Abstract
Elevated plasma lipoprotein levels play a crucial role in the development of coronary artery disease. Genetic factors strongly influence the levels of plasma lipoproteins, but the genes and sequence variations contributing to the most common forms of dyslipidemias are not known. We used GeneChip probe arrays to resequence the coding regions of 10 key genes of lipid metabolism. The sequences of these genes were analyzed in 80 dyslipidemic individuals. Fourteen nonsynonymous and twenty-two synonymous single nucleotide changes were identified that could be confirmed by conventional sequencing. Seven of the fourteen nonsynonymous sequence variants were polymorphisms with allele frequency >1% in the general population. The remaining seven were not found in normolipidemic controls (25 Caucasians and 25 African-Americans). The relationship between nonsynonymous sequence variations and various dyslipidemias was explored in association and family studies. No evidence was found for coding sequence variations in any of the 10 genes contributing to dyslipidemia. Only a single sequence variation, a missense mutation in the low density lipoprotein receptor gene, co-segregated with hyperlipidemia in the proband's family. This study illustrates some of the difficulties associated with identifying sequence variations contributing to complex traits.
Genetic factors contribute importantly to many of the common diseases afflicting Western societies, such as coronary artery disease, hypertension, diabetes, and cancer. Although these disorders cluster in families, they are not inherited in a Mendelian fashion; thus it has been difficult to identify the sequence variations that confer susceptibility. Most sequence variations within coding regions of genes result in synonymous substitutions or conservative amino acid replacements that have little or no apparent effect on protein function (Cargill et al. 1999). Only 10% to 20% of single nucleotide polymorphisms (SNPs) lead to a nonconservative amino acid replacement (Cargill et al. 1999). It has been proposed that a subset of common sequence variants contribute to common diseases in the population (Lander 1996; Risch and Merikangas 1996). The common disease–common variant (CD–CV) hypothesis is supported by a few examples of well-documented associations between sequence variants and medical conditions: F5 1691G→A (Factor V Leiden) and thrombophilia (Dahlback 1997), APOE*E4 and late-onset Alzheimer disease (Strittmatter et al. 1993), and C-C chemokine receptor 5 (CCR5) Δ32 and resistance to the human immunodeficiency virus (Liu et al. 1996).
Considerable effort is now focused on identifying genetic polymorphisms that contribute to other common diseases, including coronary artery disease (Cambien et al. 1999). Elevated plasma levels of cholesterol play a central role in the development of atherosclerosis. Most of the cholesterol circulating in plasma is transported as a constituent of low density lipoproteins (LDLs) or high density lipoproteins (HDLs). The risk of coronary artery disease is directly related to the plasma level of LDL-cholesterol and inversely related to plasma HDL-cholesterol levels (Kannel et al. 1979). Twin and family studies are consistent with ∼50% of the interindividual variation in the levels of these plasma lipoproteins being a result of genetic factors (Austin et al. 1987; Heller et al. 1993; Perusse et al. 1989). Mutations in the LDL receptor (LDLR) and apolipoprotein (apo) B-100 genes are responsible for the two most common Mendelian forms of hypercholesterolemia, familial hypercholesterolemia (FH; MIM143890) and familial defective apoB-100 (MIM 144010; Innerarity et al. 1990; Goldstein et al. 1995); however, sequence variations in these two genes do not contribute significantly to the moderate elevations in plasma lipoproteins that are most frequent in subjects with coronary artery disease (Wang et al. 1998).
Many of the genes encoding proteins involved in cholesterol metabolism have not been screened systematically for sequence variants. The present study was undertaken to determine whether sequence variations in the coding regions of 10 key genes in lipid metabolism contribute to variations in plasma levels of cholesterol and triglycerides. The genes analyzed in this study include those encoding sterol regulatory element binding proteins (SREBPs) 1 and 2 (SREBF1 and SREBF2; Hua et al. 1995; Brown and Goldstein 1997; Miserez et al. 1997), SREBP cleavage-activating protein (SCAP; Hua et al. 1996; Nakajima et al. 1999), 3-hydroxy-3-methylglutaryl coenzyme A (HMG CoA) reductase (HMGCR; Reynolds et al. 1984), acyl coenzyme A acyltransferase2 (ACAT-2; Chang et al. 1997; Cases et al. 1998), cholesterol 25-hydroxylase (CH25H; Lund et al. 1998), LDLR (LDLR; Sudhof et al. 1985), scavenger receptor, type B, class 1 (SR-BI; human gene name CLA-1; Calvo and Vega 1993; Acton et al. 1996; Cao et al. 1997), site-1 protease (S1P) (S1P; Sakai et al. 1998a,b; Nakajima et al. 2000), LXRα (NR1H3; Willy et al. 1995) and 40 bp encoding the LDLR binding domain of apolipoproteinB-100 (APOB; Yang et al. 1986). We also screened 29 bp of the known regulatory sequences of the LDLR.
These genes were selected because of the crucial role they play in the regulation, biosynthesis, and clearance of cholesterol. SREBP-1 and SREBP-2 are structurally related transcription factors that regulate many genes involved in lipid metabolism (for review, see Brown and Goldstein 1997). These two transcription factors are oriented in a hairpin configuration in the membrane of the endoplasmic reticulum in association with SCAP, the putative cholesterol sensor. Sterol depletion is associated with the transport of the SREBP–SCAP complex to the Golgi (DeBose-Boyd et al. 1999; Nohturfft et al. 1999). S1P resides in the Golgi and mediates the cleavage of SREBP within its luminal loop, which ultimately results in the release of a fragment from the amino terminus that contains a basic helix–loop–helix zipper domain. This ∼68 kD fragment enters the nucleus and activates transcription of multiple genes involved in cholesterol and fatty acid metabolism (Brown and Goldstein 1997), including the genes encoding the LDLR, which is the major vehicle for the import of lipoprotein-derived cholesterol into cells (Brown and Goldstein 1986) and HMGCR, which encodes a rate-limiting enzyme in cholesterol biosynthesis. A potent down-regulator of SREBP cleavage is 25-hydroxycholesterol, which is generated by the hydroxylation of cholesterol at position 25 by cholesterol 25-hydroxylase (CH25H; Lund et al. 1998). Excess intracellular cholesterol is converted to cholesteryl esters by two other membrane-associated proteins of the endoplasmic reticulum, ACAT-1 and ACAT-2 (Chang et al. 1997; Cases et al. 1998). The major route of clearance of cholesterol from the body is via the biliary system (Russell and Setchell 1992). SR-BI has been implicated as the major conduit by which cholesterol is transported from HDL to the bile (Ji et al. 1999). Hepatic cholesterol can be secreted directly into the bile or first converted to bile acids prior to secretion (Russell and Setchell 1992). The rate-limiting enzyme for bile acid synthesis, cholesterol 7α hydroxylase, is regulated by LXRα (gene name, NR1H3), an orphan nuclear hormone receptor (Peet et al. 1998).
We used high density GeneChip probe arrays (Chee et al. 1996; Wang et al. 1998) to screen for sequence variations in ∼25 kb of sequence from these 10 genes. GeneChip probe arrays allow for the simultaneous determination of sequences in multiple genes in a rapid and comprehensive fashion, which makes the method ideally suited for well-characterized metabolic pathways. Because we cannot predict the specific phenotype that would result from sequence variations in these genes, we screened 80 unrelated individuals with a wide array of lipid disorders.
RESULTS
Characterization of Subjects
GeneChip probe arrays (Affymetrix) were used to screen 25,298 bp of genomic sequence (24,675 bp of coding sequence, 29 bp of LDLR promoter sequence, and 594 bp of intron sequence) from 10 candidate genes in 80 unrelated individuals with various disorders of lipid metabolism and/or fat distribution (see Methods). Subjects selected for the study had a high likelihood of having a major genetic component to their lipid disorder. The major inclusion criteria were: (1) a clinical picture consistent with homozygous FH without a reduction in LDLR function in cultured fibroblasts; (2) markedly abnormal plasma lipid levels or abnormal response to lipid-lowering agents or diet; or (3) a lipoprotein disorder and at least two first-degree relatives with a similar phenotype.
The Sensitivity of the Genechip Probe Array Method
The GeneChip probe array method detects sequence variants by hybridizing labeled PCR products to chips containing arrays of corresponding oligonucleotides (for review, see Chee et al. 1996; Wang et al. 1998). For each nucleotide position being queried, four oligonucleotides that differ only in the base at the central position are arrayed in rows on the chip. For our study, these oligonucleotide arrays were designed to detect all exon sequences and 2–3 bp of adjacent intron sequences.
Putative SNPs were reported when the hybridization pattern of the sample DNA did not match the canonical sequence. Each individual GeneChip probe array-detected SNP was assigned a graded level of certainty: certain (C), likely (L), or possible (P). In some instances in which the same SNP was identified in multiple samples, some samples were scored as C whereas others were scored as L; in this paper these SNPs were classified as CL (or LP for SNPs that were scored as likely or possible).
A total of 8, 13, 12, and 96 SNPs were classified as CL, L, LP and P, respectively (Table 1). No SNP site in this study was classified as C in all samples containing the polymorphic sequence. Because subjects with rare disorders as well as common phenotypes were included in the sample population, we anticipated identifying so-called private, unique sequence variations. Therefore, SNPs classified as P (n = 96) were included in our analysis.
Table 1.
Frequency of Confirmed SNPs in Coding Sequences
| Genes | Basepairs screened | Number SNP | Frequency of SNP | θ (×10−4) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| total | non-syn | syn | total | non-syn | syn | total | non-syn | syn | ||
| ACAT2 | 1558 | 3 | 1 | 2 | 1/519 | 1/1558 | 1/779 | 3.41 | 1.136 | 2.274 |
| APOB | 40 | 0 | 0 | 0 | 0/40 | 0/40 | 0/40 | — | — | — |
| CH25H | 819 | 2 | 0 | 2 | 1/410 | 0/819 | 1/410 | 4.317 | — | 4.317 |
| CLA1 | 1650 | 3 | 2 | 1 | 1/550 | 1/825 | 1/1650 | 3.218 | 2.145 | 1.072 |
| HMGCR | 2667 | 1 | 1 | 0 | 1/2667 | 1/2667 | 0/2667 | 0.663 | 0.663 | — |
| LDLR | 2583 | 10 | 3 | 7 | 1/258 | 1/861 | 1/369 | 6.861 | 2.055 | 4.797 |
| NR1H2 | 1344 | 1 | 0 | 1 | 1/1344 | 0/1344 | 1/1344 | 1.317 | — | 1.317 |
| S1P | 3159 | 4 | 2a | 2 | 1/790 | 1/1580 | 1/1580 | 2.24 | 1.12 | 1.12 |
| SCAP | 3840 | 3 | 1 | 2 | 1/1280 | 1/3840 | 1/1920 | 1.382 | 0.46 | 0.921 |
| SREBF1 | 3575 | 1 | 1 | 0 | 1/3575 | 1/3575 | 0/3575 | 0.495 | 0.495 | — |
| SREBF2 | 3440 | 7 | 3 | 4 | 1/491 | 1/1147 | 1/860 | 3.605 | 1.543 | 2.058 |
Includes one nonsense mutation.
Average frequency of SNP.
Each sequence polymorphism detected by GeneChip probe arrays was subjected to verification by use of a combination of SSCP analysis and sequencing. Some SNPs detected on the chip in multiple individuals could be confirmed only in a subset of samples. Nevertheless, we classified the SNP site as confirmed even if the nucleotide change was verified in only a single individual. Most of the eight SNPs classified as CL were confirmed (75%). The rate of confirmation of SNPs in the other categories was lower: 38% for L, 67% for LP, and 18% for the SNPs classified as P. Although the false positive rate was very high for the possible SNPs, the absolute number of confirmed SNPs in the P category (n = 17) was nearly as high as in all the other categories combined (n = 19).
GeneChip probe array analysis also classified each sample as homozygous, heterozygous or indeterminate for each SNP. These designations were tested for true SNP frequency by SSCP and sequencing for all SNPs identified in SREBF2; the confirmation rates for these categories were 48% for the sites designated as homozygous, and 31% and 14% for those called heterozygous and indeterminate, respectively (data not shown).
We did not determine systematically the false negative rate for GeneChip probe array analysis, but several SNPs, as well as a deletion and an insertion, were not detected by this method. Four sequence variants that resulted in changes in the amino acid sequence were missed by GeneChip probe arrays: (1) An insertion in exon 2 of SREBF2 in an expanded repeat found by E. Forgacs (pers. comm.), which was present in 1 of the 80 subjects; (2) an LDLR mutation (Asp147His; Hobbs et al. 1992; Leitersdorf et al. 1993); (3) a sequencing error in the original sequence of SREBF2 (resulting in Gly1045Ala); and (4) a nonsynonymous SNP detected in SREBF2 (Table 2) that was not identified in three of the eight samples heterozygous for the polymorphism.
Table 2.
Confirmation of SNP Sites Detected by GeneChip Probe Arrays in 10 Genes of 80 Probands
| Bases screened | Confirmed/reported SNPs | Total | Confirmed | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Coding | NCa | Total | CLbc | L | LPd | P | ||
| ACAT2 | 1558 | 54 | 1612 | 0/0 | 1/3 | 0/1 | 2/6 | 3/10 | 30% |
| APOB | 40 | 0 | 40 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0% |
| CH25H | 819 | 0 | 819 | 1/2 | 1/1 | 0/0 | 0/2 | 2/5 | 40% |
| CLA1 | 1650 | 48 | 1698 | 1/1 | 1/1 | 0/0 | 1/3 | 3/5 | 60% |
| HMGCR | 2667 | 72 | 2739 | 0/0 | 0/0 | 1/1 | 0/8 | 1/9 | 11% |
| LDLR | 2583 | 97 | 2680 | 2/3 | 1/1 | 3/4 | 4/13 | 10/21 | 48% |
| NR1H2 | 1344 | 32 | 1376 | 0/0 | 0/0 | 0/0 | 1/4 | 1/4 | 25% |
| S1P | 3159 | 84 | 3243 | 2/2 | 0/2 | 0/0 | 2/6 | 4/10 | 40% |
| SCAP | 3840 | 84 | 3924 | 0/0 | 1/2 | 1/1 | 1/9 | 3/12 | 25% |
| SREBF1 | 3575 | 82 | 3657 | 0/0 | 0/2 | 1/2 | 1/18 | 2/22 | 9% |
| SREBF2 | 3440 | 70 | 3510 | 0/0 | 0/1 | 2/3 | 5/27 | 7/31 | 23% |
| Total | 24675 | 623 | 25298 | 6/8 | 5/13 | 8/12 | 17/96 | 36/129 | 28% |
| Confirmed | 75% | 38% | 67% | 18% | 28% | ||||
Noncoding.
Certain.
Likely.
Possible.
We also did not determine systematically the accuracy of the allele genotypes in the sample for each SNP. Only one SNP in exon 10 of SREBF2 that was classified as LP was reanalyzed in the entire sample by use of SSCP. Reanalysis revealed that 25 samples reported as wild type were heterozygous, and 16 samples were homozygous for the SNP.
A total of 36 SNP sites was confirmed in at least 1 of the 1600 different alleles assayed. No sequence variants were identified in the 40-bp segment of APOB that was screened. An average of 1 site per 705 bp varied in the 24,675 bp coding sequences. The 10 genes differed more than 10-fold in their overall frequency of sequence variations (Table 3). The LDLR gene had the highest nucleotide diversity and SREBF1 the lowest. The average value for the normalized number of variant sites (θ) was 2.51 × 10−4.
Table 3.
Confirmed Common and Rare Nonsynonymous Single Nucleotide Polymorphisms (SNP)
| Gene | Exon | Codon | Amino acid | Ethnicity | Disordera | Frequency A-A whitesb | Comments | |
|---|---|---|---|---|---|---|---|---|
| Common (>1%)c | ||||||||
| ACAT2 | 7 | ACC→ATC | T254Id | Multiple | — | 0.20 | 0.25 | |
| CHA1 | 1 | GGC→AGC | G2S | Multiple | — | — | — | Acton et al. 1999 |
| 3 | GTC→ATC | V135I | Multiple | — | — | — | Acton et al. 1999 | |
| SCAP | 15 | GTC→ATC | V796I | Multiple | — | 0.15 | 0.48 | Iwaki et al. 1999 |
| SREBF-2 | 10 | GCC→GGC | A595G | Multiple | — | 0.39 | 0.75 | |
| 10 | GTG→ATG | V623M | A-A | — | 0.04 | 0 | ||
| 14 | AGG→AGC | R860S | Multiple | — | 0.02 | 0.05 | ||
| Rare (<1%)c | ||||||||
| HMGCR | 16 | ATA→GTA | I638V | Caucasian | Atypical Type III | 0 | 0 | Insufficient datae |
| Caucasian | FCHL | Insufficient data | ||||||
| Caucasian | FCHL | No co-segregation | ||||||
| Caucasian | Low HDL-C (AD) | No co-segregation | ||||||
| LDLR | 3 | GAC→GTC | D79V | Caucasian | Atypical Type III | 0 | 0 | See Fig. 1A. Not detected by SSCP. |
| 4 | TCG→TTG | S156L | Puerto Rican | FH | 0 | 0 | Hobbs et al. 1992 | |
| 6 | GAG→AAG | E256K | Caucasian | Diet-induced | 0 | 0 | Ekstrom et al. 1995 | |
| HCL | Family not available. | |||||||
| S1P | 13 | AAC→AGC | N544S | Greek | FCHL | 0 | 0 | No co-segregation. |
| 20 | CAG→TAG | Q868X | Caucasian | Polygenic HCL | 0 | 0 | See Fig. 1B. No co-segregation. | |
| SREBF1 | 18a | CGG→CAG | R1064Q | Caucasian | Type V, DM | 0 | 0 | No co-segregation. |
Abbreviations: FH, Familial Hypercholesterolemia; HCL, hypercholesterolemia; DM, Diabetes Mellitus; FCHL, Familial Combined Hyperlipidemia; HDL-C, HDL-cholesterol; A-A, African-Americans.
Minor allele frequency in African-Americans (A-A) and Whites.
Common (>1%) or rare (<1%) in the general population.
Isoleucine at amino acid position 254 is substituted for threonine. All amino acids are numbered according to the initiator codon (ATG) being position 1.
Not a sufficient number of segregations in the family of the proband to assess relationship between sequence variant and the hyperlipidemia.
Over 75% of the SNPs were transitions (28 of 36) and 36% of these SNPs were cytosine to thymidine transitions at CpG dimers.
SNPs Detected in 80 Dyslipidemic Subjects and Normal Controls
Of the 36 confirmed SNPs, 14 were nonsynonymous substitutions (Table 3). Seven of the nonsynonymous changes had an allele frequency >1% in the general population. All of these common SNPs resulted in conservative amino acid changes, with the exception of an arginine to serine substitution at amino acid 860 in SREBF-2. However, this residue is also serine in hamster SREBF2. The most common SNP identified in this study was Ala595Gly in SREBF2. The allele frequency of this SNP differed significantly between African-Americans and Caucasians. Another relatively common SNP found in SREBF2 was a valine to methionine substitution at amino acid 623, which was detected in an African-American with lipodystrophy. This sequence variant comprised 4% of the alleles in the normolipemic African-Americans and was not found in the Caucasian samples tested.
Mutations Detected in Dyslipidemic Subjects
Seven mutations were identified exclusively in the group of 80 probands and not in the 50 control subjects (Table 2). Only one of these mutations was found in more than one proband. Valine was substituted for isoleucine at amino acid 638 of the HMGCR in four individuals. This residue is located in the catalytic domain of HMG CoA reductase, but is not conserved across species (Istvan et al. 2000). The corresponding residues in hamster and rat are methionine and leucine, respectively, and some bacterial forms of the enzyme also have valine at this position (E. Istvan, pers. comm.). The SNP did not cosegregate with hyperlipidemia in the four families with this sequence variant (data not shown).
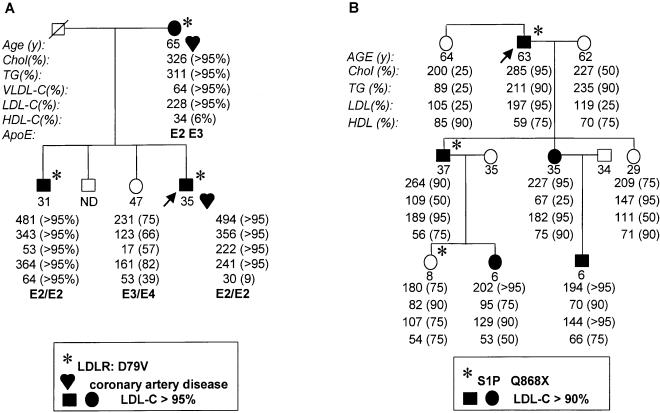
The other six missense mutations were each found in only a single subject. Three of these mutations were in LDLR, and of these mutations, one was previously known (Hobbs et al. 1989). The second LDLR mutation was identified in a 41-year-old Caucasian man whose plasma cholesterol level was extremely responsive to changes in dietary cholesterol content; the mutation had been identified previously in a patient with classic FH (Ekstrom et al. 1995), but was not associated with defective LDLR function in cultured fibroblasts (Ekstrom et a. 2000). The third LDLR mutation was identified in a 35-year-old Caucasian man who had a myocardial infarction, type III hyperlipidemia, and an elevated plasma LDL-cholesterol level. Analysis of the family revealed that the other first-degree relatives who inherited the LDLR mutation had markedly elevated plasma LDL-cholesterol levels (Fig. 1). This LDLR mutation (Asp79Val) had not been described previously in any other FH patients.
Figure 1.
Segregation of mutant LDLR (A) and S1P (B) alleles in families of probands with dyslipidemias. Fasting lipid and lipoprotein levels were measured as described in Methods. The percentiles were determined by comparison with age- and sex-matched controls (http://www.cdc.gov/nchs/about/major/nhanes/resrchact.htm).
The two nonsynonymous mutations identified in S1P were each found in only a single subject. Both mutations were reproduced by site-directed mutagenesis and the cDNA was expressed in cultured cells to assess the effect of the sequence change on enzyme activity (data not shown). Expression of S1P with the Asn544Ser substitution was associated with normal processing of SREBP, whereas no processing of SREBP was seen with expression of S1P containing the Gln868 nonsense mutation (J. Zhang, unpubl.). The proband heterozygous for the nonsense mutation had moderately elevated plasma LDL-cholesterol, but the sequence variant failed to cosegregate with the hyperlipidemia in the family (Fig. 1B).
An Arg1064Gln substitution in SREBF1 was identified in a 39-year-old Caucasian woman with noninsulin-dependent diabetes mellitus and type V hyperlipidemia. The missense mutation is located in the region of SREBP-1 that interacts with SCAP and participates in cholesterol sensing (Sakai et al. 1997). The arginine residue at this position is also present in hamster and fruit fly, suggesting that this residue may be functionally important. However, when this mutant allele was expressed in tissue culture, the SREBP-1 protein was processed normally in response to cholesterol depletion (R. DeBose-Boyd, unpubl.).
Finally, we screened 50 unrelated noninsulin-dependent diabetic, 50 hypercholesterolemic, and 50 hypertriglyceridemic subjects for each of these seven mutations. Only the Ile638Val substitution in HMGCR was found in one hypercholesterolemic and two diabetic subjects, giving on overall allele frequency of 1%.
Silent SNPs Detected in 10 Candidate Genes
Of the 25 silent polymorphisms identified in this study (Table 4), 22 were detected by GeneChip probe array sequencing, and all but one were in exons. Over 85% of the SNPs located in the coding regions were in the third position of the codon, and the remainder were in the first position. The only intronic SNP detected by GeneChip probe array was at the −3 position of the splice acceptor site of intron 18c (C→T) of SREBF1. This polymorphism occurs with a frequency of ∼50% in the population. Two other intronic SNPs were identified by SSCP and sequencing in nucleotides not queried by the GeneChip probe arrays method. Neither polymorphism segregated with the hyperlipidemia in the family.
Table 4.
SNPs That Do Not Alter Protein Sequencea
| Gene | Exon/Intron | NT change | Position | Comments |
|---|---|---|---|---|
| ACAT2 | Ex6 | CTG→CTT | 609G→Tb | |
| Ex6 | CTA→TTA | 610C→T | ||
| CH25H | Ex1 | CTG→CTC | 54G→C | |
| Ex1 | AAC→AAT | 657C→T | ||
| CLA1 | Ex8 | GCC→GCT | 1050C→T | Acton et al. 1999 |
| LDLR | Ex2 | TGT→TGC | 81T→C | Soutar et al. 1991 |
| EX2 | AAC→AAT | 90C→T | ||
| Ex10 | AGG→AGA | 1413G→A | Warnich et al. 1992 | |
| Ex11 | CCC→CCT | 1617C→T | Leren et al. 1992 | |
| Ex12 | CTC→CTT | 1725C→T | Yamakawa et al. 1993 | |
| Ex12 | AAT→AAC | 1773T→C | Chaves et al. 1991 | |
| Ex13 | GTC→GTT | 1959C→T | Hobbs et al. 1987 | |
| NR1H2 | Ex3 | AGC→AGT | 297C→T | |
| S-1-P | Ex11 | CCC→CCT | 1317C→T | |
| Ex11 | CTC→CTT | 1407C→T | ||
| In23d | G→A | IVS23+7G→Ac | ||
| SCAP | Ex15 | ATC→ATA | 2031C→A | |
| Ex20 | TCG→TCT | 3321G→T | ||
| SREBF-1 | In18ad | G→A | IVS18a-10G→A | |
| In18c | C→T | IVS18c-3C→T | ||
| SREBF-2 | Ex1 | CTG→CTA | 75G→A | |
| Ex10d | GCC→GCT | 1782C→T | Identified in A-A control sample | |
| Ex18 | CAT→CAC | 3105T→C | ||
| Ex18 | CGG→AGG | 3175C→A | ||
| Ex19 | CTG→TTG | 3316C→T |
The reference is included for SNPs that were described previously.
The A of the initiator ATG is counted +1. Only coding sequence is counted.
Intron mutation (IVS = intervening sequence) is designated by intron number and nucleotide number counted from splice donor site (positive number) or from splice acceptor site (negative number) (Antonarakis 1998).
Not detected by GeneChip probe arrays. SNPs = Single nucleotide polymorphisms.
DISCUSSION
In this study, we sequenced 25,298 bp from 160 independent alleles of 10 genes encoding key proteins in the regulatory, biosynthetic, and clearance pathways of lipid metabolism. A total of 36 SNPs was identified by GeneChip probe arrays; 1 was in an intron, and 35 were in coding sequences. Of the 35 SNPs identified in the coding sequences, 21 were synonymous, and 14 were nonsynonymous. One-half of the 14 nonsynonymous sequences were polymorphisms with a frequency of >1% in the population. All but one of the remaining seven nonsynonymous sequence variants were found in only a single individual and not in 50 unrelated controls or 100 hyperlipidemic (50 hypercholesterolemic and 50 hypertriglyceridemic) subjects. Only one of these sequence variants (LDLR: D79V) could be shown to cosegregate with hyperlipidemia in the family of the proband.
Common forms of dyslipidemia should constitute a model system for the genetic analysis of complex traits for two reasons. First, there is considerable evidence from family and twin studies that genetic variation is an important determinant of plasma lipoprotein levels (Austin et al. 1987; Perusse et al. 1989; Heller et al. 1993). Second, the metabolism of lipids and lipoproteins has been studied in considerable detail, and many of the key genes involved have been cloned and extensively characterized. Because each of the 10 genes examined in this study has a clearly defined role in lipid metabolism, and because all of the 80 individuals screened had a familial lipid disorder, it is surprising that the only DNA sequence variants identified that were clearly associated with altered plasma lipoprotein levels were rare mutations in LDLR.
The simplest interpretation of this finding is that sequence polymorphisms in coding regions of these genes are not a common cause of dyslipidemia. This interpretation is tempered by the fact that dyslipidemia is a heterogeneous entity, and that each specific type of dyslipidemia (e.g., familial combined hyperlipidemia) was present in only a minor subset of the individuals in the study. Nonetheless, at least 25 of the individuals in the study had increased plasma LDL concentrations, and at least 26 others had increased plasma triglyceride concentrations. Therefore, it is very likely (>90 %) that any allele present in 10 % or more of individuals with these characteristics would have been detected in this study.
A second possibility is that common polymorphisms conferring susceptibility to dyslipidemia are present in these genes but were either not detected by the GeneChip probe array method or were in regions of the genes (promoter or introns) not screened in this study. Few promoter polymorphisms have been associated with Mendelian dyslipidemias, but common polymorphisms in the 5′ flanking sequences of the genes encoding hepatic lipase (LIPC) and cholesterol 7α-hydroxylase have been associated with variation in the plasma concentrations of HDL-cholesterol and LDL-cholesterol, respectively (Guerra et al. 1997; Wang et al. 1998). Therefore, further screening of the noncoding regions of the candidate genes may reveal sequence polymorphisms that are associated with dyslipidemia.
A third possibility is that the polymorphisms identified in this study have little influence on plasma lipoprotein levels when considered individually, but that the inheritance of a combination of deleterious alleles confers susceptibility to dyslipidemia. Analysis of the joint effects of DNA sequence variants requires very large sample sizes and was not possible in this study. However, because conditions such as hypercholesterolemia and hypertriglyceridemia are relatively common and clinically well recognized, it is quite feasible to assemble large collections of individuals with homogeneous lipoprotein phenotypes. The application of high throughput genotyping methods to such cohorts may help to define the combinations of polymorphisms that act together to create the phenotype of complex diseases such as hyperlipidemia and atherosclerosis.
The least polymorphic of the coding sequences screened was SREBF1, a transcription factor that plays a central role in the hepatic regulation of cholesterol and triglyceride metabolism (Brown and Goldstein 1997) and adipocyte differentiation (Tontonoz et al. 1993; Shimomura et al. 1999). The gene with the second lowest frequency of sequence variation was HMGCR, which encodes a rate-limiting enzyme of cholesterol biosynthesis. The low frequency of sequence polymorphism in these two genes may be related to selection pressure, although we cannot exclude the possibility that regional differences in sequence variation and/or genetic drift are contributing factors.
The most polymorphic coding sequence was found in LDLR. Selection bias may have contributed to the high frequency of sequence variation in LDLR as mutations abolishing LDLR function almost invariably cause hypercholesterolemia (Goldstein et al. 1995). Accordingly, it is likely that mutations causing more subtle impairment of LDLR function may confer susceptibility to increased plasma LDL concentrations. Although linkage studies have indicated that polymorphisms in LDLR account for little of the variation in LDL-cholesterol concentrations in the general population (Wang et al. 1998), such polymorphisms may be more prevalent in a sample selected for high plasma cholesterol levels, like the one analyzed in this study.
Two new subjects with FH were identified here. In one subject, previous screening of the LDLR gene by SSCP did not reveal mutations. After the mutation was identified by GeneChip probe arrays and confirmed by conventional sequencing, it was reassessed by SSCP, and, again, the sequence variant was not detected. Although it was not uncommon for a sequence variant to be detected under one SSCP condition and not the other, this particular LDLR mutation was the only variant identified in the GeneChip probe array analysis that did not generate an abnormally migrating band in either of the two SSCP conditions employed in this study.
The high density GeneChip probe array method is very efficient at the detection of more common sequence variations. These polymorphisms, which were often classified as C or L, had confirmation rates that were similar to the two other larger studies that used high density GeneChip probe arrays for SNP detection (Cargill et al. 1999; Halushka et al. 1999). A major difference between earlier studies and the current study was that we included the SNPs classified as P in our analysis. Sequence variations classified by GeneChip probe arrays as P were much more common than those classified as C or L, but were also much less likely to be confirmed. The SNPs classified as P comprised almost half of the confirmed SNPs in this study, including all of the rare nonsynonymous mutations. Therefore, SNP detection by GeneChip probe arrays may be less reliable for detecting rare mutations in individual samples than for detecting common polymorphic sites.
METHODS
Subjects
A primary population that consisted of 80 unrelated subjects was screened. Then, the sequence variants identified in these subjects were analyzed in genomic DNA from 50 unrelated normolipidemic controls (25 Caucasians and 25 African-Americans), and a subset was screened in 50 individuals with hypercholesterolemia (plasma cholesterol >90th percentile compared with age- and sex-matched controls), in 50 individuals with hypertriglyceridemia (plasma triglyceride level >350 mg/dL), and in 50 individuals with noninsulin-dependent diabetes. The 80 primary subjects included probands from 61 families identified in the Lipid Clinic at Parkland Memorial Hospital or as part of a large family study of dyslipidemias. All available relatives of the probands completed a detailed medical questionnaire and were examined for the presence of tendon or cutaneous xanthomas. After obtaining of informed consent, fasting venous blood was collected and kept at 4°C until the plasma was isolated. The plasma cholesterol and triglyceride levels were measured by use of enzymatic assays (Guerra et al. 1997) and the lipoprotein levels were determined by β-quantification (S. Grundy, Univ. of Texas Southwestern Medical Center). Age- and sex- specific percentile values for plasma lipid and lipoprotein concentrations were calculated with data from the National Health and Nutrition Education Survey (NHANES) III survey (http://www.cdc.gov/nchs/about/major/nhanes/resrchact.htm). The dyslipidemias were classified into categories on the basis of the lipoprotein profile of the proband and, when available, family members. The criteria for diagnosis of familial combined hyperlipidemia (n = 13), familial hypertriglyceridemia (n = 8), polygenic hypercholesterolemia (n = 10), and familial type V (n = 5) are as described (Goldstein et al. 1972). The descriptive term familial was used in cases where at least three first-degree relatives had similar lipoprotein patterns. The terms hypoalphalipoproteinemia (n = 4), hyperalphalipoproteinemia (n = 5), and hypobetalipoproteinemia (n = 5) were used to describe lipoprotein levels that were less than the 10th or greater than the 90th percentile when compared with age- and sex-matched controls. Genomic DNA was isolated from either the white blood cells or transformed lymphocytes (Louie and King 1991).
GeneChip probe array analysis was also performed on 19 genomic DNA samples isolated from cultured skin fibroblasts donated to the Department of Molecular Genetics (J. Goldstein, M. Brown, and H.H. Hobbs). Twelve of these samples were from probands with presumed autosomal recessive hypercholesterolemia, who had marked elevations in plasma cholesterol, tendon xanthomas, and normolipidemic parents (Khachadurian and Uthman 1973; Sirtori et al. 1991; Zuliani et al. 1995). In addition, subjects with each of the following disorders were analyzed: (1) severe hypercholesterolemia (∼2000 mg/dL), bony lesions, and liver disease of unknown etiology (n = 1); (2) hypolipidemia (cholesterol level of 100 mg/dL, triglyceride level of 45 mg/dL), and xanthomatosis (n = 1); (3) type III hyperlipidemia that does not cosegregate with apoE (Giroux et al. 1997) or that has an atypical lipoprotein profile (n = 7); (4) nonalcoholic steatohepatitis (n = 1); (5) FH that does not respond to HMG-CoA reductase inhibitors (n = 1); (6) congenital generalized lipodystrophy (n = 1); and (7) multiple symmetric lipomatosis (n = 1; Enzi 1984). A 41-yr-old man with diet-responsive hypercholesterolemia was also included in the study. His plasma LDL-cholesterol level fell dramatically after eating a low-fat diet for 1 month (from 596 mg/dL to 247 mg/dL). His LDL-cholesterol level continued to fall on a low-fat diet, and then stabilized between 86 and 114 mg/dL. Upon challenge with a high-fat diet for 5 days per week for 4 weeks, his LDL-cholesterol rose to 267 mg/dL. The patient was homozygous for the ApoE3 allele and had no detectable circulating β-sitosterol (P. Barton Duell, pers. comm.).
The ethnic composition of the probands was as follows: 10 Hispanics, 1 Asian Indian, 1 American Indian, 1 African-American, 1 Ashkenazi Jews, 1 Sephardic Jew, 1 Arab, and the remainder of the sample were non-Jewish Caucasians.
PCR Amplification of Coding Regions in Candidate Genes
The sequences of most genes included were available from GenBank. If the intron sequences were not available, we obtained plasmid or bacterial artificial chromosome clones and used PCR to amplify the introns. Sufficient intron sequence was obtained to design primers for the amplification of the exons with flanking dinucleotide consensus sequences. Oppositely oriented 25-base amplification primers complementary to the flanking intron sequences of the candidate genes were used to amplify the exons and adjacent splice junction sites of the candidate genes. Oligonucleotide sequences are available on request. After optimization of the PCR conditions for each amplicon, the exons of each gene were amplified for all 80 genomic DNA samples. Each amplification product was subjected to agarose gel electrophoresis and visualized after ethidium bromide staining to assess the integrity and concentration of the fragment. Aliquots containing ∼0.3 pmoles of each fragment from a given subject were pooled and sent to Affymetrix for GeneChip probe array analysis.
Hybridization to 80 GeneChip Probe Arrays
For sequence analysis by GeneChip probe arrays, 1–2 μg of the pooled PCR products from each subject was purified by use of Qiaquick Purification kits (Qiagen) and then fragmented by use of DNase (Promega). The fragments were biotinylated and denatured prior to hybridization to the chip for 15 h at 44°C in a buffer containing tetramethylammonium chloride as described (Wang et al. 1998). Hybridization patterns were analyzed by Affymetrix software to detect SNPs (Chee et al. 1996).
Each detected SNP was assigned a graded level of certainty: C, L, or P. When the same SNP scored as C in some samples but L in other samples, the SNP was classified as CL (or LP for SNPs that were scored as likely or possible).
Verification of SNPs Detected by GeneChip Probe Array Analysis
Each SNP identified by GeneChip probe array sequencing was examined by SSCP (Orita et al. 1989). 32P-labeled PCR products containing the putative SNPs were denatured, and the single-stranded fragments were electrophoresed for 20 h at 350 V on two different nondenaturing gels: (1) 7% polyacrylamide in 2× Tris Borate EDTA buffer (TBE) and 10% (vol/vol) glycerol, and (2) 0.5× MDE (BioWhittaker), 0.6× TBE. If an abnormally migrating band was identified by SSCP, the corresponding PCR product was sequenced by use of an ABI 377 automated sequencer (Applied Biosystems). If no aberrantly migrating band was detected using SSCP, a fragment containing the putative SNP was PCR amplified from a representative individual and sequenced. SSCP or restriction endonuclease analysis was used to evaluate the segregation of the polymorphism in the families of the probands and to screen normolipidemic and hyperlipidemic subjects.
Calculations
To obtain an estimate of nucleotide diversity, the normalized numbers of variant sites (θ) was calculated by dividing the number of observed nucleotide changes by the length (basepairs) of the sequence and correcting for sample size (n) as described (Li 1997).
Electronic Database Information
The following URLs were important for this study: NHANES III data, http://www.cdc.gov/nchs/about/major/nhanes/resrchact.htm; Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/omim.htm.
Acknowledgments
We thank the following physicians who contributed patient samples for this study: Drs. Giovanni Zuliani, Renato Fellin, Marcello Arca, A.K. Khachadurian, C. Sertori, James Crockett, Vera Rose, William Lee, Paul Benke, J.L. de Gennes, F. Dariou, P. Barton Duell, Jan Breslow, Eran Leitersdorf, Jean Davignon, and Abimanyu Garg. We especially thank Michael S. Brown and Joseph L. Goldstein for providing patient samples and helpful discussions. We also thank David W. Russell and Aravinda Chakravarti for helpful discussions. This work was supported by National Institutes of Health (NIH) HL20948 (H.H.H.), the Perot Family Fund, NIH-HL53917 (J.C.C.), and the D.W. Reynolds Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL helen.hobbs@emailswmed.edu; FAX (214) 648-7539.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.172301.
REFERENCES
- Acton S, Rigotti A, Landschulz KT, Xu S, Hobbs HH, Krieger M. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science. 1996;271:518–520. doi: 10.1126/science.271.5248.518. [DOI] [PubMed] [Google Scholar]
- Austin MA, King MC, Bawol RD, Hulley SB, Friedman GD. Risk factors for coronary heart disease in adult female twins. Genetic heritability and shared environmental influences. Am J Epidemiol. 1987;125:308–318. doi: 10.1093/oxfordjournals.aje.a114531. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- ————— The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- Calvo D, Vega MA. Identification, primary structure, and distribution of CLA-1, a novel member of the CD36/LIMPII gene family. J Biol Chem. 1993;268:18929–18935. [PubMed] [Google Scholar]
- Cambien F, Poirier O, Nicaud V, Herrmann SM, Mallet C, Ricard S, Behague I, Hallet V, Blanc H, Loukaci V, et al. Sequence diversity in 36 candidate genes for cardiovascular disorders. Am J Hum Genet. 1999;65:183–191. doi: 10.1086/302448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G, Garcia CK, Wyne KL, Schultz RA, Parker KL, Hobbs HH. Structure and localization of the human gene encoding SR-BI/CLA-1. Evidence for transcriptional control by steroidogenic factor 1. J Biol Chem. 1997;272:33068–33076. doi: 10.1074/jbc.272.52.33068. [DOI] [PubMed] [Google Scholar]
- Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalyanaraman N, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nature Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- Cases S, Novak S, Zheng YW, Myers HM, Lear SR, Sande E, Welch CB, Lusis AJ, Speiner TA, Krouse BR, et al. ACAT-2, a second mammalian acyl-CoA:cholesterol acyltransferase: Its cloning, expression, and characterization. J Biol Chem. 1998;273:26755–26764. doi: 10.1074/jbc.273.41.26755. [DOI] [PubMed] [Google Scholar]
- Chakravarti A. Population genetics—making sense out of sequence. Nature Genet. 1999;21:56–60. doi: 10.1038/4482. [DOI] [PubMed] [Google Scholar]
- Chang TY, Chang CCY, Cheng D. Acyl-coenzyme A: Cholesterol acyltransferase. Annu Rev Biochem. 1997;66:613–638. doi: 10.1146/annurev.biochem.66.1.613. [DOI] [PubMed] [Google Scholar]
- Chee M, Yang R, Hubbell E, Berno A, Huang XC, Stern D, Winkler J, Lockhart DJ, Morris MS, Fodor SP, et al. Accessing genetic information with high-density DNA arrays. Science. 1996;274:610–614. doi: 10.1126/science.274.5287.610. [DOI] [PubMed] [Google Scholar]
- Dahlback B. Resistance to activated protein C as risk factor for thrombosis: Molecular mechanisms, laboratory investigation, and clinical management. Semin Hematol. 1997;34:217–234. [PubMed] [Google Scholar]
- DeBose-Boyd RA, Brown MS, Li WP, Nohturfft A, Goldstein JL, Espenshade PJ. Transport-dependent proteolysis of SREBP: Relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell. 1999;99:703–712. doi: 10.1016/s0092-8674(00)81668-2. [DOI] [PubMed] [Google Scholar]
- Ekstrom U, Abrahamson M, Sveger T, Lombardi P, Nilsson-Ehle P. An efficient screening procedure detecting six novel mutations in the LDL receptor gene in Swedish children with hypercholesterolemia. Hum Genet. 1995;96:147–150. doi: 10.1007/BF00207370. [DOI] [PubMed] [Google Scholar]
- Ekstrom U, Abrahamson M, Sveger T, Sun X-M, Soutar AK, Nilsson-Ehle P. Expression of an LDL receptor allele with two different mutations (E256K and I402T) J Clin Pathol Mol Pathol. 1999;2000:31–36. doi: 10.1136/mp.53.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enzi G. Multiple symmetric lipomatosis. Medicine (Baltimore) 1984;63:56–64. doi: 10.1097/00005792-198401000-00004. [DOI] [PubMed] [Google Scholar]
- Giroux LM, Cohn JS, LaMarre J, Davignon J. Pseudo type III dyslipoproteinemia is associated with normal fibroblast lipoprotein receptor activity. Atherosclerosis. 1997;132:85–94. doi: 10.1016/s0021-9150(97)00072-5. [DOI] [PubMed] [Google Scholar]
- Goldstein JL, Hazzard WR, Schrott HG, Bierman EL, Motulsky AG. Genetics of hyperlipidemia in coronary heart disease. Trans Assoc Am Physicians. 1972;85:120–138. [PubMed] [Google Scholar]
- Goldstein J, Hobbs H, Brown M. Familial hypercholesterolemia. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The metabolic and molecular bases of inherited disease. NY: McGraw Hill; 1995. pp. 1981–2030. [Google Scholar]
- Guerra R, Wang J, Grundy SM, Cohen JC. A hepatic lipase (LIPC) allele associated with high plasma concentrations of high density lipoprotein cholesterol. Proc Natl Acad Sci. 1997;94:4532–4537. doi: 10.1073/pnas.94.9.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halushka MK, Fan JB, Bentley K, Hsie L, Shen N, Weder A, Cooper R, et al. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nature Genet. 1999;22:239–247. doi: 10.1038/10297. [DOI] [PubMed] [Google Scholar]
- Heller DA, de Faire U, Pedersen NL, Dahlen G, McClearn GE. Genetic and environmental influences on serum lipid levels in twins. N Engl J Med. 1993;328:1150–1156. doi: 10.1056/NEJM199304223281603. [DOI] [PubMed] [Google Scholar]
- Hobbs HH, Leitersdorf E, Leffert CC, Cryer DR, Brown MS, Goldstein JL. Evidence for a dominant gene that suppresses hypercholesterolemia in a family with defective low density lipoprotein receptors. J Clin Invest. 1989;84:656–664. doi: 10.1172/JCI114212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs HH, Brown MS, Goldstein JL. Molecular genetics of the LDL receptor gene in familial hypercholesterolemia. Hum Mutat. 1992;1:445–466. doi: 10.1002/humu.1380010602. [DOI] [PubMed] [Google Scholar]
- Hua X, Wu J, Goldstein JL, Brown MS, Hobbs HH. Structure of the human gene encoding sterol regulatory element binding protein-1 (SREBF1) and localization of SREBF1 and SREBF2 to chromosomes 17p11.2 and 22q13. Genomics. 1995;25:667–673. doi: 10.1016/0888-7543(95)80009-b. [DOI] [PubMed] [Google Scholar]
- Hua X, Nohturfft A, Goldstein JL, Brown MS. Sterol resistance in CHO cells traced to point mutation in SREBP cleavage-activating protein. Cell. 1996;87:415–426. doi: 10.1016/s0092-8674(00)81362-8. [DOI] [PubMed] [Google Scholar]
- Innerarity TL, Mahley RW, Weisgraber KH, Bersot TP, Krauss RM, Vega GL, Grundy SM, Friedl W, Davignon J, McCarthy BJ, et al. Familial defective apolipoprotein B-100: A mutation of apolipoprotein B that causes hypercholesterolemia. J Lipid Res. 1990;31:1337–1349. [PubMed] [Google Scholar]
- Istvan ES, Palnitkar M, Buchanan SK, Deisenhofer J. Crystal structure of the catalytic portion of human HMG-CoA reductase: Insights into regulation of activity and catalysis. EMBO J. 2000;19:819–830. doi: 10.1093/emboj/19.5.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaki K, Nakajima T, Ota N, Emi M. A common Ile796Val polymorphism of the human SREBP cleavage-activating protein (SCAP) gene. J Hum Genet. 1999;44:421–422. doi: 10.1007/s100380050193. [DOI] [PubMed] [Google Scholar]
- Ji Y, Wang N, Ramakrishnan R, Sehayek E, Huszar D, Breslow JL, Tall AR. Hepatic scavenger receptor BI promotes rapid clearance of high density lipoprotein free cholesterol and its transport into bile. J Biol Chem. 1999;274:33398–33402. doi: 10.1074/jbc.274.47.33398. [DOI] [PubMed] [Google Scholar]
- Kannel WB, Castelli WP, Gordon T. Cholesterol in the prediction of atherosclerotic disease. New perspectives based on the Framingham study. Ann Intern Med. 1979;90:85–91. doi: 10.7326/0003-4819-90-1-85. [DOI] [PubMed] [Google Scholar]
- Khachadurian AK, Uthman SM. Experiences with the homozygous cases of familial hypercholesterolemia. A report of 52 patients. Nutr Metab. 1973;15:132–140. doi: 10.1159/000175431. [DOI] [PubMed] [Google Scholar]
- Lander ES. The new genomics: Global views of biology. Science. 1996;274:536–539. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- Leitersdorf E, Reshef A, Meiner V, Dann EJ, Beigel Y, van Roggen FG, van der Westhuyzen DR, Coetzee GA, et al. A missense mutation in the low density lipoprotein receptor gene causes familial hypercholesterolemia in Sephardic Jews. Hum Genet. 1993;91:141–147. doi: 10.1007/BF00222714. [DOI] [PubMed] [Google Scholar]
- Li W. Molecular evolution. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- Liu R, Paxton WA, Choe S, Ceradini D, Martin SR, Horuk R, MacDonald ME, Stuhlmann H, Koup RA, Landau NR, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- Lund EG, Kerr TA, Sakai J, Li WP, Russell DW. cDNA cloning of mouse and human cholesterol 25-hydroxylases, polytopic membrane proteins that synthesize a potent oxysterol regulator of lipid metabolism. J Biol Chem. 1998;273:34316–34327. doi: 10.1074/jbc.273.51.34316. [DOI] [PubMed] [Google Scholar]
- Miserez AR, Cao G, Probst LC, Hobbs HH. Structure of the human gene encoding sterol regulatory element binding protein 2 (SREBF2) Genomics. 1997;40:31–40. doi: 10.1006/geno.1996.4525. [DOI] [PubMed] [Google Scholar]
- Nakajima T, Hamakubo T, Kodama T, Inazawa J, Emi M. Genomic structure and chromosomal mapping of the human sterol regulatory element binding protein (SREBP) cleavage-activating protein (SCAP) gene. J Hum Genet. 1999;44:402–407. doi: 10.1007/s100380050187. [DOI] [PubMed] [Google Scholar]
- Nakajima T, Iwaki K, Kodama T, Inazawa J, Emi M. Genomic structure and chromosomal mapping of the human Site-1 protease (S1P) gene. J Hum Genet. 2000;45:212–217. doi: 10.1007/s100380070029. [DOI] [PubMed] [Google Scholar]
- Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci. 1989;86:2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peet DJ, Turley SD, Ma W, Janowski BA, Lobaccaro JM, Hammer RE, Mangelsdorf DJ. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell. 1998;93:693–704. doi: 10.1016/s0092-8674(00)81432-4. [DOI] [PubMed] [Google Scholar]
- Perusse L, Despres JP, Tremblay A, Leblanc C, Talbot J, Allard C, Bouchard C. Genetic and environmental determinants of serum lipids and lipoproteins in French Canadian families. Arteriosclerosis. 1989;9:308–318. doi: 10.1161/01.atv.9.3.308. [DOI] [PubMed] [Google Scholar]
- Reynolds GA, Basu SK, Osborne TF, Chin DJ, Gil G, Brown MS, Goldstein JL, Luskey KL, et al. HMG CoA reductase: A negatively regulated gene with unusual promoter and 5' untranslated regions. Cell. 1984;38:275–285. doi: 10.1016/0092-8674(84)90549-x. [DOI] [PubMed] [Google Scholar]
- Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Russell DW, Setchell KD. Bile acid biosynthesis. Biochemistry. 1992;31:4737–4749. doi: 10.1021/bi00135a001. [DOI] [PubMed] [Google Scholar]
- Sakai J, Nohturfft A, Cheng D, Ho YK, Brown MS, Goldstein JL. Identification of complexes between the COOH-terminal domains of sterol regulatory element-binding proteins (SREBPs) and SREBP cleavage-activating protein. J Biol Chem. 1997;272:20213–20221. doi: 10.1074/jbc.272.32.20213. [DOI] [PubMed] [Google Scholar]
- Sakai J, Rawson RB, Espenshade PJ, Cheng D, Seegmiller AC, Goldstein JL, Brown MS. Molecular identification of the sterol-regulated luminal protease that cleaves SREBPs and controls lipid composition of animal cells. Mol Cell. 1998a;2:505–514. doi: 10.1016/s1097-2765(00)80150-1. [DOI] [PubMed] [Google Scholar]
- Sakai J, Nohturfft A, Goldstein JL, Brown MS. Cleavage of sterol regulatory element-binding proteins (SREBPs) at site-1 requires interaction with SREBP cleavage-activating protein: Evidence from in vivo competition studies. J Biol Chem. 1998b;273:5785–5793. doi: 10.1074/jbc.273.10.5785. [DOI] [PubMed] [Google Scholar]
- Shimomura I, Hammer RE, Ikemoto S, Brown MS, Goldstein JL. Leptin reverses insulin resistance and diabetes mellitus in mice with congenital lipodystrophy. Nature. 1999;401:73–76. doi: 10.1038/43448. [DOI] [PubMed] [Google Scholar]
- Sirtori CR, Catapano AL, Franceschini G, Corsini A, Noseda G, Fragiacomo C, Panzeri E, Vaccarino V, Guenzi S, Casari J, et al. Aortic and coronary atheromatosis in a woman with severe hypercholesterolaemia without LDL receptor alterations. Eur Heart J. 1991;12:818–824. [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: High-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC, Goldstein JL, Brown MS, Russell DW. The LDL receptor gene: A mosaic of exons shared with different proteins. Science. 1985;228:815–822. doi: 10.1126/science.2988123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tontonoz P, Kim JB, Graves RA, Spiegelman BM. ADD1: A novel helix-loop-helix transcription factor associated with adipocyte determination and differentiation. Mol Cell Biol. 1993;13:4753–4759. doi: 10.1128/mcb.13.8.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Freeman DJ, Grundy SM, Levine DM, Guerra R, Cohen JC. Linkage between cholesterol 7alpha-hydroxylase and high plasma low-density lipoprotein cholesterol concentrations. J Clin Invest. 1998;101:1283–1291. doi: 10.1172/JCI1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DG, Fan JB, Siao CJ, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- Willy PJ, Umesono K, Ong ES, Evans RM, Heyman RA, Mangelsdorf DJ. LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes & Dev. 1995;9:1033–1045. doi: 10.1101/gad.9.9.1033. [DOI] [PubMed] [Google Scholar]
- Yang CY, Chen SH, Gianturco SH, Bradley WA, Sparrow JT, Tanimura M, Li WH, Sparrow DA, DeLoof H, Rosseneu M, et al. Sequence, structure, receptor-binding domains and internal repeats of human apolipoprotein B-100. Nature. 1986;323:738–742. doi: 10.1038/323738a0. [DOI] [PubMed] [Google Scholar]
- Zuliani G, Vigna GB, Corsini A, Maioli M, Romagnoni F, Fellin R. Severe hypercholesterolaemia: Unusual inheritance in an Italian pedigree. Eur J Clin Invest. 1995;25:322–331. doi: 10.1111/j.1365-2362.1995.tb01709.x. [DOI] [PubMed] [Google Scholar]