Abstract
Differences in the fertilization behavior of Xenopus borealis from X. laevis and X. tropicalis suggest differences in the glycosylation of the egg jellies. To test this assumption, O-linked glycans were chemically released from the egg jelly coat glycoproteins of X. borealis. Over 50 major neutral glycans were observed, and no anionic glycans were detected from the released O-glycan pool. Preliminary structures of ∼30 neutral oligosaccharides were determined using matrix-assisted laser desorption/ionization (MALDI) infrared multiphoton dissociation tandem mass spectrometry (MS). The mass fingerprint of a group of peaks for the core-2 structure of O-glycans was conserved in the tandem mass spectra and was instrumental in rapid and efficient structure determination. Among the 29 O-glycans, 22 glycans contain the typical core-2 structure, 3 glycans have the core-1 structure and 2 glycans contained a previously unobserved core structure with hexose at the reducing end. There were seven pairs of structural isomers observed in the major O-linked oligosaccharides. To further elucidate the structures of a dozen O-linked glycans, specific and targeted exoglycosidase digestions were carried out and the products were monitored with MALDI-MS. Reported here are the elucidated structures of O-linked oligosaccharides from glycoproteins of X. borealis egg jelly coats. The structural differences in O-glycans from jelly coats of X. borealis and its close relatives may provide a better understanding of the structure–function relationships and the role of glycans in the fertilization process within Xenopodinae.
Keywords: exoglycosidase, MALDI-IRMPD mass spectrometry, O-glycan structure, tandem MS, X. borealis
Introduction
Glycosylation is one of the most common forms of protein post-translational modifications in eukaryotic systems, with over 50% of all proteins being glycosylated (Ohtsubo and Marth 2006; Agard and Bertozzi 2009). The oligosaccharide moieties of glycoproteins are implicated in a wide range of cell–cell and cell–matrix recognition events that are required for diverse biological processes ranging from immune recognition to cancer development (Apweiler et al. 1999; Szymanski and Wren 2005; Brooks 2006; Freeze 2006; Ohtsubo and Marth 2006; Lauc et al. 2010). Not only can a glycan chain reach further away from the bulk surface of a globular protein for a specific interaction (Cyster et al. 1991), but more importantly, many biologically vital proteins would not fold properly without the glycan(s) attached to the peptide backbone (Helenius and Aebi 2001).
Xenopus laevis, together with the mouse and the chick, have been the vertebrate species used for research into the cellular and molecular mechanisms of reproduction and developmental biology. The extracellular matrix surrounding amphibian eggs includes an outer thick glycoprotein jelly coat composed of several morphologically and biochemical different layers and an inner thinner vitelline envelope composed of five or more glycoproteins. The glycoproteins in the extracellular matrix play essential roles in the fertilization process and early embryo development (Lindsay et al. 2002). Functionally, sperm must bind to glycans on glycoproteins in the extracellular matrix, involving a multifaceted and complex series of interactions, and then penetrate the matrix in order to reach the egg plasma membrane for fusion (McLeskey et al. 1998; Dell et al. 1999; Wassarman 1999; Easton et al. 2000; Horrocks et al. 2000; Prasad et al. 2000). Thus, structurally elucidating carbohydrate moieties of the jelly coat glycoproteins would add to the understanding of the biomolecules involved in fertilization and early embryo development.
The oligosaccharide structures from jelly coats of X. laevis and its relative, X. tropicalis, have been elucidated in this laboratory and elsewhere (Plancke et al. 1995; Strecker et al. 1995; Tseng et al. 1997, 1999, 2001; Guerardel et al. 2000; Xie et al. 2001, 2004; Zhang, Lindsay, et al. 2004; Zhang, Xie, et al. 2004; Zhang et al. 2005). Xenopus borealis, also called Kenya clawed frog, is a closer relative to X. laevis than X. tropicalis (Evans et al. 2004). Although the cross-fertilizations between X. laevis and X. tropicalis were successful in both directions (Lindsay et al. 2003), Brun and Kobel (1977) found the one-direction fertilization between X. laevis and X. borealis. Specifically, X. borealis eggs usually cannot be fertilized by X. laevis sperm, whereas X. laevis eggs do not exhibit such a fertilization block to borealis sperm and therefore can be cross fertilized. However, if X. borealis eggs are obtained directly from the ovaries and coated with X. laevis jelly, then fertilization by X. laevis sperm can occur. Surprisingly, X. laevis eggs coated with X. borealis egg jelly cannot be fertilized by X. laevis sperm but still can be fertilized by X. borealis sperm. This evidence suggests that X. laevis sperm is blocked by X. borealis egg jelly and cannot effectively penetrate into the innermost jelly layer of X. borealis eggs thus no fertilization occurs (Brun and Kobel 1977). These observations suggest that there must be some structural features in glycoproteins of X. borealis egg jelly coats that exert the mono-directional fertilization between the two species. Furthermore, because these proteins are mucins and are highly glycosylated, the role of the glycans may be key to understanding these observations.
In the present study, the O-linked glycans of jelly coat glycoproteins in X. borealis eggs were targeted by chemically releasing them from glycoproteins. O-Linked oligosaccharide structures were elucidated via matrix-assisted laser desorption/ionization (MALDI) Fourier transform ion cyclotron resonance (FTICR) infrared multiphoton dissociation (IRMPD) mass spectrometry (MS) analysis, a well-developed approach in this laboratory (Zhang et al. 2005; Lancaster et al. 2006; Kirmiz et al. 2007; Li et al. 2009b), coupled with exoglycosidase digestions. Previously reported mass fingerprints for O-linked glycan core structures were used to further aid structural elucidation (Xie et al. 2001; Zhang et al. 2005). We believe this is the first report with a relatively complete determination of O-glycan structures from egg jelly coats of X. borealis.
Results and discussion
MS analysis of total glycans released from egg jelly glycoproteins
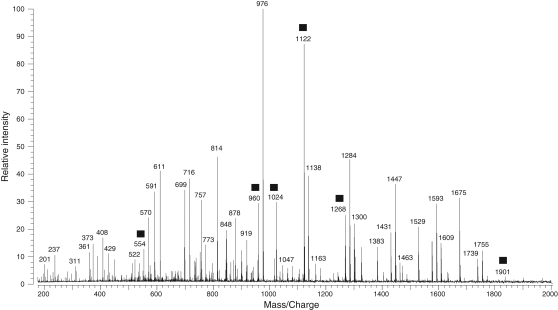
The positive ion MALDI mass spectrum of the neutral O-linked oligosaccharide mixture after porous graphitized carbon (PGC) purification is shown in Figure 1. The mass range spans 200–2000 Da, and the base peak is m/z 976. There are over 100 mass peaks above a 5% intensity threshold. Most of the glycans have masses at m/z 350–1800 Da. It is speculated that there are even more minor glycan peaks that were suppressed by the abundant glycans. In Figure 1, some peaks, m/z 554, 960, 1024, 1122, 1268 and 1901, marked with filled squares, are neutral glycan examples chosen for structure elucidation using the current approach in the subsequent description. The neutral glycan mixture was also subjected to MALDI mass analysis in negative ion mode, but there were the less peaks observed with lower abundance (data not shown).
Fig. 1.
MALDI mass spectrum of the neutral oligosaccharide mixture released from glycoproteins of X. borealis egg jelly coats after PGC purification. The peaks marked with filled squares are neutral glycan examples chosen for structure elucidation using the current approach described in the text.
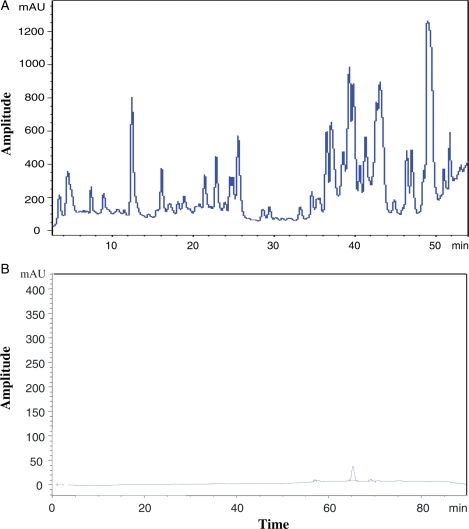
To elucidate the monosaccharide sequence in each of the glycans by tandem MS, the glycan mixture was separated into 250 µL fractions using HPLC (high-performance liquid chromatography or high-pressure liquid chromatography) with a PGC column over an 80 min elution time. The chromatograms for neutral and anionic oligosaccharide mixtures from the PGC purification are shown in Figure 2A and B under different elution gradients, respectively (see Materials and methods section). It was surprisingly observed that there were many intense peaks in the neutral glycan HPLC; in contrast, almost no noticeable absorption was found in the anionic mixture (from 40% fraction) under these conditions. Different elution gradient conditions were tried with the anionic glycan mixture, but no signal improvement was observed. These results suggest that there are no appreciable anionic oligosaccharides in the jelly coat glycoproteins of X. borealis eggs. The 40% fraction was examined extensively with both MALDI and nanoLC MS in both negative and positive modes. It was found that the 40% fraction contained no anionic oligosaccharide, unlike X. laevis. For this reason, we focused primarily on the neutral component. Each fraction of HPLC was subjected to MALDI mass analysis. It was found that the neutral oligosaccharide mixture was well separated and only a few major glycans were in each fraction. After 55 min of elution, there was a broad absorption (not shown in Figure 2A) and MALDI mass analysis showed that no glycans were in those fractions.
Fig. 2.
HPLC separation of O-glycans released from glycoproteins of egg jelly coats of X. borealis for (A) neutral (50 μL) and (B) anionic (50 μL) oligosaccharides. The separation conditions are detailed in the Materials and methods section.
The experimental mass, theoretical mass and monosaccharide components of each of the major glycan peaks are summarized in Table I. It was shown that MALDI-FTMS produced high mass accuracy with the absolute average mass error for all glycans in the list at 4.4 ppm. The peaks were determined to be O-linked oligosaccharides by an accurate mass calculation using an in-house program, Oligosaccharide Calculator. The released oligosaccharides comprise 0–5 Hexoses (Hex), 0–4 Fucoses (Fuc) and 1–5 N-acetylhexosamines (HexNAc). The largest oligosaccharide observed was in F34 with a m/z 1999 [M + Na]+. Among the 57 glycans listed in Table I, there were at least six pairs of isomers and one triple of isomers, such as F12/F33 (m/z 1122), F39/F46 (m/z 1268), F17/F34 (m/z 1325), F42/F47 (m/z 1431), F43/F48 (m/z 1593), F24/F45 (m/z 1755) and F18/F21/F36 (m/z 1577). Fortunately, within each group of isomers, the retention times were quite different in the HPLC separation, which highlights the advantage of using a PGC column for oligosaccharide separations.
Table I.
Neutral oligosaccharide components in HPLC fractions from egg jelly glycoproteins of X. borealis (all mass ions are sodiated, [M + Na]+)
| Fraction | Expt (m/z) | Theor (m/z) | Hex | Fuc | HexNAc |
|---|---|---|---|---|---|
| F4 | 960.3646 | 960.3644 | 1 | 1 | 3 |
| F7 | 554.2065 | 554.2056 | 1 | 1 | 1 |
| F7 | 595.2378 | 595.2322 | 0 | 1 | 2 |
| F8 | 862.3106 | 862.3164 | 2 | 2 | 1 |
| F9 | 814.3095 | 814.3065 | 1 | 0 | 3 |
| F10 | 903.3343 | 903.3430 | 1 | 2 | 2 |
| F10 | 1017.3951 | 1017.3860 | 1 | 0 | 4 |
| F10 | 1633.6180 | 1633.6070 | 3 | 2 | 4 |
| F12 | 1024.3752 | 1024.3690 | 3 | 2 | 1 |
| F12 | 1122.4210 | 1122.4170 | 2 | 1 | 3 |
| F14 | 1065.4050 | 1065.3960 | 2 | 2 | 2 |
| F15 | 1414.5350 | 1414.5330 | 2 | 3 | 3 |
| F15 | 1138.4110 | 1138.4120 | 3 | 0 | 3 |
| F16 | 757.2824 | 757.2850 | 1 | 1 | 2 |
| F16 | 919.3372 | 919.3379 | 2 | 1 | 2 |
| F17 | 1227.4480 | 1227.4490 | 3 | 2 | 2 |
| F17 | 1325.4946 | 1325.4970 | 2 | 1 | 4 |
| F18 | 798.3199 | 798.3116 | 0 | 1 | 3 |
| F18 | 1300.4750 | 1300.4650 | 4 | 0 | 3 |
| F18 | 1576.5910 | 1576.5860 | 3 | 3 | 3 |
| F19 | 716.2595 | 716.2585 | 2 | 1 | 1 |
| F20 | 1763.6489 | 1763.6700 | 2 | 4 | 4 |
| F21 | 1576.5976 | 1576.5860 | 3 | 3 | 3 |
| F22 | 1738.6505 | 1738.6390 | 4 | 3 | 3 |
| F24 | 1462.5143 | 1462.5180 | 5 | 0 | 3 |
| F24 | 1608.5764 | 1608.5760 | 5 | 1 | 3 |
| F24 | 1754.6373 | 1754.6340 | 5 | 2 | 3 |
| F25 | 1674.6437 | 1674.6340 | 2 | 2 | 5 |
| F28 | 1722.6519 | 1722.6440 | 3 | 4 | 3 |
| F28 | 1836.6996 | 1836.6870 | 3 | 2 | 5 |
| F28 | 1884.6964 | 1884.6970 | 4 | 4 | 3 |
| F31 | 976.3600 | 976.3593 | 2 | 0 | 3 |
| F31 | 1341.4860 | 1341.4920 | 3 | 0 | 4 |
| F32 | 878.3111 | 878.3113 | 3 | 1 | 1 |
| F32 | 1056.3620 | 1056.3590 | 5 | 0 | 1 |
| F32 | 1179.4310 | 1179.4390 | 2 | 0 | 4 |
| F33 | 1122.4095 | 1122.4170 | 2 | 1 | 3 |
| F34 | 1325.4986 | 1325.4970 | 2 | 1 | 4 |
| F34 | 1382.5141 | 1382.5180 | 2 | 0 | 5 |
| F34 | 1998.7270 | 1998.7400 | 4 | 2 | 5 |
| F35 | 1284.4680 | 1284.4700 | 3 | 1 | 3 |
| F36 | 1576.5847 | 1576.5860 | 3 | 3 | 3 |
| F37 | 1900.6790 | 1900.6920 | 5 | 3 | 3 |
| F38 | 1106.4214 | 1106.4220 | 1 | 2 | 3 |
| F39 | 1268.4754 | 1268.4750 | 2 | 2 | 3 |
| F39 | 716.2630 | 716.2585 | 2 | 1 | 1 |
| F41 | 1471.5557 | 1471.5550 | 2 | 2 | 4 |
| F42 | 1430.5296 | 1430.5280 | 3 | 2 | 3 |
| F43 | 1446.5209 | 1446.5230 | 4 | 1 | 3 |
| F43 | 1592.5810 | 1592.5810 | 4 | 2 | 3 |
| F45 | 1754.6287 | 1754.6340 | 5 | 2 | 3 |
| F46 | 1268.4671 | 1268.4750 | 2 | 2 | 3 |
| F46 | 1389.4900 | 1389.5010 | 4 | 2 | 2 |
| F47 | 1430.5397 | 1430.5280 | 3 | 2 | 3 |
| F48 | 1592.5683 | 1592.5810 | 4 | 2 | 3 |
| F52 | 1617.5950 | 1617.6130 | 2 | 3 | 4 |
| F54 | 408.1443 | 408.1477 | 1 | 0 | 1 |
Determination of O-linked oligosaccharide structures by IRMPD tandem MS
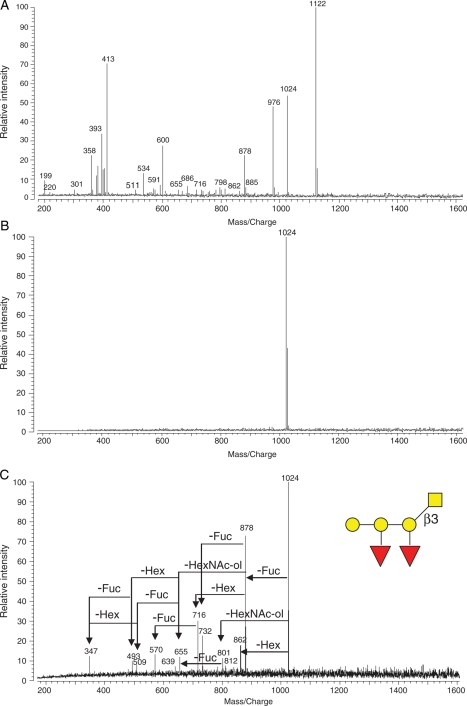
The applications of IRMPD for the elucidation of glycan structures have been reported previously (Zhang et al. 2005; Goldberg et al. 2006; Kirmiz et al. 2006; Lancaster et al. 2006). With IRMPD tandem MS, an individual ion peak is selectively isolated, followed by IR laser irradiation resulting in precursor ion fragmentation. The mass spectrum of HPLC F12 is shown in Figure 3A with many peaks. An ion (m/z 1024) was mass selected and isolated, resulting in a clean mass spectrum (Figure 3B). After the absorption of IR photons, the quasimolecular ion fragmented into product ions by the loss of monosaccharide residues (Figure 3C). Based on an accurate mass calculation, the sodiated ion, m/z 1024, was composed of three Hex, two Fuc and one HexNAc. The precursor ion lost simultaneously either one Fuc (146 Da) to m/z 878, one Hex (162 Da) to m/z 862 or one HexNAc in alditol (HexNAc-ol, 223 Da) to m/z 801, which suggested that the three residues were located in termini and HexNAc-ol was at the reducing end, being N-acetylgalactosamine (GalNAc)-ol as reserved core for O-glycans. The m/z 862 ion lost one Fuc to m/z 716 and one GalNAc-ol to m/z 639, but no loss of one Hex to m/z 700 from the ion was observed. The ion of m/z 878 lost any one of the three residues, which means only one Hex and both Fuc were at termini. Ions of m/z 347 and m/z 509 were observed in Figure 3C, which belong to Hex–Hex and Hex–Hex–Hex, respectively. This core was most similar to that of a core-1 structure [Galβ(1-3)GalNAc-ol; Brockhausen 1999], which has been reported previously (Tseng et al. 2001; Zhang et al. 2005). These results lead to the sequence of the m/z 1024 being assigned and shown in Figure 3C. The remaining peaks in the spectrum further confirmed this structure (Figure 3C). Based on the extensive experience in this laboratory on O-glycan structures, especially in egg jelly coat of Xenopus frog family, where so far all Hex residues in O-glycans have been galactose (Gal) (Tseng et al. 1997, 2001; Xie et al. 2001, 2004; Zhang, Lindsay, et al. 2004; Zhang et al. 2005; Li et al. 2009a, 2009b), we have assigned all Hex residues in this study putatively as Gal residues. Furthermore, all monosaccharide residues at the reducing end take the alditol form under conditions used in this work.
Fig. 3.
IRMPD tandem MS was applied to elucidate O-linked oligosaccharide structures. (A) The mass spectrum of one HPLC fraction (F12) from X. borealis egg jelly, (B) glycan ion (m/z 1024, [M + Na]+) peak isolated in the ICR cell, and (C) IRMPD mass spectrum yielding fragments from the precursor ion (m/z 1024), enabling structural elucidation of the oligosaccharide.
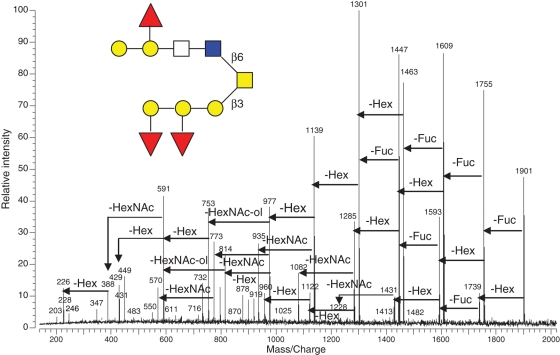
The sequence of a larger oligosaccharide ion from F37 (m/z 1901) was similarly determined based on its IRMPD fragmentation pattern (Figure 4). The quasimolecular ion consisted of five Hex, three Fuc and three HexNAc. There was a group of peaks including m/z 611, 449, 431, 408 and 246, which have been reported as the mass fingerprint of core-2 structure (Brockhausen 1999; Tseng et al. 2001; Zhang et al. 2005) [Galβ(1-3)(GlcNAcβ(1-6))GalNAc-ol] in O-glycans. The mass fingerprinting of typical core-2 motif in O-glycans is instrumental in structure elucidation and can be used as a signature of O-glycans containing the core-2 motif. The presence of m/z 428 (HexNAc–HexNAc) and m/z 591(Hex–HexNAc–HexNAc) indicated that the target glycan contained the residual linkage, Hex–HexNAc–GlcNAc–GalNAc-ol. The m/z 1901 ion only lost Fuc or Hex subsequently in early fragmentation events until the loss of (2Hex + 1Fuc) to m/z 1431, which then allowed for the first HexNAc loss to m/z 1228. This suggested that the 2Hex + 1Fuc extended from the HexNAc nonreducing end. All of fragmentations supported a single structural assignment of ion m/z 1901 shown in the inset of Figure 4. The remaining tandem mass peaks further confirmed the proposed structure.
Fig. 4.
IRMPD mass spectrum of a larger oligosaccharide (F37, m/z 1901, [M + Na]+) released from egg jelly glycoproteins of X. borealis. The fragmentation pattern and assigned preliminary structure of the glycan are shown.
Observation and structural determination of novel O-glycans by IRMPD
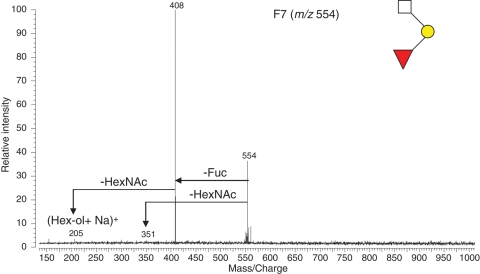
IRMPD was used to rapidly and efficiently identify novel glycans with sub-picomoles of material from jelly coats of X. borealis. For example, a glycan in HPLC F7 (m/z 554) was subjected to IRMPD MS analysis and the resulting spectrum is shown in Figure 5. The quasimolecular ion consisted of one Hex, one Fuc and one HexNAc based on an accurate mass calculation. The quasimolecular ion lost one Fuc to m/z 408 and one HexNAc to m/z 351. Both of which were then fragmented to produce an ion m/z 205. These results indicate that one Fuc and one HexNAc were at the two termini with one Hex located between them. The m/z 205 was assigned to Hex-ol. Thus, the sequence structure was readily assigned as Fuc(HexNAc)Hex-ol and shown in the inset of Figure 5. Two more glycans, m/z 821 and m/z 1122 (the F33 of HPLC) were similarly observed with Hex-ol as the reducing end. It has been confirmed in this laboratory that the presence of Hex-ol as the reducing end of O-glycans is the direct product from O-glycan release but not a peeling product under conditions used in the study (Tseng et al. 2001; Xie et al. 2001; Zhang, Xie, et al. 2004). Although it has been widely reported that a Hex residue can be linked to serine or threonine residues of O-glycoprotein of eukaryotes and some prokaryotes (Nishimura et al. 1992; Gerwig et al. 1993; Mann et al. 1996; Endo 1999; Busch and Aktories 2000; Spiro 2002), the O-glycans observed may represent the first report to date that Hex as a glycosylating residue is bonded to an amino acid of proteins in egg jelly coats of the X. borealis species.
Fig. 5.
A small O-linked glycan from egg jelly glycoproteins of X. borealis (F4, m/z 554, [M + Na]+) with a novel reducing end was identified by IRMPD. The assigned preliminary structure of the glycan is shown.
Differentiation of O-glycan isomers
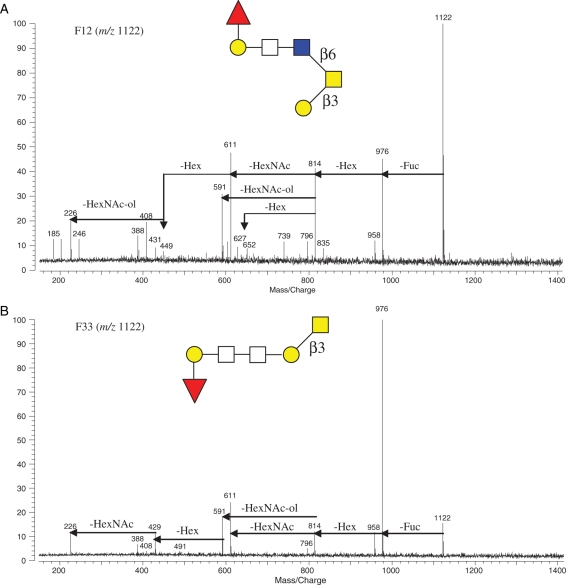
It was found that there were several pairs of glycan isomers among the oligosaccharides released from X. borealis egg jelly glycoproteins (Table I). The individual structures of each pair of isomers were distinguished by IRMPD MS analysis. For example, both F12 and F33 included an m/z 1122 glycan mass ion that consisted of two Hex, one Fuc and three HexNAc. They produced very different characteristic tandem mass spectra shown in Figure 6A and B, respectively. At first glance, the relative intensity ratios of mass peaks in two mass spectra are different; for example, the quasimolecular ion (m/z 1122) is the base peak and other major product ions are evenly distributed for F12, whereas the m/z 976 ion is the base peak and all other ion peaks are of low intensity, which is indicative of the distinctive fragmentation behavior of the two isomers. Indeed, in F12, the tandem mass spectrum of m/z 1122 produced the core-2 fingerprint peaks of O-glycans, m/z 611, 449, 431, 408 and 246, whereas in F33 m/z 1122 lacked this fingerprint. The quasimolecular ion m/z 1122 of F12 subsequently lost one Fuc and one Hex to m/z 814, which then had three fragmentation pathways, loss of one HexNAc, one HexNAc-ol or one Hex. Conversely, in F33, m/z 814 only had two fragmentation pathways and no loss of one Hex from the ion was observed (Figure 6). Furthermore, there was a fragment ion peak at m/z 449 in the IRMPD spectrum of F12 belonging to HexNAc–HexNAc-ol, but it was absent in the IRMPD spectrum of F33. Based on this information, the structural sequences of the two isomers were assigned and shown in Figure 6. Although both isomers contain a piece of HexNAc–HexNAc (m/z 429), F12 m/z 1122 ion does not retain it in order to keep core-2 fingerprint that normally is relatively stable and F33 m/z 1122 can produce and retain the ion (m/z 429) as detected. The rest of ion peaks in both spectra have the structural assignments accordingly.
Fig. 6.
A pair of glycan isomers (m/z 1122, [M + Na]+) from HPLC (A) F12 and (B) F33 was structurally differentiated by IRMPD tandem MS. The fragmentation patterns and assigned preliminary structures of the isomers are shown.
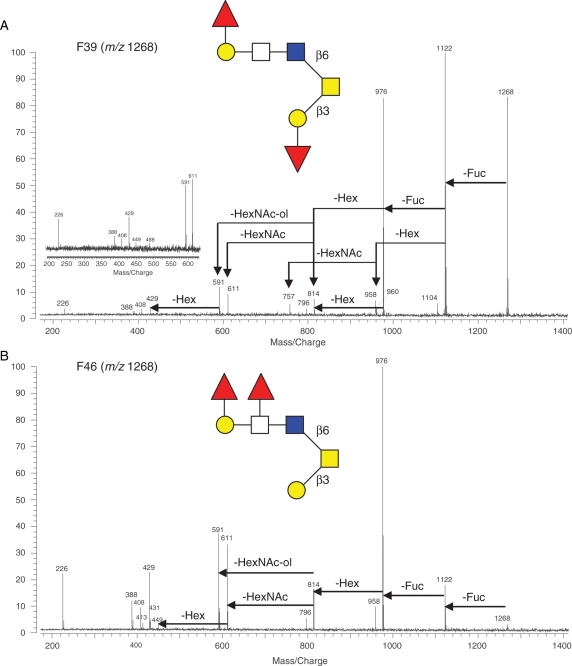
Another pair of isomers with m/z 1268 was present in F39 and F46, respectively, and also had distinctive fragmentation patterns (Figure 7). Both produced core-2 fingerprint peaks (m/z 611, 449, 431 and 408), but the former lost one Hex to m/z 960 after one Fuc was lost (Figure 7A), whereas the latter lost two Fuc prior to one Hex, thus lacking the peak of m/z 960 (Figure 7B). There was also a unique peak at m/z 757 in Figure 7A for F39, which came from a HexNAc loss from m/z 960. The structures of both isomers were assigned and presented in Figure 7. A retrospective check for mass spectra with corresponding structures helps explain the fragmentation patterns. In Figure 7A, the first lost Fuc should be the one linked to Hex residue of the noncore end so that there is one HexNAc loss to m/z 757 right after losing one Hex following the first Fuc loss. Although in Figure 7B, the first lost Fuc should be the one connected to HexNAc but not the other, otherwise there would be a Hex loss after the first Fuc off. Once the two Fuc get lost, either Hex can be cutoff as Fuc is labile residue and usually loses before the core Hex.
Fig. 7.
Another pair of glycan isomers (m/z 1268, [M + Na]+) from HPLC (A) F39 and (B) F46 was structurally differentiated by IRMPD tandem MS. The fragmentation patterns and assigned structures of the isomers are shown. The inset spectrum in (A) shows the core-2 fingerprint from another shot of IRMPD of the sample.
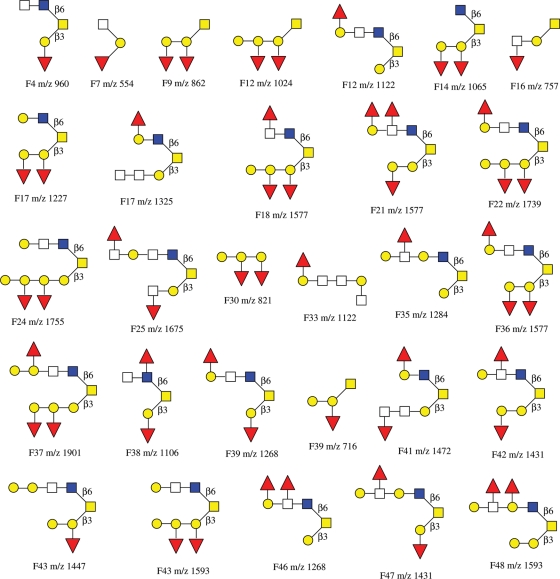
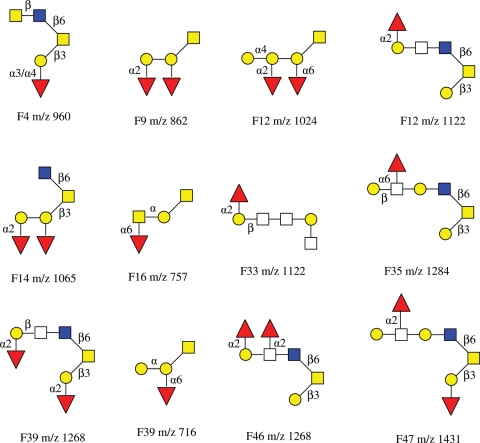
Twenty nine of the major glycans in the HPLC fractions were subjected to IRMPD tandem MS analysis, and their structure sequences are summarized in Chart 1. Some glycans did not produce high quality tandem mass spectra due to limited sample amounts. Among the 29 primary structures of O-glycans elucidated, the linkages between monosaccharide residues were not determined except those containing core-2 structure detected in the IRMPD mass spectra. The linkages were to be further elucidated by exoglycosidase digestions discussed in the following section.
Chart 1.
A summary of preliminary structures of O-linked neutral oligosaccharides elucidated based on IRMPD tandem MS from egg jelly glycoproteins of X. borealis.
Structure determination of O-glycans with exoglycosidase digestions
The specific glycosidic linkages within O-glycans can be determined by exoglycosidase digestions. The enzymes for this purpose are available and their working digestion conditions have been reported (Xie et al. 2001; Zhang, Lindsay, et al. 2004). Exoglycosidases cleave monosaccharide residues with high specificity from the nonreducing end of oligosaccharides.
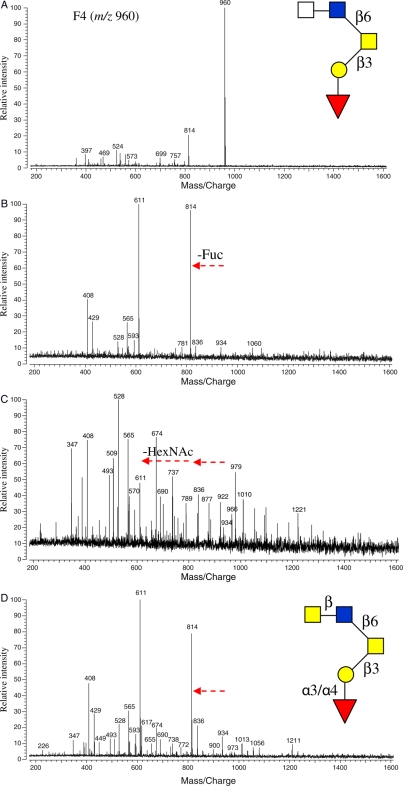
Figure 8 shows an oligosaccharide from F4 (m/z 960) with its primary sequence elucidated by IRMPD. This oligosaccharide was subjected to subsequent exoglycosidase digestions, and its structure was further determined. As shown in the inset of Figure 8A, the glycan had a Fuc linked to a Gal residue of the core with an unknown linkage. The quasimolecular ion (m/z 960) was the sole major peak in the fraction. After incubation with α(1-3,4) fucosidase at 37°C for 20 h, the m/z 960 peak was absent and m/z 814 peak greatly gained intensity due to the complete loss of Fuc (Figure 8B). This suggested that the Fuc was α(1-3,4) linked to Gal. Since no individual α(1-3) or α(1-4) fucosidase was commercially available, the linkage could not be fully determined. When another enzyme, N-acetyl-β-hexosaminidase, was added to the α(1-3,4) fucosidase digestion and incubated for another 20 h at 37°C, the m/z 814 ion disappeared due to the complete loss of its nonreducing end HexNAc residue to produce m/z 611 (Figure 8C). This result indicated that the HexNAc was β-linked to the GlcNAc residue. To determine the identity of the HexNAc residue, an alternative second enzyme, N-acetyl-β-glucosaminidase, was added and the digestion mixture was incubated at 40°C for 20 h. The resulting mass spectrum is shown in Figure 8D. It was found that this enzyme did not cleave the HexNAc as the ion m/z 814 had no change in intensity. Although N-acetyl-β-galactosaminidase was not commercially available to confirm the HexNAc identity, it was tentatively assigned GalNAc with a β-linkage. This tentative assignment was based on the observation that when m/z 814 (Figure 8D) was subjected to IRMPD, the typical core-2 fingerprint of peaks was observed (data not shown). The detailed structure of the glycan (m/z 960) is shown in Figure 8D.
Fig. 8.
(A) The MALDI mass spectra of HPLC F4 (m/z 960, [M + Na]+) before exoglycosidase digestion and (B) after digestions with α(1-3,4) fucosidase at 37°C for 20 h, (C) with N-acetyl β-hexosaminidase added to α(1-3,4) fucosidase digestion at 37°C for 20 h and (D) with N-acetyl β-glucosaminidase added to α(1-3,4) fucosidase digestion at 40°C for 20 h. Arrows represent residue loss after enzyme digestions. The preliminary structure of the glycan based on IRMPD is shown in the inset of (A) and the further elucidated structure with exoglycosidase digestions in the inset of (D).
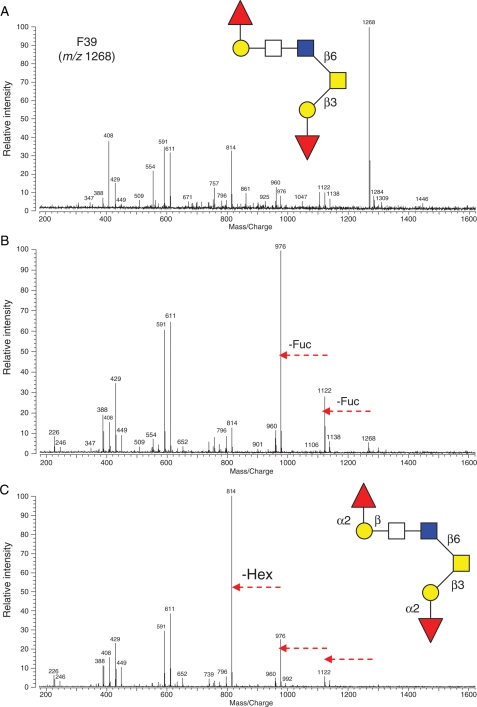
F39 (m/z 1268) was also subjected to exoglycosidase digestions to further elucidate its structure. The quasimolecular ion displayed dominant intensity in the mass spectrum prior to digestion (Figure 9A). Following incubation with α(1-2) fucosidase at 37°C for 20 h, two Fuc residues were cleaved leaving a low-intensity m/z 1268 ion and resulting in a minor m/z 1122 and a strong base peak of m/z 976, as shown in Figure 9B. When the nonspecific β-galactosidase was added to the α(1-2) fucosidase digestion mixture, the m/z 976 lost one Hex residue resulting a product ion at m/z 814 (Figure 9C). Since β(1-3) galactosidase, the only one commercially available in the β-galactosidase family, showed no effect on the Hex residue (data not shown), the residue was tentatively assigned as Gal β-linked to HexNAc. The updated structure of F39 (m/z 1268) is shown in Figure 9C.
Fig. 9.
The mass spectra of O-linked oligosaccharide (F39, m/z 1268, [M + Na]+) (A) before exoglycosidase digestion, (B) after reaction with α(1-2) fucosidase at 37°C for 20 h and (C) with β-galactosidase added to the α(1-2) fucosidase digestion at 37°C for 20 h. Arrows represent residue loss after enzyme digestions. The preliminary structure of the glycan based on IRMPD is shown in the inset of (A) and the further elucidated structure with exoglycosidase digestion in the inset of (C).
Ten other O-linked glycans were further structurally elucidated with exoglycosidase digestions based on sequences determined by IRMPD tandem mass analysis and are summarized in Chart 2. Among the structures, some individual linkage information remains to be determined due to the lack of the necessary exoglycosidases at present. It is noted in the schematic structure of glycans with symbol and color that the representation with known anomericity but unknown linkage position is displayed with only Greek letter.
Chart 2.
The structures of 12 O-linked neutral oligosaccharides that were further elucidated by exoglycosidase digestions coupled with IRMPD tandem MS from egg jelly glycoproteins of X. borealis.
Oligosaccharide comparison among three members of Xenopus family: X. borealis, X. laevis and X. tropicalis
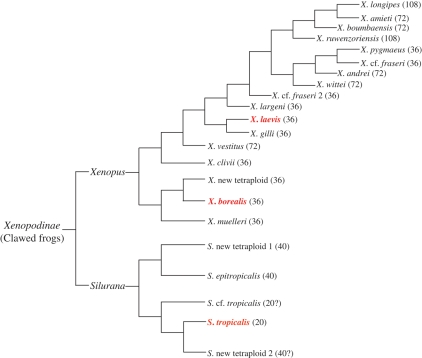
From a phylogenetic point of view, the three clawed frogs belong to two sister taxa in the subfamily Xenopodinae: laevis and borealis are in the Xenopus taxon, both are tetraploid species and contain 36 chromosomes, whereas tropicalis, a diploid species, is in the Silurana taxon and has 20 chromosomes (Figure 10; Evans et al. 2004).
Fig. 10.
The phylogenetic tree of clawed frogs. The number of chromosomes of each species is indicated in parentheses (Evans et al. 2004).
Oligosaccharide structures of jelly coats of the three species have been extensively studied in this laboratory with high-resolution MALDI-FTMS coupled with specific exoglycosidases (Tseng et al. 1997, 1999, 2001; Xie et al. 2001, 2004; Zhang, Lindsay, et al. 2004; Zhang, Xie, et al. 2004; Zhang et al. 2005). The three species displayed different oligosaccharide profiles in their egg jelly coats.
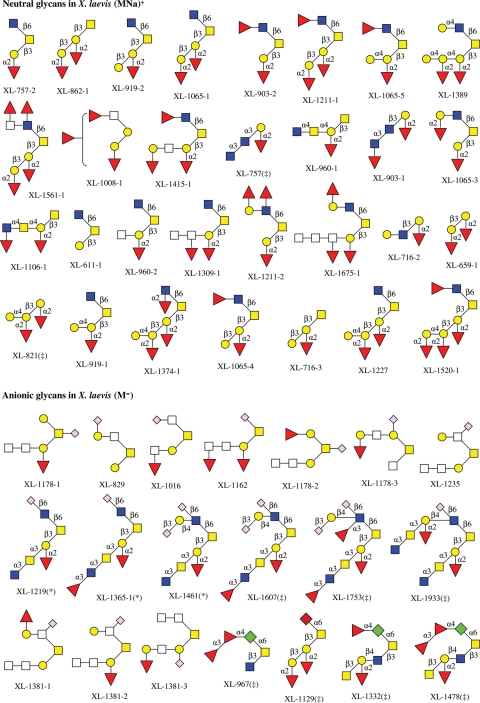
In X. laevis, 48 neutral and 47 anionic O-linked glycans have been observed by MS analysis, with the structures of 30 neutral and 20 anionic glycans are determined (Tseng et al. 2001; Zhang, Xie, et al. 2004). Although there are four sialylated glycans [three of them contain 2-keto-3-deoxynonulosonic acid (Kdn), one has 5-N-acetyl neuraminic acid (Neu5Ac)] in jelly coats of the species, which were structurally elucidated by Strecker et al. using NMR spectroscopy (Plancke et al. 1995), most of anionic glycans found in X. laevis are sulfated (Strecker et al. 1995), with some of them being doubly sulfated (Guerardel et al. 2000), as shown in Chart 3.
Chart 3.
The structures of neutral and anionic glycans observed in egg jelly coats of X. laevis. Structures with asterisk are also confirmed and those with double-dagger are only determined using NMR by Strecker et al. (1995; Plancke et al. 1995; Guerardel et al. 2000).
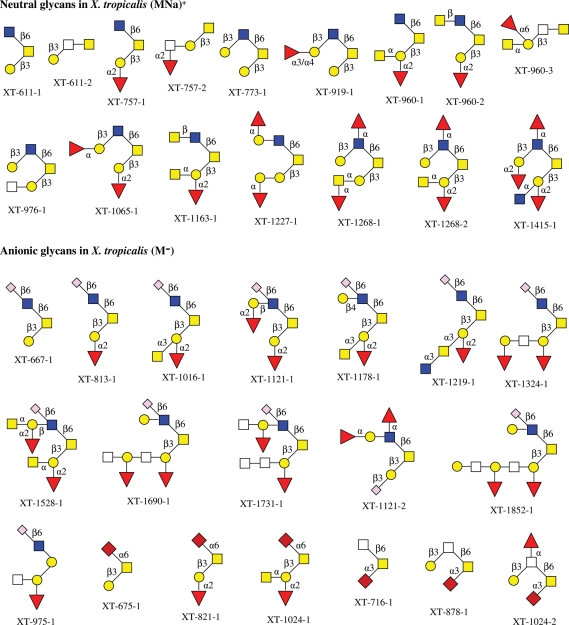
Although, in X. tropicalis, only about 16 neutral and 23 anionic O-glycans have been detected, all of neutral and 19 anionic oligosaccharides were structurally elucidated in this laboratory (Zhang, Lindsay, et al. 2004). Among the 19 anionic glycans, 13 are sulfated and 6 contain Neu5Ac. The O-glycan structures from egg jelly coats of X. tropicalis are summarized in Chart 4.
Chart 4.
The structures of neutral and anionic glycans observed in egg jelly coats of X. tropicalis.
In contrast, X. borealis exclusively contains neutral O-glycans in its egg jelly and has no detectable anionic glycans observed to date (Charts 1 and 2). It has been known that anionic glycans, mainly sulfated and sialylated, are important in the fertilization process in amphibians and other organisms (Hedrick et al. 1974; Wyrick et al. 1974; Hirohashi et al. 2008; Vilela-Silva et al. 2008). The observation of the absence of anionic glycans in the egg jelly of X. borealis in this study in contrast to glycans in X. laevis and X. tropicalis, with reports of the cross-fertilization between X. laevis and X. borealis gametes (Brun and Kobel 1977) requires an expansion of our understanding as to the role of glycans in the fertilization process in amphibians.
Based on Charts 1–4 for the elucidated O-glycans in the three frogs, no single O-glycan structure has so far been observed common to all three, although there are one neutral O-glycan (m/z 757) and one anionic O-glycan (m/z 1219) that both were detected in X. laevis and X. tropicalis. There are, however, structure motifs, such as the typical O-glycan core-2, that exist in many O-glycan structures of all three animals.
At this stage, with more structures to be elucidated from those observed and even more O-glycans to be detected of lower abundances, the absence of anionic O-glycans in egg jelly coat of X. borealis may be the major important difference that distinguishes it from the other two.
Conclusions
O-Linked glycans were chemically released from the jelly coat glycoproteins of X. borealis eggs. Surprisingly, only neutral oligosaccharides were detected with high-resolution MALDI-FTICR MS. These findings contrast those from its two close relatives, X. laevis and X. tropicalis, both of which have the acidic glycans in addition to neutral ones in their jelly coats. Structures of 29 neutral glycans of X. borealis were preliminarily determined by IRMPD tandem MS. Specific and targeted exoglycosidase digestions coupled with IRMPD were effectively applied to further elucidate 12 of the 29 glycan structures. Structural determination by IRMPD was greatly enhanced by the recognition of conserved mass fingerprint peaks for core structures (Tseng et al. 1999; Zhang, Lindsay, et al. 2004). Given that there were distinctive features of cross fertilization of X. borealis–X. laevis compared with X. laevis–X. tropicalis, the findings reported here that the O-glycan profile from X. borealis was significantly different from those of its two phylogenetic relatives X. laevis and X. tropicalis may provide for a better understanding of the structure–function relationships and the role of glycans in the fertilization process within Xenopodinae.
Materials and methods
Reagents
Carbograph solid-phase extract (SPE) cartridges (150 mg, 4 mL) packed with PGC were purchased from Alltech Associates, Inc. (Deerfield, IL). A Hypercarb PGC HPLC column (100 × 2.1 mm) was obtained from Hypersil, Inc. (Bellfonte, PA). Sodium borohydride (NaBH4, powder, 98%+ in purity) was purchased from ACROS (New Jersey). MALDI matrix [2,5-dihydroxybenzoic acid (DHB), 98%] was obtained from Sigma-Aldrich, Inc. (St Louis, MO).
Release and purification of O-linked oligosaccharides
Xenopus borealis eggs were obtained by injection with 35 IU of pregnant mare serum gonadotropin, by 1000 IU of human chorionic gonadotropin 4 days later. Eggs were collected into DeBoers solution (DB: 110 mM NaCl, 1.3 mM KCl, 1.3 mM CaCl2, to pH 7.2 with NaHCO3; Lindsay et al. 2003). The jelly coat surrounding the eggs was solubilized by 0.3% (v/v) β-mercaptoethanol, and the resulting solution of jelly coat was lyophilized. Glycoproteins were obtained from the jelly coat by a procedure described in previous publications (Xie et al. 2001, 2004; Zhang, Lindsay, et al. 2004; Zhang, Xie, et al. 2004; Zhang et al. 2005). The O-linked oligosaccharides were released from glycoproteins by a β-elimination reaction (Li et al. 2009a). Briefly, a certain amount of glycoprotein was incubated in 1.0 M NaBH4 and 0.1 M NaOH at 42°C for 16 h followed by neutralization with pre-cooled 1.0 M hydrochloric in an ice bath.
The resulting O-linked oligosaccharide mixture was desalted and purified by the SPE chromatography. The cartridge was washed with 18 MΩ water then with 0.1% (v/v) trifluoroacetic acid (TFA) in 80% (v/v) acetonitrile (ACN) followed by 18 MΩ water (all washings were ×3). The oligosaccharide sample was then loaded into the cartridge and allowed to flow slowly through the absorbent bed. Salts and other inorganic compounds were washed away with 18 MΩ water. The neutral oligosaccharides were eluted off sequentially with 10%, 20% (v/v) ACN. Anionic oligosaccharides were eluted with 40% ACN–0.05% (v/v) TFA. The effluents were collected into 1.5 mL of microcentrifuge tubes in fractions with each fraction tested for oligosaccharide components by MALDI-FTMS. The resulting neutral and anionic oligosaccharide mixtures were subjected to HPLC for further separation.
HPLC separation of oligosaccharide components
Oligosaccharide mixtures purified using the SPE chromatography were further separated by HPLC using a Hypercarb PGC column (2.1 × 100 mm, 5 µm, ThermoQ Hypersil Division). For the neutral oligosaccharide separation, the solvent A was H2O and solvent B was ACN containing 0.05% (v/v) TFA. The gradient elution system was 5–16% of B during the first 44 min, 16–28% of B during the following 12 min (44–56 min), 28–32% of B during the next 13 min (56–69 min), keeping 32% of B during the 69–80 min, finally followed by 5 min of elution with the solvent B down to 5%. For the separation of anionic glycans, solvent A was 0.05% TFA in H2O (v/v) and solvent B was 40% ACN with 0.05% TFA in H2O (v/v). The gradient elution was 5–40% of B for first 44 min, 40–62% of B for 44–62 min, 62–75% of B for 62–67 min, then keeping 75% of B for next 3 min followed by elution with 75–80% of B for 1 min. The flow rate was set up at 250 µL/min for the HPLC separation. The eluant was monitored at 206 nm by a photodiode array detector and collected into 1.5 mL microcentrifuge tubes at 1 min intervals. Each fraction was analyzed for oligosaccharides by MALDI-FTICR MS.
MALDI-FTICR MS analyses of oligosaccharides
A commercial MALDI-FT mass spectrometer (IonSpec, Irvine, CA) with an external ion source was used to perform the analysis. The instrument is equipped with a 7.0 T shielded, superconducting magnet and an air-cooled Nd:YAG laser (355 nm). MALDI samples were prepared by loading 1–5 µL of analyte, 1 µL of 0.4 M matrix (DHB in 50:50 H2O:ACN) on a stainless steel target plate. For the positive mode, 1 µL of 0.1 M NaCl in 50:50 H2O:ACN was applied to the spot to enrich the Na+ concentration and thus increase the primarily sodiated signals. The plate was left at ambient temperature for spot drying before insertion into the ion source.
IRMPD tandem MS analysis of oligosaccharides
A continuous-wave, turn-key 25 W CO2 laser, PLX25-s, from Parallax Technology, Inc. (Waltham, MA) was installed at the rear of the superconducting magnet and served to provide infrared photons for IRMPD. The laser has a working wavelength at 10.6 µm (0.1 eV/photon) and a beam diameter of 6 mm as specified by the manufacturer. A 2× beam expander from Synrad, Inc. (Mukilteo, WA) was attached to expand the laser beam to 12 mm for the complete irradiation of the ion cloud of interest. To perform IRMPD experiments, some modifications were made on the ICR cell and the vacuum chamber, which are described in detail elsewhere (Xie and Lebrilla 2003). The infrared laser was aimed directly toward the center of the analyzer cell.
Exoglycosidase digestions of O-linked oligosaccharides
Enzymes used in the study and working conditions were similar to those described previously (Zhang, Lindsay, et al. 2004). The exoglycosidases, α(1-2) fucosidase and N-acetyl-β-hexosaminidase, were purchased from New England Biolabs (Beverly, MA). The α(1-3,4) fucosidase was purchased from ProZyme (San Leandro, CA). The β-galactosidase and N-acetyl-β-glucosaminidase were purchased from Boerhinger Mannheim (Indianapolis, IN). Enzymes were dialyzed against pure water (0.025 µm membrane, Millipore Corporation, Billerica, MA) for 30 min before use. Ammonium acetate–acetic acid (NH4OAc-HOAc) solution (0.1 M) with different pHs was used as buffers for enzyme digestions (18 MΩ water was used for the digestions at pH = 7) for better results. Amounts of exoglycosidases used in the enzymatic digestions depended on the specificity were 20 U for α(1-2) fucosidase, 10 U for N-acetyl-β-hexosaminidase, 5 μU for α(1-3,4) fucosidase, 50 mU for β-galactosidase and 600 mU for N-acetyl-β-glucosaminidase. A 10–15 µL aliquot of the HPLC fraction that contained the target oligosaccharide(s) was dried by a speed-vacuum centrifuge, and the residue was resuspended in 4 µL of deionized water or 0.1 M buffer solution depending on the requirement of the exoglycosidase. The dialyzed enzyme was added to the sample solution and incubated at specified temperature for 20 h. Digestion products were monitored periodically via MALDI-FTMS to ensure digestion completion. Since the enzymes retain the activity and specificity in the presence of other exoglycosidases used in previous digestion steps, a second or third exoglycosidase was sequentially added into the reaction mixture if necessary. This allowed further MALDI-FTICR MS analyses of subsequent digestion products without any sample purification.
Funding
This work was supported by the National Institutes of Health (NIH, R01GM049077).
Conflict of interest
None declared.
Abbreviations
ACN, acetonitrile; DHB, 2,5-dihydroxybenzoic acid; Fuc, fucose; Gal, galactose; GalNAc, N-acetylgalactosamine; GlcNAc, N-acetylglucosamine; Hex, hexose; HexNAc, N-acetylhexosamine; HPLC, high-performance liquid chromatography (or high-pressure liquid chromatography); IRMPD, infrared multiphoton dissociation; Kdn/KDN, 2-keto-3-deoxynonulosonic acid; MALDI-FTICR MS, matrix-assisted laser desorption/ionization Fourier transform ion cyclotron resonance mass spectrometry; Neu5Ac, 5-N-acetyl neuraminic acid; PGC, porous graphitized carbon; SPE, solid-phase extract; TFA, trifluoroacetic acid.
The monosaccharide symbols used in the manuscript are listed as follow. (It is noted that all monosaccharide residues at reducing end are in alditol form). 
References
- Agard NJ, Bertozzi CR. Chemical approaches to perturb, profile, and perceive glycans. Acc Chem Res. 2009;42:788–797. doi: 10.1021/ar800267j. doi:10.1021/ar800267j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta. 1999;1473:4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- Brockhausen I. Pathways of O-glycan biosynthesis in cancer cells. Biochim Biophys Acta. 1999;1473:67–95. doi: 10.1016/s0304-4165(99)00170-1. [DOI] [PubMed] [Google Scholar]
- Brooks SA. Protein glycosylation in diverse cell systems: Implications for modification and analysis of recombinant proteins. Expert Rev Proteomics. 2006;3:345–359. doi: 10.1586/14789450.3.3.345. doi:10.1586/14789450.3.3.345. [DOI] [PubMed] [Google Scholar]
- Brun R, Kobel HR. Observations on fertilization block between Xenopus borealis and Xenopus laevis laevis. J Exp Zool. 1977;201:135–137. doi:10.1002/jez.1402010116. [Google Scholar]
- Busch C, Aktories K. Microbial toxins and the glycosylation of rho family GTPases. Curr Opin Struct Biol. 2000;10:528–535. doi: 10.1016/s0959-440x(00)00126-3. doi:10.1016/S0959-440X(00)00126-3. [DOI] [PubMed] [Google Scholar]
- Cyster JG, Shotton DM, Williams AF. The dimensions of the T lymphocyte glycoprotein leukosialin and identification of linear protein epitopes that can be modified by glycosylation. EMBO J. 1991;10:893–902. doi: 10.1002/j.1460-2075.1991.tb08022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell A, Morris HR, Easton RL, Patankar M, Clark GF. The glycobiology of gametes and fertilization. Biochim Biophys Acta. 1999;1473:196–205. doi: 10.1016/s0304-4165(99)00179-8. [DOI] [PubMed] [Google Scholar]
- Easton RL, Patankar MS, Lattanzio FA, Leaven TH, Morris HR, Clark GF, Dell A. Structural analysis of murine zona pellucida glycans. Evidence for the expression of core 2-type O-glycans and the Sd(a) antigen. J Biol Chem. 2000;275:7731–7742. doi: 10.1074/jbc.275.11.7731. [DOI] [PubMed] [Google Scholar]
- Endo T. O-Mannosyl glycans in mammals. Biochim Biophys Acta. 1999;1473:237–246. doi: 10.1016/s0304-4165(99)00182-8. [DOI] [PubMed] [Google Scholar]
- Evans BJ, Kelley DB, Tinsley RC, Melnick DJ, Cannatella DC. A mitochondrial DNA phylogeny of African clawed frogs: Phylogeography and implications for polyploid evolution. Mol Phylog Evol. 2004;33:197–213. doi: 10.1016/j.ympev.2004.04.018. doi:10.1016/j.ympev.2004.04.018. [DOI] [PubMed] [Google Scholar]
- Freeze HH. Genetic defects in the human glycome. Nat Rev. 2006;7:537–551. doi: 10.1038/nrg1894. doi:10.1038/nrg1894. [DOI] [PubMed] [Google Scholar]
- Gerwig GJ, Kamerling JP, Vliegenthart JF, Morag E, Lamed R, Bayer EA. The nature of the carbohydrate-peptide linkage region in glycoproteins from the cellulosomes of Clostridium thermocellum and Bacteroides cellulosolvens. J Biol Chem. 1993;268:26956–26960. [PubMed] [Google Scholar]
- Goldberg D, Bern M, Li B, Lebrilla CB. Automatic determination of O-glycan structure from fragmentation spectra. J Proteome Res. 2006;5:1429–1434. doi: 10.1021/pr060035j. doi:10.1021/pr060035j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerardel Y, Kol O, Maes E, Lefebvre T, Boilly B, Davril M, Strecker G. O-glycan variability of egg-jelly mucins from Xenopus laevis: Characterization of four phenotypes that differ by the terminal glycosylation of their mucins. Biochem J. 2000;352(Pt 2):449–463. doi:10.1042/0264-6021:3520449. [PMC free article] [PubMed] [Google Scholar]
- Hedrick JL, Smith AJ, Yurewicz EC, Oliphant G, Wolf DP. The incorporation and fate of [35S]-sulfate in the jelly coat of Xenopus laevis eggs. Biol Reprod. 1974;11:534–542. doi: 10.1095/biolreprod11.5.534. doi:10.1095/biolreprod11.5.534. [DOI] [PubMed] [Google Scholar]
- Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. doi:10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- Hirohashi N, Kamei N, Kubo H, Sawada H, Matsumoto M, Hoshi M. Egg and sperm recognition systems during fertilization. Dev Growth Differ. 2008;50(Suppl. 1):S221–S238. doi: 10.1111/j.1440-169X.2008.01017.x. doi:10.1111/j.1440-169X.2008.01017.x. [DOI] [PubMed] [Google Scholar]
- Horrocks AJ, Stewart S, Jackson L, Wishart GJ. Induction of acrosomal exocytosis in chicken spermatozoa by inner perivitelline-derived N-linked glycans. Biochem Biophys Res Commun. 2000;278:84–89. doi: 10.1006/bbrc.2000.3766. doi:10.1006/bbrc.2000.3766. [DOI] [PubMed] [Google Scholar]
- Kirmiz C, Li B, An HJ, Clowers BH, Chew HK, Lam KS, Ferrige A, Alecio R, Borowsky AD, Sulaimon S., et al. A serum glycomics approach to breast cancer biomarkers. Mol Cell Proteomics. 2007;6:43–55. doi: 10.1074/mcp.M600171-MCP200. [DOI] [PubMed] [Google Scholar]
- Lancaster KS, An HJ, Li B, Lebrilla CB. Interrogation of N-Linked oligosaccharides using infrared multiphoton dissociation in FT-ICR mass spectrometry. Anal Chem. 2006;78:4990–4997. doi: 10.1021/ac0600656. doi:10.1021/ac0600656. [DOI] [PubMed] [Google Scholar]
- Lauc G, Rudan I, Campbell H, Rudd PM. Complex genetic regulation of protein glycosylation. Mol Biosyst. 2010;6:329–335. doi: 10.1039/b910377e. doi:10.1039/b910377e. [DOI] [PubMed] [Google Scholar]
- Li B, An HJ, Hedrick JL, Lebrilla CB. Collision-induced dissociation tandem mass spectrometry for structural elucidation of glycans. Methods Mol Biol. 2009a;534:133–145. doi: 10.1007/978-1-59745-022-5_10. [DOI] [PubMed] [Google Scholar]
- Li B, An HJ, Hedrick JL, Lebrilla CB. Infrared multiphoton dissociation mass spectrometry for structural elucidation of oligosaccharides. Methods Mol Biol. 2009b;534:23–35. doi: 10.1007/978-1-59745-022-5_2. [DOI] [PubMed] [Google Scholar]
- Lindsay LL, Peavy TR, Lejano RS, Hedrick JL. Cross-fertilization and structural comparison of egg extracellular matrix glycoproteins from Xenopus laevis and Xenopus tropicalis. Comp Biochem Physiol. 2003;136:343–352. doi: 10.1016/s1095-6433(03)00169-7. [DOI] [PubMed] [Google Scholar]
- Lindsay LL, Yang JC, Hedrick JL. Identification and characterization of a unique Xenopus laevis egg envelope component, ZPD. Dev Growth Differ. 2002;44:205–212. doi: 10.1046/j.1440-169x.2002.00635.x. doi:10.1046/j.1440-169X.2002.00635.x. [DOI] [PubMed] [Google Scholar]
- Mann K, Mechling DE, Bachinger HP, Eckerskorn C, Gaill F, Timpl R. Glycosylated threonine but not 4-hydroxyproline dominates the triple helix stabilizing positions in the sequence of a hydrothermal vent worm cuticle collagen. J Mol Biol. 1996;261:255–266. doi: 10.1006/jmbi.1996.0457. doi:10.1006/jmbi.1996.0457. [DOI] [PubMed] [Google Scholar]
- McLeskey SB, Dowds C, Carballada R, White RR, Saling PM. Molecules involved in mammalian sperm-egg interaction. Int Rev Cytol. 1998;177:57–113. doi: 10.1016/s0074-7696(08)62231-7. doi:10.1016/S0074-7696(08)62231-7. [DOI] [PubMed] [Google Scholar]
- Nishimura H, Yamashita S, Zeng Z, Walz DA, Iwanaga S. Evidence for the existence of O-linked sugar chains consisting of glucose and xylose in bovine thrombospondin. J Biochem. 1992;111:460–464. doi: 10.1093/oxfordjournals.jbchem.a123780. [DOI] [PubMed] [Google Scholar]
- Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. doi:10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- Plancke Y, Wieruszeski JM, Alonso C, Boilly B, Strecker G. Structure of four acidic oligosaccharides from the jelly coat surrounding the eggs of Xenopus laevis. Eur J Biochem. 1995;231:434–439. doi: 10.1111/j.1432-1033.1995.tb20716.x. doi:10.1111/j.1432-1033.1995.tb20716.x. [DOI] [PubMed] [Google Scholar]
- Prasad SV, Skinner SM, Carino C, Wang N, Cartwright J, Dunbar BS. Structure and function of the proteins of the mammalian zona pellucida. Cells Tissues Organs. 2000;166:148–164. doi: 10.1159/000016730. doi:10.1159/000016730. [DOI] [PubMed] [Google Scholar]
- Spiro RG. Protein glycosylation: Nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology. 2002;12:43R–56R. doi: 10.1093/glycob/12.4.43r. doi:10.1093/glycob/12.4.43R. [DOI] [PubMed] [Google Scholar]
- Strecker G, Wieruszeski JM, Plancke Y, Boilly B. Primary structure of 12 neutral oligosaccharide-alditols released from the jelly coats of the anuran Xenopus laevis by reductive β-elimination. Glycobiology. 1995;5:137–146. doi: 10.1093/glycob/5.1.137. doi:10.1093/glycob/5.1.137. [DOI] [PubMed] [Google Scholar]
- Szymanski CM, Wren BW. Protein glycosylation in bacterial mucosal pathogens. Nat Rev. 2005;3:225–237. doi: 10.1038/nrmicro1100. doi:10.1038/nrmicro1100. [DOI] [PubMed] [Google Scholar]
- Tseng K, Hedrick JL, Lebrilla CB. Catalog-library approach for the rapid and sensitive structural elucidation of oligosaccharides. Anal Chem. 1999;71:3747–3754. doi: 10.1021/ac990095r. doi:10.1021/ac990095r. [DOI] [PubMed] [Google Scholar]
- Tseng K, Lindsay LL, Penn S, Hedrick JL, Lebrilla CB. Characterization of neutral oligosaccharide-alditols from Xenopus laevis egg jelly coats by matrix-assisted laser desorption Fourier transform mass spectrometry. Anal Biochem. 1997;250:18–28. doi: 10.1006/abio.1997.2193. doi:10.1006/abio.1997.2193. [DOI] [PubMed] [Google Scholar]
- Tseng K, Xie Y, Seeley J, Hedrick JL, Lebrilla CB. Profiling with structural elucidation of the neutral and anionic O-linked oligosaccharides in the egg jelly coat of Xenopus laevis by Fourier transform mass spectrometry. Glycoconj J. 2001;18:309–320. doi: 10.1023/a:1013665031668. doi:10.1023/A:1013665031668. [DOI] [PubMed] [Google Scholar]
- Vilela-Silva AC, Hirohashi N, Mourao PA. The structure of sulfated polysaccharides ensures a carbohydrate-based mechanism for species recognition during sea urchin fertilization. Int J Dev Biol. 2008;52:551–559. doi: 10.1387/ijdb.072531av. doi:10.1387/ijdb.072531av. [DOI] [PubMed] [Google Scholar]
- Wassarman PM. Mammalian fertilization: Molecular aspects of gamete adhesion, exocytosis, and fusion. Cell. 1999;96:175–183. doi: 10.1016/s0092-8674(00)80558-9. doi:10.1016/S0092-8674(00)80558-9. [DOI] [PubMed] [Google Scholar]
- Wyrick RE, Nishihara T, Hedrick JL. Agglutination of jelly coat and cortical granule components and the block to polyspermy in the amphibian Xenopus laevis. Proc Natl Acad Sci USA. 1974;71:2067–2071. doi: 10.1073/pnas.71.5.2067. doi:10.1073/pnas.71.5.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Lebrilla CB. Infrared multiphoton dissociation of alkali metal-coordinated oligosaccharides. Anal Chem. 2003;75:1590–1598. doi: 10.1021/ac026009w. doi:10.1021/ac026009w. [DOI] [PubMed] [Google Scholar]
- Xie Y, Liu J, Zhang J, Hedrick JL, Lebrilla CB. Method for the comparative glycomic analyses of O-linked, mucin-type oligosaccharides. Anal Chem. 2004;76:5186–5197. doi: 10.1021/ac0496953. doi:10.1021/ac0496953. [DOI] [PubMed] [Google Scholar]
- Xie Y, Tseng K, Lebrilla CB, Hedrick JL. Targeted use of exoglycosidase digestion for the structural elucidation of neutral O-linked oligosaccharides. J Am Soc Mass Spectrom. 2001;12:877–884. doi: 10.1016/S1044-0305(01)00267-7. doi:10.1016/S1044-0305(01)00267-7. [DOI] [PubMed] [Google Scholar]
- Zhang J, Lindsay LL, Hedrick JL, Lebrilla CB. Strategy for profiling and structure elucidation of mucin-type oligosaccharides by mass spectrometry. Anal Chem. 2004;76:5990–6001. doi: 10.1021/ac049666s. doi:10.1021/ac049666s. [DOI] [PubMed] [Google Scholar]
- Zhang J, Schubothe K, Li B, Russell S, Lebrilla CB. Infrared multiphoton dissociation of O-linked mucin-type oligosaccharides. Anal Chem. 2005;77:208–214. doi: 10.1021/ac0489824. doi:10.1021/ac0489824. [DOI] [PubMed] [Google Scholar]
- Zhang J, Xie Y, Hedrick JL, Lebrilla CB. Profiling the morphological distribution of O-linked oligosaccharides. Anal Biochem. 2004;334:20–35. doi: 10.1016/j.ab.2004.07.012. doi:10.1016/j.ab.2004.07.012. [DOI] [PubMed] [Google Scholar]