Abstract
The major function of sperm is the delivery of the paternal genome to the metaphase II oocyte, ensuring transmission of the genetic information to the next generation. For successful fertilization and healthy offspring, sperm DNA must be protected from exogenous insults. This is achieved by packaging the sperm DNA into a condensed protamine-bound form, preceded by the precisely orchestrated removal of histones and intermittent insertion and removal of transition proteins. This remodeling process requires relaxation of supercoiled DNA by transient formation of physiological strand breaks that spermatids, being haploid, cannot repair by homologous recombination. In somatic cells, the presence of DNA strand breaks rapidly induces the formation of poly(ADP-ribose) by nuclear poly(ADP-ribose) polymerases, which in turn facilitates DNA strand break signaling and assembly of DNA repair complexes. We reported earlier that chromatin remodeling steps during spermiogenesis trigger poly(ADP-ribose) (PAR) formation. Here, we show that knockout mice deficient in PARP1, PARG (110-kDa isoform), or both display morphological and functional sperm abnormalities that are dependent on the individual genotypes, including residual DNA strand breaks associated with varying degrees of subfertility. The data presented highlight the importance of PAR metabolism, particularly PARG function, as a prerequisite of proper sperm chromatin quality.
Keywords: chromatin condensation, chromatin remodeling, condensation, gametogenesis, PARG, PARP, PARP1, PARP2, poly(ADP-ribose), poly(ADP-ribose) glycohydrolase, poly(ADP-ribose) polymerase, residual DNA strand breaks, sonication-resistant spermatid nuclei, sperm, spermatid, spermatogenesis, spermiogenesis
Poly(ADP-ribose) metabolism, in particular polymer degradation by poly(ADP-ribose) glycohydrolase, is required for proper sperm chromatin quality.
INTRODUCTION
In humans, incomplete chromatin condensation and the presence of persistent DNA strand breaks indicate a severe reduction of the fertilization potential in mature spermatozoa [1]. Sperm DNA is normally shielded from potentially compromising environmental effects by extremely tight packaging into transcriptionally inactive chromatin associated with protamines linked by covalent disulfite bonds. The chromatin structure of sperm cells is therefore completely different from that of somatic cells [2, 3].
The complex chromatin reorganization during spermiogenesis is dramatic, and the steps that lead to packaging of the paternal genome into the compact transport form are not entirely understood. Histones are first replaced by the transition proteins TNP1 and TNP2, which are subsequently replaced by protamines. Along with the exchange of proteins, the DNA structure changes from the supercoiled nucleosomal structure to a relaxed state [4–6]. This requires the occurrence of controlled transient DNA strand breaks [7–9], particularly during the exchange of histones by transition proteins in elongating spermatids such as spermatids of steps 9–12 in mice [10]. Such DNA strand breaks are apparently mostly afforded by the activity of enzymes such as topoisomerase II β (TOP2B) [11, 12] and are therefore presumably mostly protein bound. However, at a certain frequency, open DNA double-strand breaks appear to be also formed in this process, posing a threat to the developing spermatid cell.
Compared with somatic cells, the presence of DNA strand breaks carries a greater risk for haploid spermatid cells, as they are not able to undergo homologous recombination, the favored and most accurate way of repairing DNA double-strand breaks in diploid cells. Cellular DNA damage repair is facilitated by two members of the poly(ADP-ribose) polymerase family, poly(ADP-ribose) polymerase 1 (PARP1) and poly(ADP-ribose) polymerase 2 (PARP2). DNA strand breaks directly and immediately recruit the ubiquitous PARP1 and PARP2. On activation, PARP1 forms a large branched biopolymer poly(ADP-ribose) (PAR) by cleaving NAD+ into nicotinamide and ADP-ribose, which becomes polymerized and covalently attached to acceptor proteins such as PARP1 itself in an intermolecular automodification reaction. Examples of other target proteins for this posttranslational modification are histones, p53, and DNA ligase III. Breakdown of the unique polymer is facilitated by poly(ADP-ribose) glycohydrolase (PARG), with PAR turnover being a rapid and dynamic process mediated by the interplay of PARP1 and PARG [13–18]. Automodified PARP1 is catalytically inactive and is released from the DNA strand break. PARP2, another member of the PARP protein family, has been shown to respond similarly and as a backup. The ability of PARP1 and PARP2 to bind to DNA strand breaks and to become activated again is restored on removal of PAR by PARG, allowing for repeated cycles of PAR turnover until the DNA strand break is repaired. Digestion of PAR by PARG is essential to cellular survival, and a complete knockout of Parg leads to an early embryonic lethal phenotype [19].
In humans, incomplete chromatin condensation and the presence of persistent DNA strand breaks in mature spermatozoa indicate a severe reduction of the fertilization potential [1]. Lack of sperm nuclear integrity often results from incomplete execution of preceding chromatin remodeling steps and from faulty DNA strand break management in early steps of spermatid development. We showed previously that the nuclear elongation phase of spermatid development involves formation of PAR concomitantly with the presence of DNA strand breaks [20].
In the present study, we tested the hypothesis that PAR metabolism may be required for efficient DNA strand break-mediated chromatin reorganization during postmeiotic germ cell maturation using Parp1−/− and Parg(110)−/− mouse models, as well as a novel double gene-disrupted Parp1−/−/Parg(110)−/− strain. Our studies revealed that mice with a perturbation in PAR metabolism display sperm nuclear abnormalities such as alterations in shaping of the nucleus and in the degree and course of nuclear condensation, as well as an increase in the amount of residual DNA strand breaks in epididymal sperm.
MATERIALS AND METHODS
Mouse Strains and Tissue Harvesting
Parp1 gene-disrupted mice (Parp1tm1Zqw [Parp1−/−] [21]) and Parg gene-disrupted mice (Pargtm1Zqw [Parg(110)−/−] [22]) were maintained according to the guidelines of the University of Pennsylvania Institutional Animal Care and Use Committee. Parg(110)−/− mice carry a targeted deletion of exons 2 and 3 of the Parg gene, which leads to abrogation of the three large PARG protein isoforms of 110 kDa, 102 kDa, and 98 kDa, but the two smaller ones of 63 kDa (ubiquitous) and 58 kDa (mitochondrial) [23, 24] are still expressed. These animals exhibit increased PAR formation with increased NAD+ depletion after genotoxic insults, but at the same time steady-state levels of the biopolymer are altered, with an overall depression of cellular PARP1 function [25]. The novel Parp1−/−/Parg(110)−/− double gene-disrupted mice were generated by conventional crossbreeding of the Parg(110)−/− with Parp1−/− mice that were originally in a 129/S1 background (The Jackson Laboratory). Parp1−/−/Parg(110)−/− double-knockout mice were then crossbred with 129/S6/SvEvTac mice (Taconics) to obtain Parp1−/− and Parg(110)−/− mice and wild-type control animals with a comparable background. All animals used in the investigations were routinely genotyped by PCR using primer sequences for Parp1 published by Wang et al. [21] and according to The Jackson Laboratory protocol and using primer sequences for Parg(110) published by Cortes et al. [22]. Wherever possible, sibling wild-type control animals were used in the experiments; in all other cases, 129/S6/SvEvTac (Taconics) mice were utilized as appropriate controls. Experimental procedures were performed by simultaneously analyzing tissue from age-matched animals of all four genotypes.
Computer-Assisted Sperm Analyses
Computer-assisted sperm analyses (CASAs) were performed using an automated system (Hamilton-Thorne Research Systems). For comparison of mouse strains, siblings or age-matched (within 2–3 days) sets of 1–5 animals among 60- to 120-day-old mice were simultaneously used in each experiment. Cauda epididymes were quickly prepared from euthanized mice, and sperm were collected by gently swirling sliced epididymes for 5 min in TYH medium (120 mM NaCl, 4.7 mM KCl, 1.7 mM CaCl2, 1.2 mM KH2PO4, 1.2 mM MgSO4, 25 mM NaHCO3, 5.5 mM glucose, 0.5 mM sodium pyruvate, and 4 mg/ml bovine serum albumin [BSA]) (Sigma Aldrich). Measurements were repeated six times per sample, and all experiments were performed at least in triplicates.
Morphometric Analyses of Sperm Nuclei
Morphometric analyses of cauda epididymal sperm were performed on air-dried sperm smears that were fixed in methanol:glacial acetic acid (3:1) and then embedded in Vectashield mounting medium (Vector Laboratories) with 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) (Sigma Aldrich). Fluorescence microscopy was performed on an inverted fluorescence microscope (Nikon). Random photographs were taken in the DAPI channel using identical exposure conditions, and data were collected using automated and manual data collection modes provided by Image-Pro plus 5.1 software (Media Cybernetics). Minor and major axes, area, perimeter, and mean density of at least 750 sperm nuclei per data set per mouse strain were measured using densitometric functions of this software. First, a polygonic line following the exact perimeter of each nucleus, including the hooked tip, was drawn automatically based on standardized background correction and signal threshold parameters that were kept constant throughout the experiments. Then, minor and major axes were defined as the longest and shortest possible diagonal lines (Axismin and Axismaj, respectively) through each marked nucleus. The elongation of nuclei was computed as the quotient of minor axis / major axis (Axismin / Axismaj), where Axismin / Axismaj = 1 describes a perfectly round nucleus. Wild-type mouse sperm were typically measured as having an elongation of ∼0.5. To provide an additional control, roundness of nuclei was also determined by dividing the expected perimeter (Pexpected) of a nucleus with a given area by the actual perimeter (Pactual). If Pexpected / Pactual = 1, a nucleus would be perfectly round. Typical mean values in wild-type mice were ∼1.32. The mean density, which describes the overall brightness of fluorescence of a given nucleus, was also determined. The result of this measurement reflects the absolute value of optical extinction over a defined area. The mean density was measured as an additional estimate of nuclear compaction. Investigations were designed so that samples from all mouse strains were collected, processed, and evaluated in parallel using age-matched animals (as in all other investigations presented herein) to minimize experimental error. Statistical data analyses were calculated using Excel (Microsoft).
Immunoblot Analyses
Protein extracts used for SDS-PAGE were obtained by collecting testes from several mice each per strain, where all mice of all strains used for a given experiment were of the same age. For SDS protein extraction of whole testes, decapsulated testes were weighed and then homogenized in an equal volume of 2× Laemmli sample buffer. Protein extracts from sonication-resistant nuclei for SDS-PAGE were prepared by homogenization of four testes in 5 ml SMTTP buffer (0.31 M sucrose, 5 mM MgCl2, 10 mM Tris/HCl [pH 7.5], 0.05% Triton X-100, and 0.5 mM PMSF) in a tight-fitting 15-ml glass homogenizer. After filtration through a 0.1-mm nylon mesh and centrifugation at 3000 × g, pellets were resuspended in 10 ml MTMP buffer (5 mM MgCl2, 10 mM Tris/HCl [pH 7.5], 0.2% beta-mercaptoethanol, and 0.5 mM PMSF) and centrifuged. Nuclei were resuspended in 8 ml water and were sonicated twice for 1 min at amplitude 100, with intermittent cooling on ice for 1 min using an ultrasonic processor (Cole-Parmer). They were centrifuged, resuspended in 2 ml water, layered over 2 ml SMP (5% sucrose, 5 mM MgCl2, and 10 mM Tris/HCl [pH 6.5]), and centrifuged at 740 × g for 5 min as previously described [26]. After resuspending the pellet in 2 ml water, this purification step was repeated, and then nuclei resistant to ultrasonic shear were diluted and counted using a hemacytometer. Samples were lysed in an appropriate volume of 2× Laemmli sample buffer to give a final concentration of 6.7 × 104 nuclei/μl. Cauda epididymes were obtained by necropsy, and sperm cells were allowed to swim out of the sliced tissue in 0.75 ml/epididymis prewarmed PBS. Spermatozoa were washed by centrifugation (700 × g), resuspended in PBS, and then pelleted and lysed in 2× SDS Laemmli sample buffer at a final concentration of 2 × 104 cells/μl. An equivalent of 2 × 105 cells/lane was separated by 8% or 10% SDS-PAGE, electrotransferred to polyvinylidene fluoride membranes (Immobilon-P; Millipore), and subjected to antibody detection. Primary antibodies used for immunoblot detection were rabbit anti-PARP1 (SA-253, 1:500; Biomol), rabbit anti-PAR (LP96–10, 1:2000; BD Bioscience), rabbit anti-PARG (C-terminal antigen, 1:500) [23], rabbit anti-PARP2 (Yuc unpurified, 1:2000; Alexis), mouse anti-phospho-H2AFX (1:500; Upstate), mouse anti-H2AFX (1:1000; R&D Systems), and mouse anti-beta-actin (1:5000; Sigma Aldrich). For Western blot detection, horseradish peroxidase-coupled donkey anti-rabbit, donkey anti-mouse, or donkey anti-goat secondary antibodies (Jackson Immunolabs) were used and were detected in a standard enhanced chemiluminescence reaction. Signals were quantified using ImageJ software (Wayne Rasband, National Institutes of Health; http://rsb.info.nih.gov/ij/). In general, quantification was performed by normalizing the signal intensities observed for samples of different genotypes to the signal intensity observed for the respective wild-type sample. In Figure 3, PAR signal intensities were compared between wild-type and Parg(110)−/− animals, as well as between Parp1−/− and Parp1−/−/Parg(110)−/− animals. This procedure adjusts for the different composition in nuclear PARP enzymes in these animals (being PARP1 and PARP2 in the first data set and only PARP2 in the second data set) and allows for comparison of PAR accumulation that is dependent on PARG activity only.
Flow Cytometry
Sperm chromatin integrity assays [27] were used as an estimate of sperm nuclear integrity in direct comparisons of the investigated mouse genotypes. This test is based on the metachromatic features of acridine orange (AO), where fluorescence is green after intercalation into double-stranded DNA chromatin but is red in a complex with single-stranded DNA or RNA. If DNA integrity is compromised (e.g., by the presence of strand breaks), sperm nuclei become sensitive to acid denaturation, causing red fluorescence. A total of 20 000 sperm cells per sample was analyzed using a FACScan flow cytometer (BD Biosciences) in combination with Cell Quest Pro software 5.2 for raw data analysis. Results were expressed as DNA fragmentation index (i.e., the ratio of red:[red + green] fluorescence) and as the percentage of cells with intense green fluorescence or high DNA stainability as an indication of low sperm DNA compaction [27].
RESULTS
Parg(110)−/− Mice Have Reduced Fertility
Parg(110)−/−, but not Parp1−/−, mice exhibited a mild but highly significant (P < 0.0001) subfertility phenotype, with mean ± SD reduced litter sizes of 4.2 ± 2.1 compared with 5.7 ± 2.1 in 129SVE wild-type animals of (Table 1). All animals were routinely removed from the breeding program at age 6 mo, after which fertility may decline naturally in mice, and were replaced by 7- to 8-wk-old animals. Unexpectedly, in Parp1−/−/Parg(110)−/− double gene-disrupted mice, reduced fecundity was not observed in the F1 and F2 generations, and breeding data averages were statistically no different from those of the wild type.
TABLE 1.
Fecundity of wild-type (Wt), Parp1−/−, Parg(110)−/−, and Parp1−/−/Parg(110)−/− mice.
Parg(110)−/− Animals Exhibit No Reduction in Sperm Motility
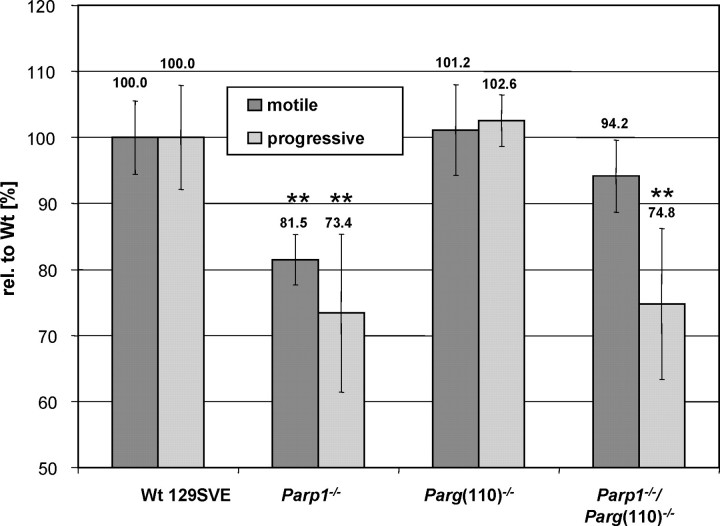
To investigate the reason for reduced fertility of Parg(110)−/− mice, physiological sperm parameters of mouse strains were tested using a CASA system (Fig. 1). While the mean ± SEM percentages of motility spermatozoa and progressive motility spermatozoa isolated from the cauda epididymis were comparable to those of the wild type (with typical motile sperm fractions being ∼55%–60% in individual experimental days) in Parg(110)−/− mice (101.2% ± 6.9% relative to the wild type), Parp1−/− mice had consistently and significantly (P < 0.001) lower percentages of motile sperm (81.5% ± 3.8% relative to the wild type) and fewer progressively motile sperm among those (73.4% ± 11.9% relative to the wild type). Sperm from Parp1−/−/Parg(110)−/− double gene-disrupted animals had almost normal mean ± SEM fractions of motile cells (94.2% ± 5.5% relative to the wild type), but progressiveness of motile sperm was impaired at a level that was statistically comparable to that of Parp1−/− mice (74.8% ± 11.4%). This result is puzzling and indicates that PARP1 may also have a previously unrecognized role in sperm cell progressive motility.
FIG. 1.
Reduction of sperm motility in Parp1−/− mice. Cauda epididymal spermatozoa from age-matched sets of animals were measured by CASA. Motile sperm characterized as progressive were each recorded as percentage of the motile population, and overall motility and progressiveness were expressed relative to the wild type (rel. to wt) in each experiment. Data represent five independent experiments involving 3–5 animals each. Error bars represent the SEM of three independent experiments. ** P < 0.001; wt, wild type.
Expression of Short PARG Isoforms and Cleavage of PARP2 and PARP1 Are Hallmarks of Late Spermiogenesis
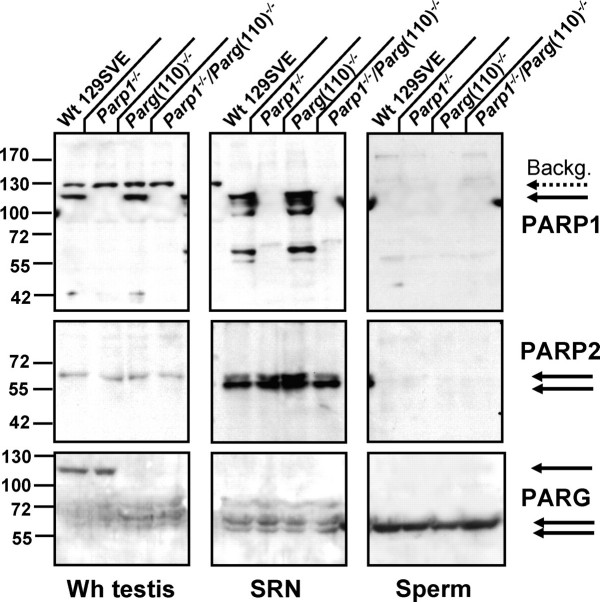
The expression of three major enzymes involved in PAR metabolism in spermatogenesis (PARP1, PARP2, and PARG) was compared in lysates of whole testes, lysates of sonication-resistant nuclei (equivalent to the fraction of spermatids in late steps of spermiogenesis), or lysates of cauda epididymal sperm (Fig. 2). Immunoblotting of SDS-soluble proteins confirmed the absence of PARP1 expression in Parp1−/− and Parp1−/−/Parg(110)−/− double-knockout mice (Fig. 2, upper panel). (Note that the upper band is an unspecific signal always produced by the primary antibody recognizing an unidentified protein that is lost during spermiogenesis.) It also revealed that in wild-type control animals PARP1 is still present in the sonication-resistant nuclei (SRN). The SRN contains testicular spermatids that should have completed nuclear elongation but are not yet fully condensed. At this stage, a large portion of the PARP1 enzyme is present in a cleaved form, reminiscent of the caspase 3-dependent PARP1 cleavage during apoptosis [28] (Fig. 2, top middle panel). However, there was no indication of ongoing apoptosis in spermatid cells as analyzed by TUNEL assays (data not shown). Apoptotic PARP1 cleavage yields fragments of 89 kDa and 24 kDa, and only the 89-kDa fragment is recognized by the antibody used. Epididymal sperm seemed to be devoid of PARP1 enzyme (Fig. 2, top right panel).
FIG. 2.
PARP1, PARP2, and PARG expression in testis. The SDS extracts of whole testis (Wh testis, left column), enriched SRN (middle column), and sperm (right column) were analyzed in Parp1−/−, Parg(110)−/−, and Parp1−/−/Parg(110)−/− double gene-disrupted mice and in wild-type littermates (Wt 129SVE) using identical starting tissue fresh weight (Wh testis), identical numbers of isolated nuclei (SRN), or sperm cells. Note that the 130-kDa band visible in PARP1 (113 kDa) immunoblots is a background artifact of the primary antibody (dotted arrow, Backg.). The three PARG bands represent at least five isoforms expressed from alternatively spliced transcripts of the Parg gene as follows: an unresolved triple band cluster of full-length PARG(110), PARG(101), and PARG(98) (upper arrow); PARG(63) (middle arrow); and PARG(58) (lower arrow).
PARP2 was detected at low levels in whole-testis lysates but seemed to be highly expressed in SRN (Fig. 2, middle panel). A double band (arrows) indicates that PARP2 is modified or cleaved in a fashion similar to PARP1. The observed size differences resulting in a double band could be due to acetylation, as has been recently described for PARP2 [29]. Acetylation leads to a slight upward shift of the PARP2 protein band. This indicates that the majority of PARP2 protein in the whole-testis lysates, as well as the PARP2 comprising the upper part of the double band in SRN, is modified, while the lower band represents unmodified protein. Alternatively, the upper band could consist of unmodified PARP2 protein, while the lower band represents an as yet undescribed cleavage product. Again, spermatozoa seem to possess either nondetectable amounts or no PARP2 (Fig. 2, middle right panel).
As expected, expression of at least one large PARG isoform (110 kDa, 101 kDa, or 98 kDa) was detected in wild-type control and Parp1−/− whole-testis lysates but not in Parg(110)−/− or Parp1−/−/Parg(110)−/− mice (Fig. 2, bottom panel). While all genotypes are expected to express endogenous 63-kDa and 58-kDa isoforms of PARG, extracts from Parg(110)−/− testes seem to have higher amounts of these smaller proteins, possibly compensating for the loss of PARG(110). Most important, no large PARG isoforms were detectable in SRN (Fig. 2, bottom right panel) of wild-type control and Parp1−/− mice, indicating that the processes leading to the observed phenotypic effects in Parg(110) deletion mutants occur before nuclei become sonication resistant. Arguably, PARG(110) could have been selectively lost or degraded through the sonication procedure; however, reprobing of the blot using a PARG N-terminus-specific polyclonal serum did not show any fragments that would support this view (data not shown), so that short-enzyme isoforms that were observed likely originate in alternative splicing of Parg transcripts. Surprisingly, a single small PARG isoform remains as an unexpected component of epididymal sperm (Fig. 2, bottom right panel) in all mouse strains tested.
Poly(ADP-Ribosyl)ation of Testicular Proteins Varies Among the Parp1 and Parg Mutant Mice
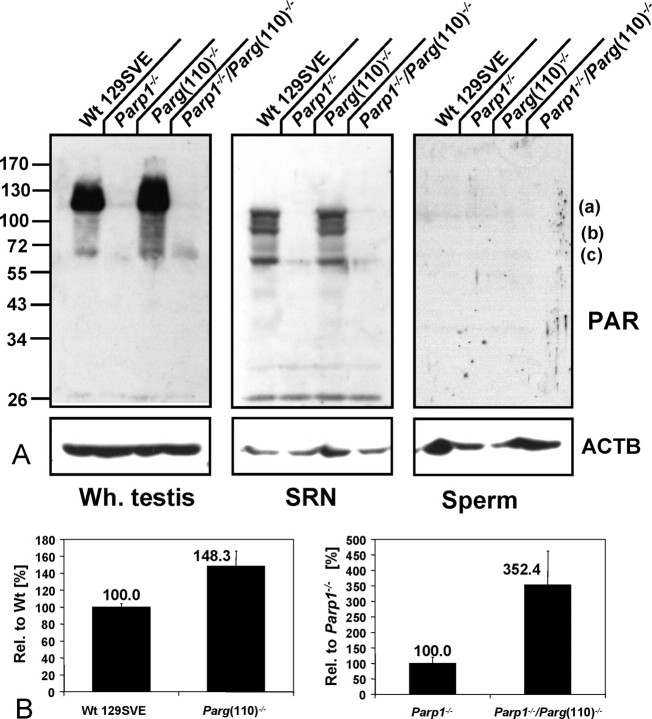
PAR was detected in the immunoblot analyses as a polymer presumably attached to target proteins, mainly PARP1 and PARP2 (Fig. 3A). A significant amount of testicular proteins seemed to be poly(ADP-ribosyl)ated in the wild type, with an overall ∼48% increase in the Parg(110)−/− mouse (Fig. 3, A and B, left panels). In contrast, in Parp1−/− testes very little PAR was detected, presumably produced by PARP2 (lane 2), as judged from the molecular weight of the observed band. This PARP2-dependent PAR formation was increased ∼3.5-fold in Parp1−/−/Parg(110)−/− double-knockout mice (Fig. 3, A [left panel, upward “smear” in lane 4] and B [right panel]). This indicates that PARP2 activity is significantly altered in Parg(110)−/− and Parp1−/−/Parg(110)−/− double-knockout mice compared with the wild type. In SRN, immunoblotting PAR signals mainly seemed to overlap with the PARP1 protein pattern and therefore point to short polymer lengths. The fact that PAR formation appeared to be comparable in SRN of the wild type and the Parg(110)−/− mutant supports the notion that large PARG isoforms may indeed already be absent at this stage. Faint PAR signals between 55 kDa and 72 kDa indicate the presence of automodified PARP2 in Parp1−/− and Parp1−/−/Parg(110)−/− SRN (Fig. 3A, middle panel, lanes 2 and 4). Redetection of the blot using PARP1- and PARP2-specific antibodies (similar to Fig. 2) show that labeled bands (a, b, and c) all correspond to PARP1 isoforms absent in the Parp1−/− mouse. Band c comprises both PARP1 and PARP2 proteins in wild-type and Parg(110)−/− mice but PARP2 only in Parp1−/− and Parp1−/−/Parg(110)−/− mice (data not shown). PAR was not detectable in epididymal sperm samples of either genotype (Fig. 3A, right panel).
FIG. 3.
Parg gene disruption results in increased PAR accumulation. A) Immunoblot analyses of whole testis (Wh. testis), SRN, and sperm using a PAR-specific antibody to detect poly(ADP-ribosyl)ated proteins. Parg(110)−/− mice showed increased amounts of PARP1 and PARP2 automodification and other poly(ADP-ribosyl)ated proteins in whole-testis lysates. Parp1−/− and Parp1−/−/Parg(110)−/− showed increased PARP2 automodification only. B) Quantification of PAR bands in blotted lysates of whole testes (such as on the left side of A) showing the mean ± SD of three independent experiments. To allow for comparison of PAR accumulation that is dependent on PARG activity, PAR signal intensities in genotypes containing the same set of PARP enzymes were quantified and compared. In the left panel, PAR signals seen in wild-type and Parg(110)−/− animals (both contain PARP1 and PARP2 enzyme) are compared. In the right panel, PAR signal intensities observed in Parp1−/− and Parp1−/−/Parg(110)−/− (both are devoid of PARP1 and contain only PARP2 as a DNA damage-dependent PARP enzyme) are compared. Overall, Parg(110)−/− showed ∼1.5-fold increase in PAR formation compared with the wild type (left), and PARP2-dependent PAR formation was increased ∼3.5-fold in Parp1−/−/Parg(110)−/− mice compared with Parp1−/− (right). Units for the data to the left of the gels are kilodaltons. Error bars represent the SEM of three independent experiments.
Abnormally Reduced Nuclear Elongation and Chromatin Condensation in Parp1−/−, Parg(110)−/−, and Parp1−/−/Parg(110)−/− Epididymal Spermatozoa
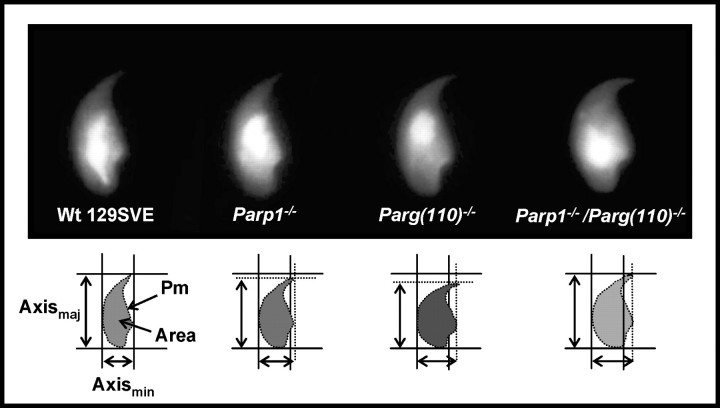
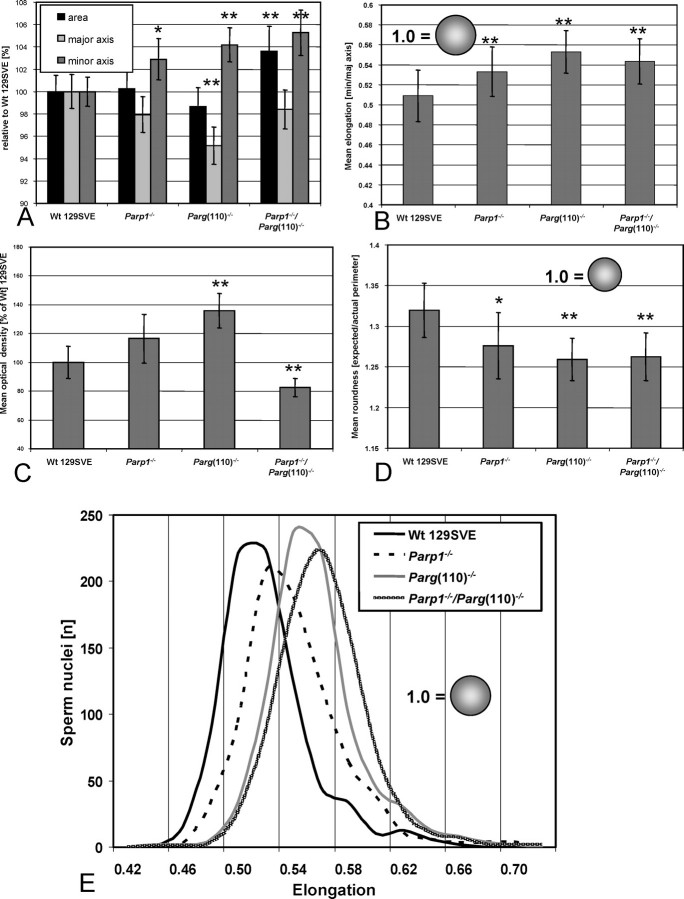
Because motility measurements of sperm cells in the CASA system initially also indicated an elongation defect in live Parp1−/− and Parg(110)−/− sperm heads (data not shown), further systematic morphometric measurements of fixed sperm nuclei were performed. These analyses revealed that Parp1−/−, Parg(110)−/−, and Parp1−/−/Parg(110)−/− mice all had a varying degree of abnormal shaping of sperm nuclei (Figs. 4 and 5). While some individual variability of sperm nuclei within a semen sample can be anticipated, analyses showed a subtle but highly significant progressive loss of nuclear elongation (i.e., increasing roundness) in Parp1−/−, Parg(110)−/−, and Parp1−/−/Parg(110)−/− sperm (Fig. 4, lower panel) compared with the wild type. Measurement of the minimal and maximal axes and the two-dimensionally projected area of large numbers of nuclei confirmed the notion that Parp1−/−, Parg(110)−/−, and Parp1−/−/Parg(110)−/− nuclei were altogether shorter and wider than those in the wild type (Fig. 5A) with high significance (P < 0.0001). Compared with the wild type, the mean area was measurably smaller in Parg(110)−/− nuclei but was larger in the Parp1−/−/Parg(110)−/− double gene-disrupted genotype (Fig. 5A). When calculated as the mean nuclear elongation, sperm nuclei from all genotypes were indeed found to be rounder than those in the wild type (Fig. 5B), suggesting a defect in spermatid elongation during developmental steps 9–12. The mean optical density is increased in Parp1−/− and Parg(110)−/− nuclei, which is consistent with the two-dimensional optical projection of more spherical objects (Fig. 5C), similar to the area measurements. In contrast, combined data from measurements of the mean density (Fig. 5C) and optical area (Fig. 5A) revealed that the Parp1−/−/Parg(110)−/− double gene disruption also leads to the formation of altogether larger, less condensed, and rounder nuclei, indicative of interference of the gene deletions with chromatin condensation. A different method of determining nuclear elongation based on area and perimeter measurements (“roundness” [Fig. 5D]) consistently supported the results obtained from elongation measurements. Histogram analyses further confirmed that the observed highly significant (P < 0.0001) differences in elongation in Parp1−/−, Parg(110)−/−, and double-knockout mice were not due to the presence of extremely aberrant subpopulations of nuclei (Fig. 5E).
FIG. 4.
Shaping and condensation of average epididymal sperm are progressively abnormal from Parp1−/− to Parg(110)−/− and particularly Parp1−/−/Parg(110)−/− gene-deleted mice with increased sperm nuclear width (major axes). DAPI-stained nuclei of random images taken from sperm smears of age-matched animals were analyzed for nuclear area, exact perimeter (Pm), length (Axismaj), and width (Axismin). Deviations from normal sperm nuclei shape are indicated by broken lines in the lower panel sketches.
FIG. 5.
Morphometric analyses of increasingly abnormal nuclear shaping in Parp1−/−, Parg(110)−/−, and Parp1−/−/Parg(110)−/− sperm. Parp1−/− sperm were the least different from the wild type but had a significant increase of the nuclear minor axis (A) and resulting greater mean elongation (B) (i.e., overall rounder shape). D) Comparisons of area and perimeter (see Materials and Methods) confirm this notion. C) The mean optical density is significantly increased in Parp1−/− and Parg(110)−/− sperm, which is consistent with the two-dimensional projection of a three-dimensional more rounded object. However, Parp1−/−/Parg(110)−/− nuclei were the exception, with a decreased mean density compared with the wild type. Parg(110)−/− and Parp1−/−/Parg(110)−/− sperm nuclei had significantly shorter major and longer minor axes than the wild type (P < 0.0001), resulting in rounder nuclei. E) Histogram plotting of the individual elongation data, pooled from three independent experiments that each comprised a minimum of 750 nuclei per genotype, demonstrates normal distributions, indicating that the observed differences were not due to extremely abnormal sperm subpopulations. Error bars represent the SD. * Significantly different from Wt 129SVE control; ** highly significantly different from WT 129SVE control.
PAR Metabolism Affects H2AFX Phosphorylation During Spermiogenesis
Because PARP1 and PARG are intimately involved in DNA strand break signaling and repair, DNA integrity was evaluated by studying H2AFX phosphorylation as a marker for the presence of DNA strand breaks using immunoblot analyses. H2AFX is a variant histone of the H2A group that is phosphorylated in the presence of DNA strand breaks and as part of the heterochromatic XY body during meiosis [30]. Owing to its functions in sex chromatin silencing and consequently its high abundance during meiosis, no major differences in H2AFX content or phosphorylation between genotypes were detectable in whole-testis extracts (Fig. 6, top two panels). We isolated and extracted SRN, which normally correspond to testicular spermatids of developmental steps 12 and later. Testicular spermatids isolated in such a way showed a small increase of γ-H2AFX detected in Parg(110)−/− compared with the wild type (Fig. 6, middle panels). Overall H2AFX protein content was reduced to 50%–60% in the Parg(110)−/−, suggesting that a comparatively greater portion of the H2AFX present in SRN of these mice was phosphorylated, which in turn indicates significantly elevated levels of DNA strand breaks. Comparative immunofluorescence analyses of testes paraffin sections did not show any significant differences of γ-H2AFX signals in stage 12 tubules of the various mouse genotypes (data not shown). While the amount of detectable pan H2AFX was not different in sperm of the different genotypes, Parg(110)−/− sperm showed approximately a 2-fold increase in γ-H2AFX compared with the wild type; the mean ± SEM amounts of γ-H2AFX in sperm from animals with Parp1−/− and Parp1−/−/Parg(110)−/− genotypes were reduced to 52% ± 8% and 88% ± 6% (n = 3 for both) of those in the wild type, respectively. PARP1, PAR, and ATM physically interact to mediate H2AFX phosphorylation, and it has been shown that γ-H2AFX formation is delayed and reduced in Parp1−/− fibroblasts [31]. This notion is supported by the results shown in Figure 6, where H2AFX phosphorylation is decreased in Parp1−/− and Parp1−/−/Parg(110)−/− SRN. Therefore, using H2AFX phosphorylation as an indicator for the presence of DNA double-strand breaks could produce false-negative results and underestimate the extent of DNA strand breaks in Parp1−/− and Parp1−/−/Parg(110)−/− double gene-disrupted mice. Therefore, we performed sperm chromatin integrity assays as a sensitive method to test for DNA integrity in sperm cells.
FIG. 6.
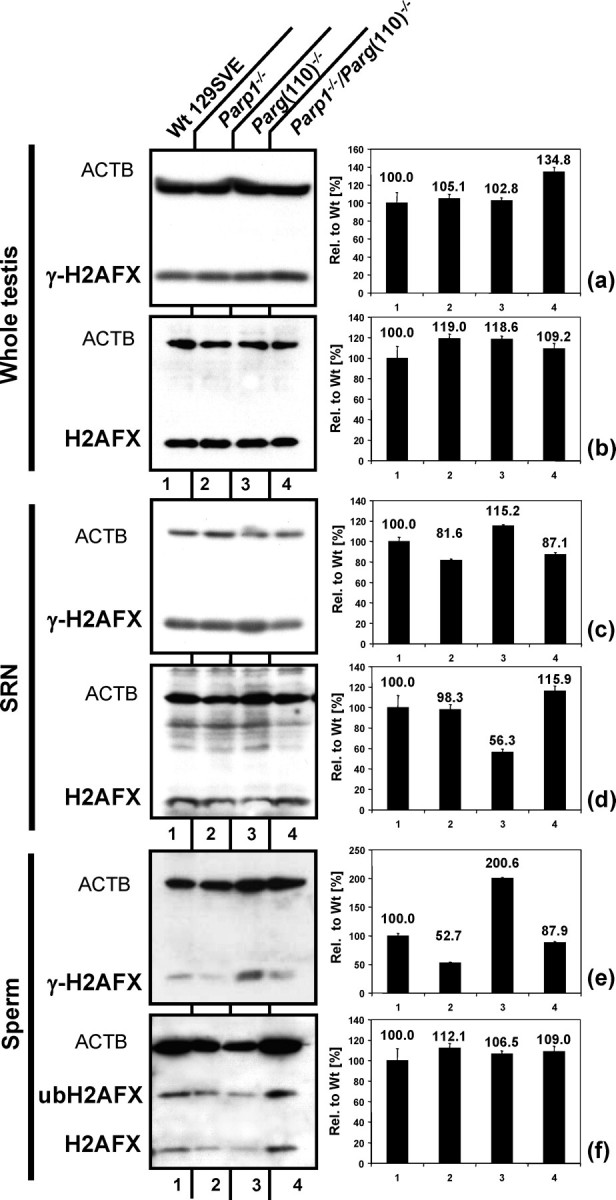
Elevated H2AFX phosphorylation in late developmental steps of Parg(110)−/−spermatids and in sperm. a and b) No major differences in H2AX content or phosphorylation between genotypes were detectable in whole-testis extracts. In SRN, total H2AFX was significantly (P < 0.001 [n = 3]) decreased in Parg(110)−/− (d), indicating that nuclei perhaps become sonication resistant later in development compared with the wild type, but γ-H2AFX was not reduced (c), indicating that a larger portion of H2AFX was phosphorylated in this strain in SRN. This was reflected by the situation in sperm, where H2AFX contents in all genotypes were similar (f), but the amount of γ-H2AFX was ∼2-fold in Parg(110)−/− (e). f) In sperm, 56%–68% of the total H2AFX was shifted in size, indicative of ubiquitination, and only nonubiquitinated H2AFX was in part phosphorylated. Bars represent the SEM of three independent experiments. ubH2AFX, ubiquitinated H2AFX.
Abrogation of PARG(110) Expression Results in Reduced Sperm Chromatin Integrity
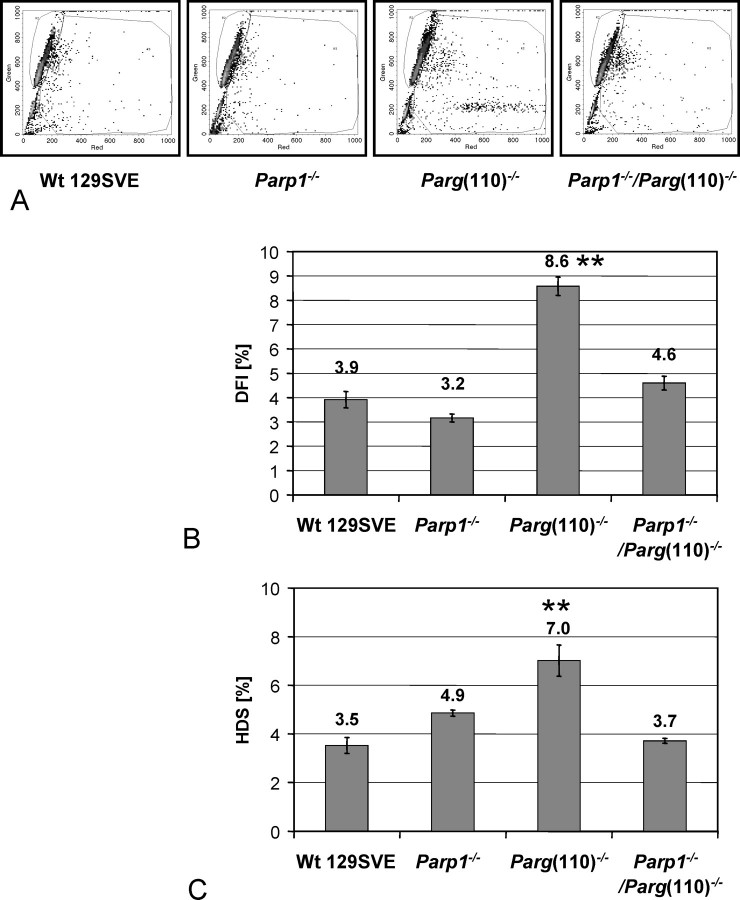
To investigate if abnormal H2AFX phosphorylation caused by the genetic alteration of PAR metabolism indeed correlates with reduced chromatin integrity, we performed flow cytometry-based sperm chromatin integrity assays. As predicted by the finding that γ-H2AFX is increased in Parg(110)−/− spermatozoa, an elevated sensitivity of sperm nuclei toward acid denaturation was observed in this strain. The mean ± SD increase in DNA fragmentation index was mild but significant (8.6% ± 0.4%) compared with that in wild-type littermates (3.9% ± 0.3%) (P < 0.001) (Fig. 7, A and B). The denaturing acid sensitivity of Parp1−/−/Parg(110)−/− sperm nuclei (mean ± SD, 4.6% ± 0.3%) was intermediate between Parg(110)−/− nuclei and the wild-type or Parp1−/− nuclei (mean ± SD, 2.8% ± 0.2%), with the wild-type and Parp1−/− nuclei being statistically indistinguishable from each other (Fig. 7B). This result indicated that H2AFX phosphorylation (Fig. 6) correlated well with actual DNA strand break frequencies. Therefore, γ-H2AFX formation was likely not selectively impaired in the Parp1-deleted animals, and Western blot results shown in Figure 2 suggest that PARP1 may be absent at the spermatid developmental stage, at which the residual DNA strand breaks originate. Genotype-specific concentrations of nuclei with high DNA stainability are similar to the individual DNA fragmentation index values, with Parg(110)−/− values being approximately twice as high as those in the wild type (Fig. 7C). Intense green fluorescence of AO-stained nuclei, as measured by high DNA stainability, indicates low chromatin compaction in the absence of DNA strand breaks.
FIG. 7.
Sperm chromatin integrity assays indicate poor sperm chromatin quality in Parg(110)−/− mice. A) Acridine orange-based sperm chromatin integrity assays [27] indicate residual DNA strand breaks (red fluorescence after acid denaturation) in Parg(110)−/− but not in Parp1−/−, Parp1−/−/Parg(110)−/−, or wild-type 129SVE siblings (R3 region). B) Data were compared as DNA fragmentation index (DFI). C) A region R2 subpopulation with extremely high green fluorescent stainability (high DNA stainability [HDS]) was computed from the raw data and compared. This subpopulation is characterized by overall low or immature chromatin condensation, allowing for intercalation of the dye. Representative data are shown (n = 4). Bars represent the SD of triplicates in one representative experiment. ** Highly significantly different from Wt 129SVE control.
DISCUSSION
An estimated 15% of all couples are affected by a clinical degree of infertility. In approximately 50% of these couples, successful conception is affected by the male factor [32]. Crucial prerequisites for successful fertilization in healthy individuals and in infertility patients are the degree of chromatin integrity and the frequency of residual DNA strand breaks in mature sperm cells [33–42]. In this study, we show that deletion of large PARG isoforms (110-kDa, 101-kDa, and 98-kDa enzymes) leads to increased testicular levels of PAR, abnormal sperm nuclear shaping that is likely indicative of interference with the spermatid elongation process, a 2-fold increase in sperm DNA strand breaks, and finally a mild subfertility phenotype with significantly reduced litter sizes in the Parg(110)−/− mouse model.
Differentiating spermatids undergo a dramatic change in DNA topology from bulky histone-bound chromatin to compact and metabolically inert protamine-bound DNA. This condensation process is preceded and partially accompanied by the reshaping of the nucleus from the round early spermatid nucleus to the elongated final shape. The elongation phase of differentiating spermatids is marked by a massive occurrence of physiological DNA strand breaks [12], which likely allow for the necessary relaxation of the superspiralized DNA during the DNA topology change. These physiological DNA strand breaks lead to H2AFX phosphorylation, which has been observed in elongating and early condensing spermatids [20, 43]. While the nature of the DNA activity that repairs the occurring DNA strand breaks correctly and to completeness is not yet well understood, we observed that extensive poly(ADP-ribosyl)ation takes place concomitantly with these very steps, indicating that PARsylation participates in the DNA break management [20].
Results of the present study indicate that Parg(110)−/− and the Parp1−/−/Parg(110)−/− mice retain DNA strand breaks in mature sperm, suggesting that PAR metabolism may be necessary for proper safeguarding of sperm cell chromatin integrity during spermiogenesis. Theoretically, the observed DNA strand breaks can be residual strand breaks remaining from the physiologically introduced strand breaks during the extensive DNA remodeling steps during spermiogenesis, or they can be acquired during the passage of the sperm to the epididymis and the subsequent epididymal maturation process, indicating that altered PARsylation leads to more damage-prone sperm with inferior chromatin quality.
Sperm nuclear morphology analyses demonstrated that Parp1−/− mice have less elongated nuclei than the wild type, but this observation was more pronounced in the Parg(110)−/− and Parp1−/−/Parg(110)−/− double gene-disrupted mice (Figs. 4 and 5). This result suggests that chromatin packaging may be impaired by a varying degree among the three mutant mouse strains, with the lowest degree of DNA compaction being present in the double gene-disrupted mouse. PAR metabolism, involving interplay of the PARP1, PARP2, and PARG enzymes, mediates cell recovery from genotoxic stress by a variety of mechanisms [44–46]. Phosphorylation of H2AFX is mediated by ATR and ATM and has an important role in DNA double-strand break signaling involving cofactors such as BRCA1 [47]. Recent findings revealed an involvement of PAR formed by PARP1 and PARP2 in DNA double-strand break-dependent phosphorylation of H2AFX [31]. In that study, downregulation of acute DNA damage-dependent γ-H2AFX formation was demonstrated in Parp1−/− mouse embryonic fibroblasts that could be simulated by treatment with PJ34, a potent inhibitor of PARP enzymes. Consistent with this finding, mouse embryonic fibroblasts derived from Parg(110)−/− mice also show such defects [25]. H2AFX phosphorylation and the percentage of acid-denatured nuclei in the sperm chromatin integrity assay (Fig. 7, A and B) indicate that Parg(110)−/− mice are most severely affected by the presence of residual strand breaks. However, it cannot be ruled out that H2AFX phosphorylation as a response to double-strand breaks may still be regulated independently from PARP1-dependent PAR signaling triggered by DNA single-strand breaks. The morphometry results indicated that, to a certain degree, all genotypes have mildly atypical sperm nuclei that are more round shaped (Figs. 4 and 5), indicating that they may have suboptimal chromatin packaging and quality. Poor chromatin integrity and residual strand breaks, as measured by the sperm chromatin integrity assay, are predictive of lower reproductive performance and specifically of reduced embryonic survival [48, 49], which is consistent with reduced litter sizes observed in the Parg(110)−/− mouse.
H2AFX phosphorylation observed in Parg(110)−/− sperm (Fig. 6e) is another indication of residual DNA strand breaks, but it cannot be determined based on the data presented whether these were newly acquired after the sperm had left the testis or represent unrepaired DNA lesions that stem from DNA remodeling during spermatid elongation. These chromatin reorganization events are accompanied by controlled physiological DNA strand breaks mediated by DNA topoisomerases, with a probable predominant role of TOP2B [12, 43]. Putative lesions caused by stalled topoisomerases comprise DNA double- and single-strand breaks. Most of the DNA single-strand breaks are repaired using the base excision pathway, which has been commonly thought to involve PARP1; however, its exact function there is still under debate [45, 50]. DNA double-strand breaks are repaired by homologous recombination repair or by nonhomologous end joining in somatic cells. However, because spermatids, being haploid, are unable to perform homologous recombination repair, double strands must be repaired by nonhomologous end joining. The latter consists of two alternatively used variant pathways, one involving DNA-dependent protein kinase (default pathway) and another involving the homolog of the yeast protein RAD18 and PARP1 (backup pathway) [51, 52]. Therefore, it is conceivable that DNA damage found in mature sperm may be in the form of residual DNA strand breaks that remained unrepaired owing to functional PARP1 inhibition caused by abnormal automodification of the enzyme in the PARG(110) deletion mutant (Fig. 3A, left panel). The increased levels of poly(ADP-ribosyl)ated PARP are likely caused by the reduced PARG activity in Parg(110)−/− mice [22] in combination with altered control of PARG activity [25] due to the loss of the regulatory A-domain in the mutant. Western blot analyses in the present study showed that elevated levels of poly(ADP-ribosyl)ated proteins in whole-testis extracts of Parg(110)−/− mice (Fig. 3) were not due to increased apoptosis, as indicated by in situ TUNEL assays (data not shown) and by the absence of caspase-dependent PARP1 cleavage. The suggestion that the observed DNA strand breaks are unrepaired residual lesions rather than newly acquired lesions is supported by the finding that SRN and sperm from wild-type mice do not contain PARG(110). This suggests that the chromatin defects resulting from abnormal PARP1/PARG protein composition in sperm from Parg(110)−/− and Parp1−/−/Parg(110)−/− animals must arise at earlier time points (e.g., during or before the elongation phase before chromatin condensation). Because γ-H2AFX formation was only moderately changed in SRN of Parg(110)−/− mice, it seems reasonable to suggest that DNA strand breaks first exist as unrepaired single-strand lesions that, depending on their frequency and spacing along the DNA, later develop into double-strand breaks during further condensation of spermatid nuclei, causing H2AFX phosphorylation in mature spermatozoa.
Arguably, the deletion of PARP1 may have a similarly detrimental effect on DNA repair as its inhibition, but recent evidence shows that inhibition of PARP enzymes (such as by using a small-molecule inhibitor) results in a delay or defect in DNA single-strand break repair that is different from, and often more severe than, the effect caused by genetic ablation of PARP1 enzyme [50]. In addition, PARP2, which has substantially overlapping functions with PARP1, was still expressed in all genetic strains tested in this study, which may also account for the mild phenotypes observed in the investigations presented herein. In a similarly mild fashion, residual DNA strand breaks, compromised chromatin integrity, and decreased embryonic survival were reported after single deletion of either transition protein 1 (TNP1) or TNP2 [26, 53], but if deleted together, the effect was much more dramatic [54]. However, deletion of both Parp1 and Parp2, which are ubiquitously expressed in the mammalian body, was shown to be inconsistent with embryonic survival [55].
In summary, the results of this study strongly indicate that PAR metabolism is involved in proper execution of spermatid maturation. The study demonstrates for the first time (to our knowledge) that partial deficiency in PAR metabolism resulting from deletion of the Parp1 and Parg genes will negatively affect chromatin integrity in spermatid nuclei, as measured by abnormal sperm nuclear shaping, altered H2AFX phosphorylation, and reduced resistance to acid denaturation, particularly in Parg gene-disrupted mice.
Footnotes
1Supported by grants from the National Institutes of Health (NIH R01 HD048837 to R.G.M.) and the Mari Lowe Center for Comparative Oncology at the University of Pennsylvania. Z.-Q.W. was supported by Association pour la Recherche sur le Cancer (ARC) France.
REFERENCES
- Leduc F, Nkoma GB, Boissonneault G. Spermiogenesis and DNA repair: a possible etiology of human infertility and genetic disorders. Syst Biol Reprod Med 2008; 54: 3 10 [DOI] [PubMed] [Google Scholar]
- Braun RE. Packaging paternal chromosomes with protamine. Nat Genet 2001; 28: 10 12 [DOI] [PubMed] [Google Scholar]
- Rousseaux S, Faure AK, Caron C, Lestrat C, Govin J, Hennebicq S, Sele B, Khochbin S. Organizing the sperm nucleus. Gynecol Obstet Fertil 2004; 32: 785 791 [DOI] [PubMed] [Google Scholar]
- Caron C, Govin J, Rousseaux S, Khochbin S. How to pack the genome for a safe trip. Prog Mol Subcell Biol 2005; 38: 65 89 [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi P. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science 2002; 296: 2176 2178 [DOI] [PubMed] [Google Scholar]
- Lewis JD, Abbott DW, Ausio J. A haploid affair: core histone transitions during spermatogenesis. Biochem Cell Biol 2003; 81: 131 140 [DOI] [PubMed] [Google Scholar]
- Boissonneault G. Chromatin remodeling during spermiogenesis: a possible role for the transition proteins in DNA strand break repair. FEBS Lett 2002; 514: 111 114 [DOI] [PubMed] [Google Scholar]
- Risley MS, Einheber S, Bumcrot DA. Changes in DNA topology during spermatogenesis. Chromosoma 1986; 94: 217 227 [DOI] [PubMed] [Google Scholar]
- McPherson S, Longo FJ. Chromatin structure-function alterations during mammalian spermatogenesis: DNA nicking and repair in elongating spermatids. Eur J Histochem 1993; 37: 109 128 [PubMed] [Google Scholar]
- Marcon L, Boissonneault G. Transient DNA strand breaks during mouse and human spermiogenesis: new insights in stage specificity and link to chromatin remodeling. Biol Reprod 2004; 70: 910 918 [DOI] [PubMed] [Google Scholar]
- Laberge RM, Boissonneault G. Chromatin remodeling in spermatids: a sensitive step for the genetic integrity of the male gamete. Arch Androl 2005; 51: 125 133 [DOI] [PubMed] [Google Scholar]
- Laberge RM, Boissonneault G. On the nature and origin of DNA strand breaks in elongating spermatids. Biol Reprod 2005; 73: 289 296 [DOI] [PubMed] [Google Scholar]
- Meyer-Ficca ML, Meyer RG, Jacobson EL, Jacobson MK. Poly(ADP-ribose) polymerases: managing genome stability. Int J Biochem Cell Biol 2005; 37: 920 926 [DOI] [PubMed] [Google Scholar]
- Woodhouse BC, Dianov GL. Poly ADP-ribose polymerase-1: an international molecule of mystery. DNA Repair (Amst) 2008; 7: 1077 1086 [DOI] [PubMed] [Google Scholar]
- Gagne JP, Hendzel MJ, Droit A, Poirier GG. The expanding role of poly(ADP-ribose) metabolism: current challenges and new perspectives. Curr Opin Cell Biol 2006; 18: 145 151 [DOI] [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol 2006; 7: 517 528 [DOI] [PubMed] [Google Scholar]
- Burkle A. Poly(ADP-ribose): the most elaborate metabolite of NAD+. FEBS J 2005; 272: 4576 4589 [DOI] [PubMed] [Google Scholar]
- Meyer RG, Meyer-Ficca ML, Jacobson EL, Jacobson MK. Enzymes in poly(ADP-ribose) metabolism. Burkle A. Poly(ADP-ribosyl)ation. Austin, TX: Landes Bioscience; 2004. [Google Scholar]
- Koh DW, Lawler AM, Poitras MF, Sasaki M, Wattler S, Nehls MC, Stoger T, Poirier GG, Dawson VL, Dawson TM. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc Natl Acad Sci U S A 2004; 101: 17699 17704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Ficca ML, Scherthan H, Burkle A, Meyer RG. Poly(ADP-ribosyl)ation during chromatin remodeling steps in rat spermiogenesis. Chromosoma 2005; 114: 67 74 [DOI] [PubMed] [Google Scholar]
- Wang ZQ, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, Wagner EF. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev 1995; 9: 509 520 [DOI] [PubMed] [Google Scholar]
- Cortes U, Tong WM, Coyle DL, Meyer-Ficca ML, Meyer RG, Petrilli V, Herceg Z, Jacobson EL, Jacobson MK, Wang ZQ. Depletion of the 110-kilodalton isoform of poly(ADP-ribose) glycohydrolase increases sensitivity to genotoxic and endotoxic stress in mice. Mol Cell Biol 2004; 24: 7163 7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Ficca ML, Meyer RG, Coyle DL, Jacobson EL, Jacobson MK. Human poly(ADP-ribose) glycohydrolase is expressed in alternative splice variants yielding isoforms that localize to different cell compartments. Exp Cell Res 2004; 297: 521 532 [DOI] [PubMed] [Google Scholar]
- Meyer RG, Meyer-Ficca ML, Whatcott CJ, Jacobson EL, Jacobson MK. Two small enzyme isoforms mediate mammalian mitochondrial poly(ADP-ribose) glycohydrolase (PARG) activity. Exp Cell Res 2007; 313: 2920 2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Coyle DL, Meyer-Ficca ML, Meyer R, Jacobson EL, Wang ZQ, Jacobson MK. Altered poly(ADP-ribose) metabolism impairs cellular responses to genotoxic stress in a hypomorphic mutant of poly(ADP-ribose) glycohydrolase. Exp Cell Res 2007; 313: 984 996 [DOI] [PubMed] [Google Scholar]
- Yu YE, Zhang Y, Unni E, Shirley CR, Deng JM, Russell LD, Weil MM, Behringer RR, Meistrich ML. Abnormal spermatogenesis and reduced fertility in transition nuclear protein 1-deficient mice. Proc Natl Acad Sci U S A 2000; 97: 4683 4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evenson D, Jost L. Sperm chromatin structure assay is useful for fertility assessment. Methods Cell Sci 2000; 22: 169 189 [DOI] [PubMed] [Google Scholar]
- Lazebnik YA, Kaufmann SH, Desnoyers S, Poirier GG, Earnshaw WC. Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature 1994; 371: 346 347 [DOI] [PubMed] [Google Scholar]
- Haenni SS, Hassa PO, Altmeyer M, Fey M, Imhof R, Hottiger MO. Identification of lysines 36 and 37 of PARP-2 as targets for acetylation and auto-ADP-ribosylation. Int J Biochem Cell Biol 2008; 40: 2274 2283 [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Mahadevaiah SK, Celeste A, Romanienko PJ, Camerini-Otero RD, Bonner WM, Manova K, Burgoyne P, Nussenzweig A. H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev Cell 2003; 4: 497 508 [DOI] [PubMed] [Google Scholar]
- Haince JF, Kozlov S, Dawson VL, Dawson TM, Hendzel MJ, Lavin MF, Poirier GG. Ataxia telangiectasia mutated (ATM) signaling network is modulated by a novel poly(ADP-ribose)-dependent pathway in the early response to DNA-damaging agents. J Biol Chem 2007; 282: 16441 16453 [DOI] [PubMed] [Google Scholar]
- Oehninger S. Strategies for the infertile man. Semin Reprod Med 2001; 19: 231 237 [DOI] [PubMed] [Google Scholar]
- Agarwal A, Said TM. Role of sperm chromatin abnormalities and DNA damage in male infertility. Hum Reprod Update 2003; 9: 331 345 [DOI] [PubMed] [Google Scholar]
- Sakkas D, Mariethoz E, Manicardi G, Bizzaro D, Bianchi PG, Bianchi U. Origin of DNA damage in ejaculated human spermatozoa. Rev Reprod 1999; 4: 31 37 [DOI] [PubMed] [Google Scholar]
- Sakkas D, Urner F, Bizzaro D, Manicardi G, Bianchi PG, Shoukir Y, Campana A. Sperm nuclear DNA damage and altered chromatin structure: effect on fertilization and embryo development. Hum Reprod 1998; 13 (suppl 4): 11 19 [DOI] [PubMed] [Google Scholar]
- Sharma RK, Said T, Agarwal A. Sperm DNA damage and its clinical relevance in assessing reproductive outcome. Asian J Androl 2004; 6: 139 148 [PubMed] [Google Scholar]
- Avendano C, Franchi A, Taylor S, Morshedi M, Bocca S, Oehninger S. Fragmentation of DNA in morphologically normal human spermatozoa. Fertil Steril 2009; 91: 1077 1084 [DOI] [PubMed] [Google Scholar]
- Carrell DT, De Jonge C, Lamb DJ. The genetics of male infertility: a field of study whose time is now. Arch Androl 2006; 52: 269 274 [DOI] [PubMed] [Google Scholar]
- Aitken RJ, De Iuliis GN. Value of DNA integrity assays for fertility evaluation. Soc Reprod Fertil Suppl 2007; 65: 81 92 [PubMed] [Google Scholar]
- Olsen AK, Lindeman B, Wiger R, Duale N, Brunborg G. How do male germ cells handle DNA damage? Toxicol Appl Pharmacol 2005; 207: 521 531 [DOI] [PubMed] [Google Scholar]
- Spano M, Seli E, Bizzaro D, Manicardi GC, Sakkas D. The significance of sperm nuclear DNA strand breaks on reproductive outcome. Curr Opin Obstet Gynecol 2005; 17: 255 260 [DOI] [PubMed] [Google Scholar]
- Sakkas D, Moffatt O, Manicardi GC, Mariethoz E, Tarozzi N, Bizzaro D. Nature of DNA damage in ejaculated human spermatozoa and the possible involvement of apoptosis. Biol Reprod 2002; 66: 1061 1067 [DOI] [PubMed] [Google Scholar]
- Leduc F, Maquennehan V, Nkoma GB, Boissonneault G. DNA damage response during chromatin remodeling in elongating spermatids of mice. Biol Reprod 2008; 78: 324 332 [DOI] [PubMed] [Google Scholar]
- Oei SL, Keil C, Ziegler M. Poly(ADP-ribosylation) and genomic stability. Biochem Cell Biol 2005; 83: 263 269 [DOI] [PubMed] [Google Scholar]
- Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci 2008; 13: 3046 3082 [DOI] [PubMed] [Google Scholar]
- Beneke S, Burkle A. Poly(ADP-ribosyl)ation in mammalian ageing. Nucleic Acids Res 2007; 35: 7456 7465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinner A, Wu W, Staudt C, Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res 2008; 36: 5678 5694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Occhio MJ, Hengstberger KJ, Johnston SD. Biology of sperm chromatin structure and relationship to male fertility and embryonic survival. Anim Reprod Sci 2007; 101: 1 17 [DOI] [PubMed] [Google Scholar]
- Lin MH, Kuo-Kuang Lee R, Li SH, Lu CH, Sun FJ, Hwu YM. Sperm chromatin structure assay parameters are not related to fertilization rates, embryo quality, and pregnancy rates in in vitro fertilization and intracytoplasmic sperm injection, but might be related to spontaneous abortion rates. Fertil Steril 2008; 90: 352 359 [DOI] [PubMed] [Google Scholar]
- Godon C, Cordelieres FP, Biard D, Giocanti N, Megnin-Chanet F, Hall J, Favaudon V. PARP inhibition versus PARP-1 silencing: different outcomes in terms of single-strand break repair and radiation susceptibility. Nucleic Acids Res 2008; 36: 4454 4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res 2006; 34: 6170 6182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res 2008; 18: 134 147 [DOI] [PubMed] [Google Scholar]
- Zhao M, Shirley CR, Yu YE, Mohapatra B, Zhang Y, Unni E, Deng JM, Arango NA, Terry NH, Weil MM, Russell LD, Behringer RR, et al. Targeted disruption of the transition protein 2 gene affects sperm chromatin structure and reduces fertility in mice. Mol Cell Biol 2001; 21: 7243 7255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley CR, Hayashi S, Mounsey S, Yanagimachi R, Meistrich ML. Abnormalities and reduced reproductive potential of sperm from Tnp1- and Tnp2-null double mutant mice. Biol Reprod 2004; 71: 1220 1229 [DOI] [PubMed] [Google Scholar]
- Menissier de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, Schreiber V, Ame JC, Dierich A, LeMeur M, Sabatier L, Chambon P, et al. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J 2003; 22: 2255 2263 [DOI] [PMC free article] [PubMed] [Google Scholar]