Abstract
Identifying the complete transcriptional regulatory network for an organism is a major challenge. For each regulatory protein, we want to know all the genes it regulates, that is, its regulon. Examples of known binding sites can be used to estimate the binding specificity of the protein and to predict other binding sites. However, binding site predictions can be unreliable because determining the true specificity of the protein is difficult because of the considerable variability of binding sites. Because regulatory systems tend to be conserved through evolution, we can use comparisons between species to increase the reliability of binding site predictions. In this article, an approach is presented to evaluate the computational predicitions of regulatory sites. We combine the prediction of transcription units having orthologous genes with the prediction of transcription factor binding sites based on probabilistic models. We augment the sets of genes in Escherichia coli that are expected to be regulated by two transcription factors, the cAMP receptor protein and the fumarate and nitrate reduction regulatory protein, through a comparison with the Haemophilus influenzae genome. At the same time, we learned more about the regulatory networks of H. influenzae, a species with much less experimental knowledge than E. coli. By studying orthologous genes subject to regulation by the same transcription factor, we also gained understanding of the evolution of the entire regulatory systems.
The number of complete microbial genome sequences is increasing at an unprecedented rate. To date, 29 bacterial genomes have been determined, 11 more are in annotation stage, and 83 are in progress. This surge of sequence information provides an enormous amount of data for comparative genomics analysis. During the earlier stage of genomic analysis, most of the effort was devoted to analyses of protein-coding regions because, in the course of evolution, protein-coding sequences change much slower than the noncoding sequences (Koonin et al. 1997, 1998). These comparative genomics studies have proved highly informative, allowing functional assignments for many putative proteins in poorly studied organisms (Overbeek et al. 1999). One surprising result from these analyses was the lack of long-range conservation of gene order in bacterial genomes, with the exception of species within the same genus (Tatusov et al. 1996; Himmelreich et al. 1997). For species of intermediate phylogenetic distance, such as in Escherichia coli and Haemophilus influenzae, many clusters are conserved, but their orders are less conserved (Dandekar et al. 1998). However, a more recent study shows a clear conservation of pairs of orthologs to genes within an operon, as opposed to genes at the boundaries of transcription units (TU) (G. Moreno-Hagelsieb et al. 2001).
Besides knowing individual protein functions, knowledge about the transcriptional regulatory network is an indispensable prerequisite for an adequate understanding of cellular functions. The computational identification of regulatory proteins from a bacterial genome sequence is more solid given the limited number of transcription factor families and the conservation of the helix–turn–helix motif in bacteria (Perez-Rueda and Collado-Vides 2000). For the set of regulatory proteins, we want know the entire set of genes whose expression is regulated by each of these regulators, its regulon (Salgado et al. 2000a). The first step in this direction is the identification of transcription factor binding sites, which then can help to predict transcription units regulated by these proteins. Although the problem of regulatory site prediction has been studied for >20 years, it is still far from being solved (Gelfand 1995; Thieffry et al. 1998a). The major reasons for this are small training set size (often <20 sequences) and poor understanding of the biophysics of protein–DNA interaction, making it very difficult to deduce a proper set of rules for pattern recognition algorithms.
Besides identifying regulatory sites, the other key to predicting new members of a regulon is to have a good estimate of transcription units in a given genome. However, even for an organism as extensively studied as E. coli, the set of known TUs is far from complete (Salgado et al. 2000a). Also, predicting TUs is a nontrivial problem that has not been studied extensively. Recently, three groups have published new methods to predict TUs (Yada et al. 1999; Craven et al. 2000; Salgado et al. 2000b). These studies represent a promising first step toward a more accurate prediction of TUs. In this article, transcription units are defined as sets of genes (one or more) that are cotranscribed. Operons are defined as the polycistronic subset (more than one gene) of all transcripition units.
We have adopted a combined approach to identifying new members of regulons. We find that high scoring matches to binding patterns for transcription factors are likely to represent real regulatory sites based on the distribution of such sites. The predictions of lower scoring sites are less reliable, so we add evidence from a comparative analysis with other species, based on the premise that regulons tend to be conserved. If we find that orthologous genes in two or more species appear to be controlled by the same factor, that provides added confidence in the prediction of even the lower scoring sites. However, because many prokaryotic genes are transcribed as operons, the transcriptional control regions may be far removed from a particular gene. Therefore, the analysis of TUs is essential to the identification of pairs of orthologous genes belonging to common regulons. Therefore, the overall approach combines the prediction of TUs in each species, the identification of orthologous genes, and the prediction of transcription factor binding sites based on probabilistic models, such as weight matrices.
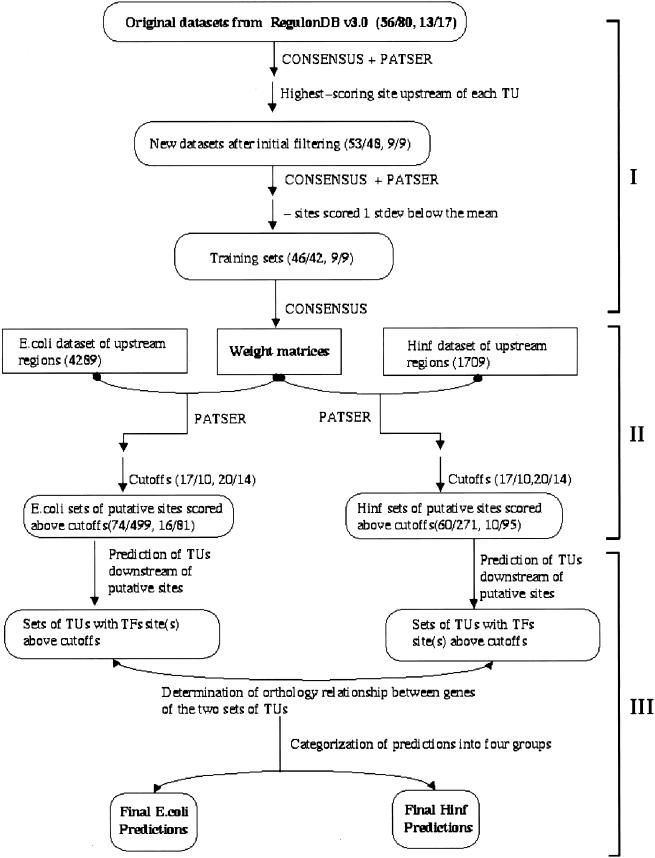
In this article, we predict new members of the cAMP receptor protein (CRP) and fumarate and nitrate reduction regulatory protein (FNR) regulons in E. coli and H. influenzae. We chose these two genomes because E. coli transcription regulation is by far the best understood among all bacterial species, and H. influenzae is the only complete genome (as of this writing) that is close enough so that many TUs are conserved. The CRP and the FNR are two global transcriptional regulators that occur in many bacteria. Genes regulated by them (CRP and FNR regulons) have a wide range of functions. Our overall strategy is shown in Figure 1. Briefly, binding patterns derived from known E. coli CRP- and FNR-binding sequences are used to predict novel binding sites for these two proteins. Predicted binding sites are combined with our knowledge of orthologous genes and predictions of TUs in both genomes. This combined information is used to predict novel members of CRP and FNR regulons.
Figure 1.
Flowchart depicting our overall strategy for predicting additional members of CRP and FNR regulons. The approach is divided into three stages. In the first stage (I), raw data sets from RegulonDB are filtered for strong binding sites, and weight matrices based on these strong sites are generated. Two pairs of numbers are shown in this part of the chart; the first pair is CRP data and the second pair FNR data. Within each number pair, the first number is the number of TUs regulated by a particular transcription factor, and the second number is the number of transcription factor binding sites. In the second stage (II), regulatory region (−400 to +50 bp) of each ORF in both genomes (4289 in E. coli and 1709 in H. influenzae) are searched by PATSER for potential transcription factor binding sites. Cutoff scores for strong (17 for CRP and 20 for FNR) and weak (10 for CRP and 14 for FNR) binding sites are chosen. Only predicted sites scored above weak site cutoffs are used for further analyses. Numbers of predicted CRP- and FNR-binding sites scored above cutoffs are shown for both genomes. The first pair of numbers represent CRP sites and the second FNR sites. In stage three (III), transcription units after predicted binding sites are predicted, and the orthology relationship between genes in E. coli and H. influenzae transcription units are determined. Finally, site scores and orthology information are used together to categorize our predictions. TU, transcription unit; TF, transcription factor.
Other groups previously have used comparative analyses to predict new sets of regulated genes. McGuire et al. (2000) recently examined 17 completely sequenced microbial genomes to identify regulatory sites for groups of related genes. They used a pattern discovery approach to find putative sites and then used various filtering techniques to diminish the number of false predictions. They used known E. coli regulons as positive controls and showed that the method worked well to identify known sites. They even showed that the method could be applied to archaebacterial species, as in another article by Gelfand et al. (2000). However, they did not use the patterns for E. coli regulatory sites to expand the set of genes likely to be regulated by specific factors, which is the main purpose of this article. Mironov et al. (1999) also used a comparative analysis to predict regulatory sites in other species for a few regulons in E. coli. In addition, they did predict a few new sites in E. coli for the PurR and ArgR regulatory proteins. Our approach in this study was similar, but by incorporating TU prediction and using two well-studied regulons, we were able to predict many more new members of the CRP and FNR regulons.
RESULTS
Conserved Recognition Patterns by CRP and FNR in Both Genomes
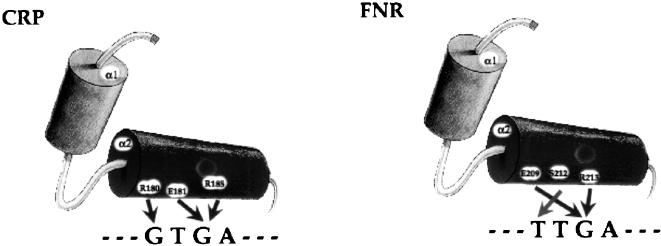
The cocrystal structure of an E. coli CRP–DNA complex has been solved at 2.5 Å resolution (Parkinson et al. 1996). The principal specificity-conferring interactions are those between the first two residues of the recognition helix, R180 and E181, and the two G : C pairs in the deduced consensus half-site TGTGA (Ebright et al. 1989; Gunasekera et al. 1992). Residue R185 in the recognition helix also contributes to binding specificity although to a lesser extent (Fig. 2). We aligned 10 CRP orthologs from various bacterial genomes (Fig. 3). The first two residues of the specificity-conferring motif, RE—R are 100% conserved across the species. The third residue in the motif is conserved except for CRP_MTB in which the second arginine is replaced by a lysine. This high level of conservation in the DNA-binding domain implies a CRP-binding pattern similar to that of E. coli. CRP also exists in these bacterial genomes with a cognate CRP protein.
Figure 2.
Schematic representation of the specificity-conferring interactions between the recognition helices (helix 2) of E. coli CRP and FNR proteins and their consensus half-site binding motifs. CRP, cAMP receptor protein; FNR, fumarate and nitrate reduction regulatory protein.
Figure 3.
Multiple sequence alignment of CRP and FNR proteins from various bacterial genomes. Only sequences around the second helix of the helix–turn–helix motif are shown. The boundaries of the second helix are labelled with a solid line. The highly conserved RE—R motif in CRP protein and E–SR motif in FNR protein are shaded. FNR_AAC, FNR_COC, and FNR_HAH are partial sequences derived from homology cloning (Hattori et al. 1996). (AAC) Actinobacillus actinomycetemcomitans; (BSU) Bacillus subtilis; (COC) Capnocytophaga ochracea; (ECO) Escherichia coli; (HAH) Haemophilus aphrophilus; (HIN) Haemophilus influenzae; (HSO) Haemophilus somnus; (KAE) Klebsiella aerogenes; (KPN) Klebsiella pneumoniae; (MTB) Mycobacterium tuberculosis; (PHA) Pasteurella haemophilus serotype 1; (PMU) Pasteurella multocida; (SDY) Shigella dysenteriae; (STM) Salmonella typhimurium; (VCH) Vibrio cholerae. CRP, cAMP receptor protein; FNR, fumarate and nitrate reduction regulatory protein.
Both CRP and FNR belong to the CRP/FNR helix–turn–helix transcription factor superfamily. In E. coli, the two proteins are 23% identical and 36% similar, with the conservation concentrated in the domain containing the HTH motif in which they have 27% identity and 43% similarity (using BLAST and BestFit). The FNR consensus half-site motif (Spiro et al. 1990), TTGAT, is analogous to that of CRP half-site (TGTGA). In fact, a common site that can bind both FNR and CRP has been reported (Jennings and Beacham 1993). In E. coli, the proposed specificity-conferring interactions for FNR are those between E209 and the G-C base pair common to both core motifs and a discriminatory interaction between S212 and the first T-A base pair in the FNR site, which replaces that between R180 and the common G-C base pair in the CRP site. Another conserved interaction involves R213 and the common G-C base pair (Fig. 2). From the multiple alignment of eight FNR orthologs (Fig. 3), we see that the first and third residues of the specificity-conferring motif, E–SR, are absolutely conserved across the species whereas the second residue is highly but not absolutely conserved. Again, this high degree of sequence conservation implies a conserved recognition pattern for FNR binding to its operators.
CRP and FNR Weight Matrices Obtained by Aligning Characterized Binding Sequences in E. coli
Using the program CONSENSUS (Hertz and Stormo 1999), we aligned the training set sequences to generate weight matrices used by the program PATSER. Specifically, a mononucleotide matrix was used to represent the binding specificity of a transcription factor. The assumption in using such a matrix is that contributions to binding specificity are additive across all positions of the site. We tested this assumption by using the program MIXY (Gutell et al. 1992) that can identify covariation(s) between any two positions across the binding sites. No significant covariation was observed between positions for CRP and FNR. Thus, we believe that a mononucleotide matrix is a valid representation of the binding specificities of CRP and FNR.
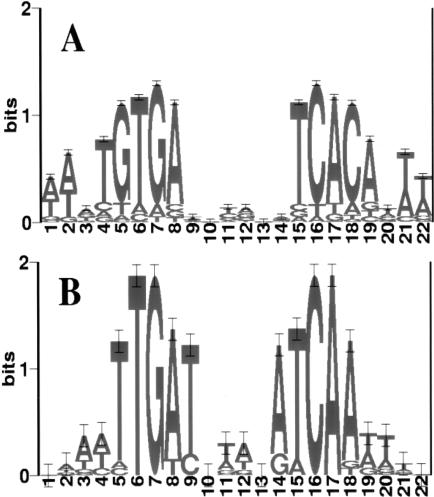
CONSENSUS calculates a P value for an ungapped multiple alignment, so different alignments can be compared and the most significant one identified (Hertz and Stormo 1999). For each protein, we compared site lengths ranging from 14 to 28 nucleotides and compared symmetric models with asymmetric ones. Symmetric models are clearly more significant than asymmetric ones, with expectation values at least 102 times lower at all even lengths tested. The expectation values for different lengths were not very different over the entire range of lengths tested, consistent with the proteins having a core conserved region of 16 or 14 bp, for CPR and FNR, respectively, surrounded by more weakly conserved sequences. We used 22 nucleotides for the length of each protein's binding site based on previous work (Kolb et al. 1993) and for consistency with previous analyses (Salgado et al. 2000a). The CRP protein has the half-site consensus of TGTGA with a separation of six nucleotides between the two half-sites. The FNR protein has a half-site consensus of TTGAT with a separation of four nucleotides between the half-sites (Fig. 4; Table 1).
Figure 4.
Sequence logos for the CRP- and FNR-binding motifs. It was generated based on the multiple sequence alignments by CONSENSUS by using the training sequences. (horizontal axis) Position in the binding motif; (vertical axis) information content in bits. The height of each letter is proportional to its prevalence at the given position.
Table 1.
Positional Weight Matrices for CRP and FNR Binding Motifs
| CRP Weight Matrix | FNR Weight Matrix | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| A | C | G | T | Cns | A | C | G | T | Cns |
| 0.97 | −2.16 | −1.70 | 0.61 | A | 0.80 | −0.55 | −0.16 | −0.55 | A |
| 1.09 | −6.57 | −1.51 | 0.61 | A | 0.97 | −0.55 | −1.09 | −0.16 | A |
| 0.80 | −1.19 | −0.14 | −0.14 | A | 1.25 | −1.09 | −1.94 | −0.16 | A |
| −1.69 | −1.19 | −1.90 | 1.58 | T | 0.80 | −0.55 | −4.26 | 0.61 | A |
| −2.46 | −3.39 | 1.74 | −1.33 | G | −1.94 | −1.94 | −4.26 | 1.78 | T |
| −1.51 | −2.16 | −4.23 | 1.75 | T | −4.26 | −4.26 | −4.26 | 1.96 | T |
| −1.51 | −6.57 | 1.80 | −2.46 | G | −4.26 | −4.26 | 1.96 | −4.26 | G |
| 1.75 | −2.16 | −2.16 | −2.46 | A | 1.78 | −4.26 | −4.26 | −1.09 | A |
| −0.35 | 0.06 | −0.61 | 0.61 | T | −4.26 | −0.55 | −4.26 | 1.70 | T |
| −0.20 | 0.52 | −0.14 | −0.35 | C | 0.39 | −0.55 | −0.16 | 0.14 | A |
| −1.06 | 0.33 | −0.71 | 0.72 | T | 0.14 | −1.94 | −1.09 | 1.12 | T |
| 0.72 | −0.71 | 0.33 | −1.06 | A | 1.12 | −1.09 | −1.94 | 0.14 | A |
| −0.35 | −0.14 | 0.52 | −0.20 | G | 0.14 | −0.16 | −0.55 | 0.39 | T |
| 0.61 | −0.61 | 0.06 | −0.35 | A | 1.70 | −4.26 | −0.55 | −4.26 | A |
| −2.46 | −2.16 | −2.16 | 1.75 | T | −1.09 | −4.26 | −4.26 | 1.78 | T |
| −2.46 | 1.80 | −6.57 | −1.51 | C | −4.26 | 1.96 | −4.26 | −4.26 | C |
| 1.75 | −4.23 | −2.16 | −1.51 | A | 1.96 | −4.26 | −4.26 | −4.26 | A |
| −1.33 | 1.74 | −3.39 | −2.46 | C | 1.78 | −4.26 | −1.94 | −1.94 | A |
| 1.58 | −1.90 | −1.19 | −1.69 | A | 0.61 | −4.26 | −0.55 | 0.80 | T |
| −0.14 | −0.14 | −1.19 | 0.80 | T | −0.16 | −1.94 | −1.09 | 1.25 | T |
| 0.61 | −1.51 | −6.57 | 1.09 | T | −0.16 | −1.09 | −0.55 | 0.97 | T |
| 0.61 | −1.70 | −2.16 | 0.97 | T | −0.55 | −0.16 | −0.55 | 0.80 | T |
| I = 12.88 bits | I = 13.74 bits | ||||||||
Each column displays the weight of the given nucleotide at that position. Highly conserved nucleotides are in bold. Cns, consensus. They are derived at each position by highest value. I, sample-size adjusted information content (in units of bits).
Determination of Cutoff Scores for CRP and FNR Sites
To determine the appropriate weight matrix for each transcription factor and the cutoff scores to be used for strong and weak predicted sites, we needed to identify a trusted set of example sites and the score distribution for those sites as well as potential sites in the genome. One of the difficulties arises because transcription factors may bind to DNA cooperatively so that a particular experimentally determined site would not, in fact, be a high-affinity site for the factor in another context without a neighboring site. To eliminate such potential artifacts, we picked only the highest scoring site for each transcription unit (see Methods), assuming that at least one of the sites should be high affinity on its own. This still may result in a few intrinsically low affinity sites in our training set but should minimize that number. We then set thresholds for high scoring sites based on the scores of the training sets and taking into account the distribution of scores in the background (i.e., genomic) sequence.
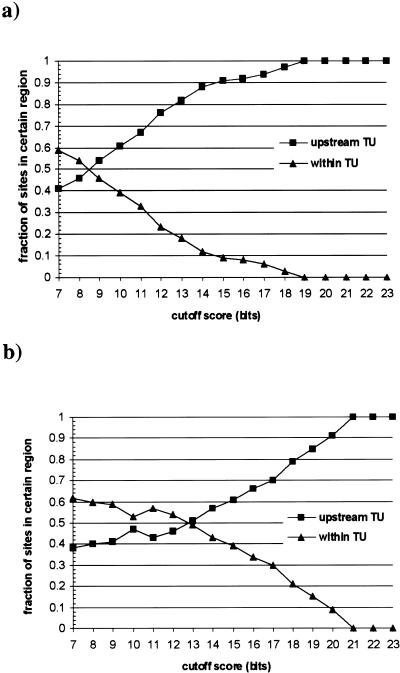
To determine cutoff scores for CRP sites, we used the following procedure. First, we determined the range and mean score for the following two sets of sequences: (1) training sequences; and (2) all 22 mers in the E. coli genome. As shown in Table 2, training sequences scored between 8.77 and 20.62 bits with a mean of 14.4 bits and a standard deviation of 3.6 bits. The mean score and the standard deviation of all 22 mers in the E. coli genome were −15.84 and 8.53 bits, respectively. Such a negative mean score is expected because most of the genomic sequence contains no CRP–binding sites. Next, for each site with a score between 7 and 23 bits from the whole-genome scan, we determined its location relative to the TUs downstream from or encompassing it. Functional regulatory sites usually are located upstream of TUs (in the regulatory region) although there are a few known cases where the sites are located within TUs (8 of 361 in RegulonDB). Given this observation, we can approximate the false-positive rate of our site predictions based on the fraction of predicted sites that are located within transcription units. Figure 5a shows the fraction of CRP-binding sites located either upstream of or within a TU in the E. coli genome. At low cutoff scores, almost all sites are located within transcription units, indicating a high false-positive rate. The size of all upstream regions is ∼1.23 Mb, ∼27% of the genome size of E. coli (4.63 Mb). Thus, sites with random localization occur ∼73% of the time within TUs. Raising the cutoff score decreases the fraction of predicted sites located within TUs and thus decreases false-positive rates. Using a cutoff of 17 bits, only 6% of all sites are located within transcription units, indicating a low false-positive rate at this cutoff. Thus, we used 17 as the cutoff score for strong sites. To increase the sensitivity of our search, we also chose a cutoff score for weak sites. We decided to use a cutoff at which greater than half of all sites are located upstream of TUs. As shown in Figure 5a, at a cutoff of 10 bits, 56% of all sites are located upstream of rather than within TUs. Thus, we chose 10 as the cutoff score for weak sites. Using this weak site cutoff, we only missed two training sequences (glnALG and rpoS; Table 3).
Table 2.
Site Score Distributions of Training Sequences and All 22-Mers in Both Genomes Scanned by PATSER
| CRP sites | FNR sites | ||||
|---|---|---|---|---|---|
| Range | Mean ± S.D. | Range | Mean ± S.D. | ||
| All training sequences | 8.77–20.62 | 14.41 ± 3.6 | All training sequences | 12.0–25.84 | 19.80 ± 4.5 |
| All 22-mers in E. coli genome | −60.97–26.65 | −15.84 ± 8.5 | All 22-mers in E. coli genome | −63.32–28.97 | −23 ± 9.3 |
| All 22-mers in H. influenzae genome | −60.96–26.67 | −16.06 ± 8.1 | All 22-mers in H. influenzae genome | −63.28–28.99 | −21.48 ± 9.1 |
Scores are in unit of bit. S.D., standard deviation.
Figure 5.
Fraction of sites located either upstream of or within TUs in E. coli. All sites in E. coli genome above certain cutoff are divided into two groups according to their locations relative to TUs: upstream of or within. (a) CRP sites; (b) FNR sites. TU, transcription unit.
Table 3.
Training Set TUs Regulated by CRP
| Operon | Site sequence | Position | Score |
|---|---|---|---|
| ansB | TTTTGTTACCTGCCTCTAACTT | −125 | 10.57 |
| araC | AAGTGTGACGCCGTGCAAATAA | −230 | 13.93 |
| araBAD | TTATTTGCACGGCGTCACACTT | −131 | 13.93 |
| araE | AATTGGAATATCCATCACATAA | −131 | 12.78 |
| araFG-araH1-araH2 | CGATGTGATATTGCTCTCCTAT | −163 | 10.0 |
| caiTABCDE | TATTGTTAAGTTCCTCACCAAT | −335 | 10.97 |
| TATTGTTTTATGGATCACCAAT | −278 | 10.88 | |
| AAATGTGATACCAATCACAGAA | −158 | 20.42 | |
| crp | GTATGCAAAGGACGTCACATTA | −137 | 11.71 |
| cyaA | AGGTGTTAAATTGATCACGTTT | −173 | 12.67 |
| dadAX | AGATGTGAGCCAGCTCACCATA | −117 | 16.48 |
| deoCABD | TTATTTGAACCAGATCGCATTA | −150 | 17.12 |
| AATTGTGATGTGTATCGAAGTG | −97 | 11.87 | |
| dsdXA | TAAAGTGAACCATATCTCAATT | −155 | 16.77 |
| epd-pgk | AAGTGTGATGTGAGTCAGATAA | −213 | 17.30 |
| focA-pf1B | AGATATGATCTATATCAATTTC | −78 | 10.14 |
| fur | AAATGTAAGCTGTGCCACGTTT | −158 | 13.81 |
| galETKM | TAATTTATTCCATGTCACACTT | −78 | 13.12 |
| galS | TGCTGTGACTCGATTCACGAAG | −95 | 10.41 |
| glgS | AAGTGTGATCGGGGACAATATA | −124 | 11.51 |
| glnALG | CTTTGTGATCGCTTTCACGGAG | −269 | 9.23 |
| glpTQ | ATGTGTGCGGCAATTCACATTT | −129 | 17.25 |
| glpD | TAATGTTATACATATCACTCTA | −117 | 15.65 |
| glpFK | TTTTATGACGAGGCACACACAT | −142 | 10.48 |
| gntT | TAATATGACCAACCTCTCATAA | −237 | 13.80 |
| lacZYA | TAATGTGAGTTAGCTCACTCAT | −110 | 18.32 |
| malEFG | TTATGTGCGCATCTCCACATTA | −161 | 11.25 |
| TTCTGTAACAGAGATCACACAA | −132 | 16.06 | |
| malI | TAGTGAGGCATAAATCACATTA | −102 | 15.70 |
| AAACGTTTTATCTGTCACATAA | −42 | 11.14 | |
| malK-lamB-malM | TTGTGTGATCTCTGTTACAGAA | −255 | 16.06 |
| TAATGTGGAGATGCGCACATAA | −226 | 11.25 | |
| malS | ATTTGAGAGTTGAATCTCAAAT | −270 | 13.97 |
| AAATGTGGGGGTTATCGCAAAA | −153 | 11.26 | |
| malT | AATTGTGACACAGTGCAAATTC | −143 | 13.48 |
| malXY | TTATGTGACAGATAAAACGTTT | −155 | 11.14 |
| TAATGTGATTTATGCCTCACTA | −95 | 15.70 | |
| nagE | TTTGGTGACAAAACTCACAAAA | −177 | 14.51 |
| ATTTGCGATACGAATTAAATTT | −143 | 12.59 | |
| nagBACD | TTTTGTGAGTTTTGTCACCAAA | −178 | 14.51 |
| AAATTTAATTCGTATCGCAAAT | −212 | 12.59 | |
| nupG | AAATGTTATCCACATCACAATT | −119 | 20.43 |
| TTATTTGCCACAGGTAACAAAA | −69 | 10.59 | |
| ompA | ATGCCTGACGGAGTTCACACTT | −188 | 12.84 |
| ppiA | TTTTGTGATCTGTTTAAATGTT | −204 | 10.23 |
| AGAGGTGATTTTGATCACGGAA | −151 | 12.33 | |
| proP | ATGTGTGAAGTTGATCACAAAT | −228 | 20.45 |
| ptsHI-crr | TTTTGTGGCCTGCTTCAAACTT | −338 | 14.43 |
| rhaBAD | AATTGTGAACATCATCACGTTC | −127 | 15.16 |
| AAATGCGGTGAGCATCACATCA | −162 | 12.41 | |
| rhaT | AGATGTGAAGCAAATCACCCAC | −145 | 13.65 |
| rpoS | AACTGCGACCACGGTCACAGCG | −270 | 8.77 |
| sdhCDAB | TATCGTGACCTGGATCACTGTT | −314 | 16.41 |
| tdcABC | ATTTGTGAGTGGTCGCACATAT | −82 | 15.0 |
| AAATGTGACATGCCGCATTATT | −384 | 10.83 | |
| tnaLAB | GATTGTGATTCGATTCACATTT | −95 | 19.61 |
| tsx | AACTGTGAAACGAAACATATTT | −129 | 12.93 |
| udp | TTATGTGATTTGCATCACTTTT | −145 | 20.62 |
| CATGGTGATGAGTATCACGAAA | −93 | 11.04 | |
| uhpT | AAGCGTGATGCATCTCACCTTT | −145 | 16.51 |
| yhfa | TAATGTGACGTCCTTTGCATAC | −187 | 11.71 |
We applied the same criteria described above to determine the cutoff scores for FNR-binding sites. The training sequences had a score range of 12 to 25.84 bits and a mean of 19.8 bits with a standard deviation of 4.5 bits (Table 2). Based on Figure 5b, we chose 20 as the cutoff for strong sites. At this cutoff, only 9% of all sites are located within TUs. As for the weak site cutoff, we chose 14 because at this cutoff greater than half of all sites (57%) are located upstream of rather than within TUs (Fig. 5b). Using the weak site cutoff, we only missed one training sequences (dmsA; Table 4).
Table 4.
Training Set TU in Escherichia coli Regulated by FNR
| Operon | Site sequence | Position | Score |
|---|---|---|---|
| ansB | TAAATTGTTTAACGTCAAATTT | −75 | 20.55 |
| dmsA | CCCTTTGATACCGAACAATAAT | −276 | 12.0 |
| fnr | AAAATTGACAAATATCAATTAC | −37 | 23.67 |
| focA | AGATATGATCTATATCAATTTC | −78 | 21.10 |
| narGHJI | ACTCTTGATCGTTATCAATTCC | −110 | 17.57 |
| narK | GGTAATGATAAATATCAATGAT | −116 | 16.41 |
| TCGTTTGATTTACATCAAATTG | −78 | 20.38 | |
| ndh | AAACTTGATTAACATCAATTTT | −155 | 25.84 |
| nirBDCcysG | GAATTTGATTTACATCAATAAG | −77 | 23.12 |
| tdcABC | TTTTTTGACAAAAATCAGGGTT | −187 | 14.03 |
This table contains top scoring site for all nine TUs, plus any additional sites with scores above 14.
New Members of the CRP Regulon
The sets of upstream sequences from both genomes were scanned by PATSER by using the CRP weight matrix. Putative sites were filtered using the two cutoffs for CRP sites described above. For each CRP site scored above 10 bits, we predicted the TU downstream from it. Orthologs (if any) to all genes in a predicted TU were identified. Based on the two cutoffs for CRP-binding sites, we first partitioned our predictions into the following two categories: (1) TUs having at least one strong site; and (2) TUs having only weak site(s). Because the cutoff for strong sites is 2.6 bits higher than the mean score of training sequences and are likely to have few false-positives (Fig. 5a), we were confident of those category I predictions even without orthology information. Predictions in category II have only weak binding sites and are less reliable than those in category I. However, for some category II predictions, additional evidence exists to support them. The first type of evidence is orthology information. If a category II TU shares orthologous member(s) with a TU from the other genome and the latter also has CRP-binding site(s) (either weak or strong), we put such a TU in category IIA. The second type of evidence is the presence of two or more weak binding sites in the regulatory region of a TU. The probability that two or more sites occur in close proximity by chance is fairly low. We examined all weak CRP sites in the E. coli genome. For all sites located upstream of TUs, 12% are within 100 nucleotides apart. Conversely, only 2% of all sites located within TUs are within 100 nucleotides apart. Thus, closely positioned tandem sites in the regulatory region are more likely to be true binding sites than a single weak site in the regulatory region. We put all category II predictions with two or more sites but without orthology information in category IIB. The rest of category II predictions, TUs having only one weak binding site and no orthology information, are labeled category IIC. This category has the least evidence to support them. Thus, we expect a high false-positive rate among category IIC predictions.
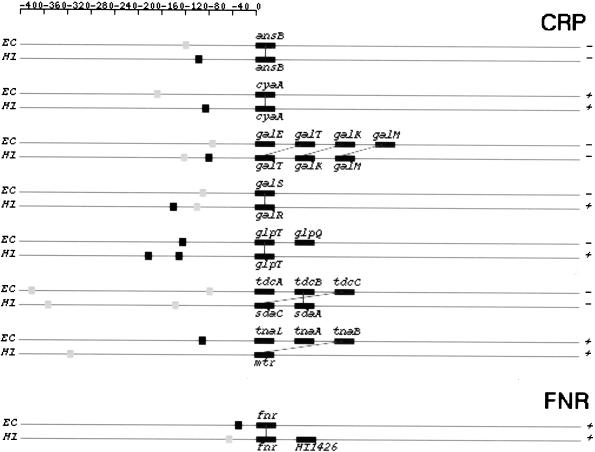
For clarity, the 46 training set TUs and H. influenzae TUs having orthologs to genes in the training set were put in a separate category. For the 46 E. coli TUs, our predictions of CRP-binding sites largely agreed with the data in RegulonDB except for a few cases in which our method predicted extra binding sites (Table 3). We identified 23 H. influenzae TUs that have orthologs to genes in the training set TUs. Of these 23 TUs, only seven contain CRP-binding sites in their upstream regions (Fig. 6; Table 5)
Figure 6.
Training set TUs regulated by CRP and FNR and H. influenzae TUs containing orthologs to genes in the training set. Genes in a TU are represented by rectangular boxes. Binding sites are represented by square boxes with gray box representing a weak site and black box representing a strong site. The distance between a binding site and the translation start is proportional to the real distance on the genomic sequence. Gene boxes and distances between genes are not in proportion. Orthology relationship is indicated by a solid line between the two genes involved. (+ and −) The strandness of a transcription unit. EC, E. coli; HI, H. influenzae; CRP, cAMP receptor protein; FNR, fumarate and nitrate reduction regulatory protein.
Table 5.
Haemophilus influenzae TUs Predicted to Belong to the CRP Regulon
| Operon | Site sequence | Position | Score |
|---|---|---|---|
| TUs Orthologous to Training Set TUs | |||
| ansB | TTATGTGATCGAGATCATAAAT | −102 | 18.78 |
| cyaA | AATTGTGATTTATGTCACATTT | −90 | 23.59 |
| galR | AACCGTGATCTTTGTCACAAAA | −145 | 17.09 |
| TTTTATGATTTAGTTCATACTT | −104 | 13.74 | |
| galTKM | TTTTGTGACAAAGATCACGGTT | −85 | 17.09 |
| AAGTATGAACTAAATCATAAAA | −126 | 13.74 | |
| glpT | TTTTGTGATATTGATCACAATA | −186 | 21.10 |
| ATTTGTGAAACACTTCACATTT | −135 | 21.58 | |
| mtr | TGATGTGAAAAATTCAATATT | −319 | 11.14 |
| sdaCA | AAATTTTAACTTGATCACAATT | −141 | 15.67 |
| TTTTTTGCTTTGATTTACAATA | −366 | 10.23 | |
| Category I | |||
| arcA | AACTATGATTTAGATCACAAAA | −130 | 17.84 |
| argR-HI1208 | TTCTATGATCTAGTTCACATTT | −125 | 18.68 |
| aspA | AAATGTGATCTTCATCAAGTTT | −71 | 18.39 |
| brnQ | TATTGTGACAAAATTCACATTT | −96 | 20.49 |
| cdd | ATAAGTGATCAAGATCACAGTT | −117 | 19.14 |
| cmkA | TTCTGTGATCCATCTCACAATC | −204 | 20.06 |
| cydAB | AAATGTGATCTATATAGCATTT | −234 | 17.77 |
| CAATTTGATCTAAGTCAATTAA | −298 | 11.86 | |
| dsbD | TTTTGTGATCTAAATCATAGTT | −84 | 17.84 |
| fdhD | TATTGTGATCTAGATCATAAAT | −73 | 19.77 |
| frdABCD | TTTTTTGAGGTAGATCACAAAA | −147 | 18.19 |
| gapdH | TGATTTGATATAGATCACAAAA | −145 | 18.14 |
| genX | TTTTGTGATCTACCTCAAAAAA | −57 | 18.19 |
| glpABC | AAATGTGAAGTGTTTCACAAAT | −180 | 21.58 |
| TATTGTGATCAATATCACAAAA | −129 | 21.10 | |
| glpR | ATCTGAGATCTAGATCACAGAA | −96 | 18.12 |
| gmk | TTTTGTGATCTATATCAAATCA | −278 | 18.14 |
| hemX-HI0602.1-hemY | AAATGTGACATAAATCACAATT | −237 | 23.59 |
| hslVU | AAATATGATCAACTTCACATTT | −129 | 19.07 |
| HI0053-HI0052-HI0051 | AACTGTGGCGTGGATCACAGTT | −127 | 20.80 |
| HI0074-HI0073 | TTTTGTGATCAATATCACAATG | −96 | 18.33 |
| TTATTAGAAAAATATCAAATTA | −150 | 11.87 | |
| HI0082-HI0083 | TTTCTTGATCCACGTCACATTA | −265 | 17.10 |
| HI0145 | AAATGAGAAGTTGATCACATTT | −184 | 20.01 |
| HI0146-HI0147 | AAATGTGATCAACTTCTCATTT | −188 | 20.01 |
| HI0310 | TAATTTGACACGCATCACAAAT | −116 | 19.12 |
| TAAAATGAAAAAAATCACGCTT | −5 | 10.80 | |
| HI0432-HI0431-hupA | TAACGTGAGATTTGTCACAATT | −4 | 17.61 |
| HI0485.1-atpBEFHAGDC | AAGTGTGATTTATATAACACTT | −95 | 18.96 |
| HI0495-HI0494 | AAATGTGAAGTTGATCATATTT | −112 | 19.07 |
| HI0521-HI0520 | AACTGTGATCTTCCTCACGTTT | −83 | 18.26 |
| HI0522 | AAACGTGAGGAAGATCACAGTT | −152 | 18.26 |
| HI0744-secB | ATTTATGATCTCGATCACATAA | −146 | 18.78 |
| HI1010-HI1011- | TTCTGTGATCTAGATCTCAGAT | −85 | 18.12 |
| HI1012-HI1013 | |||
| HI1126 | ATTTGTGACTTGTATCACATTT | −86 | 22.72 |
| HI1176 | TTTTGTGATCTTGATCACATAT | −152 | 22.64 |
| HI1315 | TTCTGTGATCCATCTCACAATC | −76 | 17.91 |
| HI1349 | AACTGTGATCTTGATCACTTAT | −179 | 19.14 |
| HI1427 | TTTTGTGATCTCGATCACAAAT | −120 | 22.06 |
| lctP | TTCTGTGATCCATCTCACAATC | −98 | 20.06 |
| mdh | AAATGTGAACTAGATCATAGAA | −104 | 18.68 |
| metR-HI1738-HI1737 | AATTGTGATCTAGTTCTCAAAA | −106 | 19.75 |
| AATTGCTAAACGGATCAAATAT | −377 | 11.55 | |
| AAATGTTTTGCACCGCACATTT | −309 | 11.36 | |
| mglBAC | ATTTGTGACATGGATCACAAAT | −92 | 22.19 |
| nhaA | AAATGTGAATTTTGTCACAATA | −114 | 20.49 |
| nrdD | CATTGTGATATTGATCACAAAA | −207 | 18.33 |
| TAATTTGATATTTTTCTAATAA | −153 | 11.87 | |
| ompP2 | AAATGTGATCTCGATCAGATTT | −161 | 20.10 |
| Operon | Site sequence | Position | Score |
|---|---|---|---|
| pckA | AAATGAGATCTACTTAACATTT | −88 | 17.54 |
| ATTTTTGCTCTATATCACAATA | −148 | 16.26 | |
| pntAB | TTTCGTGATCCCTATCACAATA | −171 | 18.29 |
| sucAB | GAGTTTGAACTAGATCACAAAT | −83 | 17.64 |
| uspA | AATTGTGATCTAGTACACAGTT | −89 | 18.58 |
| uxuRA | TTTTGTGAGCCATATCACAAAA | −5 | 20.80 |
| xylAB | AACTGTGATCCACGCCACAGTT | −121 | 17.19 |
| xylFGH | AACTGTGGCGTGGATCACAGTT | −135 | 17.19 |
| Category IIA | |||
| artPIQM | ATTCGTGTTAAAAATCTCAATT | 7 | 10.71 |
| citCDEF | ATTAGTGAAATAATTTAAAATT | −263 | 11.17 |
| cspD | TTTTGTGATCTACTTATCATTT | −146 | 15.77 |
| TATTGTAAAATGGTTCAATAAT | 11 | 11.29 | |
| folE-HI1446 | AAATTTGCAATTTTTCTCATTT | −78 | 11.28 |
| fucRIKUAP | AAGTGCGGTCGGTTTCACACCA | −170 | 10.52 |
| ndh | TAATGTAACATTTTTAACAATT | −43 | 13.07 |
| HI0017 | AATTTTAATTTAGATCAAATTT | −148 | 15.42 |
| HI0257 | AAATGAGACATAGATCATCCTT | −130 | 13.52 |
| HI1030-HI1029-HI1028 | TAATATAAAACGAATCACATTT | −44 | 15.46 |
| HI1031 | AAATAGGATCTAGATCACAAAA | −148 | 14.71 |
| HI1032 | TTTTGTGATCTAGATCCTATTT | −50 | 14.71 |
| Hi1245 | AAGTTTGCAGTTCGTCACAATT | −92 | 13.23 |
| HI1394 | TATTGTGATGAAATTTTTATTT | −188 | 10.35 |
| HI1612-sfsA | CTTTGTGGTCTCGCTCACTTTT | −117 | 12.19 |
| nifR3-fis | AAATGCGAATCGGTTCATACCA | −3 | 11.10 |
| oppABCDF | TTATTAGACACAACTCACAAAA | −132 | 13.97 |
| ribA | AACTGAAATCCCCATCACAATT | −294 | 14.71 |
| rpS6 | AATTGTGCCTTGCATCTCAATG | −400 | 12.04 |
| sodA | TTTAATGATCTAAATCAATTTT | −100 | 10.20 |
| spr | CATTGTGTAAAAGATCACAAAA | −110 | 13.16 |
| ung | AAATTTGATCTAAATTAAAATT | −132 | 15.42 |
Listed are predictions from categories I and IIA and TUs containing orthologs to genes in the training set.
In category I, we predicted 62 and 49 TUs in E. coli and H. influenzae, respectively. In category IIA, we predicted 30 and 21 TUs in E. coli and H. influenzae, respectively. For both categories, predicted CRP sites, their scores, and locations relative to the transcription start are tabulated in Table 6 (E. coli) and Table 5 (H. influenzae). Category IIB contains 25 and 12 TUs in E. coli and H. influenzae, respectively. This is a total of 117 and 82 new TUs in E. coli and H. influenzae, respectively, that we are reasonably confident belong to the CRP regulon. Category IIC contains 319 and 150 TUs in E. coli and H. influenzae, respectively. These predictions are less reliable but probably contain some true regulated TUs. Because of space limitation, we are unable to display results in categories IIB and IIC in this article. These data are available as supplementary material at http://www.genome.org.
Table 6.
Escherichia coli TUs Predicted to Belong to the CRP Regulon
| Operon | Site sequence | Position | Score |
|---|---|---|---|
| Category I | |||
| adhE | AAATTTGATTTGGATCACGTAA | −241 | 18.71 |
| aer | AATTGCGATCTAAATCAAATTA | −104 | 17.51 |
| atpIBEFHAGDC | ATATGTGATCTGAAGCACGCTT | −251 | 17.30 |
| b1458 | AATTGTGATGTAAATCACGATT | −100 | 20.51 |
| b1498-b1497 | ATAAGTGACATCCATCACATAT | −265 | 19.07 |
| b1520 | GAATGTGATCGTAATCACGTTT | −4 | 17.16 |
| b1904 | TTATGTGATACAAATCACATAA | −184 | 22.99 |
| TAATATGACAACCATCACAAAA | −214 | 15.20 | |
| b1963-yedJ | AAATGTGACTTTTATCACATAA | 25 | 21.90 |
| TGGTGTGATCAGGCGCACATTA | −269 | 15.12 | |
| b2146-yeiA | ATTTGTGAATCTTTTCACAGTT | −113 | 17.61 |
| b2343 | AAGTGTGATTTCGGTCACTTAT | −100 | 19.13 |
| AATTTTGTTTTAGATCATTTTT | −122 | 10.09 | |
| b2595-b2596-yfiA | TTATGAGATTTTCATCACACAT | −82 | 17.84 |
| b2736-b2737 | TTATGTGAATCAGATCACCATA | −91 | 18.83 |
| cdd | TAATGAGATTCAGATCACATAT | −79 | 21.29 |
| ATTTGCGATGCGTCGCGCATTT | −129 | 10.13 | |
| deaD | TACTTTGAGCCGGTTCACACTT | −48 | 17.36 |
| TTTTTTGATTGCCATCACCTTA | −105 | 13.58 | |
| fadL | ATAAGTGACCGAAATCACACTT | −264 | 19.13 |
| AAAAATGATCTAAAACAAAATT | −242 | 10.09 | |
| fixAB | ATAAGTGACCGAAATCACACTT | −300 | 20.42 |
| ATTGGTGAGGAACTTAACAATA | −123 | 10.97 | |
| ATTGGTGATCCATAAAACAATA | −180 | 10.88 | |
| flhDC | TTGTGTGATCTGCATCACGCAT | −271 | 19.67 |
| folA | AAGAGTGACGTAAATCACACTT | −190 | 19.46 |
| galP | TGATGTGATTTGCTTCACATCT | −83 | 18.25 |
| gapA | AATCGTGATGAAAATCACATTT | −227 | 19.67 |
| CACTTTAATCGTGCTCACATTA | −330 | 10.39 | |
| gcd | AATTGTGATGACGATCACACAT | −80 | 20.46 |
| glpABC | AAATGTGAATTGCCGCACACAT | −166 | 17.25 |
| gntKU1U2 | AAATTTGAAGTAGCTCACACTT | −171 | 19.84 |
| hpt | ATGTGTGATCGTCATCACAATT | −136 | 20.46 |
| idnDO | TGACGTGATCTTCATCACAAAT | −81 | 17.10 |
| idnK | ATTTGTGATGAAGATCACGTCA | −158 | 17.10 |
| kdgT | TTTTGTGATCAATTTCAAAATA | −172 | 17.07 |
| TGATGTGGTTTTGATCACTTTT | −112 | 13.87 | |
| mtlADR | AAATGTGACACTACTCACATTT | −280 | 21.14 |
| TTATGTGATTGATATCACACAA | −163 | 21.01 | |
| TTTTGTGATGAACGTCACGTCA | −207 | 15.55 | |
| TCTTGTGATTCAGATCACAAAG | −324 | 11.91 | |
| TAACATGCTGTAGATCACATCA | −367 | 11.80 | |
| mutH | ATTCGTGACCCAGGTCACACCT | −378 | 17.49 |
| serC-aroA | TTGTGTGATGCAAGCCACATTT | −156 | 18.52 |
| CATTGCGATGTGTGTCACTGAA | −114 | 10.29 | |
| tsr | ATATGTGATTCATATCACATAT | −169 | 23.71 |
| yagA | AAATTTAAGCTGGATCACATAT | −177 | 18.19 |
| yagEF | ATATGTGATCCAGCTTAAATTT | −119 | 18.19 |
| ycdZ | AAGTGTGATCTACGTCACTCAT | −56 | 19.86 |
| AAATGTGTGCTCGATCTCATTC | −5 | 13.87 | |
| ycfQ | AGATGTGATCTGGATCACATAC | −97 | 20.54 |
| ycfR | GTATGTGATCCAGATCACATCT | −88 | 20.54 |
| ychE | TTACGTGATCCAAATCAAATTT | −258 | 18.71 |
| yciCB | TAATGTGATTTAAATCAATTTT | −212 | 18.20 |
| yciD | AAAATTGATTTAAATCACATTA | −167 | 18.20 |
| ydaJ-b1337-ydaH-ogt | CTACGTGAACCGGGTCACACTT | −19 | 17.07 |
| ydaK | AAGTGTGACCCGGTTCACGTAG | −154 | 17.07 |
| ydeA | AAACGCGATCCAGATCACAAAT | −95 | 18.17 |
| yeaA-b1777 | AAATGTGATTTTCATCACGATT | −137 | 19.67 |
| yeaF | AATTGTGACCAAACTCAAACTT | −4 | 17.32 |
| ygbI | TATGGTGATCTGATTCACATAA | −97 | 18.83 |
| ygcW | CACTGTGATTACGATCACATTA | −68 | 17.12 |
| TTGTGTGAGAGTAATCACGCTT | −125 | 15.17 | |
| Operon | Site sequence | Position | Score |
|---|---|---|---|
| Category I | |||
| ygdP-pstP | AGGTGTGACCTGGGTCACGAAT | −329 | 17.49 |
| ygiG | TAATTTGATTTAGATCGCAATT | −225 | 17.51 |
| yhcN | TTTTGTGATATGGGTCACGAAA | −10 | 18.70 |
| yhcRQP | AAAAGTGATTTAGATCACATAA | −98 | 21.67 |
| yhcS | TTATGTGATCTAAATCACTTTT | −38 | 21.67 |
| yibIH | TTGTGTGATATCAATCACATAA | −396 | 21.01 |
| AAATGTGAGTAGTGTCACATTT | −279 | 21.14 | |
| TGACGTGACGTTCATCACAAAA | −352 | 15.55 | |
| CTTTGTGATCTGAATCACAAGA | −235 | 11.91 | |
| TGATGTGATCTACAGCATGTTA | −192 | 11.80 | |
| yjcB | AATTGTGATATAGTTCACAAAA | −80 | 22.28 |
| yjcC | TTTTGTGAACTATATCACAATT | −303 | 22.28 |
| yjhIHG | AAGTGTGTACAAGATCACATTT | −115 | 18.78 |
| yjiY | ATATGTGATATGAATCACATAT | −216 | 23.71 |
| yjjIW | TAATGCGATCTGGTTCAAATAA | −130 | 17.11 |
| CACTTCGATACACATCACAATT | −183 | 11.87 | |
| yohI | AAACGTGCTACCGATCACATTA | −192 | 17.07 |
| yohJK | TAATGTGATCGGTAGCACGTTT | −69 | 17.07 |
| yqcD | AATTGTGGGTTGTATCACAATA | −299 | 17.33 |
| yqcE-ygcE | TAATGTGATCGTAATCACAGTG | −198 | 17.11 |
| AAGCGTGATTACTCTCACACAA | −141 | 15.17 | |
| ysgA | AAAAGTGATGCAAATCACATAA | −176 | 20.62 |
| TTTCGTGATACTCATCACCATG | −228 | 11.04 | |
| Category IIA | |||
| aphA | TTTTGCAACAAATCTCACAATA | −58 | 11.41 |
| AAATATGCGCAAGATCACACAG | −5 | 11.01 | |
| artPIQMJ | AATCGTGATGCCCGTAACATTC | −397 | 12.39 |
| b2463 | ATGAGTGCGTTAATTCACACTT | −257 | 13.22 |
| citCDEF | CTATGTGAAATAAATCAAAATT | −96 | 16.51 |
| cspD | TAGCGTTAACTGCTTCAAATTT | −180 | 12.32 |
| ATCAGCGACATCTGTCACATTC | −209 | 10.39 | |
| fdhD | AAATGTGACAAATATCACAGGT | −72 | 13.26 |
| folE-yeiB | TTATGTGCGCCGCCTCACGCAC | −101 | 11.20 |
| fucAO | TTATGTGACTACCATCACTTTA | −361 | 16.91 |
| TTAGTTGAACCAGGTCACAAAA | −144 | 15.59 | |
| TAGTGTGAAAGGAACAACATTA | −54 | 12.46 | |
| mglBAC | ATCTGTGAGTGATTTCACAGTA | −270 | 16.65 |
| ndh | AAACTTGATTAACATCAATTTT | −155 | 11.17 |
| nrdD | TACTTTGAGCTACATCAAAAAA | −253 | 14.52 |
| oppABCDF | AAAAGAGAATTGCTTAACAATT | −338 | 11.12 |
| pckA | GAATGCGATTCCACTCACAATA | −241 | 14.03 |
| ribA | TTAGGTGAACCCCTTCTCGTTA | −67 | 11.55 |
| rpsF-priB-rpsR-rplI | AAGTGTGATGAACTTCAAATCA | −199 | 15.68 |
| sdaC | ATTTGAGATCAAGATCACTGAT | −180 | 14.46 |
| sfsA-dksA | TGCGGTGACGGAGTTCACCCTT | −105 | 10.25 |
| sodA | GTGGGTGATTTGCTTCACATCT | −162 | 13.65 |
| spr | TTTTGTGCGTTAGTCCACAGAT | −131 | 11.55 |
| ung | ATCTTTGATTTAAATCAATAAA | −202 | 11.25 |
| uxuR | AAATTTGATTAACCGCACCTAA | −36 | 10.83 |
| xylAB | TTTTGCGAGCGAGCGCACACTT | −134 | 12.26 |
| ATTTATGACCGAGATCTTACTT | −224 | 10.68 | |
| xylFGHR | AAGTGTGCGCTCGCTCGCAAAA | −254 | 12.26 |
| AAGTAAGATCTCGGTCATAAAT | −164 | 10.68 | |
| yfiD | TTTATTGATTTAAATCAAAGAT | −125 | 11.25 |
| yhdG-fis | AAGTGCGAGCAAGCTCACAAAA | −298 | 15.84 |
| AATTGAGAACTTACTCAAATTT | −213 | 14.58 | |
| yiaJ | GATCGTGAACTACGGCACACTT | −47 | 13.32 |
| yiaKL | AAGTGTGCCGTAGTTCACGATC | −176 | 13.32 |
| yiaMNO-lyxK-sgbHUE | AATTGTGGTTAAAGTCGCATTA | −155 | 13.48 |
| yidKJ | AATTCCGCTGGAGATCACATTT | −259 | 10.70 |
| yqfB | TAAGGTGAGTTTTTTCACTATC | −62 | 10.42 |
Listed are predictions from categories I and IIA.
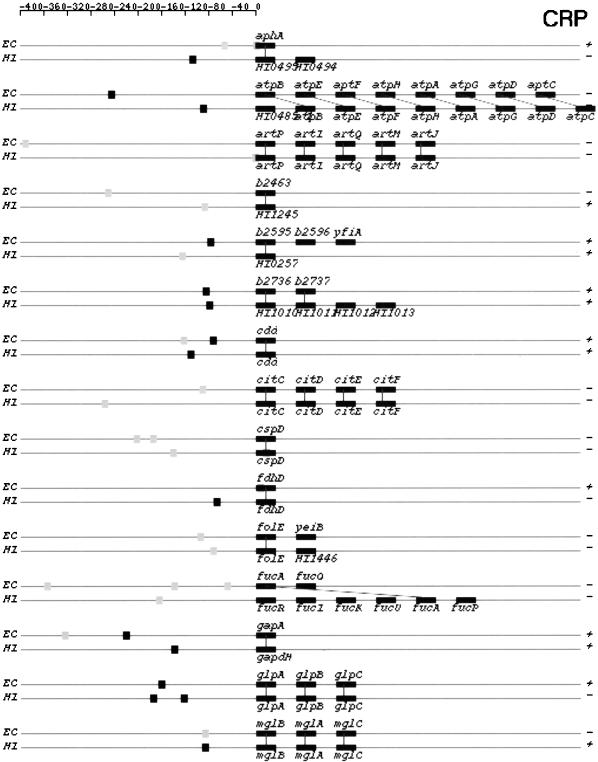
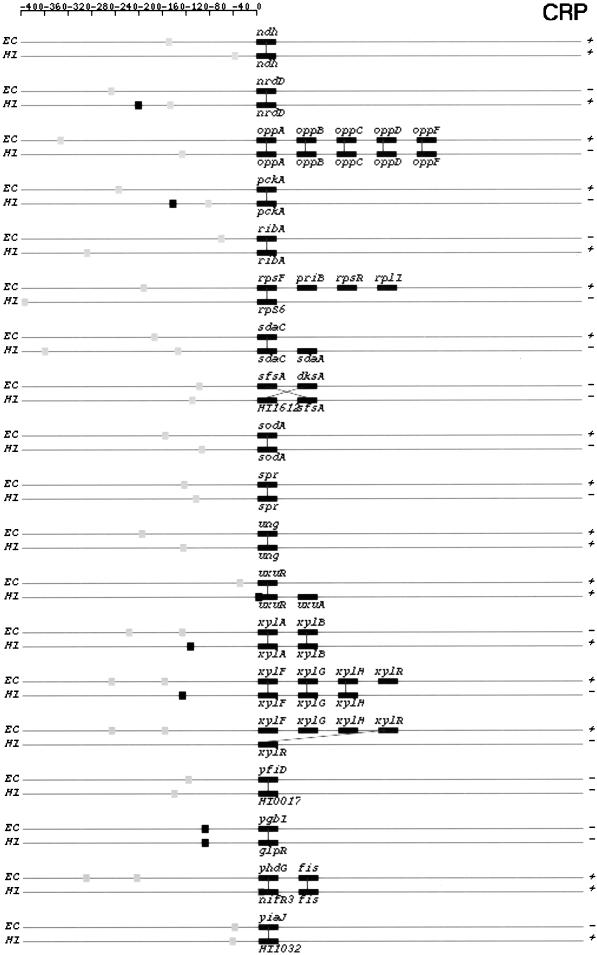
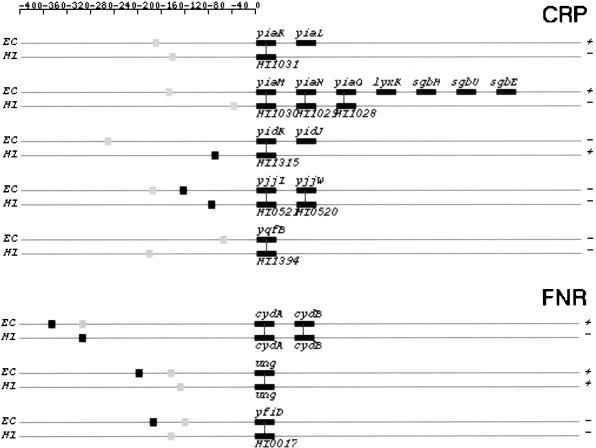
In Figure 7, we depict structures of predicted TUs that share orthologous members. They are from categories I and IIA in both genomes. Strong and weak binding sites are represented by black and gray squares, respectively. Thus, one can identify the category to which a TU belongs by the colors of binding site squares.
Figure 7.
Orthologous TU pairs from categories I and IIA that are predicted to be regulated by CRP and FNR. CRP-regulated TUs are shown first followed by FNR-regulated TUs. Symbols and drawing schemes are as described for Figure 6. CRP, cAMP receptor protein; FNR, fumarate and nitrate reduction regulatory protein.
New Members of the FNR Regulon
The same procedures (FNR-binding site predictions, predictions of downstream TUs, and categorization) were performed to identify new members of the FNR regulon. We have nine training set TUs (Table 4) and four H. influenzae TUs that have ortholgs to genes in the training set. Among these four H. influenzae TUs, only one still maintains FNR regulation (Fig. 6; Table 7). The other five E. coli TUs do not have detectable orthologs in H. influenzae.
Table 7.
Haemophilus influenzae TUs Predicted to Belong to the FNR Regulon
| Operon | Site sequence | Position | Score |
|---|---|---|---|
| TUs Orthologous to Training Set TUs | |||
| fnr-HI1426 | ATATTTGCGTTAGATCAATTTT | −53 | 14.48 |
| Category I | |||
| cpdB | GATTTTGATGAAAATCAATTAC | −165 | 22.58 |
| GAATTTGATTTTGATGAAAATC | −171 | 16.91 | |
| cydAB | CAATTTGATCTAAGTCAATTAA | −298 | 21.54 |
| moaACDE | AATACTGATTTTCATCAATATT | −200 | 20.68 |
| AATTATGATTTAAATCAATAAA | −348 | 19.29 | |
| ATAATTGATTTTTAAGAATTTA | −226 | 16.20 | |
| pepT | AATATTGTTATATATCAAGATG | −76 | 20.61 |
| potA | CATCTTGATATATAACAATATT | −215 | 20.61 |
| pyrG | TTAATTGACTTAGATCAAATTG | −376 | 21.53 |
| HI1503 | TAATTTGATTTACATCAATCAA | −319 | 21.00 |
| HI1677 | AATATTGATGAAAATCAGTATT | −267 | 20.68 |
| TTTATTGATTTAAATCATAATT | −119 | 19.29 | |
| TAAATTCTTAAAAATCAATTAT | −241 | 16.20 | |
| Category IIA | |||
| HI0017 | AATTTTAATTTAGATCAAATTT | −148 | 19.80 |
| ung | AAATTTGATCTAAATTAAAATT | −132 | 19.80 |
| Category IIB | |||
| HI0588 | TTGTTTGACGAATATCAAAAAA | −45 | 15.29 |
| TTTTTTCATATTCATCAAAAGT | −287 | 14.06 | |
| HI1129-HI1130ftsLI-murEF-mraY-murD | TATTTTGATAAAAATCAGTTGC TTATTTGTTCTACAACAAAATT | −148 −353 | 18.62 17.49 |
Listed are predictions from categories I, IIA, IIB, and TUs containing orthologs to genes in the training set.
Category I contains 10 and eight TUs in E. coli and H. influenzae, respectively, each with at least one strong site. Category IIA contains 0 and 2 TUs in E. coli and H. influenzae, respectively. For both categories, predicted FNR sites, their scores, and distances relative to the transcription start are tabulated in Table 7 (H. influenzae) and Table 8 (E. coli). We did not find any TU in category IIB in E. coli. In H. influenzae, category IIB contains 2 TUs. Thus, this is a total of 10 and 12 new TUs in E. coli and H. influenzae, respectively, that we are fairly confident belong to the FNR regulon. In category IIC, we predicted 70 E. coli and 79 H. influenzae TUs, all of which have only one weak binding site and no orthology information. Categories IIB and IIC are available as supplementary material at http://www.genome.org. Again, the structures of predicted TUs that share orthologous members are depicted in Figure 7.
Table 8.
Escherichia coli TUs Predicted to Belong to the FNR Regulon
| Operon | Site sequence | Position | Score |
|---|---|---|---|
| Category I | |||
| b1674-b1673-b1672- | TTAATTGATAACGATCAATGTT | −218 | 20.04 |
| b1671-ydhU-b1669 | |||
| b2503 | TTGTTTGATATATATCAATTGG | −145 | 20.26 |
| b2504 | CCAATTGATATATATCAAACAA | −229 | 20.26 |
| cydAB | GGAATTGATATTTATCAATGTA | −350 | 21.30 |
| TAAATTGTTCTCGATCAAATTG | −298 | 19.10 | |
| narXL | CAATTTGATGTAAATCAAACGA | −283 | 20.38 |
| ATCATTGATATTTATCATTACC | −245 | 16.41 | |
| ung | ATCTTTGATTTAAATCAATAAA | −202 | 20.93 |
| TTTATTGTTTTACATCAACTTA | −149 | 14.99 | |
| yciCB | TAATGTGATTTAAATCAATTTT | −212 | 20.88 |
| yciD | AAAATTGATTTAAATCACATTA | −167 | 20.88 |
| yehDCBA | TAATTTGTTTTAAATCAATAAA | −124 | 20.17 |
| yfiD | TTTATTGATTTAAATCAAAGAT | −125 | 20.93 |
| TAAGTTGATGTAAAACAATAAA | −178 | 14.99 | |
Listed are predictions from category I.
DISCUSSION
We have described a method to systematically search for additional members of bacterial regulons based on information both intrinsic and extrinsic to a given genome. The intrinsic information consists of transcription factor binding sites and structures of downstream TUs. The extrinsic information is the orthology relationship between TUs obtained by comparing the respective complete sets of gene products. Our comparative approach consists of the following three major steps: (1) obtaining DNA recognition pattern for a given regulatory protein; in this study, we used weight matrices to represent binding site patterns; (2) prediction of transcription factor binding sites using the recognition pattern obtained in step one; and (3) prediction of TUs downstream from binding sites from step two and identification of any orthologs to members of the predicted TUs. At low thresholds, transcription factor binding site predictions by any present-day computer algorithm are expected to have a relatively high false-positive rate due to small training set size and poor conservation of noncoding sequences. However, incorporation of orthology information in step three increases the reliability of our inferences. Another reinforcement to the prediction of regulatory sites is the use of information on TUs. The correspondence between predicted TUs and the assignment of putative regulatory sites will help to establish other means to score the predictions and make them as more reliable. Certainly, we do not have a statistical model to evaluate how much the probability of a site increases when the site is present in front of orthologous TUs. However, qualitatively, our confidence does increase in the presence of orthology information. In this way, we are at least confident that predictions in categories IIA and IIB have a lower false-positive rate compared with those in category IIC.
The sensitivity and specificity of our predictions are difficult to determine because we do not know the complete set of genes that are regulated by CRP and FNR in either species. In E. coli we have a set of genes that are known to be regulated by each protein, based on genetic and biochemical criteria, but that set is certainly incomplete. For most genes, we simply do not know whether they are regulated by CRP or FNR, and the primary purpose of this article was to identify new TUs that are likely to be regulated by these factors. We can estimate sensitivity and specificity measures by using both the known set of regulated genes and some assumptions about the distribution of sites with various scores. For example, we know that functional regulatory sites usually occur in the region we have defined as the upstream region, between −400 and +50 bp of the start of translation of the first gene in the TU. Rarely, although occasionally, functional sites occur either farther upstream of or within the TU. We also assume that if a binding site for a regulatory protein occurs within that upstream region then it is very likely to be involved in the regulation of the adjacent TU. We set the threshold based on those assumptions for strong sites to be such that >90% of the sites occur in the upstream regions, and therefore we expect very few false-positive among category I predictions. This gives us confidence in the new predicted category I CRP-regulated TUs, 62 and 49 in E coli and H. influenzae, respectively, even without additional evidence. The category I new predictions for FNR are 10 and 8. However, of the known E. coli TUs regulated by these proteins, only 9 and 6 have strong sites, so the sensitivity based on strong site cutoffs alone is only 16.1% and 46.2% for CRP and FNR, respectively.
The threshold for weak sites was chosen such that >50% occur in the upstream regions. Remember that even these weak sites have much higher scores than the average background site (Table 2), and that most upstream regions do not have them, and any randomly chosen sites would occur only 27% of the time in the upstream regions, based on the sizes of the two sequence sets. Therefore, even such weak sites, category II predictions, are likely to contain many functional sites but undoubtedly contain false-positives as well. Therefore, we look for additional evidence before considering them reliable. One type of additional evidence is if TUs in H. influenzae that contain orthologous genes also appear to be regulated by the factor with either a strong or a weak site, which we call category IIA. Another type is if there are two weak sites near each other, which we call category IIB. We know that CRP can bind cooperatively, so nearby weak sites may have a combined affinity comparable to single strong sites. Furthermore, nearby pairs of weak sites occur infrequently within TUs, but relatively frequently in the upstream regions, as is expected of functional regulatory sites. Combining categories IIA and IIB, we predict 55 and 33 new CRP regulated TUs in E. coli and H. influenzae. An additional 319 and 150 TUs are in category IIC, some of which are probably real and some false. For FNR, we predict 0 and 4 new TUs in E. coli and H. influenzae from categories IIA and IIB, and there are an additional 70 and 79 category IIC TUs.
We can estimate the sensitivity of our approach by scoring the known TUs for each regulon. For the 56 TUs in the CRP regulon that we extracted from RegulonDB, only nine of them score in category I. An additional 41 have weak sites and therefore are put in category II, resulting in a combined sensitivity of 89.3% (50/56). However, among the 41 TUs with only weak sites, only 16 are in categories IIA and IIB (six and 10, respectively), with the remaining 25 in category IIC. Therefore, our confident predictions, combining categories I, IIA, and IIB, account for only 25 known sites, a sensitivity of only 44.6%. If the same proportions exist in the whole genome, then many of the category IIC sites will be functional CRP regulatory sites; however, we cannot determine which are true and which are false from the current data. Similar results are obtained for FNR, in which categories I and II together account for 11 of the 13 known TUs, for a senstivity of 84.6%, but five of those are in category IIC.
The net result of our analysis is the prediction of 116 and 10 new CRP and FNR TUs in E. coli that we consider highly reliable because they fall into categories I, IIA, and IIB. These are clearly not all of the genes regulated by these factors because some of the known TUs are missing from such predictions. Functional sites may be missed because these factors bind cooperatively with some other factor that is not included in the analysis, or because the weight matrix is not a good enough descriptor of the proteins' binding specificity to get all of the functional sites. Many of the missing sites can be found in category IIC predictions, but those predictions probably also contain many false predictions, and we do not include them in our reliable set. Nonetheless, the computational approach we have applied in this article has greatly increased the set of TUs likely to be regulated by these factors in E. coli, with high but not perfect sensitivity. In addition, we make 82 and 12 reliable predictions of TUs regulated by CRP and FNR in H. influenzae, most of which had not been previously identified as members of those regulons.
Interestingly, one experimentally verified CRP site exists in RegulonDB for the E. coli operon glpABC. However, this is a weak site (6.2 bits). In this study, we detected another strong CRP site for this operon (17.25 bits; Table 6). Another interesting case is the E. coli operon fucAO. Before this study, only genetic evidence existed to support the regulation of this operon by CRP. In our study, we identified three CRP-binding sites upstream of fucA (Fig. 7; Table 6), providing further evidence for previous observations.
In E. coli, the gene ansB is under the dual regulation of CRP and FNR (Scott et al. 1995). This joint regulation by both transcription factors might be important in achieving optimal gene expressions. Based on our analysis, ansB may be regulated only by CRP in H. influenzae because the highest scored FNR site in the regulatory region of H. influenzae ansB was only 9.74 bits. This is much lower than the weak site cutoff for FNR but might still be a functional site. Two other TUs, ung and yfiD (its ortholog in H. influenzae is HI0017), seem to be dually reguated in both genomes. Interestingly, for both TUs, the CRP site is the same as the FNR site in both genomes with E. coli TUs having an additional FNR site. It is possible that those sites are true only for one of the regulators and are false-positives for the other regulator. Conversely, we cannot rule out the possibility that those sites are truly recognized by both regulators, because some sites that can bind CRP also can bind FNR (Sawers et al. 1997).
Negative autoregulation is quite dominant in E. coli, and it can be viewed as playing a homeostatic role for the regulatory genes (Thieffry et al. 1998b). Based on our results, CRP seems not to be autoregulated in H. influenzae (the highest scoring CRP site had a score of 4.6 bits). Conversely, FNR does seem to be autoregulated in H. influenzae.
Based on our comparative analysis of the CRP and FNR regulons in the two genomes, we noticed three types of structural changes in operons that are subject to the same mode of regulation. The first type involves insertion or deletion of individual genes in otherwise conserved operons. Examples in E. coli includes operons glpTQ (glpT in H. influenzae, Fig. 6), fnr (fnr-HI1426 in H. influenzae, Fig. 6), and b2736-b2737 (HI1010-HI1011-HI1012-HI1013 in H. influenzae, Fig 7.).
The second type of change involves breakup of an operon in one genome into several smaller ones in the other genome. Not all of the smaller operons retain their regulation by the same regulator. For instance, the E. coli xylFGHR operon is broken in H. influenzae into two operons, xylFGH and xylR (Fig. 7). Only xylFGH maintains CRP regulation in H. influenzae. The protein products of genes xylF, G, and H constitute the high-affinity xylose transport system in both genomes and that of xylR encodes a regulatory protein (Sumiya et al. 1995). In E. coli, xylR acts as a transcriptional activator for the xylFGHR operon and the expression of itself is regulated by CRP (Song and Park 1997). In H. influenzae, the regulation of xylR might be taken over by a different regulator. Alternatively, it could be autoregulated. If this is the case, it is another example of uncoupled versus coupled transcription regulations in two bacteria, an organization with different dynamic consequences (Hlavacek and Savageau 1996). Another example of this second type of change involves the E. coli galETKM operon. The same operon is broken up into two pieces in H. influenzae : galE and galTKM (Fig 6). Again, only galTKM still is regulated by CRP in H. influenzae.
Third, also the most common type of change during regulon evolution is the loss of E. coli regulon members in the H. influenzae genome. Examples include operons caiTABCDE, malEFG, and narGHJI. Tatusov et al. (1996) suggested that the common ancestor of E. coli and H. infuenzae could have a genome of intermediate size. The subsequent evolution may have proceeded in opposite directions—toward the reduction of the genome size by deletion of genes and entire transcription units in the Haemophilus lineage and toward the diversification of regulatory and transport functions via gene duplication in the E. coli lineage (Tatusov et al. 1996). As a result, the decrease in CRP and FNR regulon members may be the result of degenerative evolution of H. influenzae. The parasitic lifestyle of H. influenzae might require a less complicated metabolism to cope with enviromental changes. However, as a fraction of the total number of genes, both species appear to have similar sized regulons.
The location of regulatory sites along the genome has a clear influence on how regulation through these sites occurs (Gralla and Collado-Vides 1996). An interesting question to ask is whether regulatory sites of orthologous genes have identical or close positions, that is, whether the distance between regulatory sites and their regulated promoters remain more or less unchanged between bacterial species. To obtain such information, we would need to have a reasonably accurate method to predict promoters in those organisms. Unfortunately, current promoter prediction methods are not satisfactory in this regard. Future work is needed to address this very interesting question.
We noticed that some of our predicted TUs have quite distal binding site(s). Because we report the position of a binding site relative to the translation start of the first downstream gene, these large distances could simply result from the existence of a long 5′ untranslated region. Conversely, they could be true distal sites even if our measurement were based on transcription start. Because of the global nature of regulatory functions, CRP and FNR regulated TUs often have another local, dedicated regulator, such as LacI for the lac operon and GalR for the gal operon. Thus, we suspect TUs predicted here with distal sites will show regulation by additional proteins.
The approach we have used in this article has identified many new genes that we predict are regulated by the CRP and FNR proteins in E. coli and H. influenzae. Combined evidence from site scores and comparative analyses gives us high confidence in many of these predictions. But this is clearly just a first step. More bacterial species can be included and many more regulons can be studied, although regulons with few known members are more problematic because of the small sample size. The accurate prediction of transcription units is critical to the success of such an approach, as operons are often rearranged in evolution and common regulatory sites may be located at long and variable distances from orthologous pairs of genes. In this work, many steps were performed manually, in that careful examination of some results were used to constrain further analyses. Experience gained from this work will allow us to develop more fully automated procedures that can be applied to more regulatory systems in more species in a rapid and reliable approach.
METHODS
Sequence Data and Programs
Experimentally characterized (mostly by DNA footprinting technique) E. coli CRP- and FNR-binding sequences were extracted from the RegulonDB (Salgado et al. 2000a) database. Complete genome sequences of E. coli and H. influenzae were downloaded from GenBank (Benson et al. 1999). Weight matrices were constructed by CONSENSUS (Hertz and Stormo 1999), which generates optimal ungapped multiple sequence alignments with predefined width. In addition, the program reports the statistical significance of the generated multiple sequence alignment. Given a weight matrix, searches for transcription factor binding sites were performed using PATSER (Hertz et al. 1990). PATSER scores each possible binding site position in a sequence by using the designated weight matrix and returns the scores and positions of all sites above a user-defined threshold. Multiple alignments of protein sequences were constructed using the program CLUSTALX (Thompson et al. 1997). Protein sequence database searches were performed using the gapped BLASTP program (Altschul et al. 1997). All searches were performed against the National Center for Biotechnology Information nonredundant protein sequence database. Sequence comparisons between E. coli CRP and FNR were performed using the BestFit program (Wisconsin Package Version 10.0; Genetics Computer Group). Sequence logos were constructed using the web interface (S.E. Brenner, http://www.bio.cam.ac.uk/cgi-bin/seqlogo/logo.cgi) to the MAKELOGO program by Schneider (Schneider and Stephens 1990). The rest of the analysis was performed by using ad hoc PERL scripts (Wall et al. 1996).
Preparation of Training Set Sequences
The current version of the RegulonDB database (version 3.0) contains 80 experimentally verified E. coli CRP-binding sequences from 56 TUs (because some of these 56 TUs have multiple CRP-binding sites the number of sites exceeds the number of TUs). We expect that some of these 80 sites are weak CRP-binding sites. Presumably, CRP binds these weak sites through cooperativity with other regulatory proteins. Weak sites were filtered out from our training set by using the following procedures. In step one, we ran CONSENSUS on the 80 binding sequences and generated an initial weight matrix. PATSER then was used to score the original 80 binding sequences by using the weight matrix generated in step one. After this initial step, we only chose the highest-scoring sequence from each TU for further processing. This gave us 48 sites representing 53 TUs (all sites from three of the 56 TUs were rejected by CONSENSUS and thus not included). Because of the existence of divergent TUs, the number of sites is less than the number of TUs. The mean and standard deviation of the scores of these 48 sites were 13.1 and 3.6 bits, respectively. For our final training set, we excluded, from the 48 sites, any sites with scores that are more than one standard deviation below the mean, that is, 9.5 bits. We ended up with 42 sequences in the training set, representing 46 TUs.
The current version of RegulonDB database contains 17 experimentally verified E. coli FNR-binding sequences from 13 TUs. We applied the same procedures to these 17 sequences to generate the training set. We ended up with nine sequences in our training set, representing nine TUs. The mean and standard deviation of these nine sequences were 19.8 and 4.5 bits, respectively.
Prediction of Transcription Factor Binding Sites
During the first step of our analysis, weight matrices for both CRP and FNR binding sites were generated by CONSENSUS by using our training set sequences (42 for CRP and nine for FNR). Subsequently, the published annotations of all the open reading frames (ORFs) in E. coli (Blattner et al. 1997) and H. influenzae (Fleischmann et al. 1995) were used to generate two sets of putative regulatory sequences (one for each genome), covering 400 nt upstream of and 50 nt downstream from the beginning of each ORF. This length was chosen from the known distribution of a large collection of regulatory sites in ς70 promoters (Gralla and Collado-Vides 1996). Then, PATSER was used to scan the sets of regulatory sequences to identify potential binding sites by using the weight matrices generated in step one (Hertz and Stormo 1999; Hertz et al. 1990). Potential binding sites scored above the chosen cutoffs were reported. Eventually, binding site information was combined with orthology relationship between TUs to predict new members of the CRP and FNR regulons. We classified binding sites into two categories based on their locations relative to the TUs downstream from or encompassing it (1) sites located in the regulatory region of a TU; and (2) sites located within a TU. The latter category includes two cases: within genes of a TU and within the upstream region of an internal gene.
Determination of Orthology between E. coli and H. influenzae Genes
Fitch first introduced the term ortholog for genes derived from speciation events (Fitch 1970). At present, there is not a simple and perfect method for detecting orthology relationship because of complicating events during genome evolution, such as gene duplication, gene loss, and horizontal gene transfer (Huynen and Bork 1998). For our study, we used the minimal definition of orthology described by Huynen and Bork (1998): (1) orthologous ORFs between two genomes compared must be the most similar ORF reciprocally; (2) sequence similarity between the ORFs has to be statistically significant. In this article, sequence similarity was calculated by the BLASTP program (version 2.0; Altschul et al. 1997). Any alignment with an E-value of 1e-15 was considered significant for our purpose. and (3) sequence similarity extends to at least 60% of one of the genes.
Prediction of Transcription Units
The prediction of TUs was described for E. coli by Salgado et al. (2000b). The method is based on the differences between pairs of adjacent genes in operons and pairs of adjacent genes at the borders of TUs. The differences studied were distances between genes and their functional relationships, the latter ones being an update of the functional classification described by Monica Riley (Riley 1993; Riley and Labedan 1996). Here, to apply the method to H. influenzae, we inherited the functional classification for E. coli genes and then applied the prediction method to the whole H. influenzae genome, dividing it into putative TUs. In this way, we obtained sets of TUs that can be compared between organisms when a regulatory site was found close to orthologous genes that may in turn lie inside analogous TUs.
Acknowledgments
We thank members of the Stormo and Collado-Vides labs for insightful discussions. We thank three anonymous reviewers for their comments. This work was supported by Grant HG-00249 from National Institutes of Health (G.D.S.), Grant 0028 from Conacyt (J.C.-V.), and Grant DE-FG02-98ER62558 from U.S. Department of Energy (J.C.-V.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL stormo@ural.wustl.edu; FAX (314) 362-7855.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.149301.
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Boguski MS, Lipman DJ, Ostell J, Francis Ouellette BF, Rapp BA, Wheeler DL. GenBank. Nucleic Acids Res. 1999;27:12–17. doi: 10.1093/nar/27.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Craven M, Page D, Shavlik J, Bockhorst J, Glasner J. A probabilistic learning approach to whole-genome operon prediction. Proc Int Conf Intell Syst Mol Biol. 2000;8:116–127. .. [PubMed] [Google Scholar]
- Dandekar T, Snel B, Huynen M, Bork P. Conservation of gene order: A fingerprint of proteins that physically interact. Trends Biochem Sci. 1998;23:324–328. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- Ebright RH, Ebright YW, Gunasekera A. Consensus DNA site for the Escherichia coli catabolite gene activator protein (CAP): CAP exihibits a 450-fold higher affinity for the consensus than for the E. coli lac DNA site. Nucleic Acids Res. 1989;17:10295–10305. doi: 10.1093/nar/17.24.10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch WM. Distinguishing homologous from analogous proteins. Syst Zool. 1970;19:99–100. [PubMed] [Google Scholar]
- Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- Gelfand MS. Prediction of function in DNA sequence analysis. J Comput Biol. 1995;2:87–115. doi: 10.1089/cmb.1995.2.87. [DOI] [PubMed] [Google Scholar]
- Gelfand MS, Koonin EV, Mironov AA. Prediction of transcription regulatory sites in Archaea by a comparative genomic approach. Nucleic Acids Res. 2000;28:695–705. doi: 10.1093/nar/28.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralla JD, Collado-Vides J. Organization and function of transcription regulatory elements. In: Neidhardt FC, editor. Cellular and molecular biology: Escherichia coli and Salmonella. 2nd ed. Washington, DC.: American Society for Microbiology; 1996. pp. 1232–1245. [Google Scholar]
- Gunasekera A, Ebright YW, Ebright RH. DNA sequence determinants for binding of the Escherichia coli catabolite gene activator protein. J Biol Chem. 1992;267:14713–14720. [PubMed] [Google Scholar]
- Gutell PR, Power A, Hertz GZ, Putz E, Stormo GD. Identifying constraints on the higher-order structure of RNA: Continued development and application of comparative analysis methods. Nucleic Acids Res. 1992;20:5785–5795. doi: 10.1093/nar/20.21.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori T, Takahashi K, Nakanishi T, Ohta H, Fukui K, Taniguchi S, Takigawa M. Novel FNR homologs identified in four representative oral facultative anaerobes: Capnocytophaga Ochracea, Capnocytophaga sputigena, Haemophilus aphrophilus, and Actinobacillus actinomycetemcomitans. FEMS Microbiol Lett. 1996;137:213–220. doi: 10.1111/j.1574-6968.1996.tb08108.x. [DOI] [PubMed] [Google Scholar]
- Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- Hertz GZ, Hartzell GW, III, Stormo GD. Identification of consensus patterns in unaligned DNA sequences known to be functionally related. Comput Appl Biosci. 1990;6:81–92. doi: 10.1093/bioinformatics/6.2.81. [DOI] [PubMed] [Google Scholar]
- Himmelreich R, Plagens H, Hilbert H, Reiner B, Herrmann R. Comparative analysis of the genomes of the bacteria Mycoplasma pneumoniae and Mycoplasma genitalium. Nucleic Acids Res. 1997;25:701–712. doi: 10.1093/nar/25.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlavacek MS, Savageau MA. Rules for coupled expression of regulator and effector genes in inducible circuits. J Mol Biol. 1996;255:121–139. doi: 10.1006/jmbi.1996.0011. [DOI] [PubMed] [Google Scholar]
- Huynen MA, Bork P. Measuring genome evolution. Proc Natl Acad Sci USA. 1998;95:5849–5856. doi: 10.1073/pnas.95.11.5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings MP, Beacham IR. Co-dependent positive regulation of the ansB promoter of Escherichia coli by CRP and the FNR protein: A molecular analysis. Mol Microbiol. 1993;9:155–164. doi: 10.1111/j.1365-2958.1993.tb01677.x. [DOI] [PubMed] [Google Scholar]
- Kolb A, Busby S, Buc H, Garges S, Adhya S. Transcriptional regulation by cAMP and its receptor protein. Annu Rev Biochem. 1993;62:749–795. doi: 10.1146/annurev.bi.62.070193.003533. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Galperin MY. Prokaryotic genomes: The emerging paradigm of genome-based microbiology. Curr Opin Genet Dev. 1997;7:757–763. doi: 10.1016/s0959-437x(97)80037-8. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Tatusov RL, Galperin MY. Beyond complete genomes: From sequence to structure and function. Curr Opin Struct Biol. 1998;8:355–363. doi: 10.1016/s0959-440x(98)80070-5. [DOI] [PubMed] [Google Scholar]
- McGuire AM, Hughes JD, Church GM. Conservation of DNA regulatory motifs and discovery of new motifs in microbial genomes. Genome Res. 2000;10:744–757. doi: 10.1101/gr.10.6.744. [DOI] [PubMed] [Google Scholar]
- Mironov AA, Koonin EV, Roytberg MA, Gelfand MS. Computer analysis of transcription regulatory patterns in completely sequenced bacterial genomes. Nucleic Acids Res. 1999;27:2981–2989. doi: 10.1093/nar/27.14.2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Hagelsieb, G., Trevino, V., Perez-Rueda, E., Smith, T.F., and Collado-Vides, J. 2001. Transcription unit conservation in the three domains of life: A perspective from Escherichia coli. Trends Genet. (in press). [DOI] [PubMed]
- Overbeek R, Fonstein M, D'Souza M, Pusch GD, Maltsev N. The use of gene clusters to infer functional coupling. Proc Natl Acad Sci. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson G, Wilson C, Gunasekera A, Ebright YW, Ebright RE, Berman HM. Structure of the CAP-DNA complex at 2.5 A resolution: A complete picture of the protein–DNA interface. J Mol Biol. 1996;260:395–408. doi: 10.1006/jmbi.1996.0409. [DOI] [PubMed] [Google Scholar]
- Perez-Rueda E, Collado-Vides J. The repertoire of DNA-binding transcriptional regulators in Escherichia coli K-12. Nucleic Acids Res. 2000;28:1838–1847. doi: 10.1093/nar/28.8.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M. Functions of the gene products of Escherichia coli. Microbiol Rev. 1993;57:862–952. doi: 10.1128/mr.57.4.862-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M, Labedan B. Escherichia coli gene products: Physiological functions and common ancestries. In: Neidhardt FN, et al., editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC.: American Society for Microbiology; 1996. pp. 2118–2202. [Google Scholar]
- Salgado H, Santos-Zavaleta A, Gama-Castro S, Millán-Zárate D, Blattner FR, Collado-Vides J. RegulonDB (version 3.0): Transcriptional regulation and operon organization in Escherichia coli. Nucleic Acids Res. 2000a;28:65–67. doi: 10.1093/nar/28.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado H, Moreno-Hagelsieb G, Smith TF, Collado-Vides J. Operons in Escherichia coli: Genomic analyses and predictions. Proc Natl Acad Sci. 2000b;97:6652–6657. doi: 10.1073/pnas.110147297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawers G, Kaiser M, Sirko A, Freundlich M. Transcriptional activation by FNR and CRP: Reciprocity of binding site recognition. Mol Microbiol. 1997;23:835–845. doi: 10.1046/j.1365-2958.1997.2811637.x. [DOI] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott S, Busby S, Beacham I. Transcriptional co-activation at the ansB promoters: Involvement of the activating regions of CRP and FNR when bound in tandem. Mol Microbiol. 1995;18:521–531. doi: 10.1111/j.1365-2958.1995.mmi_18030521.x. [DOI] [PubMed] [Google Scholar]
- Song S, Park C. Organization and regulation of the D-xylose operons in Escherichia coli K-12: xylR acts as a transcriptional activator. J Bacteriol. 1997;179:7025–7032. doi: 10.1128/jb.179.22.7025-7032.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiro S, Gaston KL, Bell AI, Robers RE, Busby SJ, Guest JR. Interconvention of the DNA-binding specificities of two related transcription regulators, CRP and FNR. Mol Microbiol. 1990;4:1831–1838. doi: 10.1111/j.1365-2958.1990.tb02031.x. [DOI] [PubMed] [Google Scholar]
- Sumiya M, Davis EO, Packman LC, McDonald TP, Henderson PJF. Molecular genetics of a receptor protein for D-xylose, encoded by the gene xylF in Escherichia coli. Receptors Channels. 1995;3:117–128. [PubMed] [Google Scholar]
- Tatusov RL, Mushegian AR, Bork P, Brown NP, Hayes WS, Borodovsky M, Koonin EV. Curr Biol. 1996;6:279–291. doi: 10.1016/s0960-9822(02)00478-5. [DOI] [PubMed] [Google Scholar]
- Thieffry D, Salgado H, Huerta AM, Collado-Vides J. Prediction of transcriptional regulatory sites in the complete genome sequence of Escherichia coli K-12. Bioinformatics. 1998a;14:391–400. doi: 10.1093/bioinformatics/14.5.391. [DOI] [PubMed] [Google Scholar]
- Thieffry D, Huerta AM, Perez-Rueda E, Collado-Vides J. From specific gene regulation to genomic networks: A global analysis of transcriptional regulation in Escherichia coli. Bioessays. 1998b;20:433–440. doi: 10.1002/(SICI)1521-1878(199805)20:5<433::AID-BIES10>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall L, Christiansen T, Schwartz RL. Programming Perl. Sebastopol, CA: O'Reilly and Associates; 1996. [Google Scholar]
- Yada T, Nakao M, Totoki Y, Nakai K. Modeling and predicting transcription units of Escherichia coli genes using hidden Mokov models. Bioinformatics. 1999;15:987–993. doi: 10.1093/bioinformatics/15.12.987. [DOI] [PubMed] [Google Scholar]