Abstract
Glycan chains that terminate in sialic acid (Neu5Ac) are frequently the receptors targeted by pathogens for initial adhesion. Carbohydrate-binding proteins (lectins) with specificity for Neu5Ac are particularly useful in the detection and isolation of sialylated glycoconjugates, such as those associated with pathogen adhesion as well as those characteristic of several diseases including cancer. Structural studies of lectins are essential in order to understand the origin of their specificity, which is particularly important when employing such reagents as diagnostic tools. Here, we report a crystallographic and molecular dynamics (MD) analysis of a lectin from Polyporus squamosus (PSL) that is specific for glycans terminating with the sequence Neu5Acα2-6Galβ. Because of its importance as a histological reagent, the PSL structure was solved (to 1.7 Å) in complex with a trisaccharide, whose sequence (Neu5Acα2-6Galβ1-4GlcNAc) is exploited by influenza A hemagglutinin for viral adhesion to human tissue. The structural data illuminate the origin of the high specificity of PSL for the Neu5Acα2-6Gal sequence. Theoretical binding free energies derived from the MD data confirm the key interactions identified crystallographically and provide additional insight into the relative contributions from each amino acid, as well as estimates of the importance of entropic and enthalpic contributions to binding.
Keywords: GLYCAM, PSL, influenza viral adhesion, lectin, molecular dynamics
Introduction
The term “sialic acids” refers to a family of N- or O-substituted derivatives of neuraminic acid, a 9-carbon acidic sugar, the most common of which is 5-N-acetylneuraminic acid (Neu5Ac). They are expressed abundantly in eukaryotes and are frequently located at the termini of the glycan chains of glycoproteins and gangliosides on cell surfaces (Kelm and Schauer 1997). Because of their exposure on cell surfaces, glycans terminating in sialic acids are often involved in protein-mediated cell–cell interactions, which are essential for normal cell development (Kelm and Schauer 1997). Concomitantly, bacterial (Lehmann et al. 2006) and viral (Shinya et al. 2006) pathogens have evolved to express surface proteins [adhesins and hemagglutinins (HAs)] specific for mediating pathogen adhesion to sialylated glycans on host tissue. The ability to characterize the sialylation patterns of glycans is therefore of considerable relevance in determining the receptor specificities of pathogens. In addition to the vast diversity in glycan structures introduced by sequence and linkage, structural heterogeneity among sialylated glycans can arise from variations in the glycosidic linkage positions associated with the Neu5Ac residues, which may be α2-3 or α2-6, principally to galactose (Gal) or N-acetylgalactosamine (GalNAc) residues, or α2-8 or α2-9 to adjacent sialic acid residues (Angata and Varki 2002; Varki NM and Varki A 2007). Sialic acids may themselves be derivatized by, for example, O-acetylation or by replacement of the N-acetyl moiety with N-glycolyl. Thus, sialic acid-binding proteins can display a range of possible binding modes (Kelm and Schauer 1997).
A notable example of an interaction between sialylated glycans and a pathogen is that between the HA protein found on the surface of influenza A virus, which targets the host glycan receptor sequence Neu5Acα2-3Gal or Neu5Acα2-6Gal, or sometimes both, depending on the influenza strain. The specificity of HA largely determines species specificity, with human infective strains recognizing the Neu5Acα2-6Gal epitope, avian the α2-3 and swine both α2-3 and α2-6 (Suzuki et al. 2000). The preference of human influenza HA for the Neu5Acα2-6Gal epitope arises from mutations in the avian HA sequence (Gagneux et al. 2003). Remarkably, as few as two point mutations in avian HA may be sufficient to induce this specificity shift, as seen in the case of the 1918 “Spanish Flu” pandemic (Lewis 2006). Lectin histochemistry has been pivotal in characterizing the distribution of α2-3- and α2-6-linked sialic acids in host epithelial tissue samples, by exploiting the binding of Sambucus nigra agglutinin (SNA-I; Shibuya et al. 1987) to the α2-6 linkage and Maackia amurensis agglutinin (Wang and Cummings 1988) to the α2-3 (Baum and Paulson 1990; Shinya et al. 2006). Such data underpin our understanding of influenza transmission models.
Lectin histochemistry has also been employed to characterize the presence and distribution of aberrant host glycosylation (Boland et al. 1991; Sata et al. 1991; Benallal et al. 1995; Vierbuchen et al. 1995; Yamashita et al. 1995; Murayama et al. 1997), which can be a hallmark of several diseases, including tumor metastasis (Wang 2005). In establishing correlations between glycan modifications and disease states, it is particularly crucial to employ lectins with precisely known and appropriate specificities. In human colorectal cancer, for example, abnormal levels of Neu5Acα2-6Gal-containing glycans can result from the up-regulation of the α2-6 sialyltransferase (ST6GalI; Dall'Olio et al. 2000). In this example, a diagnostic or a histochemical reagent, such as the lectin SNA-I is not ideal as it can recognize both the ST6GalI product, as well as the Neu5Acα2-6GalNAc sequence, which is a product of sialyltransferases ST6GalNAcI or ST6GalNAcII.
The Polyporus squamosus lectin (PSL) has specificity for Neu5Acα2-6Gal over the Neu5Acα2-3 sequence, but unlike SNA-I, does not bind to the mucin-derived O-linked Neu5Acα2-6GalNAc sequence (Shibuya et al. 1987; Mo et al. 2000; Toma et al. 2001) and, therefore, has important advantages in histochemical analyses. Due to the potential importance of PSL as a tool in biomedical and cancer research (Lehmann et al. 2006) and to determine the molecular basis for its high specificity, we undertook to solve its crystal structure in complex with a human-type influenza epitope Neu5Acα2-6Galβ1-4GlcNAc (6′-SLN; Stevens et al. 2006) and to conduct a comprehensive computational analysis to quantify the per-residue binding energy contributions that determine the specificity of PSL.
Results and discussion
Molecular structure
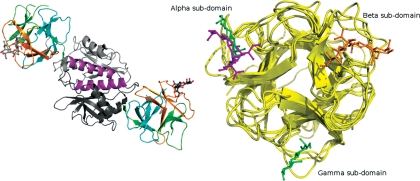
The crystallographic asymmetric unit of PSL bound to 6′-SLN contains a dimer. An analysis of protein interfaces using the Protein Interfaces, Surfaces and Assemblies (PISA) service (Krissinel and Henrick 2007) suggested that both dimeric and tetrameric quaternary structures of PSL are stable in solution. In this crystal structure, the root mean-square deviation (RMSD) of the Cα positions calculated over 275 equivalent residues in each protomer is 0.48 Å. The dimer is cylindrical in shape, and the hydrophobic interfacial contact between the two protomers takes place via a third helix in the C-terminal domain formed by residues 208–226 (Figure 1, left panel). Ten inter-domain hydrogen bonds are formed, and 12% of the solvent accessible surface of the individual protomers is buried upon dimerization.
Fig. 1.
(Left panel) Quaternary structure of PSL. The two protomers are shown in cartoon mode and the bound ligand (6′-SLN) is shown in ball-and-stick mode. (Right panel) The ricin-type β-trefoil lectins structurally related to PSL and their respective carbohydrate-binding sites. The figure shows that the N-terminal domain of PSL bound to Neu5Acα2-6Galβ1-4GlcNAc and its secondary structural overlay against SRC bound to Neu5Acα2-6Galβ1-4Glc (6′-SL, PDBID: 2DS0) and the two trefoil domains of SNA-II bound to Galβ1-4GlcNAc (PDBID: 3CA4).
Each protomer is made up of an N-terminal ricin-type B domain (residues 1–153) and a C-terminal domain (154–286) forming an α/β-fold. The B domain comprises and contains three evolutionarily conserved (Q/N-x-W)3 motifs that are found in the α (1–57), β (58–104) and γ (105–153) subdomains, respectively (Hazes 1996). The overall three-dimensional ricin-type β-trefoil fold of PSL and its related lectins is highly conserved (Figure 1, right panel). The C-terminal domain is characterized by a central five-stranded β-sheet that is flanked by three α-helices and topped by a short strand (Supplementary Material, Figure S1). It shows high fold similarity to its closest relative, the Galα1-3Gal-binding agglutinin from the mushroom Marasmius oreades agglutinin (MOA) (Grahn et al. 2007).
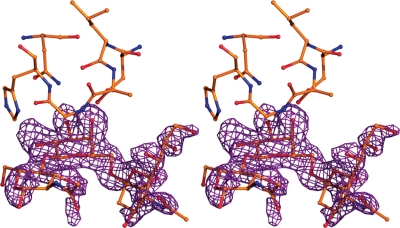
The PSL crystal structure shows well-ordered electron density for the 6'-SLN ligand in the difference density map (Fo − Fc) at the carbohydrate-binding site of the β subdomain (Figure 2), whereas no supporting ligand density was seen in the canonical α and γ subdomains.
Fig. 2.
Stereo view of the PSL β subdomain carbohydrate-binding site with bound ligand shown in ball-and-stick mode. The Fo − Fc difference density map is contoured at the 2.5 σ level. It was calculated after structure solution using MR and an initial maximum-likelihood-based refinement of the protein model but prior to the fitting of the ligands into the map.
Alignment of the sequences of PSL and MOA using Sequoia (Bruns et al. 1999), combined with the manual binding motif identification based on ligand proximity analysis, was performed to gain insight into the nature of the subdomain usage in PSL. All residues with atoms that are within 4 Å of the atoms of the Gal ring of the bound Gal residue in MOA were identified in each lectin, and the resulting motifs are highlighted in the sequences (Figure 3). In the case of MOA, binding motifs for Gal were identified that appear in each of its three subdomains. Notably, in PSL, only two such Gal-binding motifs are present, in subdomains β and γ.
Fig. 3.
Secondary structure-based sequence alignment and manual motif identification of the β-trefoil fold (residues 1–153) of PSL1a and MOA with bound Galα(1,3)[Fucα(1,2)]Gal (PDBID 3EF2) using Sequoia (Bruns et al. 1999) and Coot (Emsley and Cowtan 2004) . RMSD: 2.34 Å. Light grey: rPSL residues within 4 Å of the central Gal of the ligand from the MOA structure. Dark Grey: residues within 4 Å of the central Gal of the ligand in MOA. Bold: conserved tryptophans from the ricin B-type (Q/N-x-W) motifs. In capital letters: residues whose atomic coordinates are structurally equivalent in the alignment.
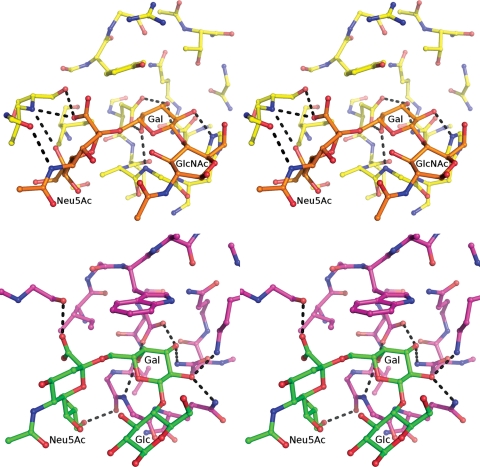
The current structural data, and the ligand proximity analysis, indicate that in the canonical β-binding site, the oligosaccharide actually interacts with segments of both the α and β subdomains (Figure 4, left panel). A similar combining of subdomains to create specificity for Neu5Ac-containing oligosaccharides has been observed in a study of the evolution of Neu5Ac-binding specificity (Yabe et al. 2007). In that work, a novel lectin (sialic acid-recognizing lectin, SRC) with specificity for the Neu5Acα2-6Gal sequence was created from a ricin-like Gal-binding lectin by introducing sequence alterations using error-prone polymerase chain reaction, followed by selection for clones with Neu5Acα2-6Gal-binding. Structural alignment of the binding sites of PSL and SRC shows remarkable conservation of the three-dimensional features and key residues (Supplementary Material, Figure S2). Both sites contain an aromatic group (Tyr87 in PSL and Trp161 in SRC) that forms a stacking interaction with the pyranose ring of the Gal residue, as well as an aspartic acid residue that hydrogen bonds with both the Gal O3 and O4 hydroxyl groups. In both PSL and SRC, affinity for the Neu5Ac residue is provided through interactions with a loop in a subdomain adjacent to the canonical Gal-binding subdomain. In both systems, a key serine residue (Ser239 in SRC and Ser32 in PSL) interacts with the carboxylic acid group of the Neu5Ac residue (Figure 4).
Fig. 4.
(Top panel) Stereo view of the hydrogen-bonding network in the 6'-SLN–PSL complex. (Bottom panel) Stereo view of the hydrogen-bonding network in the Neu5Acα2-6Galβ1-4Glc (6'-SL)–SRC complex. For a detailed interaction analysis of the hydrogen-bonding network in the 6'-SLN–PSL complex refer to Table I and Supplementary Table S1.
Details of oligosaccharide binding
The binding mode of the oligosaccharide is defined principally by hydrogen bonds between the terminal two residues (Neu5Acα2-6Gal) and the protein, with a total of eight hydrogen bonds shared between the glycosyl residues and the protein (Table I). As expected from earlier studies (Tateno et al. 2004), the majority of the interactions involve residues in the β subdomain; however, it is notable that residues Asn30 and Ser32 from the α subdomain interact with atoms N5 and O1A of the Neu5Ac residue (Figure 4, top panel). The N-acetylglucosamine (GlcNAc) residue does not interact with the lectin directly, but through mediating water molecules Wat454 and Wat542. Water molecules Wat542, Wat510, Wat467 and Wat348 hydrogen bond to both the lectin and the oligosaccharide, thus forming an extensive hydrogen bond network (Supplementary Material, Table S1). In addition to the hydrogen bond contacts, the ligand orientation is further stabilized by a hydrophobic stacking interaction between the pyranose ring of the Gal residue and the aromatic side chain of Tyr87. Stacking interactions are particularly prevalent in carbohydrate–protein complexes (Bush et al. 1999), and in the case of galactosyl, residues are seen in the binding of oligosaccharides to galectins (Ford et al. 2003; Di Lella et al. 2009).
Table I.
Key interactions between the 6′-SLN and PSL residues from the X-ray structure and MD simulation
| Interatomic distancea |
||
|---|---|---|
| X-ray | MD (10 ns) | |
| Direct H-bonds | ||
| Neu5Ac(O1A)·Ser32(Oγ) | 2.56 | 2.64 ± 0.12 (100%)b |
| Neu5Ac(O1A)·Ser32(N) | 3.07 | 3.22 ± 0.25 (87%) |
| Neu5Ac(O8)·Ser32(N) | 3.33 | 3.15 ± 0.21 (94%) |
| Neu5Ac(N5)·Asn30(O) | 2.59 | 3.00 ± 0.24 (96%) |
| Gal(O3)·Asp72(Oδ2) | 2.67 | 2.60 ± 0.09 (100%) |
| Gal(O4)·Asp72(Oδ1) | 2.64 | 2.59 ± 0.14 (100%) |
| Gal(O2)·His76(Nε2) | 2.60 | 2.86 ± 0.08 (99%) |
| Gal(O3)·Asn94(Nδ2) | 2.94 | 2.93 ± 0.16 (100%) |
| Aromatic stacking interactionsc | ||
| rGal·Tyr87 | 4.46 | 4.45 ± 0.01 |
| θGal·Tyr87 | 157.64 | 158.4 ± 4.90 |
aValues are expressed in Å. The electron density for chain A was the better ordered among the two NCS-related protomers; therefore, representative H-bond distances were calculated using this protomer.
bFor the MD data, the distances were averaged over 10 ns and H-bond length (±1 SD) and occupancy assessed for periods during which the distance between the heavy atoms was < 3.5 Å.
cDistance (r) in Å between the centroid (c) of the galactosyl ring and the tyrosine phenyl ring, and angle (θ) between the normal to the tyrosine ring plane and the vector between the centroid c, in degrees (Ford et al. 2003).
To augment the crystallographic data, a 10 ns molecular dynamics (MD) simulation in explicit water was performed on the N-terminal ricin B-type domain (residues 1–153) of PSL (ΔPSL) in complex with 6′-SLN. By monitoring the fluctuations in the inter-residue distances, data from the MD simulation provide additional insight into the strengths of these interactions. In general, the MD data (Table I) confirmed that the hydrogen bonds observed crystallographically were also populated in solution at room temperature; however, the magnitudes of the deviations in the interatomic distances, as well as the percentage occupancies of the hydrogen bonds, indicate that the four hydrogen bond interactions with the Gal residue are in general more robust than those formed with the Neu5Ac.
The three-bond α2-6 linkage is relatively flexible due to the presence of the ω-angle, which can potentially populate three distinct rotamers (Wolfe 1972), referred to as the gauche–trans (gt, ωO5-C5-C6-O6 ≈ 60°), gauche–gauche (gg, ω ≈ −60°) and trans–gauche (tg, ω ≈ 180°) rotamers (Kirschner and Woods 2001). For the Neu5Acα2-6Gal sequence in the PSL structure, as well as in the complex with SRC and in all reported complexes with influenza HA, such as PDB IDs 3HTQ (avian HA; Lin et al. 2009), 1RVZ (human HA; Gamblin et al. 2004), 1RVT (swine HA; Gamblin et al. 2004), the ω-angle adopts exclusively the gt orientation (Table II). This is also the most populated rotamer present in solution as indicated by NMR (Poppe et al. 1992), and as confirmed by 200 ns MD simulations here (Supplementary Material, Figure S3). The overall conformation of the ligand in PSL is equivalent to that seen in all reported HA crystal structures containing human flu receptor ligands (Figure 5), although the mode of binding is not equivalent. In all cases, the ψ-angles populate the trans-rotamer, whereas the φ-angle adopts only one of the two rotamers typically observed for the Neu5Acα2-6Gal linkage in solution (Poppe et al. 1992).
Table II.
Average dihedral anglesa for the glycosidic linkages in 6'-SLN bound to PSL, as well as for the same linkages in glycans bound to influenza HA, and free in solution
| Angle | PSL |
Influenza HA |
Free ligand in solution |
|||
|---|---|---|---|---|---|---|
| X-raya | MDb (10 ns) | H1c | H3d | NMR (Poppe et al. 1992) | MD (200 ns) | |
| α(2-6) glycosidic linkage | ||||||
| φ | 56.2 | 54.4 (11) | 64.2 | 54.5 | −58/−169 (0.9:0.1)e | −60/178 (0.95:0.05) |
| ψ | 161.1 | 174.8 (11) | 179.1 | −161.2 | −176/100 (0.8:0.2)e | −166/81 (0.92:0.08) |
| ω | 72.9 | 60.7 (9) | 69.8 | 69.0 | 60/−60/180 (0.71:0.20:0.08) | 69/−54/169 (0.79:0.08:0.14) |
| β(1-4) glycosidic linkage | ||||||
| φ | −87.8 | −78.8 (11) | −60.0 | – | −83e | −76/−145 (0.96:0.04) |
| ψ | −153.8 | −133.7 (17) | −123.9 | – | −126e | −124/70 (0.97:0.03) |
aValues in degrees, averaged over two NCS-related protomers in the asymmetric unit. The inter-residue torsion angles are defined as follows: for the β(1→4) linkage φ = O5-C1-O4′-C4' and ψ = C1-O4′-C4′-C5′; for the α(2→6) linkage φ = C1-C2-O6′-C6′and ψ = C2-O6′-C6′-C5′, ω = O6′-C6′-C5′-O5′.
bAverage values and standard deviations (in parentheses) computed from a 10 ns trajectory.
cAverage values from the 6'-SLN ligand conformations from the following PDB structures: 3HTQ (avian HA; Lin et al. 2009), 1RVZ (human HA; Gamblin et al. 2004), 1RVT (swine HA; Gamblin et al. 2004).
dAverage values for the ligands in PDB structure 1MQN (avian HA; Ha et al. 2003).
eAverage values and rotamer populations (in parentheses) determined from the scalar J-coupling and NOE data reported for Neu5Acα2-6Galβ1-4Glc (Poppe et al. 1992), employing a rotational isomeric state analysis (DeMarco and Woods 2009), based on states isolated from the MD simulation of the free oligosaccharide. The φ angle has been altered by −120° to comply with the φ = C1-C2-O6′-C6′ convention used here.
Fig. 5.
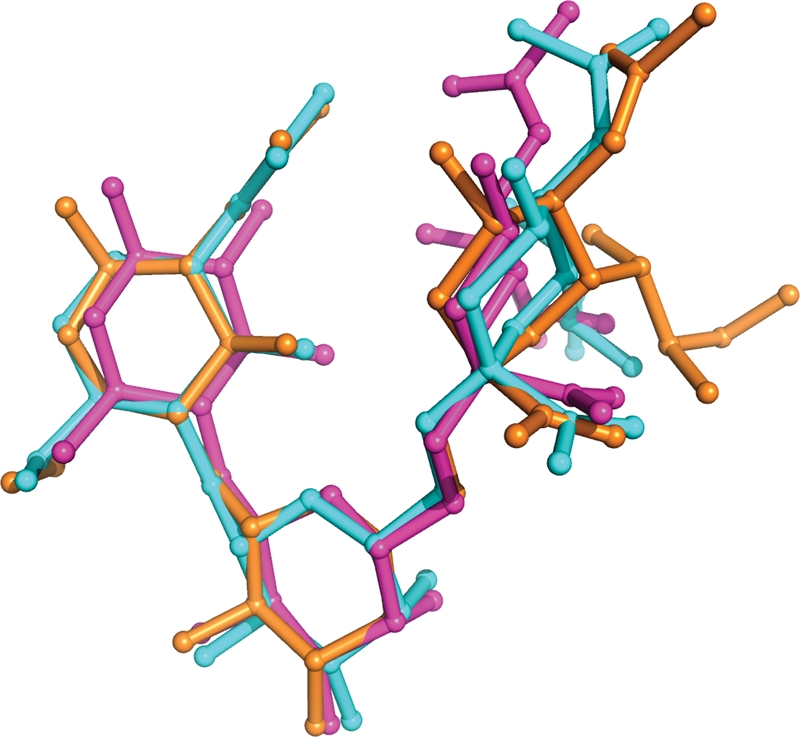
Superimposition of the ligand from PSL with that from influenza A H1 HA (PDBID:1RVZ) and SRC (Gamblin et al. 2004). Although some disorder in the Neu5Ac residues is present, all the interglycosidic torsion angles populate the same canonical rotamers (Table II).
Molecular mechanics Poisson–Boltzmann surface area analysis of binding energies
Based on the stability of the MD simulation of the 6′-SLN–ΔPSL complex, as indicated by the ability of the MD simulation to reproduce the intermolecular interactions associated with the complex (Table I), the MD data were subjected to molecular mechanics Poisson–Boltzmann surface area (MM-PBSA) energy analysis. Such an analysis enables both a global and a per-residue quantification of the energetics responsible for the specificity of the interaction (Tables III and IV). The binding energy was decomposed into contributions from direct electrostatic interactions, polar and non-polar desolvation and van der Waals contacts, employing the MM-PBSA method (Luo et al. 2002).
Table III.
Estimated average energetic contributions to ΔGBinding computed from an MM-PBSA analysis of the 10 ns MD data for the 6′-SLN–PSL complex relative to the free protein and ligand
| Average energy componenta | Energy (kcal mol−1) |
|---|---|
| <ΔEElectrostatic> | −67.1 ± 0.4b |
| <ΔGPolar desolvation> | 58.2 ± 0.4 |
| Total electrostatics <ΔGPolar + ΔEElectrostatic> | −8.9 ± 0.2 |
| <ΔEvan der Waals> | −31.4 ± 0.1 |
| <ΔGNonpolar desolvation> | −3.1 ± 0.0 |
| <ΔGBinding> | −43.5 ± 0.1 |
aAveraged over 10 ns.
bStandard error of the mean.
Table IV.
Contribution to the interaction energies from each glycan residue, as well as for all keya protein residues
| Residue | <ΔEDirect electrostatic> | <ΔGPolar desolvation> | TotalElectrostatic | <ΔEvan der Waals> | <ΔGNon-polar desolvation> | <ΔGBinding> |
|---|---|---|---|---|---|---|
| Glycan | ||||||
| GlcNAc | 2.9 | −1.5 | 1.4 | −2.3 | −0.3 | −1.2 |
| Gal | −30.2 | 19.6 | −10.6 | −5.0 | −1.3 | −16.9 |
| Neu5Ac | −6.3 | 11.5 | 5.2 | −8.4 | −1.7 | −4.9 |
| Total | −33.6 | 29.6 | −4.0 | −15.7 | −3.2 | −22.9 |
| Protein | ||||||
| Ser32 | −9.5 | 5.5 | −4.1 | −0.1 | −0.1 | −4.3 |
| Lys31 | −24.3 | 24.3 | −0.1 | −2.7 | −0.3 | −3.1 |
| Gly75 | −3.0 | 2.0 | −0.9 | −1.7 | −0.3 | −3.0 |
| Tyr87 | −1.0 | 1.8 | 0.8 | −3.1 | −0.3 | −2.6 |
| Ile85 | −1.1 | 0.9 | −0.2 | −1.8 | −0.1 | −2.1 |
| His76 | −3.4 | 2.7 | −0.6 | −1.2 | −0.2 | −2.1 |
| Ala74 | −2.2 | 1.8 | −0.3 | −1.5 | −0.1 | −2.0 |
| Asn94 | −1.3 | 0.0 | −1.3 | −0.4 | 0.0 | −1.8 |
| Asp72 | −4.0 | 1.2 | −2.8 | 2.1 | 0.0 | −0.7 |
| Asn30 | −1.2 | 2.1 | 0.8 | −1.2 | −0.2 | −0.6 |
| Asp34 | 15.8 | −12.9 | 2.9 | −0.2 | 0.0 | 2.8 |
| Subtotal | −35.2 | 29.4 | −5.8 | −11.7 | −1.9 | −19.3 |
| Percent of Totalb | 104.8 | 102.8 | 116.6 | 74.5 | 83.5 | 84.5 |
aAny residue indentified in the hydrogen-bonding analysis or that contributes >1 kcal mol−1 to <ΔGBinding>.
bRelative to the total contributions from all of the residues in the protein.
The ability to assign binding energies on a per-residue basis is a powerful feature of this type of computational analysis as it permits not only the quantification of the strengths of the crystallographically observed interactions but also objectively identifies all energetically significant interactions. In contrast to hydrogen bonds, favorable van der Waals contacts or favorable desolvation-free energies are features that are difficult to quantify from a purely structural perspective. Moreover, by computing the binding energy as the difference between the energies of the protein–oligosaccharide complex and those of the free protein and oligosaccharide, an estimate of the binding free energy is obtained that cannot be deduced from any analysis of the complex alone. Lastly, by performing the energy calculations on multiple configurations extracted from the MD simulation rather than for the single configuration observed crystallographically, the computed binding energies represent converged values that may be characterized with statistically significant error estimates.
The direct electrostatic contribution from ligand binding (−67.1 kcal mol−1) was largely offset by the energy (58.2 kcal mol−1) required to desolvate the polar residues in the binding interface. This is not unexpected given that the electrostatic interactions arise predominantly from hydrogen bonds involving hydroxyl groups in the glycan; hydrogen bonds which, depending on the polarity of the amino acid side chain involved, can be effectively equivalent in strength to those formed with water molecules (Tschampel and Woods 2003). Notably, the data in Table III lead to the conclusion that although hydrogen bonds are essential for binding specificity, a larger net contribution (−31.4 kcal mol−1) arises from van der Waals contacts. Similar relative contributions have been observed for several carbohydrate–protein complexes (Woods and Tessier 2010).
Per-residue binding energies
The overall binding energy between the protein and the carbohydrate arises from contributions from all the interacting atoms, and thus it is possible computationally to quantify the fractional contributions to the binding energy made by each residue in the protein and in the carbohydrate. Experimentally, similar per-residue affinity data could be obtained for the protein by performing alanine-scanning mutagenesis followed by affinity measurements; however, this would be extremely demanding to undertake.
As expected, all the amino acids that form hydrogen bonds with the ligand (Ser32, Asn30, Asp72, His76 and Asn94) were identified as forming strong electrostatic interactions in the MM-PBSA analysis. These interactions equate to an average hydrogen bond strength of 0.7 kcal mol−1. Notably, several residues that do not form direct interactions were also identified as key for ligand binding, and in some cases (Ile85 and Ala74), the non-specific contacts contributed more to the binding energy than that was contributed by hydrogen bond residues (Asn94, Asp72 and Asn30). In addition, the MM-PBSA analysis indicates that the aromatic stacking interaction between Tyr87 ranks this residue as fourth most critical to binding to a net binding energy of −2.6 kcal mol−1. Approximately 90% of the stacking energy comes from van der Waals contacts, with only 10% predicted to arise from desolvation. This analysis, therefore, suggests that dispersion forces (Morales et al. 2008), and not the hydrophobic effect (Di Lella et al. 2009), are the driving force behind the observed carbohydrate–aromatic stacking.
From the perspective of the carbohydrate, there is no experimental method that provides per-residue binding energy estimates. The computational per-residue binding energy data for the 6′-SLN–ΔPSL system (Table IV) indicate that ∼95% of the strength of binding comes from interactions with the disaccharide Neu5Ac-Gal, with the sialic acid accounting for 20% of the total binding affinity, with negligible contribution from the GlcNAc residue. These data are consistent with the structural observation that the GlcNAc residue does not make significant direct contacts with the protein. Further, the data indicate that although the protein might retain Gal-binding affinity, the removal of the Neu5Ac residue would be expected to decrease the affinity by almost 6 kcal mol−1. This prediction is consistent with the greatly attenuated affinity of non-sialylated ligands for PSL (Tateno et al. 2004). However, this estimate neglects any consideration of the significant entropic penalties associated with binding a flexible glycan (see following section).
Role of entropy and enthalpy in ligand binding
Given the known plasticity of glycans, it is common that carbohydrate binding is accompanied by an entropic penalty. Glycan flexibility is readily characterized by monitoring the motions of the interglycosidic torsion angles (φ, ψ and ω), and it is particularly convenient to employ changes in the torsional motions of these angles in the free and bound states to quantify any change in conformational entropy that may occur upon binding.
The conformation of 6′-SLN bound to PSL is indistinguishable from that of the predominant conformation in solution (Table II) and essentially identical to that seen in complexes with the lectin SRC and all influenza HA examples to date. This might suggest that there should be little entropic penalty incurred upon binding. However, the data for 6′-SLN indicate that an entropic penalty of ∼3.65 kcal mol−1 is incurred upon binding to PSL (Table V). Of this total penalty, 2.68 kcal mol−1 (73%) arises from stiffening of the three-bond Neu5Acα2-6Gal linkage, with the largest contribution, 1.05 kcal mol−1, arising from the restricted motion of the ω torsion angle. The remaining 0.95 kcal mol−1 comes from restrictions of the rotational degrees of freedom of the β1-4 linkage. Taking conformational entropy into account, the MM-PBSA contribution to binding, arising exclusively from the Neu5Ac group, is reduced from −5.9 kcal mol−1 to approximately −2.25 kcal mol−1.
Table V.
Contributions to the conformational-binding entropy (kcal mol−1)
| 6′-SLN | Neu5Acα2-6Gal linkage | Galβ1-4GlcNAc linkage | |
|---|---|---|---|
| −T STotal | 3.65 | — | — |
| −T Sφψω | 2.68 | — | |
| −T Sφψ | 1.10 | 0.95 | |
| −T Sφ | 0.68 | 0.35 | |
| −T Sψ | 0.73 | 0.51 | |
| −T Sω | 1.05 | — |
α2-6 vs α2-3 specificity
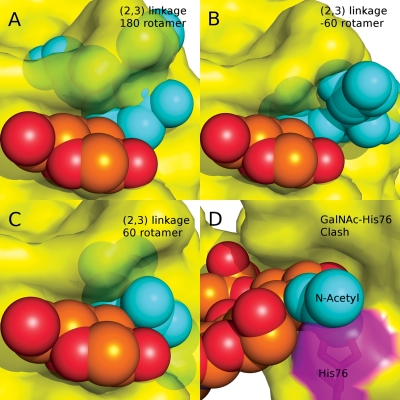
The origin of the preference of PSL for the Neu5Acα2-6Gal sequence over the α2-3 was elucidated by superimposing the ring atoms of the Gal residue in the Neu5Acα2-3Gal disaccharide, created and geometry minimized using the online carbohydrate builder Glycosides and Glycoproteins in AMBER (GLYCAM)-Web (http://www.glycam.com; Woods Group 2005–2011) on the Gal residue in the Neu5Acα2-6Gal sequence in the crystal structure. Both of the experimentally observed orientations of the φ-glycosidic angle (φ = C1-C2-O3′-C3′; Siebert et al. 1992; Kiddle and Homans 1998; Milton et al. 1998) associated with the α2-3 linkage were included in the modeling and examined for interactions with the protein surface. In each idealized case (φ = 180°, −60°, 60°), notable steric clashes with the protein surface were observed, in particular with the backbone sequence associated with residues Gly92 and Tyr87 (Figure 6A–C). As the C2-O3′ bond is perpendicular to the protein surface, it is apparent that indeed no orientation of the α2-3 linkage could fit into the PSL-binding site without requiring severe deformation of the protein surface.
Fig. 6.
(A–C) Models of the Neu5Acα2-3Galβ-1-4GlcNAc trisaccharide bound to PSL, indicating that no rotamers of the 2-3 linkage can be adopted that avoid collisions with the PSL surface. (D) Orientation of a GalNAc-containing trisaccharide in the binding site of PSL, indicating a collision with His76 (magenta).
Gal vs GalNAc (HexNAc) specificity
The molecular basis for the Gal/GalNAc specificity was identified by superimposing the ring atoms of a GalNAc residue, created and geometry minimized using the online carbohydrate builder GLYCAM-Web (Woods Group 2005–2011), onto the coordinates of the Gal residue in the binding site. In such an orientation, an obvious steric collision appeared to be present between the NAc group of the GalNAc and the side chain of His76 (Figure 6D). This side chain participates in hydrogen bonding with Gln112 and so would not be free to escape the collision with a GalNAc simply by reorienting its position. Notably, the MM-PBSA data suggest that His76 also contributes to the overall affinity for the Gal residue. By structural analogy to GalNAc, neither a 4-linked GlcNAc nor a 4-linked ManNAc residue would be predicted to be tolerated in the PSL-binding site.
Conclusions
The role of the α, β and γ subdomains of the PSL β-trefoil in ligand binding has been investigated previously using site-directed mutagenesis (Tateno et al. 2004). The results indicated that the β subdomain was the most probable carbohydrate-binding site in the N-terminal domain since it contained all the residues required both for forming the hydrophobic core of the ricin-like trefoil and those involved in binding a carbohydrate, namely, Asp72, Tyr87, Gln95 and Trp97. Sequence alignment and ligand proximity analysis of the PSL, SRC and MOA lectins (Figures 3 and 4) now indicate that the α subdomain has altered in PSL so as to provide key interactions with the Neu5Ac residue in the oligosaccharide bound in the β subdomain. This adaptation has apparently forced the abandonment of the original α subdomain-binding motif, which is consistent with conclusions drawn from the initial sequence analysis that the α subdomain in PSL lacks residues deemed essential for Gal-binding affinity (Mo et al. 2000; Tateno et al. 2004). Previously, a monomeric form of PSL was found to agglutinate rabbit erythrocytes, indicating the presence of two binding sites, which led to the conclusion that the γ domain, which still contains some of the residues essential for Gal binding, may still retain affinity for Galβ1-4GlcNAc-terminating sequences (Tateno et al. 2004). Our proximity analysis indeed suggests that the γ domain likely retains Gal-binding affinity. However, the binding data from the MM-PBSA analysis suggest that any such interactions are markedly weaker than those for 6′-SLN-terminating sequences.
A detailed computational analysis of the interaction between 6′-SLN and PSL provides a structural explanation for all the known binding specificities. In addition, the computational data lead to the conclusion that although specificity is conferred by direct hydrogen bonds between the oligosaccharide and the protein, non-specific van der Waals contacts contribute significantly to the total affinity. As in all similar analyses of carbohydrate–protein binding to date, the strength of direct electrostatic interactions is largely attenuated by desolvation free energy (Woods and Tessier 2010).
Lastly, in all co-crystal structures of influenza HAs with human-type receptor glycans, the glycan displays a characteristic three-dimensional shape (Chandrasekaran et al. 2008) equivalent to that seen in the 6′-SLN–PSL complex, suggesting that PSL will be particularly well suited to histological investigation-related influenza adhesion. Notably, this glycan conformation is also the preponderant conformation seen in solution, and yet there remains a 3.6 kcal mol−1 conformational entropic penalty to the binding of 6′-SLN to PSL, confirming that even for less-flexible glycans, such as 6′-SLN, entropy plays a significant role in determining binding affinity. This observation may be of particular relevance in the emerging area of glycomimetic drug design (Ernst and Magnani 2009).
Experimental
X-ray crystallography
A sample of 6′-SLN was purchased from V-Labs, Covington, LA, U.S.A. Recombinant PSL, with a molecular weight of 31 kDa, was cloned, expressed in Escherichia coli as an active and soluble form (Tateno et al. 2004), purified and lyophilized. The recombinant lectin was reconstituted in a buffer containing 10 mM phosphate, pH 7.2, and 150 mM NaCl, purified using gel filtration chromatography on a Sephadex G-200 column and concentrated to ∼15 mg mL−1. About 24 h prior to the crystallization trials, PSL was combined with the oligosaccharide 6'-SLN at a molar ratio of 2:1 (oligosaccharide:lectin). The microbatch-under-oil method (Chayen et al. 1992) was initially used to assess the solubility profile of the protein, choice of precipitant, pH and temperature. Small, needle-shaped crystals were obtained from polyethylene glycol 8000 (PEG 8000) precipitant solutions, and a stepwise optimization led to diffraction quality crystals. Cubic-shaped crystals were grown at 20°C from hanging drops composed of 2 μL of the lectin/ligand solution and 2 μL of precipitant. The drops were equilibrated against 22–25% PEG 8000 and 100 mM sodium 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid at pH 6.9–7.2.
The crystals were cryoprotected with PEG 400, and low-temperature diffraction data were collected on cryoprotected crystals at the IMCA-CAT beamline 17-ID at the Advanced Photon Source synchrotron ring at the Argonne National Lab, Argonne, IL. Data acquisition was accomplished using the ADSC Q210 Charge-coupled device with one crystal-rotation axis (φ) and a computer-controlled sample-to-detector distance. The data were recorded as 360 non-overlapping images with a 0.5° oscillation step and a 3 s exposure time. The HKL-2000 suite of programs (Otwinowski and Minor 1997) was used for data processing and scaling. Intensities were converted to structure factors using the program TRUNCATE from the CCP4 (Collaborative Computational Project Number 1994) suite of packages. The Matthew's coefficient was 2.86 Da Å−3, and the calculated solvent content in the asymmetric unit was 57%. The crystal parameters and data collection statistics are summarized in Table VI.
Table VI.
X-ray data and refinement statistics
| Data collection | |
| Space group | P21212 |
| Wavelength (Å) | 1.00 |
| Unit cell parameters a, b and c (Å) | 115.8, 59.4, 103.2 |
| Resolution (Å) | 38.5 to 1.7 |
| Data completeness (%) | 99.1 (92.8)a |
| Observed reflections | 451179 |
| Unique reflections | 79179 |
| Multiplicity | 5.7 |
| Rsym | 6.2 (23.8)a |
| <I/σ(I)> | 26.3 (2.7)a |
| Refinement | |
| Number of protein atoms | 4318 |
| Number of water molecules | 632 |
| Number of ligand atoms | 92 |
| RMSD bond lengthb (Å) | 0.023 |
| RMSD bond anglesb (°) | 1.98 |
| Average temperature (B) factors (Å2) | |
| All atoms | 28.9 |
| Main chain | 26.3 |
| Side chain and water molecules | 31.5 |
| Ligand | 53.2 |
| Ramachandran statistics (%) | |
| Core and additionally allowed regions | 99.4 |
| Disallowed regions | 0.6 |
| Crystallographic Rwork | 18.5 (24.2)a |
| Crystallographic Rfree | 21.7 (24.4)a |
aValues in parentheses are for the highest resolution shell (1.76–1.7 Å).
bRMSDs from restraint targets (Engh and Huber 1991).
Earlier cDNA cloning had revealed that PSL contained a ricin-like B chain (QxW)3 domain (Hazes 1996) at its N-terminus. Mutant constructs with single amino acid substitutions in the putative sugar-binding sites had also indicated that the β and γ subdomains were the most probable sugar-binding sites (Tateno et al. 2004). A list of structures was obtained for use as possible models for molecular replacement (MR) using sequences annotated by structure (Milburn et al. 1998). The highest sequence identity (38%) relative to PSL was the Galα1-3Gal-binding lectin MOA (Winter et al. 2002). The deposited crystal structure of MOA in complex with a trisaccharide (2IHO) was generously provided by Professor Ute Krengel and coworkers a few months before release by the PDB (Grahn et al. 2007). The PSL structure was solved as a single solution using a monomer of MOA as the MR model and with the program Phaser (McCoy et al. 2007) from the PHENIX (Adams et al. 2002) software suite. Model bias was reduced using density modification (Cowtan 1994) and the ReSOLVE (Terwilliger 2003) package was used for initial rebuilding. This was followed by iterative manual rebuilding and macromolecular refinement using Refmac5 (Murshudov et al. 1997) and the visualization and refinement software packages O (Jones et al. 1991) and Coot (Emsley and Cowtan 2004). The carbohydrate chains were located and refined, followed by location and refinement of water molecules. The final model consists of two polypeptide chains of 285 amino acid residues each, two 6′-SLN molecules and 535 water molecules. Loop residues 26–29 and 123–125 and Met1 were not located in electron density maps and flagged as disordered or missing residues. Glu204 is located in type IV β-turn and is an outlier in the Ramachandran plot (main chain RSCC = 0.98).
Computational simulations
MD simulations were performed on free 6′-SLN and on the N-terminal carbohydrate-binding domain (residues 1–153, ΔPSL) of the PSL–6′-SLN complex, both in explicit water, employing the assisted model building and energy refinement (AMBER) force field (Case et al. 2005) with the PARM99SB (Hornak et al. 2006) parameters selected for the protein and counter ions and the GLYCAM06 (Kirschner et al. 2008) parameters for the carbohydrate. Long-range electrostatic interactions were treated with the Particle-Mesh Ewald method (Darden et al. 1993; Sagui and Darden 1999) using a cutoff of 9 and 1.2 Å grid spacing. Methyl and acetyl groups were added to the protein structure to terminate chains with unresolved residues. The most significant impact of these missing sequences was the need to apply restraints on the Cα positions to ensure the protein was maintained in the correct overall fold during the MD simulation. Four Na+ ions were added to neutralize the net charge on the system. The PSL–6′-SLN system was then centered in a cubic box (70 Å side) of TIP3P water molecules (Jorgensen et al. 1983). Crystallographic water molecules were retained in the simulation. Water and ion positions were minimized and equilibrated for 500 ps (NVT), whereas the protein and carbohydrate non-hydrogen atoms were restrained with a force constant of 1000 kJ mol−1 nm−2. An additional 500 ps of equilibration under NPT conditions was then performed with only the non-hydrogen atoms in the protein restrained. Finally, an equilibration under NPT conditions with only the Cα atoms of the protein restrained was performed before a production run of 10 ns was collected under the same conditions. For the simulation of free 6′-SLN, one Na+ ion was added to neutralize the net charge the oligosaccharide, which was then centered in a cubic box (30 Å side) of TIP3P water molecules (Jorgensen et al. 1983). Heating and equilibration were then performed as for the protein–oligosaccharide complex, before a production run of 200 ns was collected. All MD simulations were performed with version 4.04 of gronigen machine for chemical simulations (GROMACS; Hess et al. 2008). The AMBER/GLYCAM force field parameters were imported into GROMACS by means of the amb2gmx script, written and distributed by Eric J. Sorin (www.chemistry.csulb.edu/ffamber).
MM-PBSA calculations
Employing 1000 snapshots extracted at 100 ps intervals from the MD trajectory of the 6′-SLN–ΔPSL complex, the binding energy was computed directly from the energies of the binding reaction components [Equation (1)]:
| (1) |
The approach taken follows closely that reported for other carbohydrate–protein complexes (Bryce et al. 2001; Ford et al. 2003; Kadirvelraj et al. 2006). The free energies of the components were approximated by separating the energies into molecular mechanical (electrostatic and van der Waals), polar and apolar solvation and entropic contributions [Equation (2)]:
| (2) |
Before the analysis, the water molecules were removed from the solvated trajectory. The energy contribution from solvation was then obtained through application of the Poisson–Boltzmann implicit solvation model.
Entropy calculations
Conformational entropy penalties were estimated using the Karplus–Kushick approach (Karplus and Kushick 1981), in which the relative entropies were derived from the ratios of the determinants of the covariance matrices associated with the glycosidic torsion angles for the bound (σB) and free (σF) oligosaccharides [Equation (3)]. In the case of carbohydrates, it is particularly appropriate to focus on the conformational entropy associated with the interglycosidic torsion angles (Bryce et al. 2001; Kadirvelraj et al. 2006).
| (3) |
Due to the absence of loop residues 26–29 and 123–125 in the crystallographic data, we were unable to perform an analysis of the configurational entropy change associated principally with the stiffening of the protein that occurs upon oligosaccharide binding (Ford et al. 2003).
Supplementary data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Funding
This work was supported by the National Institutes of Health (GM055230 and GM094919) and the Science Foundation of Ireland (08/IN.1/B2070).
Conflict of interest
None declared.
Atomic coordinates
The atomic coordinates and structure factor amplitudes for PSL has been deposited in the RCSB Protein Data Bank (Bernstein et al. 1977) under PDB ID 3PHZ.
Abbreviations
AMBER, assisted model building and energy refinement; RMSD, root mean-square deviation; Gal, galactose; Galβ1-4GlcNAc, lactosamine; GlcNAc, N-Acetyl glucosamine; GLYCAM, Glycosides and Glycoproteins in AMBER; GROMACS, gronigen machine for chemical simulations; HA, hemagglutinin; MD, molecular dynamics; MOA, Marasmius oreades agglutinin; MM-PBSA, molecular mechanics Poisson–Boltzmann surface area; MR, molecular replacement; Neu5Ac, sialic acid/5-N-Acetylneuraminic acid; PEG, polyethylene glycol; PSL, Polyporus squamosus lectin; SNA, Sambucus nigra agglutinin; SRC, sialic acid-recognizing lectin; ST6Gal, α2-6 sialyltransferase; 6′-SLN, Neu5Acα2-6Galβ1-4GlcNAc.
Supplementary Material
Acknowledgements
We thank the Advanced Photon Source, Argonne, IL, USA, and the staff at the IMCA-CAT beamline 17 for synchrotron time and assistance during data collection.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr Sect D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. doi:10.1107/S0907444902016657. [DOI] [PubMed] [Google Scholar]
- Angata T, Varki A. Chemical diversity in the sialic acids and related α-keto acids: An evolutionary perspective. Chem Rev. 2002;102:439–469. doi: 10.1021/cr000407m. doi:10.1021/cr000407m. [DOI] [PubMed] [Google Scholar]
- Baum LG, Paulson JC. Sialyloligosaccharides of the respiratory epithelium in the selection of human influenza virus receptor specificity. Acta Histochem Suppl. 1990;40:35–38. [PubMed] [Google Scholar]
- Benallal M, Zotter H, Anner RM, Lacotte D, Moosmayer M, Anner BM. Maackia amurensis agglutinin discriminates between normal and chronic leukemic human lymphocytes. Biochem Biophys Res Commun. 1995;209:921–929. doi: 10.1006/bbrc.1995.1586. doi:10.1006/bbrc.1995.1586. [DOI] [PubMed] [Google Scholar]
- Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank: A computer-based archival file for macromolecular structures. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. doi:10.1016/S0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Boland CR, Chen YF, Rinderle SJ, Resau JH, Luk GD, Lynch HT, Goldstein IJ. Use of the lectin from Amaranthus caudatus as a histochemical probe of proliferating colonic epithelial cells. Cancer Res. 1991;51:657–665. [PubMed] [Google Scholar]
- Bruns CM, Hubatsch I, Ridderstrom M, Mannervik B, Tainer JA. Human glutathione transferase A4–4 crystal structures and mutagenesis reveal the basis of high catalytic efficiency with toxic lipid peroxidation products. J Mol Biol. 1999;288:427–439. doi: 10.1006/jmbi.1999.2697. doi:10.1006/jmbi.1999.2697. [DOI] [PubMed] [Google Scholar]
- Bryce RA, Hillier IH, Naismith JH. Carbohydrate-protein recognition: Molecular dynamics simulations and free energy analysis of oligosaccharide binding to concanavalin A. Biophys J. 2001;81:1373–1388. doi: 10.1016/S0006-3495(01)75793-1. doi:10.1016/S0006-3495(01)75793-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush CA, Martin-Pastor M, Imberty A. Structure and conformation of complex carbohydrates of glycoproteins, glycolipids, and bacterial polysaccharides. Annu Rev Biophys Biomol Struct. 1999;28:269–293. doi: 10.1146/annurev.biophys.28.1.269. doi:10.1146/annurev.biophys.28.1.269. [DOI] [PubMed] [Google Scholar]
- Case DA, Cheatham TE, III, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. doi:10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran A, Srinivasan A, Raman R, Viswanathan K, Raguram S, Tumpey TM, Sasisekharan V, Sasisekharan R. Glycan topology determines human adaptation of avian H5N1 virus hemagglutinin. Nat Biotechnol. 2008;26:107–113. doi: 10.1038/nbt1375. doi:10.1038/nbt1375. [DOI] [PubMed] [Google Scholar]
- Chayen N, Shaw Stewart P, Blow D. Microbatch crystallization under oil—a new technique allowing many small-volume crystallization trials. J Cryst Growth. 1992;122:176–180. doi:10.1016/0022-0248(92)90241-A. [Google Scholar]
- Collaborative Computational Project Number. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr Sect D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. doi:10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Cowtan K. dm: An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994:34–38. p. [Google Scholar]
- Dall'Olio F, Chiricolo M, Ceccarelli C, Minni F, Marrano D, Santini D. β-galactoside α2,6 sialyltransferase in human colon cancer: Contribution of multiple transcripts to regulation of enzyme activity and reactivity with Sambucus nigra agglutinin. Int J Cancer. 2000;88:58–65. doi: 10.1002/1097-0215(20001001)88:1<58::aid-ijc9>3.0.co;2-q. doi:10.1002/1097-0215(20001001)88:1<58::AID-IJC9>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Darden T, York D, Pedersen L. Particle mesh Ewald: An N log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. doi:10.1063/1.464397. [Google Scholar]
- DeMarco ML, Woods RJ. Atomic-resolution conformational analysis of the GM3 ganglioside in a lipid bilayer and its implications for ganglioside-protein recognition at membrane surfaces. Glycobiology. 2009;19:344–355. doi: 10.1093/glycob/cwn137. doi:10.1093/glycob/cwn137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lella S, Ma L, Díaz Ricci JC, Rabinovich GA, Asher SA, Álvarez RMS. Critical role of the solvent environment in galectin-1 binding to the disaccharide lactose. Biochemistry. 2009;48:786–791. doi: 10.1021/bi801855g. doi:10.1021/bi801855g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. doi:10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Engh R, Huber R. Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallogr Sect A. 1991;47:392–400. doi:10.1107/S0108767391001071. [Google Scholar]
- Ernst B, Magnani JL. From carbohydrate leads to glycomimetic drugs. Nat Rev Drug Discov. 2009;8:661–677. doi: 10.1038/nrd2852. doi:10.1038/nrd2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford MG, Weimar T, Köhli T, Woods RJ. Molecular dynamics simulations of galectin-1-oligosaccharide complexes reveal the molecular basis for ligand diversity. Proteins. 2003;53:229––40.. doi: 10.1002/prot.10428. doi:10.1002/prot.10428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagneux P, Cheriyan M, Hurtado-Ziola N, van der Linden EC, Anderson D, McClure H, Varki A, Varki NM. Human-specific regulation of α 2-6-linked sialic acids. J Biol Chem. 2003;278:48245–48250. doi: 10.1074/jbc.M309813200. doi:10.1074/jbc.M309813200. [DOI] [PubMed] [Google Scholar]
- Gamblin SJ, Haire LF, Russell RJ, Stevens DJ, Xiao B, Ha Y, Vasisht N, Steinhauer DA, Daniels RS, Elliot A, et al. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science. 2004;303:1838–1842. doi: 10.1126/science.1093155. doi:10.1126/science.1093155. [DOI] [PubMed] [Google Scholar]
- Grahn E, Askarieh G, Holmner A, Tateno H, Winter HC, Goldstein IJ, Krengel U. Crystal structure of the Marasmius oreades mushroom lectin in complex with a xenotransplantation epitope. J Mol Biol. 2007;369:710–721. doi: 10.1016/j.jmb.2007.03.016. doi:10.1016/j.jmb.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Ha Y, Stevens DJ, Skehel JJ, Wiley DC. X-ray structure of the hemagglutinin of a potential H3 avian progenitor of the 1968 Hong Kong pandemic influenza virus. Virology. 2003;309:209–218. doi: 10.1016/s0042-6822(03)00068-0. doi:10.1016/S0042-6822(03)00068-0. [DOI] [PubMed] [Google Scholar]
- Hazes B. The (QxW)3 domain: A flexible lectin scaffold. Protein Sci. 1996;5:1490–1501. doi: 10.1002/pro.5560050805. doi:10.1002/pro.5560050805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B, Kutzner C, Van Der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. doi:10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. doi:10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr Sect A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. doi:10.1063/1.445869. [Google Scholar]
- Kadirvelraj R, Gonzalez-Outeiriño J, Foley BL, Beckham ML, Jennings HJ, Foote S, Ford MG, Woods RJ. Understanding the bacterial polysaccharide antigenicity of Streptococcus agalactiae versus Streptococcus pneumoniae. Proc Natl Acad Sci USA. 2006;103:8149–8154. doi: 10.1073/pnas.0602815103. doi:10.1073/pnas.0602815103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus M, Kushick JN. Method for estimating the configurational entropy of macromolecules. Macromolecules. 1981;14:325–332. doi:10.1021/ma50003a019. [Google Scholar]
- Kelm S, Schauer R. Sialic acids in molecular and cellular interactions. Int Rev Cytol. 1997;175:137–240. doi: 10.1016/S0074-7696(08)62127-0. doi:10.1016/S0074-7696(08)62127-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiddle GR, Homans SW. Residual dipolar couplings as new conformational restraints in isotopically 13C-enriched oligosaccharides. FEBS Lett. 1998;436:128–130. doi: 10.1016/s0014-5793(98)01112-0. doi:10.1016/S0014-5793(98)01112-0. [DOI] [PubMed] [Google Scholar]
- Kirschner KN, Woods RJ. Solvent interactions determine carbohydrate conformation. Proc Natl Acad Sci USA. 2001;98:10541–10545. doi: 10.1073/pnas.191362798. doi:10.1073/pnas.191362798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner KN, Yongye AB, Tschampel SM, González-Outeiriño J, Daniels CR, Foley BL, Woods RJ. GLYCAM06: A generalizable biomolecular force field. carbohydrates. J Comput Chem. 2008;29:622–655. doi: 10.1002/jcc.20820. doi:10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. doi:10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Lehmann F, Tiralongo E, Tiralongo J. Sialic acid-specific lectins: Occurrence, specificity and function. Cell Mol Life Sci. 2006;63:1331–1354. doi: 10.1007/s00018-005-5589-y. doi:10.1007/s00018-005-5589-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DB. Avian flu to human influenza. Annu Rev Med. 2006;57:139–154. doi: 10.1146/annurev.med.57.121304.131333. doi:10.1146/annurev.med.57.121304.131333. [DOI] [PubMed] [Google Scholar]
- Lin T, Wang G, Li A, Zhang Q, Wu C, Zhang R, Cai Q, Song W, Yuen KY. The hemagglutinin structure of an avian H1N1 influenza A virus. Virology. 2009;392:73–81. doi: 10.1016/j.virol.2009.06.028. doi:10.1016/j.virol.2009.06.028. [DOI] [PubMed] [Google Scholar]
- Luo R, David L, Gilson MK. Accelerated Poisson-Boltzmann calculations for static and dynamic systems. J Comput Chem. 2002;23:1244–1253. doi: 10.1002/jcc.10120. doi:10.1002/jcc.10120. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. doi:10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milburn D, Laskowski RA, Thornton JM. Sequences annotated by structure: A tool to facilitate the use of structural information in sequence analysis. Protein Eng Des Sel. 1998;11:855–859. doi: 10.1093/protein/11.10.855. doi:10.1093/protein/11.10.855. [DOI] [PubMed] [Google Scholar]
- Milton MJ, Harris R, Probert MA, Field RA, Homans SW. New conformational constraints in isotopically (13C) enriched oligosaccharides. Glycobiology. 1998;8:147–153. doi: 10.1093/glycob/8.2.147. doi:10.1093/glycob/8.2.147. [DOI] [PubMed] [Google Scholar]
- Mo H, Winter HC, Goldstein IJ. Purification and characterization of a Neu5Acα2-6Galβ1-4Glc/GlcNAc-specific lectin from the fruiting body of the polypore mushroom Polyporus squamosus. J Biol Chem. 2000;275:10623–10629. doi: 10.1074/jbc.275.14.10623. doi:10.1074/jbc.275.14.10623. [DOI] [PubMed] [Google Scholar]
- Morales JC, Reina JJ, Díaz I, Aviñó A, Nieto PM, Eritja R. Experimental measurement of carbohydrate-aromatic stacking in water by using a dangling-ended DNA model system. Chem Eur J. 2008;14:7828–7835. doi: 10.1002/chem.200800335. doi:10.1002/chem.200800335. [DOI] [PubMed] [Google Scholar]
- Murayama T, Zuber C, Seelentag WK, Li WP, Kemmner W, Heitz PU, Roth J. Colon carcinoma glycoproteins carrying α2,6-linked sialic acid reactive with Sambucus nigra agglutinin are not constitutively expressed in normal human colon mucosa and are distinct from sialyl-Tn antigen. Int J Cancer. 1997;70:575–581. doi: 10.1002/(sici)1097-0215(19970304)70:5<575::aid-ijc14>3.0.co;2-c. doi:10.1002/(SICI)1097-0215(19970304)70:5<575::AID-IJC14>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr Sect D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. doi:10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. doi:10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Poppe L, Stuike-Prill R, Meyer B, van Halbeek H. The solution conformation of sialyl-α(2-6)-lactose studied by modern NMR techniques and Monte Carlo simulations. J Biomol NMR. 1992;2:109–136. doi: 10.1007/BF01875524. doi:10.1007/BF01875524. [DOI] [PubMed] [Google Scholar]
- Sagui C, Darden TA. Molecular dynamics simulations of biomolecules: Long-range electrostatic effects. Annu Rev Biophys Biomol Struct. 1999;28:155–179. doi: 10.1146/annurev.biophys.28.1.155. doi:10.1146/annurev.biophys.28.1.155. [DOI] [PubMed] [Google Scholar]
- Sata T, Roth J, Zuber C, Stamm B, Heitz PU. Expression of α2,6-linked sialic acid residues in neoplastic but not in normal human colonic mucosa. A lectin-gold cytochemical study with Sambucus nigra and Maackia amurensis lectins. Am J Pathol. 1991;139:1435–1448. [PMC free article] [PubMed] [Google Scholar]
- Shibuya N, Goldstein IJ, Broekaert WF, Nsimba-Lubaki M, Peeters B, Peumans WJ. The elderberry (Sambucus nigra L.) bark lectin recognizes the Neu5Ac(α2-6)Gal/GalNAc sequence. J Biol Chem. 1987;262:1596–1601. [PubMed] [Google Scholar]
- Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. Avian flu: Influenza virus receptors in the human airway. Nature. 2006;440:435–436. doi: 10.1038/440435a. doi:10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- Siebert HC, Reuter G, Schauer R, Von Der Lieth CW, Dabrowski J. Solution conformations of GM3 gangliosides containing different sialic acid residues as revealed by NOE-based distance mapping, molecular mechanics, and molecular dynamics calculations. Biochemistry. 1992;31:6962–6971. doi: 10.1021/bi00145a014. doi:10.1021/bi00145a014. [DOI] [PubMed] [Google Scholar]
- Stevens J, Blixt O, Tumpey TM, Taubenberger JK, Paulson JC, Wilson IA. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006;312:404–410. doi: 10.1126/science.1124513. doi:10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Ito T, Suzuki T, Holland RE, Jr, Chambers TM, Kiso M, Ishida H, Kawaoka Y. Sialic acid species as a determinant of the host range of influenza A viruses. J Virol. 2000;74:11825–11831. doi: 10.1128/jvi.74.24.11825-11831.2000. doi:10.1128/JVI.74.24.11825-11831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateno H, Winter HC, Goldstein IJ. Cloning, expression in Escherichia coli and characterization of the recombinant Neu5Acα2,6Galβ1,4GlcNAc-specific high-affinity lectin and its mutants from the mushroom Polyporus squamosus. Biochem J. 2004;382:667–675. doi: 10.1042/BJ20040391. doi:10.1042/BJ20040391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC. SOLVE and RESOLVE: Automated structure solution and density modification. Methods Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. doi:10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- Toma V, Zuber C, Winter HC, Goldstein IJ, Roth J. Application of a lectin from the mushroom Polyporus squamosus for the histochemical detection of the NeuAcα2,6Galβ1,4Glc/GlcNAc sequence of N-linked oligosaccharides: A comparison with the Sambucus nigra lectin. Histochem Cell Biol. 2001;116:183–193. doi: 10.1007/s004180100304. [DOI] [PubMed] [Google Scholar]
- Tschampel SM, Woods RJ. Quantifying the role of water in protein–carbohydrate interactions. J Phys Chem A. 2003;107:9175–9181. doi: 10.1021/jp035027u. doi:10.1021/jp035027u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki NM, Varki A. Diversity in cell surface sialic acid presentations: Implications for biology and disease. Lab Invest. 2007;87:851–857. doi: 10.1038/labinvest.3700656. doi:10.1038/labinvest.3700656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen MJ, Fruechtnicht W, Brackrock S, Krause KT, Zienkiewicz TJ. Quantitative lectin-histochemical and immunohistochemical studies on the occurrence of α(2,3)- and α(2,6)-linked sialic acid residues in colorectal carcinomas. Relation to clinicopathologic features. Cancer. 1995;76:727–735. doi: 10.1002/1097-0142(19950901)76:5<727::aid-cncr2820760504>3.0.co;2-r. doi:10.1002/1097-0142(19950901)76:5<727::AID-CNCR2820760504>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Wang P-H. Altered glycosylation in cancer: Sialic acids and sialyltransferases. J Cancer Mol. 2005;1:73–81. [Google Scholar]
- Wang WC, Cummings RD. The immobilized leukoagglutinin from the seeds of Maackia amurensis binds with high affinity to complex-type Asn-linked oligosaccharides containing terminal sialic acid-linked α-2,3 to penultimate galactose residues. J Biol Chem. 1988;263:4576–4585. [PubMed] [Google Scholar]
- Winter HC, Mostafapour K, Goldstein IJ. The mushroom Marasmius oreades lectin is a blood group type B agglutinin that recognizes the Galα1,3Gal and Galα1,3Galβ1,4GlcNAc porcine xenotransplantation epitopes with high affinity. J Biol Chem. 2002;277:14996–15001. doi: 10.1074/jbc.M200161200. doi:10.1074/jbc.M200161200. [DOI] [PubMed] [Google Scholar]
- Wolfe S. The Gauche effect. Some stereochemical consequences of adjacent electron pairs and polar bonds. Acc Chem Res. 1972;5:102–111. doi:10.1021/ar50051a003. [Google Scholar]
- Woods Group. GLYCAM Web. Athens, GA: Complex Carbohydrate Research Center, University of Georgia; 2005–2011. http://www.glycam.com . [Google Scholar]
- Woods RJ, Tessier MB. Computational glycoscience: Characterizing the spatial and temporal properties of glycans and glycan–protein complexes. Curr Opin Struct Biol. 2010;20:575–583. doi: 10.1016/j.sbi.2010.07.005. doi:10.1016/j.sbi.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe R, Suzuki R, Kuno A, Fujimoto Z, Jigami Y, Hirabayashi J. Tailoring a novel sialic acid-binding lectin from a ricin-B chain-like galactose-binding protein by natural evolution-mimicry. Eur J Biochem. 2007;141:389–399. doi: 10.1093/jb/mvm043. [DOI] [PubMed] [Google Scholar]
- Yamashita K, Fukushima K, Sakiyama T, Murata F, Kuroki M, Matsuoka Y. Expression of Sia α2–>6Galβ1–>4GlcNAc residues on sugar chains of glycoproteins including carcinoembryonic antigens in human colon adenocarcinoma: Applications of Trichosanthes japonica agglutinin I for early diagnosis. Cancer Res. 1995;55:1675–1679. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.