Abstract
In the field of pharmacogenetics, we currently have a few markers to guide physicians as to the best course of therapy for patients. For the most part, these genetic variants are within a drug metabolizing enzyme that has a large effect on the degree or rate at which a drug is converted to its metabolites. For many drugs, response and toxicity are multi-genic traits and understanding relationships between a patient's genetic variation in drug metabolizing enzymes and the efficacy and/or toxicity of a medication offers the potential to optimize therapies. This review will focus on variants in drug metabolizing enzymes with predictable and relatively large impacts on drug efficacy and/or toxicity; some of these drug/gene variant pairs have impacted drug labels by the United States Food and Drug Administration. The challenges in identifying genetic markers and implementing clinical changes based on known markers will be discussed. In addition, the impact of next generation sequencing in identifying rare variants will be addressed.
Keywords: Adverse drug reactions, Cytochrome P450, drug metabolizing enzymes, pharmacogenetics, pharmacogenomics, single nucleotide polymorphisms
INTRODUCTION
Differences in response to medications have long been recognized by physicians, but it was not until 1957 that Arno Motulsky used previously published works on variations in drug response to propose that “...hereditary gene-controlled enzymatic factors determine why, with identical exposure, certain individuals become ‘sick,’ whereas others are not affected” [1]. Two years later, Vogel first coined the term “pharmacogenetics” to describe the relationship between genetic factors and response to medications [2]. Advances in biochemistry allowed for the discovery of drug metabolizing enzymes and characterization of the various reactions they catalyzed while advances in molecular genetics allowed for an improved understanding of both the DNA sequence responsible for the production of these enzymes and the consequence of genetic variation in that sequence on enzyme activity. The goal of pharmacogenetics is to use genetics to predict response to therapy and to tailor medications appropriately. More recently, there has been a shift from studying variants within one or more candidate genes (pharmacogenetics) towards evaluating the entire genome for associations with pharmacologic phenotypes (pharmacogenomics). This review will focus on the clinically relevant consequences of common genetic variation on drug metabolizing enzymes and the consequences of drug metabolism. Special attention will be paid to the variants that produce predictable changes in drug metabolism and have impacted clinical practice and/or regulatory labeling. Finally, newer approaches to understanding the interplay between genetic variation and drug response as well as the future of “personalized medicine” will be discussed.
DRUG METABOLISM
Drug metabolism is typically responsible for converting drugs to compounds that are more water soluble and more easily excreted but may also be involved in the conversion of prodrugs into active compounds or conversion of drugs to toxic metabolites [3]. Although there are considered two pathways of metabolism: the phase I reactions (oxidation, reduction and hydrolysis) and the phase II conjugation reactions (glucuronidation, acetylation, sulfation and methylation) [3], this classification is historical, and does not necessarily refer to the order of reactions in drug metabolism. Ultimately, all reactions serve the same general purpose of converting lipophilic drugs into hydrophilic metabolites for excretion [3].
There is a rapidly expanding list of genetic variants that affect the function of drug metabolizing enzymes and lead to altered drug responses. Clinicians are becoming increasingly aware of the impact of genetic variation on the therapeutic index of a given medication. Although there are a number of associations identified between drugs and their respective drug metabolizing enzyme, we have chosen to focus on the drug-variant combinations that should merit special consideration by clinicians at this time.
THE PHASE I ENZYMES
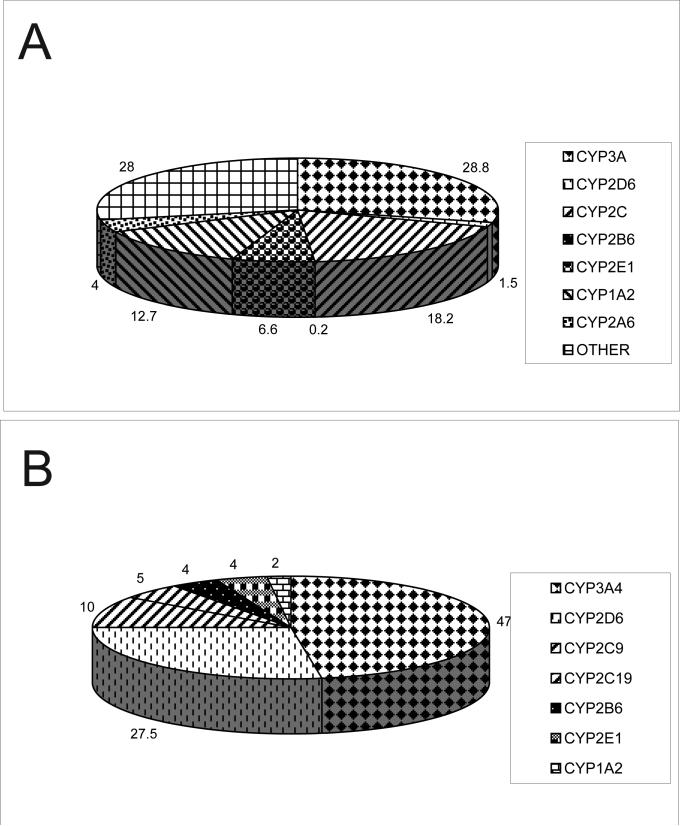
The vast majority of phase I reactions are catalyzed by the cytochrome P450 superfamily of hemoproteins. Early studies of drug metabolism demonstrated the NADPH-dependent oxidation of various compounds by liver microsomes [4,5], and the enzyme responsible for this oxidation was later described as an iron-containing molecule having an absorbance peak of 450 nm, hence cytochrome P450 [6]. Evidence for hydroxylase activity of P450 [7] as well as its role in the metabolism of commonly prescribed drugs like codeine [8] soon followed. Initially assumed to be one enzyme, evidence for multiple isoforms began to emerge in the late 1960s [9,10], and the first experiments to isolate and purify these isoforms from animal liver microsomes emerged in the late 1970s [11] and early 1980s [12]. As more and more isoforms emerged, a recommended nomenclature system was developed where gene families were represented by Roman (later changed to Arabic [13]) numerals, subfamilies represented by letters and individual genes represented by Arabic numerals [14]. The explosion of knowledge of human genetic variation from the sequencing of the human genome has necessitated another addition to the P450 nomenclature: a star (*) followed by a unique Arabic numeral is used to denote nucleotide changes in the reference sequence, which is usually denoted by -*1 [15]. Comprehensive databases of P450 variation are available online courtesy of the Human Cytochrome P450 Nomenclature Committee at http://www.cypalleles.ki.se and the Pharmacogenetics Knowledge Base at http://www.pharmgkb.org. A similar nomenclature has been adopted for the other phase I and phase II enzymes [16-21]. The P450 enzyme families involved with the majority of drug metabolism in humans are CYP3A, CYP2D, and CYP2C. The relative abundance of the P450 enzymes in the human liver and the percentage of drugs these enzymes metabolize are outlined in Fig. (1).
Fig. (1).
(a) Relative abundance of drug metabolizing enzymes in human liver microsomes, “other” includes non-P450 phase I enzymes and phase II enzymes [39]. Other includes minor phase I drug metabolizing enzymes as well as phase II enzymes. (b) Percentage of prescription drugs metabolized by each P450 enzyme [39].
CYP2D6
CYP2D6 has the largest phenotypic variation of the P450 enzymes, and some of the earliest observations of variations in drug metabolism have now been linked to polymorphisms in this gene. In the 1970's, groups investigating the metabolism of two new drugs, sparteine and debrisoquine, both found that a significant minority of individuals were unable to metabolize these drugs [22,23]. Later investigators were able to show that the inability to metabolize these drugs was a recessive trait [24]; was present in approximately 5-10% of Europeans; and that the inability to oxidize sparteine was associated with the inability to hydroxylate debrisoquine [25], suggesting that metabolism of these two drugs was by the same enzyme. It became evident that this deficiency also affected the metabolism of other drugs [26], and that it was at least partially responsible for previous observations that nortryptyline plasma steady state concentrations were influenced by genetics [27,28]. Introduction of molecular techniques in the 1990s allowed for the sequencing of patients with the 2D6 poor metabolizer phenotype and recognition of several variations in the sequence of CYP2D6 [29-31] as well as the functional consequences of these variants on gene expression [32]. Further identification of patients with an ultrarapid metabolism of CYP2D6 substrates due to duplicated or multiple extra copies of a functional CYP2D6 gene [33] and higher levels of enzyme expression led to the current allelic dosage model of CYP2D6 metabolism: where poor metabolizers are homozygous or compound heterozygotes for various loss-of-function alleles, intermediate metabolizers carry one defective allele, normal metabolizers carry zero defective alleles, and extensive metabolizers have a gain in 2D6 function due to duplicated or multiple extra copies of a functional CYP2D6 gene.
CYP2D6 is responsible for the metabolism of 25-30% of prescription medications, but represents about 2-5% of total CYP content of the liver [34]. It is also present in small amounts in the brain [35], gastrointestinal tract [36] and lungs [37]. That genetics plays such a large role in determining inter-individual variation in 2D6 metabolism may be due to the fact that it is thought to be the only non-inducible P450 in humans [38]. There are over 80 allelic variants in CYP2D6, but most are quite rare. Important variants include CYP2D6*9, CYP2D6*10, CYP2D6*17, CYP2D6*29, and CYP2D6*41 which have decreased catalytic activity (intermediate metabolizer phenotype); CYP2D6*3 through CYP2D6*8 as well as CYP2D6*36 which have no functional activity (poor metabolizer phenotype); and duplications of CYP2D6*1xN, CYP2D6*2 or CYP2D6*35 which lead to enhanced functional capacity and the ultrarapid metabolizer phenotype [39]. The distribution of poor, intermediate, extensive and ultrarapid CYP2D6 metabolizers varies among ethnic groups. For example, the reduced-function *10 allele is observed in about 38% to 50% of East Asians but only about 3% of Caucasians and 6% of Africans. Likewise, the reduced-function *17 allele is present almost exclusively in Africans (approximately 21%). The normal-function *2 allele (extensive metabolizer phenotype), in contrast, is present in about 25% of Caucasians and 31% of Africans but only 10% to 12% of East Asians [40]. These differences likely account for some of the ethnic variation in response to medications that are CYP2D6 substrates [41-43].
One of the most publicized examples in pharmacogenetics has been on the effect of variation in CYP2D6 on clinical outcome relates to the use of tamoxifen in the treatment of estrogen-receptor positive breast cancer. The metabolism of tamoxifen is complex, but CYP2D6 mediates the conversion of tamoxifen to endoxifen (4-hydroxy-N-desmethyltamoxifen), a metabolite with a much more potent estrogen receptor binding capacity than the parent compound [44]. Because 2D6 activity is so variable and is the major enzyme responsible for endoxifen production, there has been great interest in the impact of 2D6 variation on response to tamoxifen therapy in women with breast cancer. Most of these studies have been retrospective and represent a heterogeneous patient population using various tamoxifen doses for either adjuvant therapy or chemoprevention of recurrence. Although results are quite contradictory, there have been several series where patients treated with tamoxifen that have a CYP2D6 poor metabolizer phenotype have lower circulating levels of endoxifen and likely have an increased risk of relapse [45-49]. Similarly, patients on tamoxifen treated with medications that act as potent 2D6 inhibitors, like the use of paroxetine to prevent tamoxifen-induced hot flashes, may also have an increased risk of breast cancer recurrence [47,50,51]. Because of the lack of concordant, prospective data, the adoption of routine CYP2D6 genotype testing in women with early breast cancer has not been routinely adopted in clinical practice. However, direct-to-consumer genetic testing of CYP2D6 variants is now commercially available and may influence a patient's decision to start tamoxifen therapy.
A striking example of the impact of genetic variation on response to medication came with the unfortunate report of a fatal opioid overdose in a breastfeeding neonate [52]. An estimated 40% of postgestational women are prescribed codeine for the pain associated with childbirth [53], and its use is generally considered safe in breastfeeding mothers based on several studies finding only low levels of codeine excreted in breast milk [54-56]. However, in the case of the neonate described above, the child's mother was an ultrarapid CYP2D6 metabolizer, and therefore likely had a rapid conversion of codeine to its active metabolite, morphine. The infant was noted to have progressive lethargy prior to being found unresponsive on day of life 11, and a postmortem examination revealed a markedly elevated serum morphine concentration [52]. A subsequent case-control study demonstrated that breastfed infants of CYP2D6 ultrarapid metabolizers were much more likely to experience central nervous system depression when their mothers were prescribed codeine [53].
CYP2C19
The same approaches used to discover CYP2D6 polymorphisms were employed to investigate individuals who were unable to metabolize the anticonvulsant S-mephenytoin [57]. Poor metabolizers of S-mephenytoin were hypothesized to have a different defect than sparteine or debrisoquine poor metabolizers, as these two traits did not co-segregate [57]. The gene for this enzyme was cloned in 1994, and is now referred to CYP2C19 [58]. S-mephenytoin has been used extensively as a metabolic probe of CYP2C19 activity, and a marked ethnic difference in the poor metabolizer phenotype has been observed. While the poor metabolizer phenotype is only present in 2-6% of Caucasians, up to 20% of Asians are poor metabolizers [59].
Various sequencing technologies have identified nearly 30 allelic variants of CYP2C19 [60]. Some of these alleles are associated with reduced catalytic activity (CYP2C19*5 and CYP2C19*8) or no functional activity (CYP2C19*2, CYP2C19*3, CYP2C19*4, CYP2C19*6 and CYP2C19*7) and a poor metabolizer phenotype, whereas others are associated with greater catalytic activity (CYP2C19*17) and an extensive metabolizer phenotype [39]. Several pharmacologic inhibitors of 2C19 exist: cimetidine, oral contraceptives, fluoxetine among others, and inhibition occurs in a gene dose-dependent fashion such that homozygous extensive metabolizers (CYP2C19*17/*17) experience the most 2C19 inhibition with these compounds, where homozygous poor metabolizers experience little to no inhibition [61]. This phenomenon is evident in the inhibition of phenytoin metabolism by compounds like fluoxetine or cimetidine, leading to increased phenytoin exposure and toxic side effects [62].
The impact of 2C19 metabolic variation has been highlighted by several studies and a meta-analysis in patients with coronary artery disease treated with the antiplatelet agent, clopidogrel [63-66]. Clopidogrel is a prodrug which is oxidized to its active metabolite largely by CYP2C19. Patients with the CYP2C19*2 allele, a G to A polymorphism at position 681 that leads to a splicing defect and a truncated protein, are less able to activate clopidogrel and are at higher risk of serious cardiovascular events. However, even in a genetically homogenous population, the effect of CYP2C19 genotype on variability to clopidogrel response appears to be small [64]. In a separate analysis of over 1500 patients undergoing percutaneous coronary stent placement, patients with one or more CYP2C19*17 alleles had a significant increase in bleeding complications, a phenotype consistent with extensive metabolism [67]. Despite the fact that CYP2C19*2 genotype and other non-genetic factors only predict about 12% of the variability in clopidogrel response [68], in 2009 the United States Food and Drug Administration changed the label of clopidogrel to highlight the impact of CYP2C19 genotype on clopidogrel pharmacokinetics and clinical response [65]. Prospective studies on the impact of other antiplatelet agents, such as prasugrel or using a higher dose of clopidogrel in 2C19 poor metabolizers with coronary artery disease are underway [69,70]. Recently, in vitro metabolomic studies have found that paraoxonase-1 (PON1) was the enzyme responsible for the majority of clopidogrel activation, and that a variant within the gene, Q192R, accounted for the majority of variability in drug biotransformation. These findings were confirmed in a clinical cohort, where the QQ192 homozygotes had a much higher rate of coronary stent thrombosis and lower concentrations of the active metabolite of clopidogrel when compared to RR192 homozygotes [71]. This example highlights one of the main difficulties of pharmacogenetic and genomic studies, where differences in phenotype endpoints and/or populations studied can lead to variable associations.
CYP2C9
CYP2C9 is responsible for the metabolism of several drugs with narrow therapeutic indices (i.e. phenytoin and warfarin) and is another human P450 with many functionally significant polymorphisms. It is also subject to both induction and inhibition by a variety of other drugs [39]. Approximately 1-2% of Caucasians have a poor metabolizer phenotype [72]. Up to 20% of Caucasians carry one CYP2C9*2 allele, and 17% carry one CYP2C9*3 allele, both of which have reduced function [72]. Interestingly, in Chinese individuals, CYP2C9*2 is not present and CYP2C9*3 is only present in 2-5% [73]. In African Americans, CYP2C9*8 appears to be the most prevalent variant allele, and is likely the major contributor to CYP2C9 expression in this racial group [74,75].
Variations in 2C9 metabolism have had a large part in understanding the wide interpatient variability in dosing requirements of one of the most commonly prescribed anticoagulants, warfarin. Pharmacologic inhibition of 2C9 by drugs like fluconazole was known to influence the clearance of warfarin [76], and patients requiring a lower maintenance dose of warfarin to achieve therapeutic anticoagulation were 6 times more likely to have one or more CYP2C9 variant alleles [72]. In this study, poor metabolizers were also much more likely to experience a major bleeding event while on warfarin [72]. Further studies attributed between 10-20% of the variability in warfarin dose requirement to CYP2C9 genotype, and variation in a gene in the pharmacodynamic pathway of warfarin, VKORC1, was able to account for another 20-30% of the variability [77]. Two large prospective studies of pharmacogenetic-based dosing of warfarin based on CYP2C9 and VKORC1 genotype as well as other clinical factors (age, race, smoking, concomitant medications) are underway [78,79], and language regarding the impact of variation in CYP2C9 and VKORC1 on warfarin dosing has already been incorporated into the drug label [80].
CYP3A4/CYP3A5
CYP3A4 accounts for approximately 30% of hepatic P450 content and is involved in over 50% of drug metabolism, including immunosuppresants, chemotherapeutics, macrolide antibiotics, antidepressants, anxiolytics, antipsychotics, opiates, calcium channel blockers, and statins [39]. Despite considerable variation in levels of activity among individuals, CYP3A4 does not appear to have polymorphisms that result in absence of functional protein. Several polymorphisms exist within the gene, and some of these do alter the catalytic activity of the enzyme, but these variations have not impacted clinical care to date. Wide variability in CYP3A4 activity is due in part to the large number of substrates capable of inhibiting or inducing the enzyme. Classic examples of 3A4 inducers include the enzyme-inducing antiepileptics phenobarbital, phenytoin, carbamazepine and oxcarbazepine; the non-nucleoside reverse transcriptase inhibitors efavirenz, nivirapine and etravirine; the herbal antidepressant St. John's Wort (hyperforin); as well as the antituberculosis agent rifampin [39]. Inhibitors include protease inhibitors (ritonavir, indinavir, nelfinavir), macrolide antibiotics (erythromycin, clarithromycin), grapefruit juice, and azole antifungals (fluconazole, ketoconazole, voriconazole) [39]. CYP3A4 is also highly expressed in the intestinal tract, and these enzymes can be inhibited and induced by the same substrates, altering the first pass effect [81]. Another factor in the wide interpatient variability in 3A4 activity is the variable expression of CYP3A5, a related enzyme with a broad overlap in substrate specificity with CYP3A4. Only 10% of Europeans express CYP3A5, mostly because of the CYP3A5*3 variant which is an A to G polymorphism at position 6986 resulting in the creation of a new splice site and a truncated protein [82]. The proportion of CYP3A5 expressers is significantly higher in African Americans, due in large part to the rarity of the CYP3A5*3 allele in this population [83]. Other polymorphisms (CYP3A5*6 and CYP3A5*7) also result in abnormal splicing, and are the predominant polymorphisms in African Americans who do not express 3A5 [82].
Other P450 Enzymes
Several other P450 enzymes, including CYP1A2, CYP2A6, CYP2B6 and CYP2E1 are expressed in humans, but in total only account for 5-15% of drug metabolism (Fig. 1b) [39]. Variations in activity can be the result of genetic polymorphisms for several of these enzymes, but their significance in clinical practice largely remains unclear. Some emerging examples include a reduced ability of patients carrying one or more CYP2B6 variant alleles (CYP2B6*6, CYP2B6*16 and/or CYP2B6*18) to activate the antineoplastic prodrug cyclophosphamide (leading to reduced antitumor efficacy) [84] or metabolize the antiretroviral efavirenz (leading to increased systemic toxicity) [85]; CYP2A6*4 variants having slow nicotine metabolism [86,87] and an increased incidence of smoking behavior [88,89]; and CYP1A2*1F CC carriers with rheumatoid arthritis had 9.7-fold increase in toxicity with leflunomide when compared to carriers of the A allele [90]. These associations should be interpreted with caution, as they mostly represent small, single center genotype-phenotype association studies. Combination of variants like these in the pharmacokinetic pathway with variants in the pharmacodynamic pathway, similar to the warfarin and CYP2C9/VKORC1 example, will likely lead to increased predictive power of the effect of variation in these minor metabolizing enzymes on clinical practice.
Other Phase I Enzymes
One of the earliest pharmacogenetic observations involved what is now considered one of the minor phase I enzymes. In the early 1950s, acute hemolysis was noted in a subset of mostly males treated with a new antimalarial agent, primaquine [91,92]. Subsequent studies in prisoners revealed patients that developed hemolysis when exposed to primaquine lacked the enzyme glucose-6-phosphate dehydrogenase in their erythrocytes [93]. The enzyme was linked to the long arm of the X chromosome and the molecular mechanism of this deficiency was elucidated in 1988 and found to be due to two polymorphisms in the gene leading to an unstable enzyme [94,95]. This enzymatic deficiency was also linked to a disease recognized since antiquity, favism. However, a different nucleotide substitution, C to T at position 563 in exon 6, was linked to the Mediterranean form of the disease [96].
Another important pharmacogenetic association involving a minor phase I enzyme was discovered as a result of investigations into toxic deaths from the fluoropyrimidine chemotherapy, 5-fluorouracil (5-FU). Patients who experienced severe or fatal toxicity from this medicine invariably had a decreased activity of dihydropyrimidine dehydrogenase (DPD) enzyme [97]. Dihydropyrimidine dehydrogenase (DPD) is the initial and rate-limiting enzyme in the pathway that catabolizes the pyrimidines such as uracil and thymine [98]. The frequency of low DPD enzyme activity is approximately 3-5% in the general population, but varies significantly among different ethnic populations [99-103]. DYPD, the gene encoding DPD, is subject to polymorphic variation with over 30 SNPs in the gene described to date, although very few of these SNPs have functional consequences [98]. Of particular interest is a splice site variant in intron 14 (IVS14+1G>A, DYPD*2A), which is found in 40-50% of patients with partial or complete DPD deficiency [104]. However, studies attempting to link severe fluoropyrimidine toxicity with variant DYPD genotypes have had varied success, with most studies having good specificity (82-100%) but poor sensitivity (6.3-83%) to detect risk of severe toxicity [105]. Functional testing of DPD activity by various techniques likely provide a more accurate assessment than DYPD genetic testing of a patient's risk of toxicity with fluoropyrimidine therapy. Current methods to functionally test DPD deficiency include incubation of patient peripheral blood mononuclear cells with radiolabeled 5-FU and measurement of metabolite formation by liquid chromotography [106], measurement of a plasma 5,6 dihydrouracil to uracil (UH2/U) ratio prior to 5-FU treatment [107], and measurement of uracil catabolism via a uracil breath test [108]. A small single-center prospective trial of 5-FU dosing determined by DPD metabolic status measured with a plasma UH2/U ratio revealed a significant reduction in adverse effects with no impact on therapeutic efficacy [109], a finding that suggests functional DPD testing and appropriate dose adjustments in deficient patients is feasible with the use of fluoropyrimidines.
The other minor phase I drug metabolizing enzymes: flavin monooxygenases, alcohol and aldehyde dehydrogenases, and the monoamine oxidases are also subject to polymorphic variation, some with functional consequences [110-113]. However, because these enzymes only have a minor role in the metabolism of prescribed drugs, polymorphisms in these enzymes will not be discussed further in this review.
THE PHASE II ENZYMES
The phase II enzymes are responsible for conjugating various molecules resulting in a more water soluble metabolite that can be excreted in the urine or stool. The phase II enzymes are also subject to phenotypic variation.
N-Acetyltransferases
Variations in acetylation leading to adverse drug events were some of the very first pharmacogenetic observations. In the 1950's, a new antituberculosis agent, isoniazid revolutionized the treatment of this deadly disease. However, a rare adverse event, peripheral neuritis, was noted in some patients treated with isoniazid [114]. Hughes and colleagues were able to demonstrate that patients with a slow conversion of isoniazid to acetylisoniazid were more susceptible to the development of peripheral neuritis [115]. Further investigations revealed that slow acetylation was a recessive trait [116] and that there was a wide range of slow acetylators between populations, with 10% of East Asians and up to 50% of Europeans exhibiting the phenotype [117]. Cloning of the cDNA responsible for isoniazid acetylation, N-acetyltransferase 2 (NAT2), was achieved 30 years later [118] and several base substitutions creating variant alleles with absent enzyme activity made evident the molecular mechanism of slow acetylation [119,120].
Thiopurine Methyltransferase (TPMT)
To date, the only pharmacogenetic test involving a drug metabolizing enzyme that has gained widespread acceptance in clinical practice involves another phase II enzyme: thiopurine methyltransferase. The thiopurines, 6-mercaptopurine, 6-thioguanine azathioprine, were developed in the late 1940s and 1950s [121] and have been incorporated into the treatment of hematologic malignancies and autoimmune disorders as well as in the prevention of solid organ transplant rejection. The thiopurines are prodrugs that are converted by multiple enzymes into thioguanine nucleotides (TGN). TGNs are then incorporated into DNA. Inactivation of TGN occurs by two main mechanisms: oxidation by xanthine oxidase and methylation by TPMT. Xanthine oxidase activity is negligible in hematopoetic tissues, so these cells rely on TPMT for TGN inactivation.
Struck by the wide interpatient variability in both response and side effect profiles of patients treated with 6-mercaptopurine (6-MP), Weinshilboum and Sladek first described patients with undetectable erythrocyte TPMT activity [122]. Based on the population distribution of enzyme activity they identified that TPMT enzyme activity was inherited in a codominant fashion, and that 1 in every 300 patients lacked TPMT activity altogether [122]. Subsequent studies revealed that adverse events like drug induced neutropenia were directly correlated with the accumulation of the TGN, and that patients with absent erythrocyte TPMT activity had marked accumulation of these metabolites, and were much more prone to toxic side effects [123]. Conversely, patients with low TGN concentrations and high erythrocyte TPMT concentrations had an increased incidence of relapse of their leukemia when being treated with standard doses of 6-MP [123]. Localization and cloning of wild-type TPMT and 2 common alleles (TPMT*2 and TPMT*3A), each leading to amino acid substitutions and absent enzyme activity, was achieved in 1996 [124,125]. The two variant alleles as well as TPMT*3C account for 95% of intermediate or low enzyme activity cases [126]. Thiopurine dose modifications based on TPMT activity have been adopted widely in the treatment of autoimmune disease [127], and in the treatment of childhood acute lymphoblastic leukemia [128,129].
UDP Glucuronyltransferases (UGTs)
Glucuronidation represents a major pathway that enhances the elimination of many lipophilic xenobiotics and endobiotics to more water-soluble compounds. Over 35 different UGT gene products have been described from several different species. UGTs have been divided into two distinct subfamilies based on sequence identities, UGT1 and UGT2. In his classic paper reviewing early important pharmacogenetic associations, Motulsky mentioned the mild hyperbilirubinemia of Gilbert syndrome, caused by the inability to conjugate bilirubin, and hypothesized that variations in drug glucuronides may be from a similar mechanism [1]. In fact, these two phenomena were shown to be from deficiencies in UGT activity [130]. Cloning of the UGT variant responsible for Gilbert syndrome, UGT1A1*28 – a TA insertion in the promoter region of the gene leading to decreased transcription, was achieved in 1996 [131]. This variant has also been linked to severe hematologic and gastrointestinal toxicities with the antineoplastic agent irinotecan, because of absent conjugation and therefore delayed excretion of a toxic metabolite (SN-38) [132]. Several UGT1A1 genotypes were shown to be associated with the development of severe toxicity with irinotecan [133-135], and in 2005, the United States FDA updated the drug label to include a consideration of dose reduction in patients homozygous for the UGT1A1*28 allele. Studies evaluating the impact of dose modification based on UGT1A1 genotype on preventing toxicity are ongoing.
GENOMIC APPROACHES TO UNDERSTANDING GENETIC VARIATION AND DRUG RESPONSE
Most, but not all, of the examples of genotype-phenotype relationships that have resulted in an FDA label change are from candidate gene studies. These studies focus on variants within one or more genes thought to have the largest effect on metabolism, disposition or mechanism of action. The value of this approach is that it deals with a limited number of genes and variants that have been previously studied; but it is biased by our limited knowledge of the mechanism of action of drugs. Given these limitations, genome wide approaches to discover variants important in drug response and/or toxicity have emerged.
Genome wide techniques treat all interrogated variants as equal in their potential to impact the phenotype of interest. The benefit of this technique is that variants outside of those known to be involved in metabolism, disposition and/or response have the potential to emerge as important factors and may highlight new genes important in the biology of metabolism or response for a given drug. For example, prior work established that approximately 30% of the warfarin dose variance is explained by SNPs in the warfarin drug target VKORC1 and another approximately 12% by the warfarinmetabolizing gene CYP2C9 [136,137]. However, a later genome-wide association study enhanced detection of weaker effects, by conducting a multiple regression adjusting for known influences on warfarin dose (VKORC1, CYP2C9, age, gender) and identified a SNP that alters protein coding of the CYP4F2 gene [138]. Regardless of the technologies used to investigate pharmacogenetic or genomic relationships, for clinical genotype-phenotype studies, patients should be treated uniformly and phenotypes of interest (toxicity, drug response, pharmacokinetic data) should be systematically evaluated. An ideal setting for these types of studies is within the context of large multi-institution clinical trials, where comprehensive response and toxicity data (phenotype data) are kept and can be associated with genetic or genomic data extracted from patients enrolled on the trial.
Although only 2% of the human genome is genes, there has been a strong interest in studying regulatory sequences and non-coding sequences. Intergenic, intron and other non-coding regions may harbor regulators of gene expression [139]. In addition, microRNAs, short sequences encoded throughout the genome, are capable of binding to complementary sequences of mRNA, usually resulting in post-translational gene silencing [140]. Based on sequence homology, several microRNAs are thought to regulate drug metabolizing enzymes, including miR-133 and miR-137 with VKORC1 and miR-22 with MTHFR [141]. There will be even more genetic information in the form of rare SNPs from The 1000 Genomes Project, that will allow a better view of contributions to phenotypic variation [142]. Sequencing data output will be concomitant with the development of powerful tools in bioinformatics for analyzing and storing vast amounts of data that will enable continued growth in the field of personalized therapeutics.
CONCLUSIONS AND FUTURE DIRECTIONS
Common variation in the coding sequence and/or regulatory regions of genes encoding drug metabolizing enzymes has explained a great deal of interindividual variation in response and toxicity with medications. However, drug metabolism is only one aspect of drug-gene interaction, and common genetic variations in the sequence encoding drug transporters, drug receptors, target genes and other pharmacodynamic genes have also been shown to impact toxicity and response to treatment. Many formerly idiosyncratic (not predictable by drug concentration) adverse drug reactions like hypersensitivity reactions, liver injury and prolongation of the QT interval can now be at least partially explained by variation in genes outside of the metabolic or therapeutic pathway such as genes encoding for human leukocyte antigens and voltage gated ion channels [143-145]. The post-genome era and advances in microarray technology have made scanning a patient's entire genome for associations with drug response and/or toxicity much more affordable and practical.
Recognition of the potential of pharmacogenomics in eventually being able to accurately predict toxicity and response has led to the widespread collection and banking of DNA for both ongoing prospective and future genotype-phenotype association studies. Because the inherited component of drug response for a given drug is polygenic in the vast majority of cases, development of techniques for elucidating the multiple genes involved and algorithms to consider multiple alleles are of considerable interest. Using well-defined phenotypes is key to making reproducible genotype-phenotype correlations; however this presents a challenge when performing clinical studies where there are a number of confounding variables. Well designed prospective pharmacogenetic and pharmacogenomic studies will undoubtedly increase our knowledge of genetic markers predictive of drug response and toxicity, and lead to FDA label changes, and most importantly, improved efficacy and safety of current medications.
The authors have no conflicts of interest to report.
Table 1.
Overview of Common Genetic Variants in Phase I Drug Metabolising Enzymes
| Gene | Important Variants | Variation | Consequence of Variation | Effect on Enzyme Activity | Selected Medication Substrates |
|---|---|---|---|---|---|
| CYP2D6 | CYP2D6*3 | 2549 del A (rs4986774) | Frameshift mutation, premature stop | Absent activity | Amitriptyline, atomoxetine, carvedilol, chlorpheniramine, chlorpromazine, citalopram, clomipramine, clozapine, codeine, debrisoquine, desipramine, dextromethorphan, doxepin, flecainide, fluoxetine, fluvoxamine, gefitinib, haloperidol, imipramine, maprotiline, metoprolol, mexilet-ine, mianserin, morphine, nortriptyline, paroxetine, risperidone, tamoxifen, thioridazine, timolol, tolterodine, tramadol |
| CYP2D6*4 | 1846G>A (rs3892097) | Splicing defect | Absent activity | ||
| CYP2D6*5 | n/a | Whole gene deletion | Absent activity | ||
| CYP2D6*6 | 1707 del T (rs5030655) | Frameshift mutation | Absent activity | ||
| CYP2D6*9 | 2615_2617delAAG (rs5030656) | Deletion of lys281 | Reduced activity | ||
| CYP2D6*10 | 100C>T (rs1065852) | pro34ser | Reduced activity | ||
| CYP2D6*17 | 1023C>T (rs28371706) | thr107ile | Reduced activity* | ||
| CYP2D6*29 | 3183G>A (rs59421388) and 4180G>C (rs61736512) | val338met and ser486thr | Reduced activity | ||
| CYP2D6*41 | 2988G>A (rs28371725) | Splicing defect | Reduced activity | ||
| CYP2D6*UM | n/a | Additional functional copies (2-13) of gene | Increased activity | ||
| CYP2C19 | CYP2C19*2 | 681G>A (rs4244285) | Splicing defect, premature stop | Absent activity | lansoprazole, omeprazole, pantoprazole, rabeprazole, phenytoin, phenobarbitone, amitriptyline, carisoprodol, clopidogrel, citalopram, clomipramine, cyclophos-phamide, hexobarbital, imipramine, indomethacin, nelfi-navir, nilutamide, primidone, progesterone, proguanil, propranolol, teniposide, warfarin |
| CYP2C19*3 | 636G>A (rs4986893) | Premature stop | Absent activity | ||
| CYP2C9 | CYP2C9*2 | 430C>T (rs1799853) | arg144cys | Reduced activity | irbesartan, losartan, phenytoin, cyclophosphamide, tamoxifen, fluvastatin, celecoxib, diclofenac, ibuprofen, meloxicam, naproxen, glibenclamide, glimepiride, glipizide, tolbutamide, warfarin |
| CYP2C9*3 | 1075A>C (rs1057910) | ile359leu | Reduced activity | ||
| CYP3A4 | No clinically relevant variants described to date | N/A | N/A | N/A | clarithromycin, erythromycin, telithromycin, quinidine, alprazolam, diazepam, midazolam, triazolam, cyclosporine, tacrolimus, indinavir, nelfinavir, ritonavir, saquinavir, cisapride, astemizole, chlorpheniramine, terfenidine, amlodipine, diltiazem, felodipine, lercanidipine, nifedipine, nisoldipine, nitrendipine, verapamil, atorvastatin, cerivastatin, lovastatin, simvastatin, estradiol, hydrocortisone, progesterone, testosterone, alfentanil, aprepitant, aripiprazole, buspirone, cafergot, caffeine, cilostazol, cocaine, codeine, dapsone, dextromethorphan, docetaxel, domperidone, eplerenone, fentanyl, finasteride, gleevec, haloperidol, irinotecan, LAAM, lidocaine, methadone, nateglinide, ondansetron, pimozide, propranolol, quetiapine, quinine, risperidone, salmeterol, sildenafil, sirolimus, tamoxifen, taxol, terfenadine, trazodone, vincristine, zaleplon, ziprasidone, zolpidem |
| CYP3A5 | CYP3A5*3 | 6986A>G (rs776746) | Splicing defect, premature stop | Absent activity | Broad overlap with CYP3A4 |
| CYP3A5*6 | 14690G>A (rs10264272) | Splicing defect, removes exon 7 | Absent activity | ||
| CYP3A5*7 | 27131-2insT (rs41303343) | Frameshift mutation | Absent activity | ||
| CYP2B6 | CYP2B6*6 | 516G>T (rs3745274) | his172gln | Reduced activity | bupropion, cyclophosphamide, diazepam, efavirenz, ifosfamide, ketamine, methadone, MDMA, meperidine, mephenytoin, midazolam, nevirapine, nicotine, propofol, selegiline, tamoxifen |
| CYP2B6*I6 | 983T>C (rs28399499) | ile328thr | Reduced activity | ||
| CYP2B6*I8 | 1459C>T (rs3211371) | arg487cys | Reduced activity |
Data compiled from http://www.pharmgkb.org and http://www.cypalleles.ki.se. Bold type indicates drug of particular interest to the corresponding gene.
while CYP2D6*I7 is generally considered to be a reduced function allele, evidence exists that this variant has normal activity towards risperidone [146].
ACKNOWLEDGEMENT
The work performed by the authors of this manuscript is supported by Pharmacogenetic of Anticancer Agents Research (PAAR) Group (http://pharmacogenetics.org) NIH/NIGMS UO1GM61393, the University of Chicago Breast Cancer SPORE grant P50 CA125183, NIH/NCI grant CA136765 (MED) and ASCO-Young Investigator Award 43829 and St. Baldrick's Foundation Fellowship (NP).
Footnotes
CONFLICTS OF INTEREST
REFERENCES
- 1.Motulsky AG. Drug reactions enzymes, and biochemical genetics. J. Am. Med. Assoc. 1957;165:835–837. doi: 10.1001/jama.1957.72980250010016. [DOI] [PubMed] [Google Scholar]
- 2.Vogel F. Moderne Probleme der Humangenetik. Ergeb Inn Med Kinderheilkd. 1959;12:52–125. [Google Scholar]
- 3.Goodman LS, Gilman A, Brunton LL, Lazo JS, Parker K. Goodman & Gilman's the pharmacological basis of therapeutics. 11th edn McGraw-Hill; 2006. [Google Scholar]
- 4.Conney AH, Miller EC, Miller JA. Substrate-induced synthesis and other properties of benzpyrene hydroxylase in rat liver. J. Biol. Chem. 1957;228:753–766. [PubMed] [Google Scholar]
- 5.Brodie BB, Gillette JR, La Du BN. Enzymatic metabolism of drugs and other foreign compounds. Annu. Rev. Biochem. 1958;27:427–454. doi: 10.1146/annurev.bi.27.070158.002235. [DOI] [PubMed] [Google Scholar]
- 6.Omura T, Sato R. A new cytochrome in liver microsomes. J. Biol. Chem. 1962;237:1375–1376. [PubMed] [Google Scholar]
- 7.Cooper DY, Estabrook RW, Rosenthal O. The stoichiometry of C21 hydroxylation of steroids by adrenocortical microsomes. J. Biol. Chem. 1963;238:1320–1323. [PubMed] [Google Scholar]
- 8.Cooper DY, Levin S, Narasimhulu S, Rosenthal O. Photochemical Action Spectrum of the Terminal Oxidase of Mixed Function Oxidase Systems. Science. 1965;147:400–402. doi: 10.1126/science.147.3656.400. [DOI] [PubMed] [Google Scholar]
- 9.Sladek NE, Mannering GJ. Induction of drug metabolism. II. Qualitative differences in the microsomal N-demethylating systems stimulated by polycyclic hydrocarbons and by phenobarbital. Mol. Pharmacol. 1969;5:186–199. [PubMed] [Google Scholar]
- 10.Sladek NE, Mannering GJ. Induction of drug metabolism. I. Differences in the mechanisms by which polycyclic hydrocarbons and phenobarbital produce their inductive effects on microsomal N-demethylating systems. Mol. Pharmacol. 1969;5:174–185. [PubMed] [Google Scholar]
- 11.Wiebel FJ, Selkirk JK, Gelboin HV, Haugen DA, van der Hoeven TA, Coon MJ. Position-specific oxygenation of benzo(a)pyrene by different forms of purified cytochrome P-450 from rabbit liver. Proc. Natl. Acad. Sci. U S A. 1975;72:3917–3920. doi: 10.1073/pnas.72.10.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schenkman JB, Jansson I, Backes WL, Cheng KC, Smith C. Dissection of cytochrome P-450 isozymes (RLM) from fractions of untreated rat liver microsomal proteins. Biochem. Biophys. Res. Commun. 1982;107:1517–1523. doi: 10.1016/s0006-291x(82)80171-x. [DOI] [PubMed] [Google Scholar]
- 13.Nebert DW, Nelson DR, Coon MJ, Estabrook RW, Feyereisen R, Fujii-Kuriyama Y, Gonzalez FJ, Guengerich FP, Gunsalus IC, Johnson EF, et al. The P450 superfamily: update on new sequences, gene mapping, and recommended nomenclature. DNA Cell Biol. 1991;10:1–14. doi: 10.1089/dna.1991.10.1. [DOI] [PubMed] [Google Scholar]
- 14.Nebert DW, Adesnik M, Coon MJ, Estabrook RW, Gonzalez FJ, Guengerich FP, Gunsalus IC, Johnson EF, Kemper B, Levin W, et al. The P450 gene superfamily: recommended nomenclature. DNA. 1987;6:1–11. doi: 10.1089/dna.1987.6.1. [DOI] [PubMed] [Google Scholar]
- 15.Robarge JD, Li L, Desta Z, Nguyen A, Flockhart DA. The star-allele nomenclature: retooling for translational genomics. Clin. Pharmacol. Ther. 2007;82:244–248. doi: 10.1038/sj.clpt.6100284. [DOI] [PubMed] [Google Scholar]
- 16.Lawton MP, Cashman JR, Cresteil T, Dolphin CT, Elfarra AA, Hines RN, Hodgson E, Kimura T, Ozols J, Phillips IR, et al. A nomenclature for the mammalian flavin-containing monooxygenase gene family based on amino acid sequence identities. Arch. Biochem. Biophys. 1994;308:254–257. doi: 10.1006/abbi.1994.1035. [DOI] [PubMed] [Google Scholar]
- 17.Duester G, Farres J, Felder MR, Holmes RS, Hoog JO, Pares X, Plapp BV, Yin SJ, Jornvall H. Recommended nomenclature for the vertebrate alcohol dehydrogenase gene family. Biochem. Pharmacol. 1999;58:389–395. doi: 10.1016/s0006-2952(99)00065-9. [DOI] [PubMed] [Google Scholar]
- 18.Maguire JJ, Parker WA, Foord SM, Bonner TI, Neubig RR, Davenport AP. International Union of Pharmacology. LXXII. Recommendations for trace amine receptor nomenclature. Pharmacol. Rev. 2009;61:1–8. doi: 10.1124/pr.109.001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Black WJ, Stagos D, Marchitti SA, Nebert DW, Tipton KF, Bairoch A, Vasiliou V. Human aldehyde dehydrogenase genes: alternatively spliced transcriptional variants and their suggested nomenclature. Pharmacogenet. Genomics. 2009;19:893–902. doi: 10.1097/FPC.0b013e3283329023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mackenzie PI, Bock KW, Burchell B, Guillemette C, Ikushiro S, Iyanagi T, Miners JO, Owens IS, Nebert DW. Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet. Genomics. 2005;15:677–685. doi: 10.1097/01.fpc.0000173483.13689.56. [DOI] [PubMed] [Google Scholar]
- 21.Hein DW, Boukouvala S, Grant DM, Minchin RF, Sim E. Changes in consensus arylamine N-acetyltransferase gene nomenclature. Pharmacogenet. Genomics. 2008;18:367–368. doi: 10.1097/FPC.0b013e3282f60db0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahgoub A, Idle JR, Dring LG, Lancaster R, Smith RL. Polymorphic hydroxylation of Debrisoquine in man. Lancet. 1977;2:584–586. doi: 10.1016/s0140-6736(77)91430-1. [DOI] [PubMed] [Google Scholar]
- 23.Eichelbaum M, Spannbrucker N, Steincke B, Dengler HJ. Defective N-oxidation of sparteine in man: a new pharmacogenetic defect. Eur. J. Clin. Pharmacol. 1979;16:183–187. doi: 10.1007/BF00562059. [DOI] [PubMed] [Google Scholar]
- 24.Evans DA, Mahgoub A, Sloan TP, Idle JR, Smith RL. A family and population study of the genetic polymorphism of debrisoquine oxidation in a white British population. J. Med. Genet. 1980;17:102–105. doi: 10.1136/jmg.17.2.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertilsson L, Dengler HJ, Eichelbaum M, Schulz HU. Pharmacogenetic covariation of defective N-oxidation of sparteine and 4-hydroxylation of debrisoquine. Eur. J. Clin. Pharmacol. 1980;17:153–155. doi: 10.1007/BF00562624. [DOI] [PubMed] [Google Scholar]
- 26.Bertilsson L, Eichelbaum M, Mellstrom B, Sawe J, Schulz HU, Sjoqvist F. Nortriptyline and antipyrine clearance in relation to debrisoquine hydroxylation in man. Life Sci. 1980;27:1673–1677. doi: 10.1016/0024-3205(80)90642-6. [DOI] [PubMed] [Google Scholar]
- 27.Hammer W, Sjoqvist F. Plasma levels of monomethylated tricyclic antidepressants during treatment with imipramine-like compounds. Life Sci. 1967;6:1895–1903. doi: 10.1016/0024-3205(67)90218-4. [DOI] [PubMed] [Google Scholar]
- 28.Alexanderson B, Evans DA, Sjoqvist F. Steady-state plasma levels of nortriptyline in twins: influence of genetic factors and drug therapy. Br. Med. J. 1969;4:764–768. doi: 10.1136/bmj.4.5686.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heim M, Meyer UA. Genotyping of poor metabolisers of debrisoquine by allele-specific PCR amplification. Lancet. 1990;336:529–532. doi: 10.1016/0140-6736(90)92086-w. [DOI] [PubMed] [Google Scholar]
- 30.Gough AC, Miles JS, Spurr NK, Moss JE, Gaedigk A, Eichelbaum M, Wolf CR. Identification of the primary gene defect at the cytochrome P450 CYP2D locus. Nature. 1990;347:773–776. doi: 10.1038/347773a0. [DOI] [PubMed] [Google Scholar]
- 31.Gaedigk A, Blum M, Gaedigk R, Eichelbaum M, Meyer UA. Deletion of the entire cytochrome P450 CYP2D6 gene as a cause of impaired drug metabolism in poor metabolizers of the debrisoquine/sparteine polymorphism. Am. J. Hum. Genet. 1991;48:943–950. [PMC free article] [PubMed] [Google Scholar]
- 32.Hanioka N, Kimura S, Meyer UA, Gonzalez FJ. The human CYP2D locus associated with a common genetic defect in drug oxidation: a G1934----A base change in intron 3 of a mutant CYP2D6 allele results in an aberrant 3' splice recognition site. Am. J. Hum. Genet. 1990;47:994–1001. [PMC free article] [PubMed] [Google Scholar]
- 33.Johansson I, Lundqvist E, Bertilsson L, Dahl ML, Sjoqvist F, Ingelman-Sundberg M. Inherited amplification of an active gene in the cytochrome P450 CYP2D locus as a cause of ultrarapid metabolism of debrisoquine. Proc. Natl. Acad. Sci. U S A. 1993;90:11825–11829. doi: 10.1073/pnas.90.24.11825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ingelman-Sundberg M. The human genome project and novel aspects of cytochrome P450 research. Toxicol. Appl. Pharmacol. 2005;207:52–56. doi: 10.1016/j.taap.2005.01.030. [DOI] [PubMed] [Google Scholar]
- 35.Chinta SJ, Pai HV, Upadhya SC, Boyd MR, Ravindranath V. Constitutive expression and localization of the major drug metabolizing enzyme, cytochrome P4502D in human brain. Brain Res. Mol. Brain Res. 2002;103:49–61. doi: 10.1016/s0169-328x(02)00177-8. [DOI] [PubMed] [Google Scholar]
- 36.Prueksaritanont T, Dwyer LM, Cribb AE. (+)-bufuralol 1'-hydroxylation activity in human and rhesus monkey intestine and liver. Biochem. Pharmacol. 1995;50:1521–1525. doi: 10.1016/0006-2952(95)02052-7. [DOI] [PubMed] [Google Scholar]
- 37.Guidice JM, Marez D, Sabbagh N, Legrand-Andreoletti M, Spire C, Alcaide E, Lafitte JJ, Broly F. Evidence for CYP2D6 expression in human lung. Biochem. Biophys. Res. Commun. 1997;241:79–85. doi: 10.1006/bbrc.1997.7775. [DOI] [PubMed] [Google Scholar]
- 38.Ingelman-Sundberg M, Sim SC, Gomez A, Rodriguez-Antona C. Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol. Ther. 2007;116:496–526. doi: 10.1016/j.pharmthera.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 39.Boullata JI, Armenti VT. Handbook of drug-nutrient interactions. Humana Press; 2004. [Google Scholar]
- 40.Man M, Farmen M, Dumaual C, Teng CH, Moser B, Irie S, Noh GJ, Njau R, Close S, Wise S, Hockett R. Genetic variation in metabolizing enzyme and transporter genes: comprehensive assessment in 3 major East Asian subpopulations with comparison to Caucasians and Africans. J. Clin. Pharmacol. 2010;50:929–940. doi: 10.1177/0091270009355161. [DOI] [PubMed] [Google Scholar]
- 41.Rudorfer MV, Lane EA, Potter WZ. Interethnic dissociation between debrisoquine and desipramine hydroxylation. J. Clin. Psychopharmacol. 1985;5:89–92. doi: 10.1097/00004714-198504000-00006. [DOI] [PubMed] [Google Scholar]
- 42.Zhan M, Flaws JA, Gallicchio L, Tkaczuk K, Lewis LM, Royak-Schaler R. Profiles of tamoxifen-related side effects by race and smoking status in women with breast cancer. Cancer Detect. Prev. 2007;31:384–390. doi: 10.1016/j.cdp.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 43.Caraco Y, Sheller J, Wood AJ. Impact of ethnic origin and quinidine coadministration on codeine's disposition and pharmacodynamic effects. J. Pharmacol. Exp. Ther. 1999;290:413–422. [PubMed] [Google Scholar]
- 44.Wu X, Hawse JR, Subramaniam M, Goetz MP, Ingle JN, Spelsberg TC. The tamoxifen metabolite, endoxifen, is a potent antiestrogen that targets estrogen receptor alpha for degradation in breast cancer cells. Cancer Res. 2009;69:1722–1727. doi: 10.1158/0008-5472.CAN-08-3933. [DOI] [PubMed] [Google Scholar]
- 45.Goetz MP, Rae JM, Suman VJ, Safgren SL, Ames MM, Visscher DW, Reynolds C, Couch FJ, Lingle WL, Flockhart DA, Desta Z, Perez EA, Ingle JN. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J. Clin. Oncol. 2005;23:9312–9318. doi: 10.1200/JCO.2005.03.3266. [DOI] [PubMed] [Google Scholar]
- 46.Rae JM, Goetz MP, Hayes DF, Ingle JN, Li L, Storniolo AM, Stearns V, Flockhart DA. CYP2D6 genotype and tamoxifen response. Breast Cancer Res. 2005;7:E6. doi: 10.1186/bcr1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin Y, Desta Z, Stearns V, Ward B, Ho H, Lee KH, Skaar T, Storniolo AM, Li L, Araba A, Blanchard R, Nguyen A, Ullmer L, Hayden J, Lemler S, Weinshilboum RM, Rae JM, Hayes DF, Flockhart DA. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J. Natl. Cancer Inst. 2005;97:30–39. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 48.Schroth W, Goetz MP, Hamann U, Fasching PA, Schmidt M, Winter S, Fritz P, Simon W, Suman VJ, Ames MM, Safgren SL, Kuffel MJ, Ulmer HU, Bolander J, Strick R, Beckmann MW, Koelbl H, Weinshilboum RM, Ingle JN, Eichelbaum M, Schwab M, Brauch H. Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. JAMA. 2009;302:1429–1436. doi: 10.1001/jama.2009.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schroth W, Hamann U, Fasching PA, Dauser S, Winter S, Eichelbaum M, Schwab M, Brauch H. CYP2D6 polymorphisms as predictors of outcome in breast cancer patients treated with tamoxifen: expanded polymorphism coverage improves risk stratification. Clin. Cancer Res. 2010;16:4468–4477. doi: 10.1158/1078-0432.CCR-10-0478. [DOI] [PubMed] [Google Scholar]
- 50.Stearns V, Johnson MD, Rae JM, Morocho A, Novielli A, Bhargava P, Hayes DF, Desta Z, Flockhart DA. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J. Natl. Cancer Inst. 2003;95:1758–1764. doi: 10.1093/jnci/djg108. [DOI] [PubMed] [Google Scholar]
- 51.Dezentje VO, van Blijderveen NJ, Gelderblom H, Putter H, van Herk-Sukel MP, Casparie MK, Egberts AC, Nortier JW, Guchelaar HJ. Effect of concomitant CYP2D6 inhibitor use and tamoxifen adherence on breast cancer recurrence in early-stage breast cancer. J. Clin. Oncol. 2010;28:2423–2429. doi: 10.1200/JCO.2009.25.0894. [DOI] [PubMed] [Google Scholar]
- 52.Madadi P, Koren G, Cairns J, Chitayat D, Gaedigk A, Leeder JS, Teitelbaum R, Karaskov T, Aleksa K. Safety of codeine during breastfeeding: fatal morphine poisoning in the breastfed neonate of a mother prescribed codeine. Can. Fam. Physician. 2007;53:33–35. [PMC free article] [PubMed] [Google Scholar]
- 53.Madadi P, Ross CJ, Hayden MR, Carleton BC, Gaedigk A, Leeder JS, Koren G. Pharmacogenetics of neonatal opioid toxicity following maternal use of codeine during breastfeeding: a case-control study. Clin. Pharmacol. Ther. 2009;85:31–35. doi: 10.1038/clpt.2008.157. [DOI] [PubMed] [Google Scholar]
- 54.Sapeika N. The excretion of drugs in human milk; a review. J. Obstet. Gynaecol. Br. Emp. 1947;54:426–431. doi: 10.1111/j.1471-0528.1947.tb10726.x. [DOI] [PubMed] [Google Scholar]
- 55.Findlay JW, DeAngelis RL, Kearney MF, Welch RM, Findlay JM. Analgesic drugs in breast milk and plasma. Clin. Pharmacol. Ther. 1981;29:625–633. doi: 10.1038/clpt.1981.87. [DOI] [PubMed] [Google Scholar]
- 56.Meny RG, Naumburg EG, Alger LS, Brill-Miller JL, Brown S. Codeine and the breastfed neonate. J. Hum. Lact. 1993;9:237–240. doi: 10.1177/089033449300900423. [DOI] [PubMed] [Google Scholar]
- 57.Kupfer A, Preisig R. Pharmacogenetics of mephenytoin: a new drug hydroxylation polymorphism in man. Eur. J. Clin. Pharmacol. 1984;26:753–759. doi: 10.1007/BF00541938. [DOI] [PubMed] [Google Scholar]
- 58.de Morais SM, Wilkinson GR, Blaisdell J, Nakamura K, Meyer UA, Goldstein JA. The major genetic defect responsible for the polymorphism of S-mephenytoin metabolism in humans. J. Biol. Chem. 1994;269:15419–15422. [PubMed] [Google Scholar]
- 59.Flockhart DA. Drug interactions and the cytochrome P450 system. The role of cytochrome P450 2C19. Clin. Pharmacokinet. 1995;29(Suppl 1):45–52. doi: 10.2165/00003088-199500291-00008. [DOI] [PubMed] [Google Scholar]
- 60.Home Page of the Human Cytochrome P450 (CYP) Allele Nomenclature Committee. < http://www.cypalleles.ki.se/> (
- 61.Desta Z, Zhao X, Shin JG, Flockhart DA. Clinical significance of the cytochrome P450 2C19 genetic polymorphism. Clin. Pharmacokinet. 2002;41:913–958. doi: 10.2165/00003088-200241120-00002. [DOI] [PubMed] [Google Scholar]
- 62.Levy RH. Cytochrome P450 isozymes and antiepileptic drug interactions. Epilepsia. 1995;36(Suppl 5):S8–13. doi: 10.1111/j.1528-1157.1995.tb06007.x. [DOI] [PubMed] [Google Scholar]
- 63.Simon T, Verstuyft C, Mary-Krause M, Quteineh L, Drouet E, Meneveau N, Steg PG, Ferrieres J, Danchin N, Becquemont L. Genetic determinants of response to clopidogrel and cardiovascular events. N. Engl. J. Med. 2009;360:363–375. doi: 10.1056/NEJMoa0808227. [DOI] [PubMed] [Google Scholar]
- 64.Shuldiner AR, O'Connell JR, Bliden KP, Gandhi A, Ryan K, Horenstein RB, Damcott CM, Pakyz R, Tantry US, Gibson Q, Pollin TI, Post W, Parsa A, Mitchell BD, Faraday N, Herzog W, Gurbel PA. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302:849–857. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ellis KJ, Stouffer GA, McLeod HL, Lee CR. Clopidogrel pharmacogenomics and risk of inadequate platelet inhibition: US FDA recommendations. Pharmacogenomics. 2009;10:1799–1817. doi: 10.2217/pgs.09.143. [DOI] [PubMed] [Google Scholar]
- 66.Sofi F, Giusti B, Marcucci R, Gori AM, Abbate R, Gensini GF. Cytochrome P450 2C19(*)2 polymorphism and cardiovascular recurrences in patients taking clopidogrel: a meta-analysis. Pharmacogenomics J. 2010 doi: 10.1038/tpj.2010.21. [DOI] [PubMed] [Google Scholar]
- 67.Sibbing D, Koch W, Gebhard D, Schuster T, Braun S, Stegherr J, Morath T, Schomig A, von Beckerath N, Kastrati A. Cytochrome 2C19*17 allelic variant, platelet aggregation, bleeding events, and stent thrombosis in clopidogrel-treated patients with coronary stent placement. Circulation. 2010;121:512–518. doi: 10.1161/CIRCULATIONAHA.109.885194. [DOI] [PubMed] [Google Scholar]
- 68.Geisler T, Schaeffeler E, Dippon J, Winter S, Buse V, Bischofs C, Zuern C, Moerike K, Gawaz M, Schwab M. CYP2C19 and nongenetic factors predict poor responsiveness to clopidogrel loading dose after coronary stent implantation. Pharmacogenomics. 2008;9:1251–1259. doi: 10.2217/14622416.9.9.1251. [DOI] [PubMed] [Google Scholar]
- 69.Sorich MJ, Vitry A, Ward MB, Horowitz JD, McKinnon RA. Prasugrel vs. clopidogrel for cytochrome P450 2C19-genotyped subgroups: integration of the TRITON-TIMI 38 trial data. J. Thromb. Haemost. 2010;8:1678–1684. doi: 10.1111/j.1538-7836.2010.03923.x. [DOI] [PubMed] [Google Scholar]
- 70.Fontana P, Senouf D, Mach F. Biological effect of increased maintenance dose of clopidogrel in cardiovascular outpatients and influence of the cytochrome P450 2C19*2 allele on clopidogrel responsiveness. Thromb. Res. 2008;121:463–468. doi: 10.1016/j.thromres.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 71.Bouman HJ, Schomig E, van Werkum JW, Velder J, Hackeng CM, Hirschhauser C, Waldmann C, Schmalz HG, ten Berg JM, Taubert D. Paraoxonase-1 is a major determinant of clopidogrel efficacy. Nat. Med. 2011;17:110–116. doi: 10.1038/nm.2281. [DOI] [PubMed] [Google Scholar]
- 72.Aithal GP, Day CP, Kesteven PJ, Daly AK. Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. Lancet. 1999;353:717–719. doi: 10.1016/S0140-6736(98)04474-2. [DOI] [PubMed] [Google Scholar]
- 73.Xie HG, Prasad HC, Kim RB, Stein CM. CYP2C9 allelic variants: ethnic distribution and functional significance. Adv. Drug Deliv. Rev. 2002;54:1257–1270. doi: 10.1016/s0169-409x(02)00076-5. [DOI] [PubMed] [Google Scholar]
- 74.Scott SA, Jaremko M, Lubitz SA, Kornreich R, Halperin JL, Desnick RJ. CYP2C9*8 is prevalent among African-Americans: implications for pharmacogenetic dosing. Pharmacogenomics. 2009;10:1243–1255. doi: 10.2217/pgs.09.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cavallari LH, Langaee TY, Momary KM, Shapiro NL, Nutescu EA, Coty WA, Viana MA, Patel SR, Johnson JA. Genetic and clinical predictors of warfarin dose requirements in African Americans. Clin. Pharmacol. Ther. 2010;87:459–464. doi: 10.1038/clpt.2009.223. [DOI] [PubMed] [Google Scholar]
- 76.Black DJ, Kunze KL, Wienkers LC, Gidal BE, Seaton TL, McDonnell ND, Evans JS, Bauwens JE, Trager WF. Warfarin-fluconazole. II. A metabolically based drug interaction: in vivo studies. Drug Metab. Dispos. 1996;24:422–428. [PubMed] [Google Scholar]
- 77.Klein TE, Altman RB, Eriksson N, Gage BF, Kimmel SE, Lee MT, Limdi NA, Page D, Roden DM, Wagner MJ, Caldwell MD, Johnson JA. Estimation of the warfarin dose with clinical and pharmacogenetic data. N. Engl. J. Med. 2009;360:753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Joo J, Geller NL, French B, Kimmel SE, Rosenberg Y, Ellenberg JH. Prospective alpha allocation in the clarification of optimal anticoagulation through genetics (COAG) trial. Clin. Trials. 2010;7:597–604. doi: 10.1177/1740774510381285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Genetics Informatics Trial (GIFT) of Warfarin to Prevent DVT. doi: 10.1038/tpj.2011.18. < http://clinicaltrials.gov/ct2/show/NCT01006733> ( [DOI] [PMC free article] [PubMed]
- 80.Kim MJ, Huang SM, Meyer UA, Rahman A, Lesko LJ. A regulatory science perspective on warfarin therapy: a pharmacogenetic opportunity. J. Clin. Pharmacol. 2009;49:138–146. doi: 10.1177/0091270008328098. [DOI] [PubMed] [Google Scholar]
- 81.Kato M. Intestinal first-pass metabolism of CYP3A4 substrates. Drug Metab. Pharmacokinet. 2008;23:87–94. doi: 10.2133/dmpk.23.87. [DOI] [PubMed] [Google Scholar]
- 82.Daly AK. Significance of the minor cytochrome P450 3A isoforms. Clin. Pharmacokinet. 2006;45:13–31. doi: 10.2165/00003088-200645010-00002. [DOI] [PubMed] [Google Scholar]
- 83.Lamba JK, Lin YS, Schuetz EG, Thummel KE. Genetic contribution to variable human CYP3A-mediated metabolism. Adv. Drug Deliv. Rev. 2002;54:1271–1294. doi: 10.1016/s0169-409x(02)00066-2. [DOI] [PubMed] [Google Scholar]
- 84.Roy P, Waxman DJ. Activation of oxazaphosphorines by cytochrome P450: application to gene-directed enzyme prodrug therapy for cancer. Toxicol. In Vitro. 2006;20:176–186. doi: 10.1016/j.tiv.2005.06.046. [DOI] [PubMed] [Google Scholar]
- 85.Rotger M, Tegude H, Colombo S, Cavassini M, Furrer H, Decosterd L, Blievernicht J, Saussele T, Gunthard HF, Schwab M, Eichelbaum M, Telenti A, Zanger UM. Predictive value of known and novel alleles of CYP2B6 for efavirenz plasma concentrations in HIV-infected individuals. Clin. Pharmacol. Ther. 2007;81:557–566. doi: 10.1038/sj.clpt.6100072. [DOI] [PubMed] [Google Scholar]
- 86.Benowitz NL, Swan GE, Jacob P;, 3rd, Lessov-Schlaggar CN, Tyndale RF. CYP2A6 genotype and the metabolism and disposition kinetics of nicotine. Clin. Pharmacol. Ther. 2006;80:457–467. doi: 10.1016/j.clpt.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 87.Mwenifumbo JC, Tyndale RF. Genetic variability in CYP2A6 and the pharmacokinetics of nicotine. Pharmacogenomics. 2007;8:1385–1402. doi: 10.2217/14622416.8.10.1385. [DOI] [PubMed] [Google Scholar]
- 88.Ho MK, Mwenifumbo JC, Al Koudsi N, Okuyemi KS, Ahluwalia JS, Benowitz NL, Tyndale RF. Association of nicotine metabolite ratio and CYP2A6 genotype with smoking cessation treatment in African-American light smokers. Clin. Pharmacol. Ther. 2009;85:635–643. doi: 10.1038/clpt.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, Sulem P, Rafnar T, Esko T, Walter S, Gieger C, Rawal R, Mangino M, Prokopenko I, Magi R, Keskitalo K, Gudjonsdottir IH, Gretarsdottir S, Stefansson H, Thompson JR, Aulchenko YS, Nelis M, Aben KK, den Heijer M, Dirksen A, Ashraf H, Soranzo N, Valdes AM, Steves C, Uitterlinden AG, Hofman A, Tonjes A, Kovacs P, Hottenga JJ, Willemsen G, Vogelzangs N, Doring A, Dahmen N, Nitz B, Pergadia ML, Saez B, De Diego V, Lezcano V, Garcia-Prats MD, Ripatti S, Perola M, Kettunen J, Hartikainen AL, Pouta A, Laitinen J, Isohanni M, Huei-Yi S, Allen M, Krestyaninova M, Hall AS, Jones GT, van Rij AM, Mueller T, Dieplinger B, Haltmayer M, Jonsson S, Matthiasson SE, Oskarsson H, Tyrfingsson T, Kiemeney LA, Mayordomo JI, Lindholt JS, Pedersen JH, Franklin WA, Wolf H, Montgomery GW, Heath AC, Martin NG, Madden PA, Giegling I, Rujescu D, Jarvelin MR, Salomaa V, Stumvoll M, Spector TD, Wichmann HE, Metspalu A, Samani NJ, Penninx BW, Oostra BA, Boomsma DI, Tiemeier H, van Duijn CM, Kaprio J, Gulcher JR, McCarthy MI, Peltonen L, Thorsteinsdottir U, Stefansson K. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat. Genet. 2010;42:448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bohanec Grabar P, Rozman B, Tomsic M, Suput D, Logar D, Dolzan V. Genetic polymorphism of CYP1A2 and the toxicity of leflunomide treatment in rheumatoid arthritis patients. Eur. J. Clin. Pharmacol. 2008;64:871–876. doi: 10.1007/s00228-008-0498-2. [DOI] [PubMed] [Google Scholar]
- 91.Hockwald RS, Arnold J, Clayman CB, Alving AS. Toxicity of primaquine in Negroes. J. Am. Med. Assoc. 1952;149:1568–1570. doi: 10.1001/jama.1952.72930340027010c. [DOI] [PubMed] [Google Scholar]
- 92.Clayman CB, Arnold J, Hockwald RS, Yount EH, Jr., Edgcomb JH, Alving AS. Toxicity of primaquine in Caucasians. J. Am. Med. Assoc. 1952;149:1563–1568. doi: 10.1001/jama.1952.72930340022010b. [DOI] [PubMed] [Google Scholar]
- 93.Alving AS, Carson PE, Flanagan CL, Ickes CE. Enzymatic deficiency in primaquine-sensitive erythrocytes. Science. 1956;124:484–485. doi: 10.1126/science.124.3220.484-a. [DOI] [PubMed] [Google Scholar]
- 94.Hirono A, Beutler E. Molecular cloning and nucleotide sequence of cDNA for human glucose-6-phosphate dehydrogenase variant A(-). Proc. Natl. Acad. Sci. U S A. 1988;85:3951–3954. doi: 10.1073/pnas.85.11.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Town M, Bautista JM, Mason PJ, Luzzatto L. Both mutations in G6PD A- are necessary to produce the G6PD deficient phenotype. Hum. Mol. Genet. 1992;1:171–174. doi: 10.1093/hmg/1.3.171. [DOI] [PubMed] [Google Scholar]
- 96.Vives-Corrons JL, Kuhl W, Pujades MA, Beutler E. Molecular genetics of the glucose-6-phosphate dehydrogenase (G6PD) Mediterranean variant and description of a new G6PD mutant, G6PD Andalus1361A. Am. J. Hum. Genet. 1990;47:575–579. [PMC free article] [PubMed] [Google Scholar]
- 97.van Kuilenburg AB, De Abreu RA, van Gennip AH. Pharmacogenetic and clinical aspects of dihydropyrimidine dehydrogenase deficiency. Ann. Clin. Biochem. 2003;40:41–45. doi: 10.1258/000456303321016150. [DOI] [PubMed] [Google Scholar]
- 98.van Kuilenburg AB. Dihydropyrimidine dehydrogenase and the efficacy and toxicity of 5-fluorouracil. Eur. J. Cancer. 2004;40:939–950. doi: 10.1016/j.ejca.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 99.Etienne MC, Lagrange JL, Dassonville O, Fleming R, Thyss A, Renee N, Schneider M, Demard F, Milano G. Population study of dihydropyrimidine dehydrogenase in cancer patients. J. Clin. Oncol. 1994;12:2248–2253. doi: 10.1200/JCO.1994.12.11.2248. [DOI] [PubMed] [Google Scholar]
- 100.Lu Z, Zhang R, Carpenter JT, Diasio RB. Decreased dihydropyrimidine dehydrogenase activity in a population of patients with breast cancer: implication for 5-fluorouracil-based chemotherapy. Clin. Cancer Res. 1998;4:325–329. [PubMed] [Google Scholar]
- 101.Ogura K, Ohnuma T, Minamide Y, Mizuno A, Nishiyama T, Nagashima S, Kanamaru M, Hiratsuka A, Watabe T, Uematsu T. Dihydropyrimidine dehydrogenase activity in 150 healthy Japanese volunteers and identification of novel mutations. Clin. Cancer Res. 2005;11:5104–5111. doi: 10.1158/1078-0432.CCR-05-0217. [DOI] [PubMed] [Google Scholar]
- 102.Morsman JM, Sludden J, Ameyaw MM, Githang AJ, Indalo A, Ofori-Adjei D, McLeod HL. Evaluation of dihydropyrimidine dehydrogenase activity in South-west Asian, Kenyan and Ghanaian populations. Br. J. Clin. Pharmacol. 2000;50:269–272. doi: 10.1046/j.1365-2125.2000.00242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Saif MW, Mattison L, Carollo T, Ezzeldin H, Diasio RB. Dihydropyrimidine dehydrogenase deficiency in an Indian population. Cancer Chemother. Pharmacol. 2006;58:396–401. doi: 10.1007/s00280-005-0174-5. [DOI] [PubMed] [Google Scholar]
- 104.Van Kuilenburg AB, Vreken P, Abeling NG, Bakker HD, Meinsma R, Van Lenthe H, De Abreu RA, Smeitink JA, Kayserili H, Apak MY, Christensen E, Holopainen I, Pulkki K, Riva D, Botteon G, Holme E, Tulinius M, Kleijer WJ, Beemer FA, Duran M, Niezen-Koning KE, Smit GP, Jakobs C, Smit LM, Van Gennip AH, et al. Genotype and phenotype in patients with dihydropyrimidine dehydrogenase deficiency. Hum Genet. 1999;104:1–9. doi: 10.1007/pl00008711. [DOI] [PubMed] [Google Scholar]
- 105.Yen JL, McLeod HL. Should DPD analysis be required prior to prescribing fluoropyrimidines? Eur. J. Cancer. 2007;43:1011–1016. doi: 10.1016/j.ejca.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 106.Johnson MR, Yan J, Shao L, Albin N, Diasio RB. Semiautomated radioassay for determination of dihydropyrimidine dehydrogenase (DPD) activity. Screening cancer patients for DPD deficiency, a condition associated with 5-fluorouracil toxicity. J. Chromatogr. B. Biomed. Sci. Appl. 1997;696:183–191. doi: 10.1016/s0378-4347(97)00253-3. [DOI] [PubMed] [Google Scholar]
- 107.Ciccolini J, Mercier C, Evrard A, Dahan L, Boyer JC, Duffaud F, Richard K, Blanquicett C, Milano G, Blesius A, Durand A, Seitz JF, Favre R, Lacarelle B. A rapid and inexpensive method for anticipating severe toxicity to fluorouracil and fluorouracil-based chemotherapy. Ther. Drug Monit. 2006;28:678–685. doi: 10.1097/01.ftd.0000245771.82720.c7. [DOI] [PubMed] [Google Scholar]
- 108.Mattison LK, Ezzeldin H, Carpenter M, Modak A, Johnson MR, Diasio RB. Rapid identification of dihydropyrimidine dehydrogenase deficiency by using a novel 2-13C-uracil breath test. Clin. Cancer Res. 2004;10:2652–2658. doi: 10.1158/1078-0432.ccr-03-0374. [DOI] [PubMed] [Google Scholar]
- 109.Yang CG, Ciccolini J, Blesius A, Dahan L, Bagarry-Liegey D, Brunet C, Varoquaux A, Frances N, Marouani H, Giovanni A, Ferri-Dessens RM, Chefrour M, Favre R, Duffaud F, Seitz JF, Zanaret M, Lacarelle B, Mercier C. DPD-based adaptive dosing of 5-FU in patients with head and neck cancer: impact on treatment efficacy and toxicity. Cancer Chemother. Pharmacol. 2010 doi: 10.1007/s00280-010-1282-4. [DOI] [PubMed] [Google Scholar]
- 110.Koukouritaki SB, Hines RN. Flavin-containing monooxygenase genetic polymorphism: impact on chemical metabolism and drug development. Pharmacogenomics. 2005;6:807–822. doi: 10.2217/14622416.6.8.807. [DOI] [PubMed] [Google Scholar]
- 111.Peng GS, Yin SJ. Effect of the allelic variants of aldehyde dehydrogenase ALDH2*2 and alcohol dehydrogenase ADH1B*2 on blood acetaldehyde concentrations. Hum. Genomics. 2009;3:121–127. doi: 10.1186/1479-7364-3-2-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Edenberg HJ. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res. Health. 2007;30:5–13. [PMC free article] [PubMed] [Google Scholar]
- 113.Fan M, Liu B, Jiang T, Jiang X, Zhao H, Zhang J. Meta-analysis of the association between the monoamine oxidase-A gene and mood disorders. Psychiatr. Genet. 2010;20:1–7. doi: 10.1097/YPG.0b013e3283351112. [DOI] [PubMed] [Google Scholar]
- 114.Devadatta S, Gangadharam PR, Andrews RH, Fox W, Ramakrishnan CV, Selkon JB, Velu S. Peripheral neuritis due to isoniazid. Bull World Health Organ. 1960;23:587–598. [PMC free article] [PubMed] [Google Scholar]
- 115.Hughes HB, Biehl JP, Jones AP, Schmidt LH. Metabolism of isoniazid in man as related to the occurrence of peripheral neuritis. Am. Rev. Tuberc. 1954;70:266–273. doi: 10.1164/art.1954.70.2.266. [DOI] [PubMed] [Google Scholar]
- 116.Evans DA, Manley KA, Mc KV. Genetic control of isoniazid metabolism in man. Br. Med. J. 1960;2:485–491. doi: 10.1136/bmj.2.5197.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Harris HW, Knight RA, Selin MJ. Comparison of isoniazid concentrations in the blood of people of Japanese and European descent; therapeutic and genetic implications. Am. Rev. Tuberc. 1958;78:944–948. doi: 10.1164/artpd.1958.78.6.944. [DOI] [PubMed] [Google Scholar]
- 118.Blum M, Demierre A, Grant DM, Heim M, Meyer UA. Molecular mechanism of slow acetylation of drugs and carcinogens in humans. Proc. Natl. Acad. Sci. U S A. 1991;88:5237–5241. doi: 10.1073/pnas.88.12.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vatsis KP, Martell KJ, Weber WW. Diverse point mutations in the human gene for polymorphic N-acetyltransferase. Proc. Natl. Acad. Sci. U S A. 1991;88:6333–6337. doi: 10.1073/pnas.88.14.6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hickman D, Sim E. N-acetyltransferase polymorphism. Comparison of phenotype and genotype in humans. Biochem. Pharmacol. 1991;42:1007–1014. doi: 10.1016/0006-2952(91)90282-a. [DOI] [PubMed] [Google Scholar]
- 121.Hitchings GH, Elion GB, Falco EA, Russell PB, Vanderwerff H. Studies on analogs of purines and pyrimidines. Ann. N. Y. Acad. Sci. 1950;52:1318–1335. doi: 10.1111/j.1749-6632.1950.tb54032.x. [DOI] [PubMed] [Google Scholar]
- 122.Weinshilboum RM, Sladek SL. Mercaptopurine pharmacogenetics: monogenic inheritance of erythrocyte thiopurine methyltransferase activity. Am. J. Hum. Genet. 1980;32:651–662. [PMC free article] [PubMed] [Google Scholar]
- 123.Lennard L, Van Loon JA, Lilleyman JS, Weinshilboum RM. Thiopurine pharmacogenetics in leukemia: correlation of erythrocyte thiopurine methyltransferase activity and 6-thioguanine nucleotide concentrations. Clin. Pharmacol. Ther. 1987;41:18–25. doi: 10.1038/clpt.1987.4. [DOI] [PubMed] [Google Scholar]
- 124.Szumlanski C, Otterness D, Her C, Lee D, Brandriff B, Kelsell D, Spurr N, Lennard L, Wieben E, Weinshilboum R. Thiopurine methyltransferase pharmacogenetics: human gene cloning and characterization of a common polymorphism. DNA Cell Biol. 1996;15:17–30. doi: 10.1089/dna.1996.15.17. [DOI] [PubMed] [Google Scholar]
- 125.Tai HL, Krynetski EY, Yates CR, Loennechen T, Fessing MY, Krynetskaia NF, Evans WE. Thiopurine S-methyltransferase deficiency: two nucleotide transitions define the most prevalent mutant allele associated with loss of catalytic activity in Caucasians. Am. J. Hum. Genet. 1996;58:694–702. [PMC free article] [PubMed] [Google Scholar]
- 126.Otterness D, Szumlanski C, Lennard L, Klemetsdal B, Aarbakke J, Park-Hah JO, Iven H, Schmiegelow K, Branum E, O'Brien J, Weinshilboum R. Human thiopurine methyltransferase pharmacogenetics: gene sequence polymorphisms. Clin. Pharmacol. Ther. 1997;62:60–73. doi: 10.1016/S0009-9236(97)90152-1. [DOI] [PubMed] [Google Scholar]
- 127.Teml A, Schaeffeler E, Herrlinger KR, Klotz U, Schwab M. Thiopurine treatment in inflammatory bowel disease: clinical pharmacology and implication of pharmacogenetically guided dosing. Clin. Pharmacokinet. 2007;46:187–208. doi: 10.2165/00003088-200746030-00001. [DOI] [PubMed] [Google Scholar]
- 128.Karas-Kuzelicki N, Mlinaric-Rascan I. Individualization of thiopurine therapy: thiopurine S-methyltransferase and beyond. Pharmacogenomics. 2009;10:1309–1322. doi: 10.2217/pgs.09.78. [DOI] [PubMed] [Google Scholar]
- 129.Relling MV, Gardner EE, Sandborn WJ, Schmiegelow K, Pui CH, Yee SW, Stein CM, Carrillo M, Evans WE, Klein TE. Clinical Pharmacogenetics Implementation Consortium Guidelines for Thiopurine Methyltransferase Genotype and Thiopurine Dosing. Clin. Pharmacol. Ther. 2011 doi: 10.1038/clpt.2010.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Macklon AF, Savage RL, Rawlins MD. Gilbert's syndrome and drug metabolism. Clin. Pharmacokinet. 1979;4:223–232. doi: 10.2165/00003088-197904030-00004. [DOI] [PubMed] [Google Scholar]
- 131.Monaghan G, Ryan M, Seddon R, Hume R, Burchell B. Genetic variation in bilirubin UPD-glucuronosyltransferase gene promoter and Gilbert's syndrome. Lancet. 1996;347:578–581. doi: 10.1016/s0140-6736(96)91273-8. [DOI] [PubMed] [Google Scholar]
- 132.Iyer L, King CD, Whitington PF, Green MD, Roy SK, Tephly TR, Coffman BL, Ratain MJ. Genetic predisposition to the metabolism of irinotecan (CPT-11). Role of uridine diphosphate glucuronosyltransferase isoform 1A1 in the glucuronidation of its active metabolite (SN-38) in human liver microsomes. J. Clin. Invest. 1998;101:847–854. doi: 10.1172/JCI915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ando Y, Fujita K, Sasaki Y, Hasegawa Y. UGT1AI*6 and UGT1A1*27 for individualized irinotecan chemotherapy. Curr. Opin. Mol. Ther. 2007;9:258–262. [PubMed] [Google Scholar]
- 134.Jada SR, Lim R, Wong CI, Shu X, Lee SC, Zhou Q, Goh BC, Chowbay B. Role of UGT1A1*6, UGT1A1*28 and ABCG2 c.421C>A polymorphisms in irinotecan-induced neutropenia in Asian cancer patients. Cancer Sci. 2007;98:1461–1467. doi: 10.1111/j.1349-7006.2007.00541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Onoue M, Terada T, Kobayashi M, Katsura T, Matsumoto S, Yanagihara K, Nishimura T, Kanai M, Teramukai S, Shimizu A, Fukushima M, Inui K. UGT1A1*6 polymorphism is most predictive of severe neutropenia induced by irinotecan in Japanese cancer patients. Int. J. Clin. Oncol. 2009;14:136–142. doi: 10.1007/s10147-008-0821-z. [DOI] [PubMed] [Google Scholar]
- 136.Wadelius M, Chen LY, Downes K, Ghori J, Hunt S, Eriksson N, Wallerman O, Melhus H, Wadelius C, Bentley D, Deloukas P. Common VKORC1 and GGCX polymorphisms associated with warfarin dose. Pharmacogenomics J. 2005;5:262–270. doi: 10.1038/sj.tpj.6500313. [DOI] [PubMed] [Google Scholar]
- 137.Rettie AE, Tai G. The pharmocogenomics of warfarin: closing in on personalized medicine. Mol. Interv. 2006;6:223–227. doi: 10.1124/mi.6.4.8. [DOI] [PubMed] [Google Scholar]
- 138.Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, Soranzo N, Whittaker P, Ranganath V, Kumanduri V, McLaren W, Holm L, Lindh J, Rane A, Wadelius M, Deloukas P. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5:e1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Duan S, Huang RS, Zhang W, Bleibel WK, Roe CA, Clark TA, Chen TX, Schweitzer AC, Blume JE, Cox NJ, Dolan ME. Genetic architecture of transcript-level variation in humans. Am. J. Hum. Genet. 2008;82:1101–1113. doi: 10.1016/j.ajhg.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 141.Shomron N. MicroRNAs and pharmacogenomics. Pharmacogenomics. 2010;11:629–632. doi: 10.2217/pgs.10.26. [DOI] [PubMed] [Google Scholar]
- 142.Pennisi E. Genomics. 1000 Genomes Project gives new map of genetic diversity. Science. 2010;330:574–575. doi: 10.1126/science.330.6004.574. [DOI] [PubMed] [Google Scholar]
- 143.Daly AK, Day CP. Genetic association studies in drug-induced liver injury. Semin. Liver Dis. 2009;29:400–411. doi: 10.1055/s-0029-1240009. [DOI] [PubMed] [Google Scholar]
- 144.Alfirevic A, Pirmohamed M. Drug-induced hypersensitivity reactions and pharmacogenomics: past, present and future. Pharmacogenomics. 2010;11:497–499. doi: 10.2217/pgs.10.12. [DOI] [PubMed] [Google Scholar]
- 145.Nishio Y, Makiyama T, Itoh H, Sakaguchi T, Ohno S, Gong YZ, Yamamoto S, Ozawa T, Ding WG, Toyoda F, Kawamura M, Akao M, Matsuura H, Kimura T, Kita T, Horie M. D85N, a KCNE1 polymorphism, is a disease-causing gene variant in long QT syndrome. J. Am. Coll. Cardiol. 2009;54:812–819. doi: 10.1016/j.jacc.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 146.Zhou SF. Polymorphism of human cytochrome P450 2D6 and its clinical significance: Part I. Clin. Pharmacokinet. 2009;48:689–723. doi: 10.2165/11318030-000000000-00000. [DOI] [PubMed] [Google Scholar]