Abstract
The basic Helix-Loop-Helix (bHLH) proteins are transcription factors that play important roles during the development of various metazoans including fly, nematode, and vertebrates. They are also involved in human diseases, particularly in cancerogenesis. We made an extensive search for bHLH sequences in the completely sequenced genomes of Caenorhabditis elegans and of Drosophila melanogaster. We found 35 and 56 different genes, respectively, which may represent the complete set of bHLH of these organisms. A phylogenetic analysis of these genes, together with a large number (>350) of bHLH from other sources, led us to define 44 orthologous families among which 36 include bHLH from animals only, and two have representatives in both yeasts and animals. In addition, we identified two bHLH motifs present only in yeast, and four that are present only in plants; however, the latter number is certainly an underestimate. Most animal families (35/38) comprise fly, nematode, and vertebrate genes, suggesting that their common ancestor, which lived in pre-Cambrian times (600 million years ago) already owned as many as 35 different bHLH genes.
Transcription factors of the basic Helix-Loop-Helix (bHLH) family play a central role in cell proliferation, determination, and differentiation (Jan and Jan 1993; Weintraub 1993; Hassan and Bellen 2000). The bHLH domain is ∼60 amino acids long and comprises a DNA-binding basic region (b) followed by two α-helices separated by a variable loop region (HLH) (Ferre-D'Amar et al. 1993). The HLH domain promotes dimerization, allowing the formation of homodimeric or heterodimeric complexes between different family members (Murre et al. 1989a; Kadesh 1993). The two basic domains brought together through dimerization bind specific hexanucleotide sequences (Murre et al. 1989a; Van Doren et al. 1991, 1994; Ohsako et al. 1994).
The bHLH motif first was identified in the murine transcription factors E12 and E47 (Murre et al. 1989b). Numerous bHLH proteins since have been identified in animals, plants, and fungi. A phylogenetic analysis based on a sample of 122 bHLH sequences has lead to a subdivision into four monophyletic groups of proteins named A, B, C, and D (Atchley and Fitch 1997).
Group A and B include bHLH proteins that bind hexameric DNA sequences referred to as “E Boxes” (CANNTG), respectively CACCTG or CAGCTG (Group A) and CACGTG or CATGTTG (Group B) (Murre et al. 1989a; Van Doren et al. 1991; Dang et al. 1992).
Group A includes several tissue-specific bHLH proteins (e.g., MyoD, Twist, Achaete-Scute proteins; for a recent review, see Hassan and Bellen 2000) as well as the ubiquitously distributed E12/Daughterless-type bHLH proteins (Murre et al. 1989b). In many instances, the tissue-specific proteins form inactive homodimers and require the presence of a E12/Daughterless partner to form active heterodimers (Cabrera and Alonso 1991; Lassar et al. 1991; Van Doren et al. 1992). Binding of the heterodimers to an E-box usually leads to transcriptional activation of the target gene (Cabrera and Alonso 1991; Van Doren et al. 1992).
Group B includes a large number of functionally unrelated proteins (e.g., Myc, Max, USF, SREBP, MITF) involved in various developmental and cellular processes (Henriksson and Luscher 1996; Facchini and Penn 1998; Goding 2000). Some group-B proteins contain an additional motif, known as a Leucine Zipper (LZ), which also is involved in protein dimerization. Dang et al. (1992) and Atchley and Fitch (1997) included in the same group B several proteins related to the Drosophila Hairy and Enhancer of split bHLH (HER proteins; Fisher and Caudy 1998). These proteins are characterized by the presence of a proline instead of an arginine at a crucial position in the basic domain. DNA-binding site selection and in vivo studies have shown that these proteins bind preferentially to sequences referred to as “N-boxes” (CACGCG or CACG AG) and have only a low affinity for “E-boxes” (Ohsako et al. 1994; Van Doren et al. 1994). The HER proteins are characterized further by the presence of an additional motif, the 4-amino acid WRPW domain, which allows the interaction with the Groucho repressor protein (Fisher and Caudy 1998). Accordingly, the HER proteins have been shown to act as transcriptional repressors during nervous system development and segmentation (Kageyama and Nakanishi 1997; Fisher and Caudy 1998).
Group C corresponds to the family of bHLH proteins known as bHLH-PAS (Crews 1998). The characteristic feature of bHLH-PAS proteins is the PAS domain, so called for the first three proteins identified with this motif: Drosophila Period (Per), human ARNT, and Drosophila Single-minded (Sim). The PAS domain found in bHLH-PAS proteins is ∼260–310 amino acids long and allows the dimerization between PAS proteins, the binding of small molecules (e.g., dioxin), and interactions with non-PAS proteins (Crews 1998). bHLH-PAS proteins control a variety of developmental and physiological events including neurogenesis, tracheal and salivary duct formation, toxin metabolism, circadian rhythms, and response to hypoxia (Crews 1998). bHLH-PAS proteins bind to ACGTG or GCGTG core sequences.
Group D corresponds to HLH proteins that lack a basic domain and are hence unable to bind DNA. This group includes the Id and Extramacrochaete (Emc) proteins (Benezra et al. 1990; Ellis et al. 1990; Garrell and Modolell 1990), which act as antagonists of group A bHLH proteins (Van Doren et al. 1991, 1992).
An additional group of putative HLH proteins has been described more recently, the COE family (for Collier/Olf-1/EBF). This group is characterized by the presence of an additional domain involved both in dimerization and in DNA binding, the COE domain (Crozatier et al. 1996). The HLH sequences of this group are highly divergent from the other bHLH motifs, making their phyletic analysis difficult.
Other than this subdivision in a few large groups, however, little is known of the evolution and diversification of the bHLH domain. Yet, given the importance of the bHLH genes in development, it would be desirable to have a more refined classification scheme of the various types of bHLH motifs, as well as a better understanding of their evolutionary relationships both within and between organisms. We have taken advantage of the complete sequencing of the nematode's (Caenorhabditis elegans Sequencing Consortium 1998) and fly's (Adams et al. 2000) genomes to extract a large, and possibly complete, set of bHLH genes from these two organisms. We also have used the large number of bHLH genes that now have been identified in vertebrates, as well as the smaller number available in plants and fungi, to assess the evolutionary relationships within this family.
RESULTS AND DISCUSSION
Derivation of Comprehensive Sets of bHLH Sequences from Existing Databases
The completion of the nematode and fly sequencing projects provided us with an opportunity to screen whole genomes for bHLH coding regions. To collect as many such sequences as possible (hopefully, all of them) we first retrieved a large number of bHLH sequences available from the nonredundant NCBI (http://www.ncbi.nlm.nih.gov) and Sanger protein databases (http://www.sanger.ac.uk), as described in the Methods section. We used the most divergent among these sequences (as determined by preliminary phylogenetic reconstructions) to screen by BLASTP (Altschul et al. 1990) the complete genomic sequences of C. elegans and D. melanogaster. We used the retrieved sequences that were not present in our initial collection to make new BLASTP searches in both genome databases as well as in the nonredundant NCBI protein database. We also used yeast and plant sequences retrieved from our original screen to make BLASTP searches, against the Saccharomyces cerevisiae and the Arabidopsis thaliana genome databases, in order to isolate additional bHLH sequences from these organisms. These various searches generated a set of more than 350 different bHLH sequences. We did not systematically retrieve the large number of bHLH sequences from various mammals (other than mouse) that are available. Thus, it is clear that our set is far from including all the available bHLH sequences. We believe, however, that it provides a very extensive coverage of fly, nematode, and mouse genes, and a fair representation of the plant and fungal types.
We aligned these sequences using the multiple alignments software CLUSTALW (Thompson et al. 1994) and checked each alignment by hand. The verified alignments then were used to construct phylogenetic trees as described in the Methods section. The resultant trees were bootstrapped to provide information about their statistical reliability. We used these trees to define groups of orthologous sequences.
Identification of Orthologous Families
Orthologous genes in two or more organisms are homologs that evolved from the same gene in the last common ancestor (Fitch 1970). Paralogous genes are those that have resulted from within species duplication (Fitch 1970). Unfortunately, there is no absolute criterion that can be used to decide if two genes are orthologous. The criterion we used to define orthologous families was that the grouping of bHLH sequences from at least two species into one monophyletic family should be supported by different methods of analysis with bootstrap values >50%. A similar criterion has been used in other analyses of protein families (Galliot et al. 1999). This criterion was relaxed for a few families, as will be discussed later (lower bootstrap values; Table 1). The fact that congruence was observed between trees constructed by different methods suggests that our reconstruction of the bHLH phylogeny is essentially correct.
Table 1.
bHLH Genes Grouped into 44 Phylogenetically-Defined Families
| Family name | Bootstrap support | No. of worm genes | No. of fly genes | No. of mouse genes | Plants-yeast? | High-order group |
|---|---|---|---|---|---|---|
| Achaete-Scute | intermediate1 | 4 | 4 | 2 | no | A |
| Neurogenin | high | 1 | 1 | 3 | no | A |
| NeuroD | intermediate | 1 | 0 to 22 | 4 | no | A |
| Twist | high | 1 | 1 | 2 | no | A |
| MyoD | high | 1 | 1 | 4 | no | A |
| E12/E47 | high | 1 | 1 | 4 | no | A |
| Atonal | high | 1 | 3 | 2 | no | A |
| Mist | high | 0 or 13 | 1 | 1 | no | A |
| Beta3 | intermediate | 1 or 23 | 1 | 2 | no | A |
| MyoR | high | 1 | 1 | 2 | no | A |
| Mesp | low | 0 | 1 | 2 | no | A |
| Paraxis | high | 0 | 1 | 2 | no | A |
| Hand | intermediate | 1 | 1 | 2 | no | A |
| PTF1 | intermediate | 1 | 3 | 1 | no | A |
| SCL | high | 0 | 1 | 4 | no | A |
| NSCL | high | 1 | 1 | 2 | no | A |
| SRC | high | 0 | 0 | 2 | no | B |
| AHR | intermediate | 2 | 2 | 1 | no | C |
| Sim | high | 0 or 14 | 1 | 2 | no | C |
| Trh | high | 0 or 14 | 1 | 1 | no | C |
| PAS1 | high | 0 or 14 | 0 | 1 | no | C |
| HIF | high | 0 or 14 | 1 | 3 | no | C |
| Max | high | 2 | 1 | 1 | no | B |
| USF | low5 | 1 | 1 | 2 | yeast | B |
| MITF | high | 1 | 0 | 4 | no | B |
| AP4 | high | 1 | 1 | 1 | no | B |
| TF4 | high | 0 | 2 | 1 | no | B |
| Clock | high | 0 | 1 to 36 | 2 | no | C |
| ARNT | high | 1 | 1 | 2 | no | C |
| Bmal | high | 0 | 1 | 2 | no | C |
| Myc7e | intermediate | 0 | 0 | 0 | plants only | B |
| R | high | 0 | 0 | 0 | plants only | B |
| SREBP | low7 | 1 | 1 | 2 | yeast | B |
| Emc | high | 0 | 1 | 4 | no | D |
| Myc | low8 | 0 | 1 | 4 | no | B |
| Mad | high | 0 | 0 | 3 | no | B |
| COE | **9 | 1 | 1 | 5 | no | F |
| Gridlock | low10 | 0 | 2 | 2 | no | E |
| Hairy | high | 0 | 3 | 1 | no | E |
| E (spl) | low11 | 2 | 8 | 5 | no | E |
| Pho4 | high | 0 | 0 | 0 | yeast only | B |
| Sat1 | high | 0 | 0 | 0 | plants only | B |
| GBOX | high | 0 | 0 | 0 | plants only | B |
| RTG3P | intermediate | 0 | 0 | 0 | yeast only | B |
| orphans | none | 6 | 0 to 412 | 0 | yes | 0 |
Families have been named according to the name (or its common abbreviation) of the first discovered or best known member of the family. Bootstrap support has been classified as high (>75%), intermediate (50%–75%) or low (<50%). The number of members per family in worm, fly, and mouse is reported. Each family has been tentatively assigned to a high-order group using the classification of Atchley and Fitch (1997) (see text for details). Fly and worm genes that cannot be assigned to any families are categorized as “orphan” genes.
Low (∼30%) when C. elegans sequences are included, one of which (C2812.8) being most divergent but consistently found linked to the Achaete-Scute family.
delilah and CG11450, two Drosophilagenes that group together (bootstrap value 52%) to the exclusion of any other gene, are often found associated with the NeuroD family; their inclusion in this family is, however, not supported by bootstrap resampling (see text and Fig. 3).
Beta3 and Mist are closely related families; one C. elegans sequence (DY33) is almost equally related to both families.
The Hif, Sim, Trh and PAS1 families form a strongly supported monophyletic group (bootstrap value 95%) which are collectively linked to a single C. elegans gene, F38A6.3(bootstrap value 65%). Because the Hif, Sim and Trh families contain both fly and mouse genes, F38A6.3 is unlikely to be the single worm ortholog of all these families. It is more likely that F38A6.3 is the ortholog of one of these families but has been displaced at the root of these families as a result of the long branch attraction phenomenon (see text).
Low bootstrap support (∼40%) when yeast genes are included; high support (75%) when animal genes only are considered.
One Drosophila gene, known as Dm clock, is found included in this family with high bootstrap support (91%); two other closely related genes Rst(1) JH and CG6211,are found associated with this family with lower bootstrap value (45%) and as outgroup to the other clock genes from both vertebrates and fly. Rst(1) JH is involved in metamorphosis and thus might represent a divergent clock gene specific to Drosophila.
Low bootstrap support (∼45%) when yeast and worm genes are included; high support (77%) when only vertebrate and fly genes are considered.
Low bootstrap support (∼30%) when the fly gene (named diminutive, dm) is included; high support (99%) when only vertebrate, echinoderms, and retrovirus genes are considered. Functional analysis of dm supports its orthology to the Myc family (Schreiber-Agus et al. 1997).
The COE genes were not included in our analysis as their HLH was not alignable with the other ones. Support for the monophyly of this family comes from the analysis of the HLH and the COE domain within this family.
High bootstrap support (100%) when only one of the two fly genes is considered (Dm Hey); lower support (42%) for the inclusion of a second fly gene (Dm Sticky ch1) in the family, as outgroup to Hey and gridlock genes from both vertebrates and fly.
The Gridlock, Hairy, and Enhancer of split families genes form a well-supported monophyletic group (group E; see Fig. 2). Two clear families (Hairy and Gridlock families) with high bootstrap support emerge from this group. All of the remaining sequences have been grouped in a single family (named Enhancer of split) which has no real phylogenetic support. A phylogenetic tree of the group can be found on the authors' Web site.
The total number of orphan genes depends on whether we do or do not include Rst(1) JH and CG6211 in the Clock family or delilah and CG11450 in the NeuroD family.
Our analysis led us to define 44 orthologous families ( i.e., 44 ancestral types of bHLH domains). Table 1 summarizes the 44 families and some of their properties. We named each family according to its first discovered member, or in a few cases, to its best-characterized member. The complete list of all members of every family, together with database accession numbers, can be found as supplementary material at http://www.genome.org. Two types of bHLH motifs presented special problems. First, the HLH of COE family proteins were not easily alignable with other bHLH proteins. Hence, the phylogenetic analysis of this family was mostly done without other types of bHLH sequences and using the well-conserved COE domain in addition to the bHLH. Second, although Hairy/E(spl)-related (HER) proteins appear consistently monophyletic, the resolution within the group was very poor and we were unable to identify orthologous families with any confidence. Because many amino acids flanking the bHLH motif are conserved in this group, we used a larger domain for phyletic comparisons to obtain better (but still low) phylogenetic resolution (Table 1).
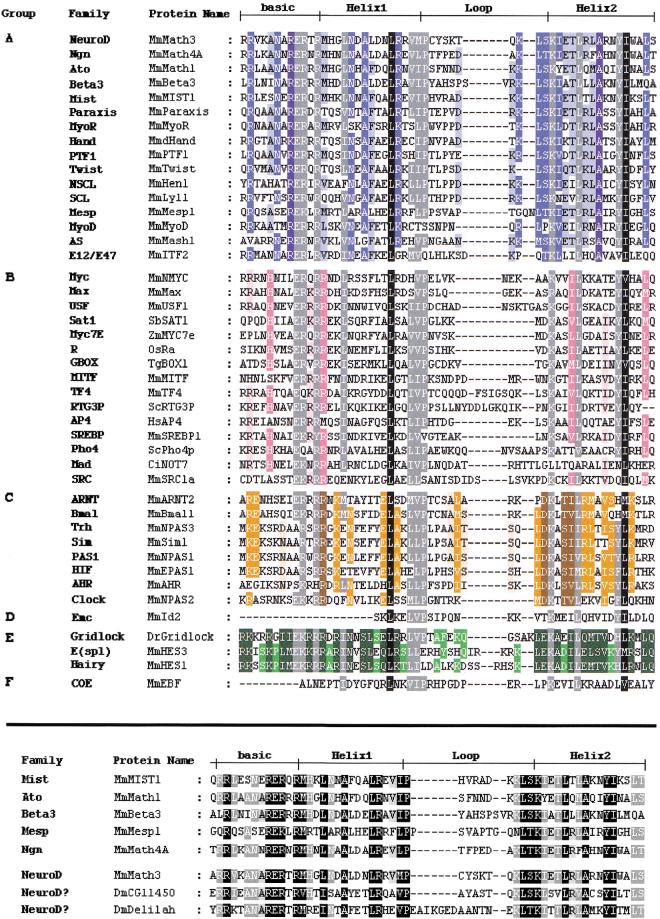
Figure 1 shows an alignment of all 44 bHLH types, based on one representative of each family. Thirty-six families comprise only animal members, four families are specific to plants, two are found only in yeasts, and two have both yeast and animal representatives. Thus, the bHLH motif appeared very early in eukaryotic history, but its expansion occurred almost entirely after the divergence between plants, fungi, and animals. The presence of only four plant families in our set is most likely a result of the fact that there were no extensive searches for bHLH genes in plants. As a consequence, most plant sequences come from one species, A. thaliana, for which an extensive genome project is conducted. Indications that more plant families are to be identified come from preliminary BLASTP searches, which revealed 30 different A. thaliana bHLH sequences, most of which are unrelated to other plant bHLH sequences. We found these sequences to form four additional “families” comprising A. thaliana sequences only. These “families” are not reported in Table 1 because we choose to consider, as significant families, only groups that contain sequences from at least two different species. Hence, most A. thaliana bHLH are considered, in our work, as orphan genes (i.e., sequences that can not be assigned to any family).
Figure 1.
(top) Alignment of the bHLH of the 44 different families listed in Table 1 (abbreviations as in Table 1). One member per family, usually from mouse, has been selected. Designation of basic, Helix1, Loop and Helix2 follows Ferre-D'Amare et al. (1993). The different families have been grouped according to the high-order group to which they belong (Atchley and Fitch 1997; see text). The evaluation of percentage conservation within each group and through the complete multiple sequence alignment was done using the Blosum62 Similarity Scoring Table. A specific background color with three intensities is attributed to each group (dark, 100% conservation; medium, 80% or greater conserved; light, 60% or greater conserved). Dark gray and black backgrounds represent conservation through all groups. Residues with black background represent 100% conservation; residues with dark gray background represent 80% or greater conservation. (bottom) Alignment of the bHLH of the constituting members of the Atonal superfamily. One member of each family plus the two orphan Drosophila genes, CG11450 and delilah are represented. The evaluation of percentage conservation was done using the Blosum62 Similarity Scoring Table. Background intensities represent conservation (dark, 100% conservation; medium 80% or greater conserved; light, 60% or greater conserved). In this and all subsequent figures, the following abbreviations for species names are used: Bb, Branchiostoma belcheri (amphioxus); Bf, Branchiostoma floridae (amphioxus); Cc, Ceratitis capitata (a lower diptera); Ce, Caenorhabditis elegans; Ci, Ciona intestinalis (an ascidian); D and Dm, Drosophila melanogaster; Dp, Drosophila pseudoobscura; Dr, (Brachy)danio rerio (zebrafish); Ds, Drosophila simulans; Dy, Drosophila yakuba; Gg, Gallus gallus (chick); Hs, Homo sapiens; Hv, Hydra vulgaris; Jc, Juonia coenia (buckeye butterfly); Mm, Mus musculus; Ol, Oryzias latipes (Japanese medaka); Os, Oryza sativa (rice); Rn, Rattus norvegicus; Sb, Soybean (Glycina max); Sc, Saccharomyces cerevisiae (yeast); Tg, Tulipa gesneriana; Tr, Takifugu rubripes (pufferfish); Xl, Xenopus laevis; Zm, Zea mays (maize).
Drosophila Genes
We found 56 bHLH sequences in D. melanogaster. Table 2 lists these sequences, the family to which they belong, their chromosomal localization, their characterization status, and their accession number. A version of this table with links to Flybase (http://flybase.bio.indiana.edu; The Flybase Consortium 1999) is available as supplementary material at http://www.genome.org.
Table 2.
The Complete List of bHLH Genes from Drosophila melanogaster
| Gene name | Localization | Family | Status | Accession no. |
|---|---|---|---|---|
| tap (biparous) | 74B1-2 | Neurogenin | 3 | emb|CAA65103.1 |
| daughterless | 31D11-E1 | E12/E47 | 4 | pir|A31641 |
| nautilus | 95B3-5 | MyoD | 3 | SW:P22816 |
| achaete | 1B1 | Achaete-Scute | 4 | gb|AAF45498.1 |
| scute | 1B1 | Achaete-Scute | 4 | gb|AAA28313.1 |
| letal of scute | 1B1 | Achaete-Scute | 4 | gb|AAF45500.1 |
| asense | 1B1 | Achaete-Scute | 4 | gb|AAF45502.1 |
| CG8667 | 39D3 | Mist | 2 | gb|AAF53991.1 |
| CG5545 | 36C6-7 | Beta3 | 2 | gb|AAF53631.1 |
| cato | 53A1-2 | Atonal | 2 | gb|AAF58026.1 |
| ato | 84F6 | Atonal | 4 | gb|AAF54209.1 |
| amos | 37A1-2 | Atonal | 3 | gb|AAF53678.1 |
| delilah | 97B1-2 | NeuroD? | 3 | gb|AAF56590.1 |
| CG11450 (net) | 21A5-B1 | NeuroD? | 3 | gb|AAF51562.1 |
| HLH54F | 54E7-9 | MyoR | 2 | gb|AAF57795.1 |
| CG12952 | 85D7-10 | Mesp | 2 | gb|AAF54351.1 |
| CG12648 | 9A4 | Paraxis | 1 | SPTREMBL:Q9W2Z5 |
| twist | 59C2-3 | Twist | 4 | emb|CAA32707.1 |
| CG6913 | 86F1-2 | PTF1 | 2 | gb|AAF54684.1 |
| CG5952 | 89B9-12 | PTF1 | 2 | gb|AAF55280.1 |
| CG10066 | 84C3-4 | PTF1 | 2 | gb|AAF54058.1 |
| CG18144 | 31D1-6 | Hand | 2 | gb|AAF52900.1 |
| HLH3b | 3B3-4 | SCL | 2 | gb|AAF45802.1 |
| HLH4C | 4C6-7 | NSCL | 2 | gb|AAF45967.1 |
| max | 76A3 | Max | 2 | gb|AAF49179.1 |
| CG17592 | 4C4 | USF | 2 | gb|AAF45953.1 |
| crp | 35F6-7 | AP4 | 1 | gb|AAF53510.1 |
| CG3350 | 97F5-6 | TF4 | 1 | gb|AAF56696.1 |
| CG18362 | 39D1-2 | TF4 | 1 | gb|AAF53989.1 |
| HLH106 | 76D1-3 | SREBP | 1 | gb|AAF49115.1 |
| diminutive | 3D3-4 | Myc | 3 | gb|AAB39842.1 |
| clock | 66A11-B1 | Clock | 4 | gb|AAD10630.1 |
| Rst(1) JH | 10C6-8 | Clock ? | 3 | gb|AAC14350.1 |
| CG6211 | 13C1 | Clock ? | 2 | gb|AAF48439.1 |
| CG12561 | 96F14-97A1 | AHR | 1 | gb|AAF56569.1 |
| spineless | 89C1-2 | AHR | 4 | gb|AAD09205.1 |
| single-minded | 87D12-13 | Sim | 4 | gb|AAF54902.1 |
| trachealess | 61C1 | Trh | 4 | gb|AAA96754.1 |
| Hif-1A | 99D5-F1 | Hif | 2 | gb|AAC47303.1 |
| tango | 85C5-7 | ARNT | 4 | gb|AAF54329.1 |
| cycle (MOP3) | 76D2-3 | BMAL | 3 | gb|AAF49107.1 |
| extramacrochaete | 61D1-2 | Emc | 4 | gb|AAF47413.1 |
| Hey | 43F9-44A1 | Gridlock | 1 | gb|AAF59152.1 |
| Sticky ch1 | 86A5-6 | Gridlock | 3 | gb|AAF24476.1 |
| hairy | 66D11-12 | Hairy | 4 | mb|CAA34018.1 |
| deadpan | 44B3-4 | Hairy | 4 | gb|AAB24149.1 |
| E(spl) m3 | 96F10-12 | E (spl) | 4 | gb|AAF56550.1 |
| E(spl) m5 | 96F10-12 | E (spl) | 4 | emb|CAA34552.1 |
| E(spl) m8 | 96F10-12 | E (spl) | 4 | sp|P13098 |
| E(spl) m7 | 96F10-12 | E (spl) | 4 | emb|CAA34553.1 |
| E(spl) mB | 96F10-12 | E (spl) | 4 | gb|AAA28910.1 |
| E(spl) mC | 96F10-12 | E (spl) | 4 | gb|AAA28911.1 |
| E(spl) mA | 96F10-12 | E (spl) | 4 | gb|AAA28909.1 |
| CG10446 | 37B9-11 | Hairy | 2 | gb|AAF53741.1 |
| CG5927 | 17A3 | E (spl) | 2 | gb|AAF48810.1 |
| collier | 51C2-5 | COE | 4 | gb|AAF58204.1 |
Sequences are listed by the family in which they are included (or stated as orphan genes), their chromosomal localization (position on the polytene chromosomes map as found in Flybase), their accession number and their characterization status (1, sequence only; 2, expression pattern known; 3, preliminary functional data exist; 4, exhaustive characterization has been done).
We believe that these sequences represent, if not the full set, at least a large proportion of the bHLH domains present in the fly genome. The repeated BLASTP searches that were used to build our original set of genes were meant to detect even very divergent types of bHLH domains. Furthermore, after we determined the 44 types of bHLH domains, we made new BLASTP screens of the complete sequence of D. melanogaster with one member of each family, without finding any new genes. On the other hand, none of these searches revealed Collier, the fly COE family representative. Therefore it is conceivable that one or more highly divergent HLH families may have escaped our screens.
The BLASTP searches detected additional sequences that we did not use in our analyses, as they did not correspond to complete HLH motifs. Such sequences were identified because they present a marked similarity with a small region of the bHLH domain, 20–30 amino acids long, often including the basic region. In all cases we checked the sequence by hand, and the decision as to whether a sequence did or did not correspond to a bona fide bHLH domain was always clearcut. We also checked the 61 “HLH DNA-binding domain” and 69 “Myc-type HLH dimerization domain” sequences recently identified in the Drosophila genome (Rubin et al. 2000), and found that only the 56 sequences listed in Table 2 correspond to complete bHLH domains. Our analysis is completely consistent with and extends that of Moore et al. (2000), who analyzed 12 previously uncharacterized bHLH from the Drosophila genome project. We also retrieved these 12 genes in our screen and our family assignment coincides with that of Moore et al. (2000).
C. elegans Genes
We found 35 bHLH sequences in C. elegans. Table 3 shows these sequences; a version of this table with links to Wormbase (http://www.wormbase.org) is available as supplementary material at http://www.genome.org. A previous report (Rubin et al. 2000) mentioned 38 “HLH DNA-binding domain” sequences and 8 “Myc-type HLH dimerization domain” sequences in the C. elegans genome. Prior analysis of the C. elegans genome revealed only 24 bHLH putative proteins (Ruvkun and Hobert 1998). Here again we checked the discrepancies between our results and the previous ones, and found that only the 35 sequences listed in Table 3 correspond to complete bHLH domains. These 35 sequences are likely to represent the full set of C. elegans bHLH.
Table 3.
The Complete List of bHLH Genes from Caenorhabditis elegans
| Gene name | Localization | Family | Status | Accession No. |
|---|---|---|---|---|
| C34E10.7 (cnd-1) | III: −2,01 | NeuroD | 3 | sp|P46581 |
| Y69A2AR | **** | Neurogenin | 1 | **** |
| HLH-2 (MO5B5.5) | I: 1,82 | E12/E47 | 3 | TR:Q17588 |
| HLH-1 (BO3O4.1) | II: −4,51 | MyoD | 4 | SW:P22980 |
| HLH-3 (T29B8.6) | II: 1 | Achaete-Scute | 1 | gb|AAB38323.1 |
| C18A3.8 | II: 1,11 | Achaete-Scute | 1 | TR:Q09961 |
| F57C12.3 | X: −19,47 | Achaete-Scute | 1 | TR:Q20941 |
| C28C12.8 | IV: 3,86 | Achaete-Scute | 1 | TR:Q18277 |
| F38C2.2 | IV: 24,06 | Beta3 | 1 | TR:O45489 |
| DY3.3 | I: 3,04 | Beta3/Misti | 1 | TR:O45320 |
| lin-32 (T14F9.5) | X: −15,13 | Atonal | 3 | TR:10574 |
| ZK682.4 | V: 1,87 | MyoR | 1 | TR:Q23579 |
| HLH-8 (CO2B8.4) | X: −0,63 | Twist | 4 | gb|AAC26105.1 |
| C44C10.8 | X: 5,8 | Hand | 1 | TR:Q18612 |
| F48D6.3 | X: −8,42 | PTF1 | 1 | TR:Q20561 |
| C43H6.8 | X: −14 | NSCL | 1 | TR:Q18590 |
| F46G10.6 | X: 12,32 | Max | 1 | TR/Q18711 |
| T19B10.11 | V: 3,05 | Max | 1 | TR:P90982 |
| F40G9.11 | III: −28,29 | USF | 1 | gb|AAC68792 |
| W02C12.3 | IV: −1,14 | MITF | 1 | TR:P91527 |
| F58A4.7 | III: 0,63 | AP4 | 1 | SW:P34474 |
| Y47D3B.7 | III: 8,9 | SREBP | 1 | TR:Q9XX00 |
| C15C8.2 | V: 4,63 | AHR | 1 | emb|CAA99775.1 |
| C41G7.5 | I: 3,75 | AHR | 1 | emb|CAB51463.1 |
| F38A6.3 | V: 27,08 | Hif/Sim/Trh | 1 | pir|T21944 |
| C25A11.1 (AHA-1) | X: 0,43 | ARNT | 1 | TR:O02219 |
| unc-3(Y16B4A.1) | X: 19,39 | COE | 4 | gb|AAC06226.1 |
| lin-22 | IV: 6,9 | E (spl) | 3 | gb|AAB68848.1 |
| C17C3.10 | II: −1,28 | E (spl) | 1 | gb|AAB52693.1 |
| Y39A3CR.6 | III: −19,16 | Orphan | 1 | gb|AAF605231 |
| T01D3.2 | V: 5,39 | Orphan | 1 | TR:P90953 |
| T15H9.3 | II: 1,51 | Orphan | 1 | emb|CAA87416.1 |
| T01E8.2 | II: 2,22 | Orphan* | 1 | emb|CAA88744.1 |
| F31A3.4 | X: 24,06 | Orphan* | 1 | TR:Q19917 |
| C17C3.8 | II: 1,28 | Orphan* | 1 | TR:Q18053 |
The localization of the genes referred to the worm recombination genetic map as found in Wormbase. Sequences marked with an asterisk form a group with good bootstrap support (70%) which is found at the root of group A. The Y69A2AR gene was not found in the databases. Its sequence comes from Hallam et al. (2000).
In contrast to the sequences from Drosophila, most of which can easily be assigned to one of the 38 animal bHLH families, 17% of the C. elegans sequences (6/35) cannot be confidently assigned to a specific family, and are therefore called “orphan”. Furthermore, several C. elegans bHLH included in families are only loosely linked to the other members (their inclusion is supported by low bootstrap values). Conversely, 40% of the animal families do not contain C. elegans members. These results are consistent with the traditional view of metazoan phylogeny, which held nematodes as very distantly related to both arthropods and vertebrates. Recent molecular phylogenies indicate that, on the contrary, arthropods and nematodes are relatives, (i.e., they group into one of the three clades of bilaterians, the ecdysozoa) (Aguinaldo et al. 1997; Adoutte et al. 2000). Many nematodes, including C. elegans, have higher mutation rates than other metazoans, not only in their rRNA genes (Aguinaldo et al. 1997), but also throughout their genome (Mushegian et al. 1998). Therefore, nematode sequences, tend to be artifactually displaced to a wrong position because they appear as being very distant from all others, and to end up at the base of the tree or even associated with the outgroup (because of chance convergence at some nucleotide positions). This phenomenon, known as “long branch attraction phenomenon” (for a recent review, see Philippe and Laurent 1998), presumably explains why our analysis led to the clustering of several C. elegans sequences at the base of the group A family, or as orphan group B genes (Table 3). Accordingly, we found that the worm bHLH sequences diverge more rapidly than those of fly and mouse (data not shown; a detailed analysis can be found on our Web site, http://www.cnrs-gif.fr/cgm/evodevo/bhlh/index.html).
Interestingly, some nematode sequences have diverged very little from their fly or mouse counterparts. These include the few functionally characterized C. elegans bHLH genes, which show overall functional conservation with their vertebrates and/or fly orthologs; for example, the C. elegans orthologs of twist and myoD are involved in muscle formation (Harfe et al. 1998a,b), and the orthologs of atonal and NeuroD (lin-32and cnd-1) play a role in nervous system development (Zhao and Emmons 1995; Hallam et al. 2000). The genetic control of developmental processes such as neurogenesis and myogenesis relies on small sets of interacting genes (syntagms Garcia-Bellido 1981). The function of syntagms crucially relies on specific molecular interactions among their members, hence imposing strong structural constraints on them and preventing structural diversification (for discussion on syntagms and evolution, see Huang 1998). This may explain why such networks are strongly conserved throughout metazoan evolution (Baylies et al. 1998; Arendt and Nübler-Jung 1999) and why nematode genes involved in such networks have been subject to special constraints.
Mouse Genes
We found a total of 90 different bHLH sequences in mouse (and related mammals). This large set of genes is the result of the extensive molecular analyses of processes such as neurogenesis, myogenesis, or oncogenesis in which bHLH are crucially involved. Therefore, it might be that in the absence of systematic bHLH searches or genome sequencing projects, only a small subset of vertebrate bHLH genes have been identified so far. Indeed, our initial searches showed that the same vertebrate bHLH genes may be reported under up to seven different names, suggesting the convergence of many research groups on small numbers of crucial genes.
However, our results show that at least 35 of the 38 vertebrate bHLH types have protostomian (fly and/or worm) orthologs (90%), and reciprocally, that all fly genes have mouse counterparts. Because we believe that our set of fly genes is close to complete, the fact that mouse counterparts have been identified for all fly genes suggests that our sample of bHLH genes in mouse is in fact quite extensive. Needless to say, a definite answer to the question of how complete is our knowledge of vertebrate bHLH genes will have to await the results of the various vertebrate sequencing projects that currently are under way.
Assessing Orthologies
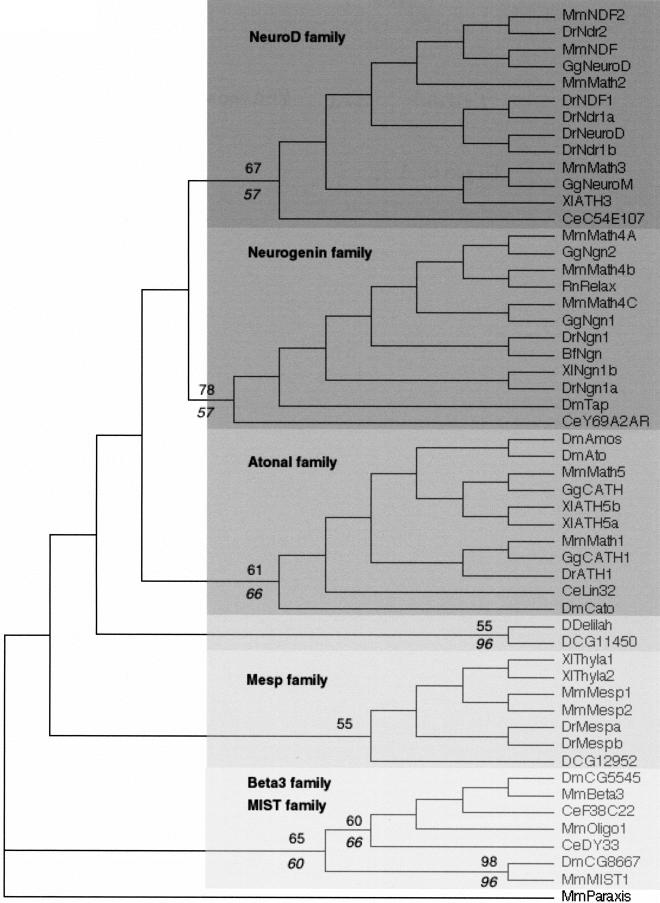
The assesment of orthologies must necessarily be based on phylogenetic reconstructions. Thus, although orthology is a very useful concept, there is no foolproof way of deciding whether two similar sequences are indeed orthologous. We will illustrate this difficult question in the case of two closely related fly genes, Delilah (dei) and CG11450 (Fig. 1 and 3). CG11450 recently has been described, based on overall similarity, as the Drosophila ortholog of the vertebrate NeuroD gene (Hassan and Bellen 2000). We similarly retrieved CG11450 as the closest Drosophila relative of NeuroD when making BLASTP searches. However, the inclusion of both genes in the NeuroD family is, not supported by the phylogenetic analyses. While both genes clearly belong to the Atonal superfamily, they cannot be associated unequivocally to either of the NeuroD, Ngn or Ato families (Fig. 2 and 3). Nevertheless, CG11450 and Dei may represent divergent NeuroD proteins as they show several residues in their bHLH typical of this family (Fig. 1; Hassan and Bellen 2000).
Figure 3.
A rooted neighbor-joining (NJ) tree showing the evolutionary relationships of Atonal superfamily members. We used the closely related paraxis sequence (see Figure 2) as outgroup. The different constituting families are pointed out. Numbers above branches indicate percent support for the nodes defining the families in distance bootstrap analyses (1000 replicates). Italicized numbers below branches indicate percent reliability in a puzzle maximum-likelihood (ML) tree. The Mesp family is not supported by ML analyses. Deep nodes are not supported by resampling methods. Note that delilah and CG11450 cluster together but are not associated to any vertebrate or nematode genes. As in Figure 2, the tree shown has branch lengths that are not proportional to distance between sequences. The alignments of which this tree is based as well as maximum parsimony (MP), ML, phylogram, and bootstrapped trees can be found at http://www.cnrs-gif.fr/cgm/evodevo/bhlh/index.html. Abbreviations are as listed in Figure 1.
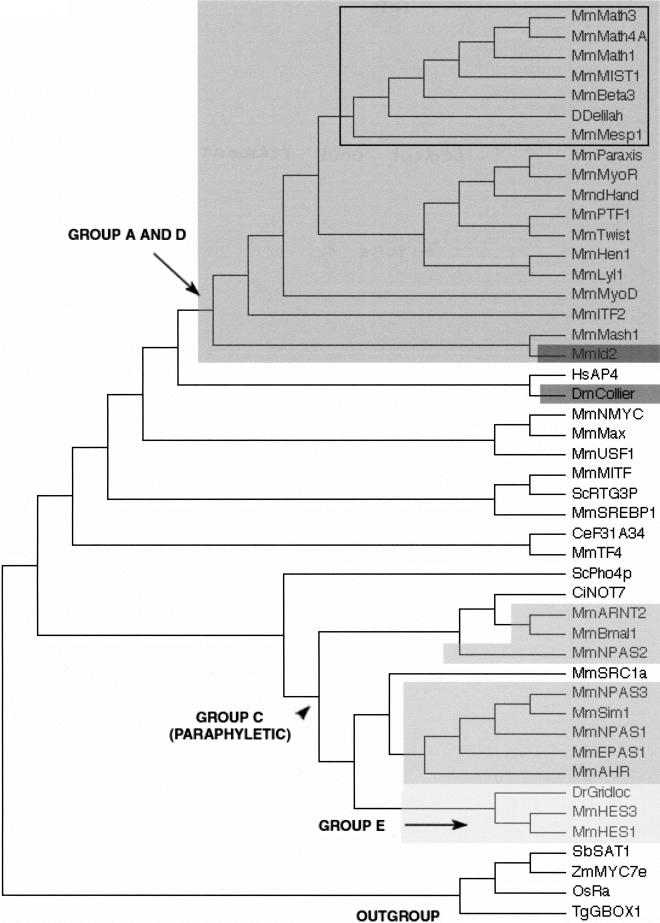
Figure 2.
A neighbor-joining (NJ) tree showing the evolutionary relationships of the 44 bHLH families listed in Table 1 as well as the orphan genes delilah (putative D. melanogaster neuroD gene) and F31A3.4 (as a representative of a group of three C. elegans genes that cluster together with high bootstrap value; see Table 3). We used one gene (usually from mouse) per family to construct this tree. Although there are strong theoretical reasons for preferring the unrooted tree, we show a rooted tree because it is easier to display compactly and more clearly represents the relationships at the tip of the branches. This tree is just a representation of an unrooted tree with rooting that should be considered arbitrary. We used the four plant bHLH families as outgroup. For similar sake of simplicity, we show a tree in which branch lengths are not proportional to distances between sequences. A tree with meaningful branch lengths can be found at http://www.cnrs-gif.fr/cgm/evodevo/bhlh/index.html. The two monophyletic high-order groups A + D (large, dark-gray box and arrows) and E (very light-gray box) are highlighted. The Emc family (the high-order group D of Atchley and Fitch 1997; represented in our tree by Mm Id2) is shown in a black box and the group F (COE family) in a dark gray box. The bHLH-PAS families (the high-order group D of Atchley and Fitch 1997) are shown in intermediate gray boxes. Their last common ancestor (arrowhead) is also that of non bHLH-PAS families and the group is hence paraphyletic. Finally, all the other families were included in the high-order group B of Atchley and Fitch (1997), a group that appears to be paraphyletic (the common ancestor of these families is that of all bHLH genes). The Atonal superfamily is pointed out (black square) and is detailed in Figure 3. Abbreviations are as listed in Figure 1. The alignment on which this tree is based and complementary phylogenetic analyses are available at http://www.cnrs-gif.fr/cgm/evodevo/bhlh/index.html.
We examined whether what is known of the function of these various genes might help us elucidate the origin of dei and CG11450. The vertebrate representatives of the Ngn, NeuroD and Ato families are mainly involved in the determination and the differentiation of neural cells (Kageyama and Nakanishi 1997; Hassan and Bellen 2000). In Drosophila, the Ato representatives ato, amos and cato are all involved in neural development (Jarman et al. 1993; Goulding et al. 2000a,b; Huang et al. 2000). The function of the neurogenin ortholog target of poxn (tap) is not known, but the gene is exclusively expressed at late stages of neural development (Bush et al. 1996; Gautier et al. 1997; Ledent et al. 1998). On the contrary, dei and CG11450 are not involved in neurogenesis. dei is required for the differentiation of specific epidermal cells as muscle attachment sites (Armand et al. 1994). CG11450 is expressed in the embryonic mesoderm in a pattern that overlaps that of twist (Moore et al. 2000). During postembryonic development, CG11450 is involved in wing vein formation (CG11450 corresponds to the net locus; Brentrup et al. 2000). Thus, one plausible interpretation of the data is that dei and CG11450 are bona fide orthologs of the NeuroD genes, and that their phylogenetic relationships have been blurred by a rapid divergence associated to the acquisition of new functions.
Comparison of Fly, Nematode and Vertebrate Families
Most families comprise one protostome (fly and/or nematode) and several (often two) vertebrate genes. The fact that most families contain both fly and vertebrate genes suggests that there was no addition of new bHLH types in the corresponding lineages, and therefore no important diversification of the ancestral repertoire. Among the few families that lack fly genes, most also lack nematode genes. These may represent the arisal of new bHLH types in the vertebrate lineage, or alternatively a loss of ancestral types in both fly and nematode. The analysis of bHLH genes from molluscs or annelids might help settle this question. It is now widely believed that bilateria (triploblastic metazoans) are composed of three main lineages: deuterostomes (which include vertebrates and echinoderms) and protostomes themselves including two large groups, the ecdysozoans (e.g., arthropods and nematodes) and the lophotrochozoans (e.g., annelids, molluscs, flatworms) (e.g., Aguinaldo et al. 1997; de Rosa et al. 1999; Adoutte et al. 2000). Therefore, the finding of ortholog genes in vertebrates and lophotrochozoans but not in fly and nematode would strongly suggest that gene loss(es) has occurred in the ecdysozoan lineage. Similarly, the case of families that contain vertebrate and either worm or fly genes is explained best by gene losses that occurred, inside the ecdysozoan clade, in either lineage after the arthropod/nematode divergence. This occurred in the fly lineage for only one family, MITF, which contains vertebrate and worm but no fly genes (the case of the NeuroD family has been discussed above). The much larger number of families that have vertebrate and fly members but no nematode representative, as well as the large number of nematode genes that cannot be clearly assigned to specific families (orphan genes) is likely because of the high divergence rate reported for nematode genes in general (Aguinaldo et al. 1997; Mushegian et al. 1998) and that we found within our specific data set (data not shown; for details, see our Web site at http://www.cnrs-gif.fr/cgm/evodevo/bhlh/index.html).
Gene and Genome Duplications
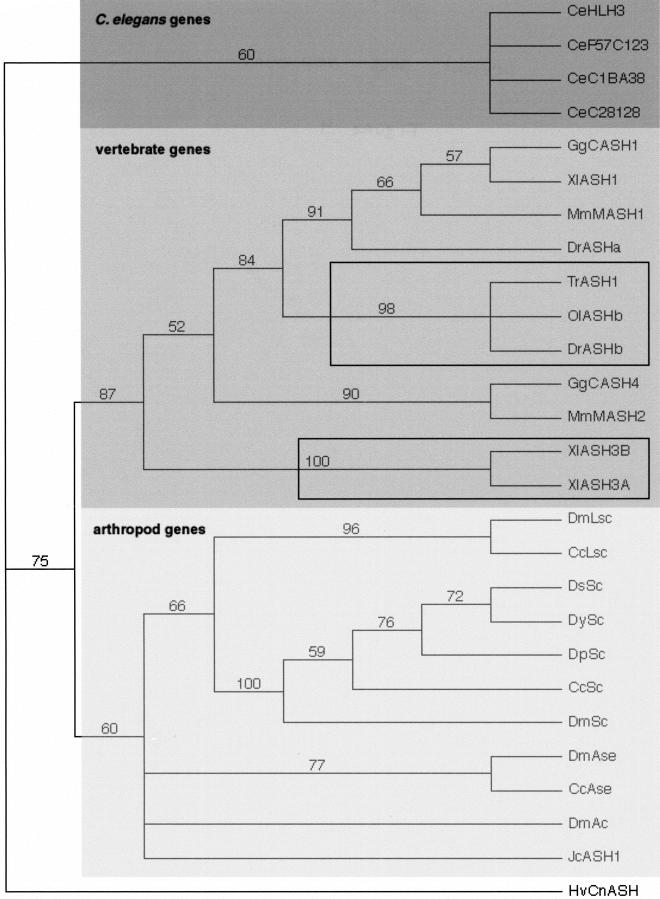
Most bHLH families, as other gene families, comprise more members in vertebrates than in other phyla (Table 1). It has been proposed that this may reflect the occurrence of two rounds of genome duplication during the early vertebrate evolution (Sidow 1996; Meyer and Schartl 1999), but this idea, mainly based on mapping of gene clusters, remains controversial (Skrabanek and Wolfe 1998; Hughes 1999; Smith et al. 1999; Martin 2001). Many gene families in vertebrates have less than four genes (Skrabanek and Wolfe 1998; Smith et al. 1999). However, this might result from gene loss during or after the rounds of duplication (Meyer and Schartl 1999). Within our set of bHLH genes, the most usual case was two mouse genes per family, but we know this set is likely to be incomplete because the entire genomic sequence of the mouse is not available. Even within this incomplete set, we observe that up to one-fourth of the families comprise four or more members (Table 1). As pointed out by Hughes (1999), the presence of four vertebrate members, by itself, does not support the genome duplication hypothesis. Support only may come from families whose phylogenetic tree shows a topology of the (AB) (CD) form (i.e., two pairs of two closely related paralogs) (Hughes 1999). Hughes (1999) discussed the phylogenies of 13 protein families important in development and found that only one of them shows a (AB) (CD) topology. We constructed individual trees for each bHLH family (available at http://www.cnrs-gif.fr/cgm/evodevo/bhlh/index.html) and often found one or two duplication(s) during vertebrates radiation (e.g., the Achaete-Scute family; Fig. 4). We checked the topology of the trees of families with four or more vertebrate members (nine families, see Table 1) and observed that none of the five families showing a reliable phylogeny, has a (AB) (CD) topology (data not shown; see http://www.cnrs-gif.fr/cgm/evodevo/bhlh/index.html). Hence, our data set does not support the hypothesis of two rounds of genome duplication. Figure 4 also shows a feature we observed in several families: the existence of extra closely related genes in the tetraploid Xenopus and in ray-finned fishes such as the zebrafish Brachydanio rerio (actinopterygia). The latter observation is consistent with the hypothesis than actinopterygia genome underwent a duplication, which took place after actinopterygian-sarcopterygian lineage divergence (the sarcopterygian lineage include coelacanths, lungfishes, and all tetrapods) (reviewed in Wittbrod et al. 1998; Meyer and Schartl 1999).
Figure 4.
A neighbor-joining (NJ) tree showing the evolutionary relationships among Achaete-Scute family members. Numbers above branches indicate percent support in bootstrap analyses (1000 replicates). This tree is rooted using the single cnidarian (Hv CNASH) member as outgroup. As in Figure 3, the rooting should be considered arbitrary. The four fly achaete-scute genes are collectively orthologous to the four worm genes and to two subfamilies of vertebrate genes, the MASH-1 and MASH-2 related genes, respectively. The three closely related actinopterygians sequences (TrASH1, OlASHB, and DrASHb) and the two closely related Xenopus sequences are pointed out (black squares). Note also the basal position of the Juonia coenia sequences (Jc ASH1), inside the arthropod clade, that indicates that this gene is most probably the single ortholog of the three or four achaete-scute genes found in diptera. The alignments of which this tree is based as well as maximum parsimony (MP), maximum likelihood (ML), phylogram, and bootstrapped trees can be found on our Web site. Abbreviations are as listed in Figure 1.
Duplications of Ecdysozoan bHLH
A few families contain more than one gene in fly and/or nematode, and in some cases, more genes than in vertebrates: the Achaete-Scute, Atonal, PTF1, Enhancer of split, Hairy, AHR, and TF4 families in Drosophila and AHR, Enhancer of split, and Max families in C. elegans (Table 1). The different protostome members of these families arose by duplications that occurred after the arthropod/nematode split within the ecdisozoan clade: For example, the four Drosophila achaete-scute genes are collectively orthologous to the two vertebrate genes and to the four nematode genes (Fig. 4); the three Drosophila Atonal genes are collectively orthologous to the two vertebrate and the single nematode genes (Fig. 3). We retrieved the chromosomal localizations of these genes from Flybase and Wormbase and observed that, in most cases, the members of a given family have very different localizations, often on different chromosomes (Table 2 and 3). The three Drosophila Atonal genes, for example, are found on three different chromosomes arms (2L, 3L and 3R; see Table 2). These localizations suggest that the duplications that gave rise to the paralogs are rather ancient events. However, in some cases, the duplications might have occurred more recently, as the paralogs are localized close to each other in the genome: this is, for example, the case of the four achaete-scutegenes and the seven Enhancer of split genes which are known for long time to form gene complexes in Drosophila. We found one similar case in C. elegans: C17C3.10 and C17C3.8 are adjacent genes and are on the same DNA strand. In addition, two worm members of the Achaete-Scute family are found at a similar chromosomal localization (Table 3), although separated by several unrelated genes. Information about the timing of duplication events may come from evolutive comparisons with increasingly distantly related species. For example, clear orthologs of three of the four achaete-scute genes have been found in another dipteran, Ceratitis capitata (Wülbeck and Simpson 2000), while a single ortholog to the four achaete-scute genes is found in the buckeye butterfly, Juonia coenia, a lepidoptera, and in the flour beetle, Tribolium castaneum, a coleoptera (Figure 4; Galant et al. 1998). Duplication, in this case, probably has occurred after the divergence of diptera from other insects.
Phylogenetic Relationships of bHLH Families: A Reappraisal of High-Order
Although the bHLH motif has good resolving power to delimit families of proteins and describe their evolutionary relationships at the tips of the clades, the very early evolutionary history of the motif is more problematic (Atchley and Fitch 1997). Deep nodes usually have a low statistical support (small bootstrap values). This is mainly a result of the small size of the conserved sequence and the existence of numerous ancient paralogs. Nevertheless, we found recurrent topologies when constructing trees with different sequences sets and different tree reconstruction procedures [maximum parsimony (MP), distance, and maximum likelihood (ML)]. The congruence between trees obtained with different methods and different data sets is usually considered in phylogenetic reconstructions as a good argument in favor of the validity of a given phylogeny (Adoutte et al. 2000); however, it is not a demonstration of its reliability. A representative tree of the different bHLH families is shown in Figure 2. Our results agree largely with those of Atchley and Fitch (1997) who described the four high-order groups (A–D) found in a neighbor-joining (NJ) tree and subsequent work of Atchley and collaborators (Atchley et al. 1999; Morgenstern and Atchley 1999). Although the high-order groups were supported only by low bootstrap values, their validity was confirmed by MP analyses of particular sites at different positions in the bHLH (Atchley and Fitch 1997), analyses of bHLH flanking regions (Morgenstern and Atchley 1999) and mathematical modeling (Atchley et al. 1999). The inclusion of the 44 orthologous families in the high-order groups is shown in Table 1.
Our results diverge from the previous analyses in a few points, however. First, we have had to revise the relationship between groups A and D, and to include group D within group A. Second, our analysis suggests that group B is paraphyletic and closest to the ancestral bHLH motif. Third, we have evidence that group C is not monophyletic but includes several independent occurrences of the bHLH-PAS association. Finally, the more extensive data set used in the present study led us to define two additional groups, E and F.
Our phylogenetic analysis (Fig. 2) reveals a large monophyletic group that corresponds to the group A defined by Atchley and Fitch (1997). This group includes the E12/E47 family genes and several other families whose members are able to heterodimerize with the E12/E47 proteins (Cabrera and Alonso 1991; Lassar et al. 1991; Van Doren et al. 1992). The phylogenetic analysis (Fig.2) clearly shows that the Emc family is deeply embedded into the group A family. Furthermore, although group D proteins lack the DNA-binding motif, they are able to dimerize with several group A proteins (Benerzra et al. 1990; Ellis et al. 1990; Garrell and Modolell 1990; Van Doren et al. 1991, 1992), but not with other types of bHLH motifs. Therefore, our results indicate that the Emc family, previously considered to define group D, should also be considered as belonging to group A.
We believe that group B is paraphyletic rather than monophyletic (Fig. 2). This group is probably closest to the ancestral bHLH type from which groups A, C, D, E, and F bHLH arise. The distribution of these proteins in various groups of organisms stongly supports this suggestion: Group B proteins are found in plants, yeast, and animals, whereas the other groups (A, C, D, E, and F) are found only in animals. Likewise, we did not find the group C of Atchley and Fitch (1997) to form a monophyletic group (Fig. 2). As this group comprises the bHLH-PAS genes, one obvious explanation for its paraphyly is that the association between the bHLH and the PAS domains occurred several times independently, consistent with the hypothesis of a modular evolution of the bHLH proteins by domain shuffling (for discussion, see Morgenstern and Atchley 1999).
We found that all Hairy and Enhancer of split-related proteins form a well-supported monophyletic group that we named group E in accordance with Atchley and Fitch nomenclature (Fig. 2). The monophyly of this group is confirmed by the presence of several conserved amino acids flanking the bHLH and the presence of the WRPW peptide (Fisher and Caudy 1998).
Similarly, the HLH domain of the COE proteins appears well conserved among them and much divergent with respect to other bHLH families. Furthermore, all COE proteins contain a highly conserved domain, the COE domain, not found in any other proteins. Taken together, this strongly indicates that the COE proteins form a clearly distinct monophyletic group, which we named group F.
Conclusions: An Overview of bHLH Evolution
We have not been able to identify procaryotic genes that would match our bHLH sequences. Therefore, it seems that the bHLH motif has been established in early eukaryote evolution. The bHLH genes of yeast are involved in general transcriptional enhancement and cell cycle control, suggesting that this may have been the original function of the bHLH genes in primitive eukaryotes. An important diversification occurred independently in the animal and plant lineages, as seen by the 36 different families found exclusively in animals and 30 different bHLH genes found in A. thaliana, compared to the five genes found in yeast.
In animals, bHLH genes generally are involved in development and in tissue-specific gene regulation. The 38 families have representatives in the two major subdivisions of the animal kingdom, protostomes and deuterostomes, and must therefore have been represented in their common ancestor prior to the Cambrian radiation, which saw the emergence of all present-day phyla and many extinct ones. Morphologically, these ancestors (also called Urbilateria; De Robertis and Sasai 1996) probably were coelomates with antero-posterior and dorso-ventral polarity, rudimentary appendages, some form of metamerism, a heart, sense organs such as photoreceptors, and a complex nervous system (Knoll and Carroll 1999). Genetically, they possessed numerous Hox genes (at least seven; de Rosa et al. 1999) as well as other homeobox genes, several intercellular signaling pathways (TGF-Β, Hedgehog, Notch, EGF), and several Pax genes (Galliot et al. 1999). Our analysis indicates that their genome contained at least 35 different bHLH genes. The functional conservation that often is observed between fly and vertebrate bHLH orthologs indicates that some of the developmental functions associated with present-day bHLH genes already were established in these ancestral organisms, further indicating the genomic and developmental complexity of this ancient ancestor.
METHODS
Protein sequences were obtained mostly by BLASTP search (Altschul et al. 1990) at the National Center for Biotechnology (NCBI) and the Sanger center, as well as from Swissprot, GenPept, and TrEMBL through SRS (LION Bioscience AG) and Nentrez (NCBI) software. A table containing all sequences and their accession numbers is available on our Web site (http://www.cnrs-gif.fr/cgm/evodevo/bhlh/index.html). Protein alignments were carried out using CLUSTALW (Thompson et al. 1994) with no adjustment of the default parameters, and were subsequently edited and manually improved in Genedoc Multiple Sequence Alignment Editor and Shading Utility, Version 2.6.001 (Nicholas et al. 1997). The evaluation of percentage conservation of residues in multiple sequence alignments was done using the Blosum62 Similarity Scoring Table (Henikoff and Henikoff 1992). Only the bHLH motif (determined as in Ferre-D'Amar et al. 1993), plus a few flanking amino acids, was used in most of our analyses because the remaining part of proteins from independent clades are either not homologous or have so diverged that the alignments are meaningless. The facilities of the Belgian EMBnet Node (http://be.embnet.org) were used for all database searches through SRS and sequence analysis using Genedoc software, and for most of the protein alignments using CLUSTALW. Trees were built using unweighted maximum parsimony (MP) and neighbor-joining (NJ) algorithms with the PAUP 4.0 program (Swofford 1993). The MP analysis was performed with the following settings: heuristic search over 100 bootstrap replicates, MAXTREES set up to 1000 because of computer limitations, other parameters set to default values. When large numbers of sequences (>150) were handled, as a result of computer limitations, bootstraps were made by “fast” stepwise-additions (1000 replicates) in PAUP 4.0. Extensive computer simulations have shown that such fast algorithms are as efficient as more extensive search algorithms when a large number of sequences is used (Takahashi and Nei 2000). Distance trees were constructed with the NJ algorithm (Saitou and Nei 1987) using PAUP 4.0 based on a Dayhoff's PAM 250 distance matrix (Dayhoff et al. 1978). Bootstrap replicates of the NJ trees (1000) also were made with PAUP 4.0, parameters set to default values.
Some alignments also were analyzed by maximum likelihood (ML) using Puzzle 4.0.2 (Strimmer and Von Haeseler 1996). The ML was performed using the quartet puzzling tree search procedure with 10000 puzzling steps, using the Jones-Taylor-Thornton (JTT) model of substitution (Jones et al. 1992), the frequencies of amino acids being estimated from the data set (Strimmer and Von Haeseler 1996).
The trees were displayed with the TreeView (Version 1.5) (Page 1996), saved as PICT files, converted into JPEG files using Graphic Converter, and then annotated using Adobe Photoshop.
Acknowledgments
We thank Robert Herzog, Marc Colet, and André Adoutte for support. We are especially grateful to Alain Ghysen for his help in the writing of this article. We thank Daniel Van Belle for comments on protein structure. We also thank André Adoutte, Robert Herzog, Nicolas Lartillot, Michel Milinkovitch, and two anonymous referees for helpful comments on the manuscript. This work has been supported by the Federal Office for Scientific, Technical, and Cultural Affairs (V.L.) and Centre National de la Recherche Scientifique and Université de Paris-Sud (M.V.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL vervoort@cgm.cnrs-gif.fr; FAX 33 169 823160.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.177001.
REFERENCES
- Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Adoutte A, Balavoine G, Lartillot N, Lespinet O, Prud'homme B, de Rosa R. The new animal phylogeny: Reliability and implications. Proc Natl Acad Sci. 2000;97:4453–4456. doi: 10.1073/pnas.97.9.4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguinaldo AMA, Turbeville JM, Linford LS, Rivera MC, Garey JR, Raff RA, Lake JA. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature. 1997;387:489–493. doi: 10.1038/387489a0. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arendt D, Nübler-Jung K. Comparisons of early nerve cord development in insects and vertebrates. Development. 1999;126:2309–2325. doi: 10.1242/dev.126.11.2309. [DOI] [PubMed] [Google Scholar]
- Armand P, Knapp AC, Hirsch AJ, Wieschaus EF, Cole MD. A novel basic helix-loop-helix protein is expressed in muscle attachment sites of the Drosophilaepidermis. Mol Cell Biol. 1994;14:4145–4154. doi: 10.1128/mcb.14.6.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchley WR, Fitch WM. A natural classification of the basic helix-loop-helix class of transcription factors. Proc Natl Acad Sci. 1997;94:5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchley WR, Terhalle W, Dress A. Positional dependence, cliques, and predictive motifs in the bHLH protein domain. J Mol Evol. 1999;48:501–516. doi: 10.1007/pl00006494. [DOI] [PubMed] [Google Scholar]
- Baylies M K, Bate M, Gomez M R. Myogenesis: A view from Drosophila. Cell. 1998;93:921–927. doi: 10.1016/s0092-8674(00)81198-8. [DOI] [PubMed] [Google Scholar]
- Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: A negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- Brentrup D, Lerch H-P, Jäckle H, Noll M. Regulation of Drosophila wing vein patterning: net encodes a bHLH protein repressing rhomboidand is repressed by Rhomboid-dependent Egfr signalling. Development. 2000;127:4729–4741. doi: 10.1242/dev.127.21.4729. [DOI] [PubMed] [Google Scholar]
- Bush A, Hiromi Y, Cole M. biparous: A novel bHLH gene expressed in neuronal and glial precursors in Drosophila. Dev Biol. 1996;180:759–772. doi: 10.1006/dbio.1996.0344. [DOI] [PubMed] [Google Scholar]
- Cabrera CV, Alonso MC. Transcriptional activation by heterodimers of the achaete-scute and daughterless gene product of Drosophila. EMBO J. 1991;10:2965–2973. doi: 10.1002/j.1460-2075.1991.tb07847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Crews ST. Control of cell lineage-specific development and transcription by bHLH-PAS proteins. Genes & Dev. 1998;12:607–620. doi: 10.1101/gad.12.5.607. [DOI] [PubMed] [Google Scholar]
- Crozatier M, Valle D, Dubois L, Ibnsouda S, Vincent A. collier, a novel regulator of Drosophilahead development, is expressed in a single mitotic domain. Curr Biol. 1996;6:707–718. doi: 10.1016/s0960-9822(09)00452-7. [DOI] [PubMed] [Google Scholar]
- Dang CV, Dolde D, Gillison ML, Kato GJ. Discrimination between related DNA sites by a single amino acid residue of myc-related basic-helix-loop-helix proteins. Proc Natl Acad Sci. 1992;89:599–602. doi: 10.1073/pnas.89.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayhoff MO, Schwartz RM, Orcutt BC. A model of evolutionary change in proteins. In: Dayhoff MO, editor. Atlas of Protein Sequence Structure. 5, Suppl. 3. Washington DC.: National Biomedical Research Foundation; 1978. pp. 345–352. [Google Scholar]
- De Robertis DM, Sasai Y. A common plan for dorsoventral patterning in Bilateria. Nature. 1996;380:37–40. doi: 10.1038/380037a0. [DOI] [PubMed] [Google Scholar]
- de Rosa R, Grenier JK, Andreeva T, Cook CE, Adoutte A, Akam M, Carroll SB, Balavoine G. Hox genes in brachiopods and priapulids and protostome evolution. Nature. 1999;399:772–776. doi: 10.1038/21631. [DOI] [PubMed] [Google Scholar]
- Ellis HM, Spann DR, Posakony JW. Extramacrochaete, a negative regulator of sensory organ development in Drosophila, defines a new class of helix-loop-helix proteins. Cell. 1990;61:27–38. doi: 10.1016/0092-8674(90)90212-w. [DOI] [PubMed] [Google Scholar]
- Facchini LM, Penn LZ. The molecular role of Myc in growth and transformation: Recent discoveries lead to new insights. FASEB J. 1998;12:633–651. [PubMed] [Google Scholar]
- Ferre-D'Amare AR, Prendergast GC, Ziff EB, Burley SK. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature. 1993;363:38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- Fisher A, Caudy M. The function of hairy-related bHLH repressors proteins in cell fate decisions. BioEssays. 1998;20:298–306. doi: 10.1002/(SICI)1521-1878(199804)20:4<298::AID-BIES6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Fitch WM. Distinguishing homologous from analogous proteins. Syst Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- The Flybase Consortium. The FlyBase database of the DrosophilaGenome Projects and community literature. Nucleic Acids Res. 1999;27:85–88. doi: 10.1093/nar/27.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galant R, Skeath JB, Paddock S, Lewis D, Carroll SB. Expression pattern of a butterfly achaete-scutehomolog reveals the homology of butterfly wing scales and insect sensory bristles. Curr Biol. 1998;8:807–813. doi: 10.1016/s0960-9822(98)70322-7. [DOI] [PubMed] [Google Scholar]
- Galliot B, de Vargas C, Miller D. Evolution of homeobox genes: Q50Paired-like genes founded the Paired class. Dev Genes Evol. 1999;209:186–197. doi: 10.1007/s004270050243. [DOI] [PubMed] [Google Scholar]
- Garcia-Bellido A. The bithorax syntagma. In: Lakovaara S, editor. Advances in genetics, development, and evolution of Drosophila. VII European Drosophila Research Conference, Plenum Press; 1981. pp. 135–148. [Google Scholar]
- Garrell J, Modolell J. The Drosophila extramacrochaetelocus, an antagonist of proneural genes that, like these genes, encodes a helix-loop-helix protein. Cell. 1990;61:39–48. doi: 10.1016/0092-8674(90)90213-x. [DOI] [PubMed] [Google Scholar]
- Gautier P, Ledent V, Massaer M, Dambly-Chaudière C, Ghysen A. tap, a DrosophilabHLH expressed in chemosensory organs. Gene. 1997;191:15–21. doi: 10.1016/s0378-1119(97)00021-8. [DOI] [PubMed] [Google Scholar]
- Goding CR. Mitf from neural cest to melanoma: Signal transduction and transcription in the melanocyte lineage. Genes & Dev. 2000;14:1712–1728. [PubMed] [Google Scholar]
- Goulding SE, zur Lage P, Jarman AP. amos, a proneural gene for Drosophila olfactory sense organs that is regulated by lozenge. Neuron. 2000a;25:69–78. doi: 10.1016/s0896-6273(00)80872-7. [DOI] [PubMed] [Google Scholar]
- Goulding SE, White NM, Jarman AP. cato encodes a basic helix-loop-helix transcription factor implicated in the correct differentiation of Drosophilasense organs. Dev Biol. 2000b;221:120–131. doi: 10.1006/dbio.2000.9677. [DOI] [PubMed] [Google Scholar]
- Hallam S, Singer E, Waring D, Jin Y. The C. elegans NeuroD homolog cnd-1functions in multiple aspects of motor neuron fate specification. Development. 2000;127:4239–4252. doi: 10.1242/dev.127.19.4239. [DOI] [PubMed] [Google Scholar]
- Harfe B D, Branda C S, Krause M, Stern M J, Fire A. MyoD and the specification of muscle and non-muscle fates during postembryonic development of the C. elegansmesoderm. Development. 1998a;125:2479–2488. doi: 10.1242/dev.125.13.2479. [DOI] [PubMed] [Google Scholar]
- Harfe B D, Vaz Gomes A, Kenyon C, Liu J, Krause M, Fire A. Analysis of a Caenorhabditis elegansTwist homolog identifies conserved and divergent aspects of mesodermal patterning. Genes & Dev. 1998b;12:2623–2635. doi: 10.1101/gad.12.16.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan BA, Bellen HJ. Doing the MATH: Is the mouse a good model for fly development? Genes & Dev. 2000;14:1852–1865. [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson M, Luscher B. Proteins of the Myc network: Essential regulators of cell growth and differentiation. Adv Cancer Res. 1996;68:109–182. doi: 10.1016/s0065-230x(08)60353-x. [DOI] [PubMed] [Google Scholar]
- Holland PWH, Garcia-Fernandez JD. Hoxgenes and chordate evolution. Dev Biol. 1996;173:382–395. doi: 10.1006/dbio.1996.0034. [DOI] [PubMed] [Google Scholar]
- Huang F. Syntagms in development and evolution. Int J Dev Biol. 1998;42:487–494. [PubMed] [Google Scholar]
- Huang M-L, Hsu C-H, Chien C-T. The proneural gene amos promotes multiple dendritic neuron formation in the Drosophilaperipheral nervous system. Neuron. 2000;25:57–67. doi: 10.1016/s0896-6273(00)80871-5. [DOI] [PubMed] [Google Scholar]
- Hughes A. Phylogenies of developmentally important proteins do not support the hypothesis of two rounds of genome duplication early in vertebrate history. J Mol Evol. 1999;48:565–576. doi: 10.1007/pl00006499. [DOI] [PubMed] [Google Scholar]
- Jan YN, Jan LY. HLH proteins, fly neurogenesis, and vertebrate myogenesis. Cell. 1993;75:827–830. doi: 10.1016/0092-8674(93)90525-u. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Grau Y, Jan LY, Jan YN. atonal is a proneural gene that directs chordotonal development in the Drosophilaperipheral nervous system. Cell. 1993;73:1307–1321. doi: 10.1016/0092-8674(93)90358-w. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. CABIOS. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kadesch T. Consequences of heteromeric interactions among helix-loop-helix proteins. Cell Growth Differ. 1993;4:49–55. [PubMed] [Google Scholar]
- Kageyama R, Nakanishi S. Helix-loop-helix factors in growth and differentiation of the vertebrate nervous system. Curr Opin Genet Dev. 1997;7:659–665. doi: 10.1016/s0959-437x(97)80014-7. [DOI] [PubMed] [Google Scholar]
- Knoll AH, Carroll SB. Early animal evolution: Emerging views from comparative biology and geology. Science. 1999;284:2129–2137. doi: 10.1126/science.284.5423.2129. [DOI] [PubMed] [Google Scholar]
- Lassar A B, Davis R L, Wright W E, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H. Functional activity of myogenic HLH proteins requires hetero-oligodimerization with E12/E47-like proteins in vivo. Cell. 1991;66:305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- Ledent V, Gaillard F, Gautier P, Ghysen A, Dambly-ChaudiÈre C. Expression and function of tap in the gustatory and olfactory organs of Drosophila. Int J Dev Biol. 1998;42:163–170. [PubMed] [Google Scholar]
- Martin A. Is tetralogy true? Lack of support for the “one-to-four rule.”. Mol Biol Evol. 2001;18:89–93. doi: 10.1093/oxfordjournals.molbev.a003723. [DOI] [PubMed] [Google Scholar]
- Meyer A, Schartl M. Gene and genome duplications in vertebrates: the one-to-four (to-eight in fish) rule and the evolution of novel gene functions. Curr Opin Cell Biol. 1999;11:699–704. doi: 10.1016/s0955-0674(99)00039-3. [DOI] [PubMed] [Google Scholar]
- Moore AW, Barbel S, Jan LY, Jan YN. A genomewide survey of basic helix-loop-helix factors in Drosophila. Proc Natl Acad Sci. 2000;97:10436–10441. doi: 10.1073/pnas.170301897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern B, Atchley WR. Evolution of bHLH transcription factors: Modular evolution by domain shuffling. Mol Biol Evol. 1999;16:1654–1663. doi: 10.1093/oxfordjournals.molbev.a026079. [DOI] [PubMed] [Google Scholar]
- Murre C, Mc Caw PS, Vaessin H, Caudy M, Jan L Y, Cabrera C V, Buskin J N, Hauschka S D, Lassar A B, Weintraub H, et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989a;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Murre C, Mc Caw PS, Baltimore D. A new DNA binding and dimerizing motif in Immunoglobulin enhancer binding, Daugtherless, MyoD, and Myc proteins. Cell. 1989b;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Mushegian AR, Garey JR, Martin J, Liu LX. Large-scale taxonomic profiling of eukaryotic model organisms: A comparison of orthologous proteins encoded by the human, fly, nematode, and yeast genomes. Genome Res. 1998;8:590–598. doi: 10.1101/gr.8.6.590. [DOI] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB, Jr, Deerfield DWII. Genedoc: Analysis and visualization of Genetic Variation/ Embnew. News. 1997;4:14. [Google Scholar]
- Ohsako S, Hyer J, Panganiban, Olivier I, Caudy M. Hairy function as a DNA-binding helix-loop-helix repressor of Drosophilasensory organ formation. Genes & Dev. 1994;8:2743–2755. doi: 10.1101/gad.8.22.2743. [DOI] [PubMed] [Google Scholar]
- Page RD. TreeView: An application to display phylogenetic trees on personal computers. Comput Appl Biosc. 1995;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Philippe H, Laurent J. How good are deep phylogenetic trees? Curr Opin Gen Dev. 1998;8:616–623. doi: 10.1016/s0959-437x(98)80028-2. [DOI] [PubMed] [Google Scholar]
- Rubin GM, et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvkun G, Hobert O. The taxonomy of developmental control in Caenorhabditis elegans. Science. 1998;282:2033–2041. doi: 10.1126/science.282.5396.2033. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schreiber-Agus N, Stein D, Chen K, Goltz JS, Stevens L, DePinho RA. Drosophila Myc is oncogenic in mammalian cells and plays a role in the diminutivephenotype. Proc Natl Acad Sci. 1997;94:1235–1240. doi: 10.1073/pnas.94.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidow A. Gen(om)e duplications in the evolution of early vertebrates. Curr Opin Genet Dev. 1996;6:715–722. doi: 10.1016/s0959-437x(96)80026-8. [DOI] [PubMed] [Google Scholar]
- Skrabanek L, Wolfe KH. Eukaryote genome duplication-where's the evidence? Cur Opin Genet Dev. 1998;8:694–700. doi: 10.1016/s0959-437x(98)80039-7. [DOI] [PubMed] [Google Scholar]
- Smith NGC, Knight R, Hurst LD. Vertebrate genome evolution: A slow shuffle or a big bang? BioEssays. 1999;21:697–703. doi: 10.1002/(SICI)1521-1878(199908)21:8<697::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Strimmer K, von Haeseler A. Quartet puzzling: A quartet maximum likelihood method for reconstructing tree topologies. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- Swofford D L. PAUP* Phylogenetic Analysis Using Parsimony, Version 4. 1998. (Sinauer, Sunderland, MA). [Google Scholar]
- Takahashi K, Nei M. Efficiencies of fast algorithms of phylogenetic inference under the criteria of maximum parsimony, minimum evolution, and maximum likelihood when a large number of sequences are used. Mol Biol Evol. 2000;17:1251–1258. doi: 10.1093/oxfordjournals.molbev.a026408. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins JD, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doren M, Ellis HM, Posakony JW. The DrosophilaExtramacrochaetae protein antagonizes sequence-specific DNA binding by Daughterless/Achaete-Scute protein complexes. Development. 1991;113:245–255. doi: 10.1242/dev.113.1.245. [DOI] [PubMed] [Google Scholar]
- Van Doren M, Powell PA, Pasternak D, Singson A, Posakony JW. Spatial regulation of proneural gene activity: Auto- and cross-activation of achaete is antagonized by extramacrochaete. Genes & Dev. 1992;6:2592–2605. doi: 10.1101/gad.6.12b.2592. [DOI] [PubMed] [Google Scholar]
- Van Doren M, Bayley AM, Esnayra J, Ede K, Posakony JW. Negative regulation of proneural gene activity: Hairy is a direct transcriptional repressor of achaete. Genes & Dev. 1994;8:2729–2742. doi: 10.1101/gad.8.22.2729. [DOI] [PubMed] [Google Scholar]
- Weintraub H. The MyoD family and myogenesis: Redundancy, networks, and thresholds. Cell. 1993;75:1241–1244. doi: 10.1016/0092-8674(93)90610-3. [DOI] [PubMed] [Google Scholar]
- Wittbrodt J, Meyer A, Schartl M. More genes in fish? BioEssays. 1998;20:511–515. [Google Scholar]
- Wülbeck C, Simpson P. Expression of achaete-scute homologues in discrete proneural clusters on the developing notum of the medfly Ceratitis capitata, suggests a common origin for the stereotyped bristle patterns of higher Diptera. Development. 2000;127:1411–1420. doi: 10.1242/dev.127.7.1411. [DOI] [PubMed] [Google Scholar]
- Zhao C, Emmons SW. A transcription factor controlling development of peripheral sense organs in C. elegans. Nature. 1995;373:74–78. doi: 10.1038/373074a0. [DOI] [PubMed] [Google Scholar]