Abstract
Bioreducible cationic polymers (p(DAHa-R/APIb)s) composed of different ratios (a:b = 2:1, 1:1, 1:2) between arginine-grafted diaminohexane (DAH-R) (cell penetrating functionality) and 1-(3-aminopropyl) imidazole (API) (endosome buffering functionality) monomers were synthesized by Michael reaction of N,N’-cystaminebisacrylamide (CBA) with them, in order to study the effect of endosome buffering moiety on arginine-grafted bioreducible polymeric gene carriers. Several experiments displayed a distinct correlation between monomer composition ratios of p(DAH-R/API)s and the polymer features. Increased endosome buffering capacities proportional to API portions was evaluated for p(DAH-R/API)s due to the imidazole group (pKa=6) of API. Increased portions of API non-ionized at physiological pH and resultant decrease of arginine residues also reduced cytotoxicities of the polymers due to less interaction of cellular compartments with less positively charged polymers, but decreased pDNA condensing abilities, Zeta-potential values, cellular uptakes of polyplexes, and finally transfection efficiencies as well. Thus, the predominance of arginine residues over endosome buffering moieties was revealed regarding efficient gene delivery for p(DAH-R/API)s. From transfection results with chloroquine or nigericin, it can be deduced that the endosomal escape of p(DAH-R/API) polyplexes occurs by direct endsome membrane penetration of arginine moieties as well as endosome buffering abilities of the polymers after cellular uptake, which emphasizes the importance of arginine moieties for polymeric gene delivery systems.
Keywords: Gene delivery, bioreducible polymer, arginine-graft, endosome buffer, membrane penetration
1. Introduction
Polymeric gene delivery carriers can deliver genetic materials (pDNAs, antisense oligonucleotides, or siRNAs) for gene therapy by formation of nano-sized polyplexes via electrostatic interaction. They have gained great interest from a lot of researchers in gene delivery field because of many advantages over viral gene delivery carriers, such as non-immunogenicity, no integration of exogeneous genes into host chromosomes, simple and less expensive good manufacturing products (GMP), convenience of handling, and modifiable multiple functionality for efficient gene delivery [1–3]. However, their low transfection efficiency, some cytotoxicity, or in vivo instability are still major drawbacks to limit the applications for human gene therapy [4–6].
As one of the strategies for solving those problems, biodegradable polymeric gene carriers have been synthesized, which contains ester [7,8], phosphoester [9], acetal [10] bonds, etc. Biodegradability refers to functionality for the degradation of polymers with response to physiological stimulus such as pH change. They have been reported to show high transfection efficiency and low cytotoxicity, which may be due to the efficient release of genetic materials from polyplexes after cell uptake and degradation of polymers into non-toxic and easily excreted small molecules by hydrolysis [11]. However, handling of hydrolysable polymers requires careful efforts because of their considerable instability in aqueous solution. Contrary to them, bioreducible polymers with disulfide bonds have higher stability in extracellular aqueous environment and can be degraded easily in cytoplasm by high concentration (0.5~10 mM) of glutathione, intracellular reducing agents after cellular uptake [12]. Many bioreducible polymers with various chemical structures have been developed for efficient gene delivery systems [13–16].
Recently, we synthesized arginine-grafted bioreducible polymer (ABP) and reported that ABP is a promising gene carrier showing much higher transfection ability than PEI25k, which is known as one of the most efficient polymeric gene carriers, and negligible cytotoxicity in mammalian cells [17]. High transfection efficiency of ABP is thought to be contributed by arginine moieties which were known to play an important role for cellular membrane penetration in cell penetrating peptides (CPPs) or protein transduction domains (PTDs) such as HIV-1 Tat sequence, penetratin, or oligoarginine [18,19]. Low cytotoxicity of ABP is thought to come from the bioreducibility of internal disulfide bonds within ABP because ABP polyplexes can be degraded in cytoplasm.
Generally, it is known that cellular uptake of polyplexes is mediated by endocytosis and endosomal escape of polyplexes before digestion in lysosome is an important step for efficient transfection [3]. Many studies reported that modulating endosome buffering capacity of polymers by introducing amine moieties protonatable in acidic endosomal pH could improve endosomal escape of polyplexes via “proton sponge effect” and consequently, increase the transfection efficiency [20–22].
Therefore, we aimed to study the effect of endosome buffering moieties on biophysical properties of arginine (cell penetrating moiety)-grafted bioreducible polymers for development of efficient polymeric gene carriers. 1-(3-aminopropyl) imidazole (API) was chosen as a monomer because its imidazole group (pKa = ~6) can function as an endosome buffer.
Here, ABP containing various portions of API moieties were synthesized to tune the endosome buffering capacity of the polymers and their structure-activity relationships were characterized for gene delivery systems. Furthermore, specific endosomal escape mechanism of the polyplexes was also proposed based on transfection results. Until now, within our knowledge it will be the first report about them.
2. Materials and methods
2.1. Materials
Hyperbranched poly(ethylenimine) (bPEI25k), N-Boc-1,6-diaminohexane (N-Boc-DAH), 1-(3-aminopropyl) imidazole (API), trifluoroacetic acid (TFA), triisopropylsilane, triisobutylsilane, dithiothreitol (DTT), N,N-diisopropylethylamine (DIPEA), piperidine, 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT), chloroquine diphosphate salt, and nigericin sodium salt were purchased from Sigma-Aldrich (St. Louis, MO). N,N’-cystaminebisacrylamide (CBA) was purchased from PolySciences, Inc. (Warrington, PA). 2-(1H-benzotriazole-1-yl)-1, 1, 3, 3-tetramethyluronium hexafluorophosphate (HBTU) was purchased from Novabiochem (San Diego, CA). Fmoc-L-Arg(pbf)-OH was purchased from Anaspec, Inc. (San Jose, CA). The plasmid pCMV-Luc, containing a firefly luciferase reporter gene was amplified in E.coli DH5α and isolated by standard Maxiprep kit (Invitrogen, Carlsbad, CA). Luciferase assay system and reporter lysis buffer were purchased from Promega (Madison, WI). SYBR safe DNA gel stain (10,000X in DMSO), fetal bovine serum (FBS), Dulbecco’s phosphate buffered saline (DPBS), and Dulbecco’s modified Eagle’s medium (DMEM) were purchased from Invitrogen (Carlsbad, CA). BCA™ protein assay kit was purchased from PIERCE (Rockford, Il). YOYO-1 iodide (1 mM solution in DMSO) was purchased from Molecular Probes (Eugene, OR). All other chemicals were purchased and used without any further purification.
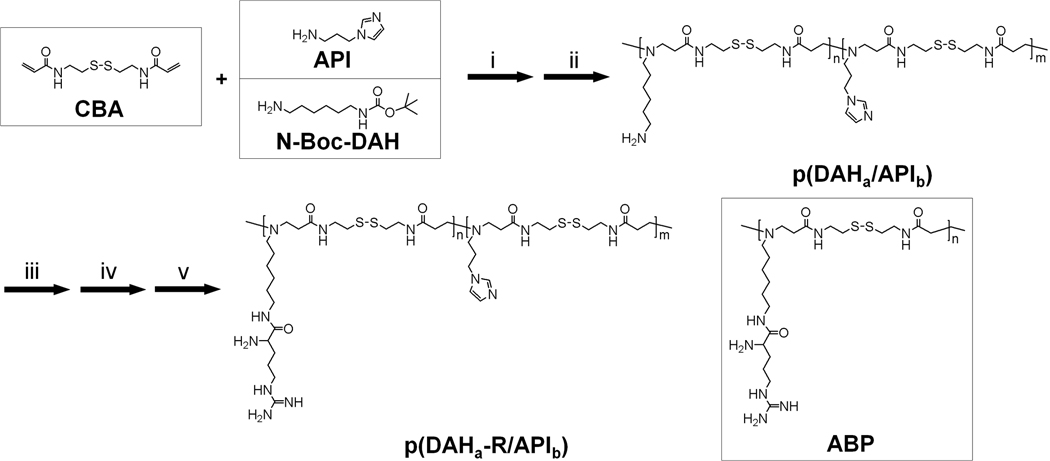
2.2. Syntheses of p(DAH/API)s
Syntheses of the polymers were conducted in two steps as shown in scheme 1. First, the bioreducible backbone polymers, p(DAHa/APIb)s were synthesized (a and b are relative molar ratio between DAH and API in the synthesized polymers, which were determined by 1H NMR analysis) in MeOH/H2O solution (9:1, v/v) by Michael reaction of acrylamide monomer (CBA) and two amine monomers (N-Boc-DAH and API). In order to obtain maximized polymer chain elongation, equal molar amounts of acrylamide monomer and amine monomers were used. The molar ratios between two amine monomers, N-Boc-DAH and API were varied as followings:
n(CBA) : n[(N-Boc-DAH) + (API)] = 1 : 1
n(N-Boc-DAH): n(API) = 1:1, 1:2, 1:4, 0:1.
Scheme 1.
Synthesis scheme of p(DAH-R/API)s and molecular structure of ABP. i) MeOH : H2O = 9 : 1, v/v, 50 °C, 4 days, ii) TFA : triisobutylsilane : H2O = 95 : 2.5 : 2.5, v/v/v, 0 °C, 30 min, iii) Fmoc-Arg(pbf)-OH, HBTU, DIPEA/DMF, r.t., 2 days, iv) 15% piperidine/DMF, r.t., 30 min, v) TFA: triisopropylsilane: H2O = 95: 2.5: 2.5, v/v/v, r.t., 30 min.
The reactions were maintained for 4 days at 50 °C under nitrogen atmosphere in the dark. Subsequently, 10 % mole of N-Boc-DAH was added to the reaction mixture to consume unreacted acrylamide groups. When only API was used as an amine monomer (n(N-Boc-DAH): n(API) = 0:1, pAPI), 10% mole of API was added for the saturation of unreacted acrylamide groups. This reaction was carried out for further two days under same conditions. The polymer products were precipitated in anhydrous diethyl ether and dried under vacuum conditions. The N-Boc groups of products were removed by using a reagent solution (TFA : triisobutylsilane : H2O = 95 : 2.5 : 2.5, v/v/v) for 30 min at 0 °C in order to expose primary amines for arginine-graft reaction. The reaction mixtures were then precipitated in diethyl ether and were dialyzed against ultrapure water overnight with dialysis membrane (MWCO=1000, Spectrum Laboratories, Inc., Rancho Dominguez, CA). After subsequent lyophilization, the backbone polymers, p(DAH/API)s were obtained. The syntheses of p(DAH/API)s were confirmed by 1H NMR (400MHz, D2O) as followings. Because of its poor water solubility, 1H NMR of pAPI was performed in CD3OD.
p(DAH/API)s → 1H NMR (D2O): δDAH (H2NCH2CH2CH2CH2CH2CH2N) = 1.21–1.27; δDAH (H2NCH2CH2CH2CH2CH2CH2N) = 1.51–1.61; δAPI (imidazole-NCH2CH2CH2N) = 1.79–2.20; δCBA (NCH2CH2CONH) = 2.53–2.62; δCBA (CH2SSCH2) = 2.69–2.78; δDAH (H2NCH2CH2CH2CH2CH2CH2N, H2NCH2CH2CH2CH2CH2CH2N), δCBA (NCH2CH2CONH), δAPI (imidazole-NCH2CH2CH2N) = 2.79–3.20; δCBA (NCH2CH2CONHCH2) = 3.35–3.39; δAPI (imidazole-NCH2CH2CH2N) = 3.92–4.12; δ imidazole of API (NCHCHN), δ imidazole of API (NCHCHN)= 7.02–7.28; δ imidazole of API (NCHN) = 7.85–8.18 pAPI →1H NMR (CD3OD): δAPI (imidazole-NCH2CH2CH2N) = 1.91–1.98; δCBA (NCH2CH2CONH) = 2.31–2.44; δCBA (CH2SSCH2) = 2.52–2.56; δCBA (NCH2CH2CONH), δAPI (imidazole-NCH2CH2CH2N) = 2.71–2.85; δCBA (NCH2CH2CONHCH2) = 3.46–3.56; δAPI (imidazole-NCH2CH2CH2N) = 4.00–4.10; δimidazole of API (NCHCHN), δimidazole of API (NCHCHN)= 6.95–7.15; δimidazole of API (NCHN) = 7.65
After confirmation of syntheses of p(DAH/API)s, arginine residues were grafted onto the primary amines of p(DAH/API)s in DMF by using four equivalents of Fmoc-Arg(pbf)-OH and HBTU, and eight equivalents of DIPEA according to the amount of the primary amines for 2 days at room temperature. Reaction mixtures were precipitated in diethyl ether for removal of unreacted reagents and Ninhydrin assay was conducted for proof of primary amine-absence. Fmoc groups of arginine residues were then removed in 15% piperidine solution (DMF, v/v) for 30 min at room temperature. After precipitation with diethyl ether again, the pbf groups of arginine residues were removed by using a reagent solution (TFA : triisopropylsilane : H2O = 95 : 2.5 : 2.5, v/v/v) for 30 min at room temperature. The final products, p(DAH-R/API)s were obtained after additional precipitation in diethyl ether and dialysis, followed by filtration with 0.2 µm membrane and lyophilization. The syntheses were also confirmed by 1H NMR.
p(DAH-R/API)s →1H NMR (D2O): δDAH (CONHCH2CH2CH2CH2CH2CH2N) = 1.21–1.27; δDAH (CONHCH2CH2CH2CH2CH2CH2N), δarginine (NHCH2CH2CH2CHNH2) = 1.48–1.57; δarginine (NHCH2CH2CH2CHNH2), δAPI (imidazole-NCH2CH2CH2N) = 1.71–2.20; δCBA (NCH2CH2CONH) = 2.53–2.62; δCBA (CH2SSCH2) = 2.69–2.78; δDAH (CONHCH2CH2CH2CH2CH2CH2N), δCBA (NCH2CH2CONH), δAPI (imidazole-NCH2CH2CH2N), δarginine (NHCH2CH2CH2CHNH2), δDAH (CONHCH2CH2CH2CH2CH2CH2N) = 2.79–3.25; δCBA (NCH2CH2CONHCH2) = 3.35–3.39; δarginine (NHCH2CH2CH2CHNH2) = 3.71–3.77; δAPI (imidazole-NCH2CH2CH2N) = 3.92–4.16; δimidazole of API (NCHCHN), δimidazole of API (NCHCHN)= 7.02–7.28; δimidazole of API (NCHN) = 7.85–8.18
2.3. Characterization of p(DAH-R/API)s
2.3.1. Molecular weight measurements
The molecular weights of p(DAH-R/API)s were determined by multi-angle light scattering (MALS, miniDAWN TREOS, Wyatt Technology Corporation, Santa Barbara, CA) at 658 nm after size exclusion chromatography (SEC) using an AKTA FPLC system. The assay was run on a Superdex 75 column with 0.1 M acetate buffer (30 % acetonitrile, v/v, pH 6.5) as an eluent. The concentration of the polymer solutions was set to 3 mg/mL, the flow rate to 0.4 mL/min.
2.3.2. Endosome buffering capacity measurements
The endosome buffering capacities of p(DAH-R/API)s were measured by slightly modified acid-base titration method compared to the previous report [23]. Each 10 mg of polymer was dissolved in 10 mL of 0.1 M aqueous NaCl solution and the solution was set to pH 11 using 1 M NaOH. The solutions were titrated to pH 3 with 0.1 M HCl. pH changes of the solutions were measured by a pH meter (Corning 340, Mettler-Toledo, Inc, Columbus, OH). Titratio of 0.1 M NaCl and bPEI25k were also performed in the same manner as controls. Endosome buffering capacity was calculated as percentage of amine groups becoming protonated within the pH-range from 7.4 to 5.1, according to the following equation:
Here, ΔVHCl is the volume of HCl solution (0.1 M) which brought the pH values of polymer solutions from 7.4 to 5.1 and N mol is the total moles of protonable amine groups in 10 mg of each polymer.
2.4. Agarose gel electrophoresis
Agarose gel electrophoresis assay was performed to assess pDNA condensation ability of p(DAH-R/API)s. Agarose gel (0.7%, w/v) containing 0.1 µL/mL SYBR Safe DNA gel stain was prepared in Tris-Acetate-EDTA (TAE) buffer. The polyplexes (0.5 µg pDNA) were prepared in Hepes buffered saline (10 mM Hepes, 1 mM NaCl, pH 7.4) at various weight ratios (polymer/pDNA). After 30 minutes of incubation at room temperature, the electrophoresis was run for 15 min at 120 V. In order to examine the pDNA condensation ability under reducing condition, the identically prepared polyplexes were additionally incubated for 1 h in the presence of 2.5 mM DTT at 37 °C before being electrophoresed. The locations of pDNA bands were visualized by UV illuminator (Gel Documentation Systems, Bio-Rad, Hercules, CA).
2.5. Average particle size and Zeta-potential measurements
Average particle sizes and Zeta-potential values of p(DAH-R/API) polyplexes were measured by Zetasizer 3000 (Malvern Instruments, USA) with a He-Ne Laser beam (633 nm, fixed scattering angle of 90°) at 25°C. 0.5 mL of the polyplex solutions (10 µg pDNA) were prepared in Hepes buffered saline at various weight ratios ranging from 1 to 40. After 30 minutes of incubation, the polyplex solutions were diluted with Hepes buffered saline to 4 mL before measurements. Average particle sizes and Zeta-potential values were measured ten times and three times, respectively.
2.6. MTT assay
MTT assays were performed to examine the cytotoxicity of p(DAH-R/API)s. C2C12 mouse myoblast cells were seeded in a 96-well plate in 90 µL DMEM (10 % FBS) at a density of 1 × 104 cells/well. Having achieved 70–80 % confluence after 24 h, the cells were exposed to 100 µL polymer solutions (serum-free DMEM) having different concentrations (5–500 µg/mL) for 4 h. PEI25k was used as a control. Subsequently, the media was changed with fresh DMEM (10% FBS). After 24 h, the cells were treated with 25 µL of MTT stock solution (2 mg/mL in DPBS) and incubated for 2 h at 37°C. After removing each medium carefully, 150 µL of DMSO was added to each well to dissolve the formazan crystal formed by proliferating cells. The absorbance was measured at 570 nm using a UV/VIS photometry microplate reader (Model 680, Bio-Rad Lab, Hercules, CA). Results were presented as relative cell viabilities (RCV, percentage values relative to value of untreated control cells). All experiments were performed in quadruplicate.
2.7. Flow cytometry
C2C12 cells were seeded at a density of 1×105 cells/well in a 12-well plate in DMEM (10 % FBS) and were grown to reach 70–80% confluence. Prior to transfection with YOYO-1 iodide labeled pDNA (1 molecule dye / 50 nucleotide base pairs), the media were exchanged with serum-free DMEM. For transfection, the cells were treated with polyplex solutions (1 µg pDNA) at weight ratio of 80 for 4 h at 37 °C. PEI25k polyplex (weight ratio = 1) was used as a control. Subsequently, medium was aspirated off from the wells and the cells were rinsed two times with ice-cold DPBS. After trypsinization, the cells were suspended in 1 mL DPBS for assay.
The cellular uptake of fluorescence-labeled polyplexes was examined by using the BD FACScan analyzer (Becton Dickinson, San Jose, CA) at a minimum of 1 × 104 cells gated per sample. Analysis was performed by using Becton Dickinson CellQuest software. Data were processed by using Windows Multiple Document interface software (WinMDI).
2.8. In vitro transfection experiments
2.8.1. Transfection in serum or without serum condition
The transfection efficiencies of p(DAH-R/API)s were examined by measuring luciferase reporter gene expression in non-serum or serum conditions in C2C12 cells. Cells were seeded in a 24-well plate at a density of 5 × 104 cells/well in DMEM (10% FBS) and were grown to reach 70–80% confluence. Before transfection, the media were exchanged with serum-free DMEM for the assay in non-serum condition but not for the assay in serum condition. The cells were treated with polyplex solutions (0.5 µg pDNA) with various weight ratios ranging from 20 to 80. PEI25k polyplex was used as a control (weight ratio = 1). After 4 h of incubation, the media were exchanged with fresh DMEM (10% FBS) and the cells were incubated for further 2 days at 37 °C in 5% CO2. Subsequently, media were removed and the cells were rinsed with 200 µL of DPBS and shaken for 30 min at room temperature in 100 µL of Reporter Lysis Buffer. The luciferase activities of 25 µL cell lysates were measured by using 100 µL of luciferase assay reagent on a luminometer (Dynex Technologies Inc., Chantilly, CA). A protein quantification assay was performed using a BCA™ Protein Assay Reagent Kit to measure total amount of cellular proteins. The final results were presented in terms of RLU/mg cellular protein. All experiments were performed in triplicate.
2.8.2. Transfection with chloroquine or nigericin
Transfection experiments were carried out using two kinds of chemical reagents, chloroquine or nigericin in order to investigate the endosomal escape mechanism of p(DAH-R/API) polyplexes. All experiments were performed with the same methods as section 2.8.1, except that seeded cells were pre-incubated for 30 min in serum-free DMEM containing 100 µM chloroquine or 10 µM nigericin before polyplexes treatment. Chloroquine and nigericin stock solutions were prepared in DPBS buffer and DMSO, respectively. ABP polyplex was used as a control with a weight ratio of 40. All the other p(DAH-R/API) polyplexes were prepared at a weight ratio of 80. The cells were transfected for 4h in the presence of the mentioned reagents. Subsequently, the media were replaced with fresh DMEM (10% FBS). After further 2 days of incubation, luciferase assay and protein quantification assay were fulfilled. Transfection efficiencies in reagent-treated condition were presented as relative ratios to transfection efficiencies in untreated condition.
3. Results and discussion
3.1. Syntheses of polymers
First, bioreducible backbone polymers, p(DAH/API)s were synthesized by Michael reaction of CBA and the amine monomers, composed of various mole ratios (N-Boc-DAH/API : 1/1, 1/2, 1/4, 0/1). API was chosen as a endosome buffering amine monomer because pKa value (~6) of its imidazole group lies within endosomal pH range (7.4~5.1). The endosome buffering capacity of final products, p(DAH-R/API)s can be tuned by variation of API portions in similar with Lin et al.’s report which used low-basicity amine monomer, histamine as a regulator for buffering capacity [24]. N-Boc-DAH served as a linkage unit for arginine-graft due to its modifiable primary amine. After removal of N-Boc groups by strong acid mixture for the exposure of primary amines, p(DAH/API)s were obtained via dialysis followed by lyophilization. The structures of synthesized p(DAH/API)s were confirmed by 1H NMR.
Interestingly, it was found that the calculated molar ratios between API and DAH based on NMR results were always nearly 2 fold lower than expected values for all p(DAH/API)s. This might be based on low reactivity of API amine because of steric hindrance from short linkage hydrocarbon chain (3 carbons) between API amine and bulky imidazole group. In this work, p(DAHa/APIb)s and p(DAHa-R/APIb)s were named in accordance with their relative composition ratios calculated from 1H NMR analysis, as shown in Table 1.
Table 1.
Characterization of p(DAH-R/API)s.
| Expected DAH/API ratio (%)a |
Calculated DAH/API ratio (%)b |
Mw (kDa) | Mn (kDa) | PDI | Buffer capacity (%) | |
|---|---|---|---|---|---|---|
| p(DAH2-R/API1) | 50/50 | 66/34 | 6.49 | 5.64 | 1.15 | 35.4 |
| p(DAH1-R/API1) | 33/67 | 53/47 | 6.58 | 5.85 | 1.13 | 39.1 |
| p(DAH1-R/API2) | 20/80 | 37/63 | 6.27 | 5.98 | 1.05 | 42.9 |
Relative molar ratios in p(DAHa/APIb)s of DAH/API added initially for polymerization
Calculated molar ratios in p(DAHa/APIb)s of DAH/API after NMR analysis of synthesized polymers
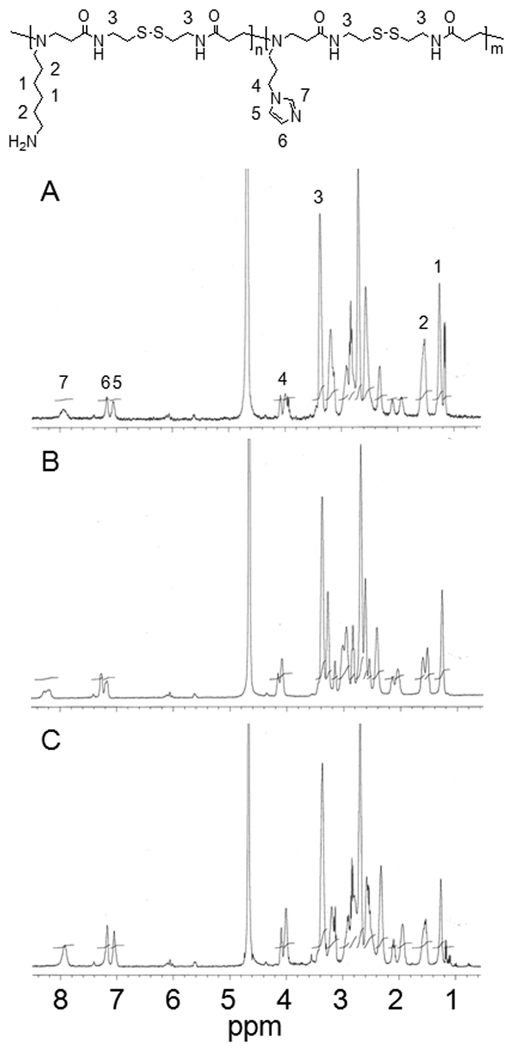
Figure 1 displays 1H NMR results of p(DAH/API)s. It can be seen that distinct proton peaks from API (peak # 5, 6, and 7) augment and proton peaks from DAH hydrocarbons (peak # 1, 2) are scaled down as API portions increase in p(DAH/API)s, when integrations of CBA proton peaks (peak # 3) are fixed. However, pAPI (N-Boc-DAH/API = 0/1) was not soluble in buffer (pH 7.4) due to hydrophobicity of API imidazoles and rigidity of CBA backbone, while being only soluble in acidic solutions. Hence, no more assays were performed with pAPI except 1H NMR in CD3OD. Subsequently, arginine-graft reaction was conducted by coupling primary amines of DAH to carboxylic acids of Fmoc-Arg(pbf)-OH using HBTU and DIPEA in DMF. After removal of Fmoc and pbf protecting groups of arginine moieties, final polymer products were obtained after dialysis followed by lyophilization. The final polymer structures were also confirmed by 1H NMR. Degree of the arginine-graft reaction was found to be almost 100% from the calculation between the integration of arginine proton peaks and the integration of polymer backbone proton peaks. Therefore, p(DAH2-R/API1), p(DAH1-R/API1), and p(DAH1-R/API2) were successfully synthesized from p(DAH2/API1), p(DAH1/API1), and p(DAH1/API2), respectively.
Figure 1.
1H NMR spectra (400 MHz, D2O) of p(DAH/API)s. (A) p(DAH2/API1), (B) p(DAH1/API1), (C) p(DAH1/API2).
3.2. Characterization of polymers
3.2.1. Molecular weight measurements
The molecular weights of p(DAH-R/API)s were determined by multi-angle light scattering (MALS) at 658 nm after SEC using an AKTA FPLC system. MALS is known to be able to measure the absolute molecular weights of polymers without the reference standard polymers [25].
As shown in Table 1, all p(DAH-R/API)s were found to possess similar Mw, which range from 6.27 kDa to 6.58 kDa and very narrow molecular weight distributions (PDI : 1.05 ~ 1.15). These low PDIs of the polymers means a good control of the Michael-type polyaddition and lack of gel formation by using CBA (N,N’-cystaminebisacrylamide) as an acrylamide monomer. dn/dc value of polymers was determined to be 0.145 mL/g. Considering the experimentally found ratios of DAH and API, about halves of initially added API moles have not participated in polymerization. These amounts of API are presumed to remain unreacted or react with equivalent moles of CBA in a marginal degree, generating oligomers. These small molecules are expected to be removed during purification, causing decrease of synthesis yield. This can be supported by low synthesis yields (9~21 %) for p(DAH-R/API)s in comparison with ABP (> 60%) containing no API.
3.2.2. Endosome Buffering capacity measurements
Acid-base titrations were performed to assess endosome buffering capacities of p(DAH-R/API)s. Several works demonstrated the important role of the endosome buffering capacities of polymeric gene delivery carriers regarding efficient endosomal escape and transfection. However, high transfection efficiency can’t be guaranteed by adjusting buffering capacities of polymers because of change in hydrophobicity and pDNA condensing ability.
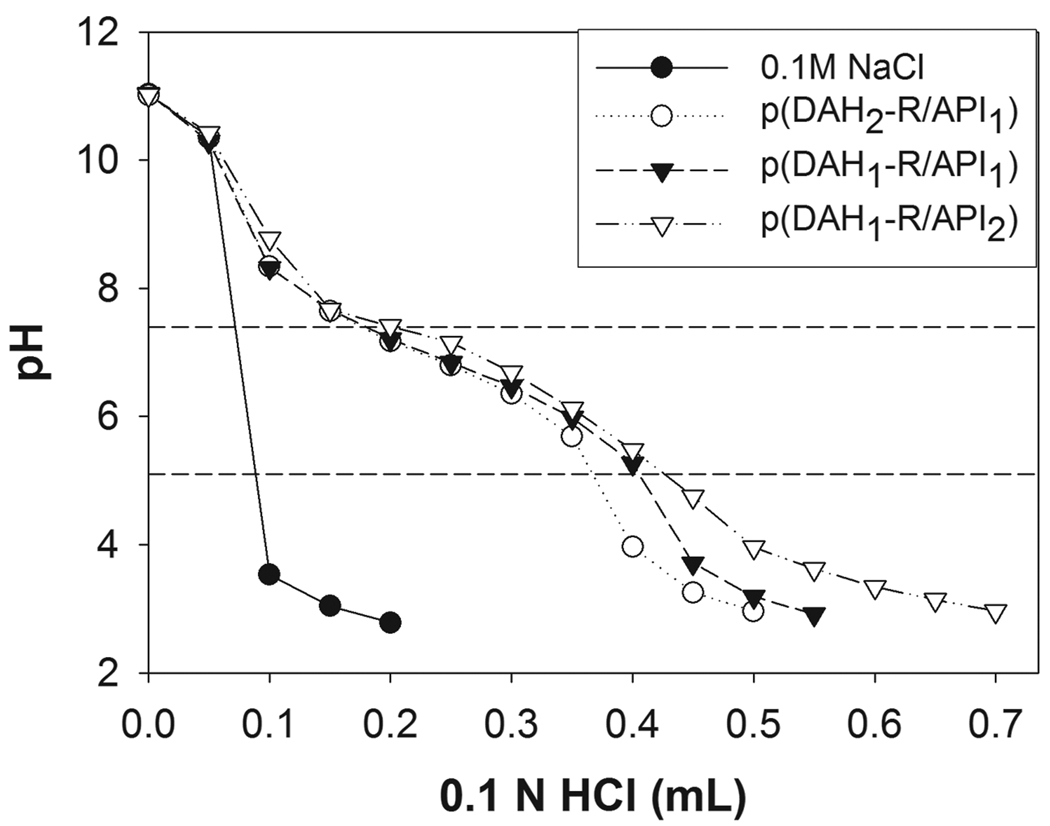
Figure 2 presents the acid-base titration curve of p(DAH-R/API)s. Endosome buffering capacities of polymers were obtained from this curve by using equation in Section 2.3.2. Table 1 shows increasing endosome buffering capacities of p(DAH-R/API)s with the increase of API portions due to its imidazole group as following order; (p(DAH2-R/API1) < p(DAH1-R/API1) < p(DAH1-R/API2). The buffering capacities of polymers were found to be higher than that of PEI25k (18.7 %, curve not shown).
Figure 2.
Acid-base titration curve of p(DAH-R/API)s. 10 mg of each polymer was dissolved in 10 mL of 0.1 M NaCl solution, which was subsequently set to pH 11 using 1 M NaOH. The solutions were titrated to pH 3 with 0.1 M HCl. Titration curve of 0.1 M NaCl is presented as a control. Dashed lines indicate endosomal pH range (5.1 and 7.4).
3.3. Agarose gel electrophoresis
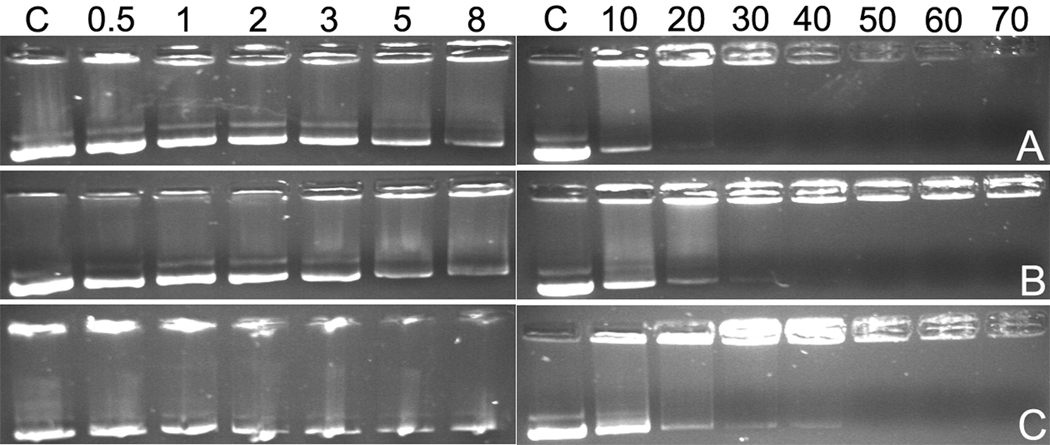
pDNA condensing ability of the p(DAH-R/API)s were examined by agarose gel electrophoresis. As shown in Figure 3, p(DAH2-R/API1) and p(DAH1-R/API1) retarded pDNA from a weight ratio of 2 completely after 30 min of polyplex formation while p(DAH1-R/API2) condensed pDNA completely at a weight ratio of 5. This data clearly displays a sufficient pDNA condensing abilities of p(DAH-R/API)s as well as their dependency on the composition ratio (DAH-R:API) which is based on low pKa value of imidazole group of API remaining electrically neutral at pH 7.4.
Figure 3.
Agarose gel electrophoresis results in the absence of DTT. (A) p(DAH2-R/API1), (B) p(DAH1-R/API1), (C) p(DAH1-R/API2). C : pDNA only. Numbers above indicate weight ratios of the polyplexes.
In addition, the polyplexes (same as above) were electrophoresed after 1 h of incubation at 37 °C in the presence of 2.5 mM DTT (reducing agent) in order to examine the pDNA condensing abilities of p(DAH-R/API)s in reducing condition.
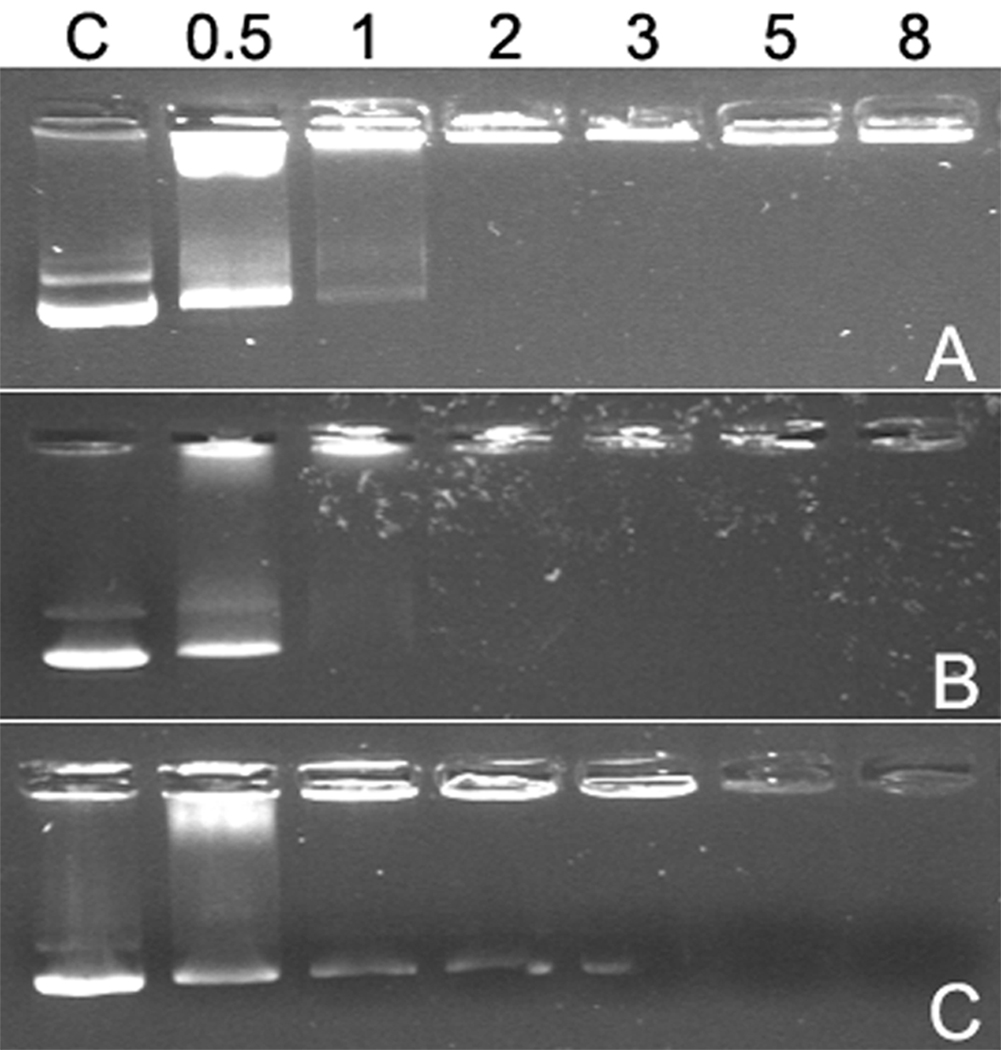
Figure 4 shows that pDNA band released from polyplexes were detected at weight ratios less than 8 for all polyplexes, which indicates all p(DAH-R/API)s are degradable and could not condense pDNA in reducing condition such as cytoplasm. However, quite different gel electrophoresis profiles were detected for polyplexes formed at higher weight ratios. It was observed that pDNA retardation started at different weight ratios according to polymers as followings; p(DAH2-R/API1) : 30, p(DAH1-R/API1) : 40, p(DAH1-R/API2) : 50. This result also demonstrates the effect of varying the composition ratio of monomers possessing different pKa on pDNA condensing abilities of p(DAH-R/API)s. It is concluded that pDNA condensing abilities of p(DAH-R/API)s are tunable by modulating the monomer composition ratio.
Figure 4.
Agarose gel electrophoresis results after incubation in 2.5 mM DTT for 1h at 37 °C. (A) p(DAH2-R/API1), (B) p(DAH1-R/API1), (C) p(DAH1-R/API2). C : pDNA only. Numbers above indicate weight ratios of the polyplexes.
3.4. Average size and Zeta-potential measurements of polyplexes
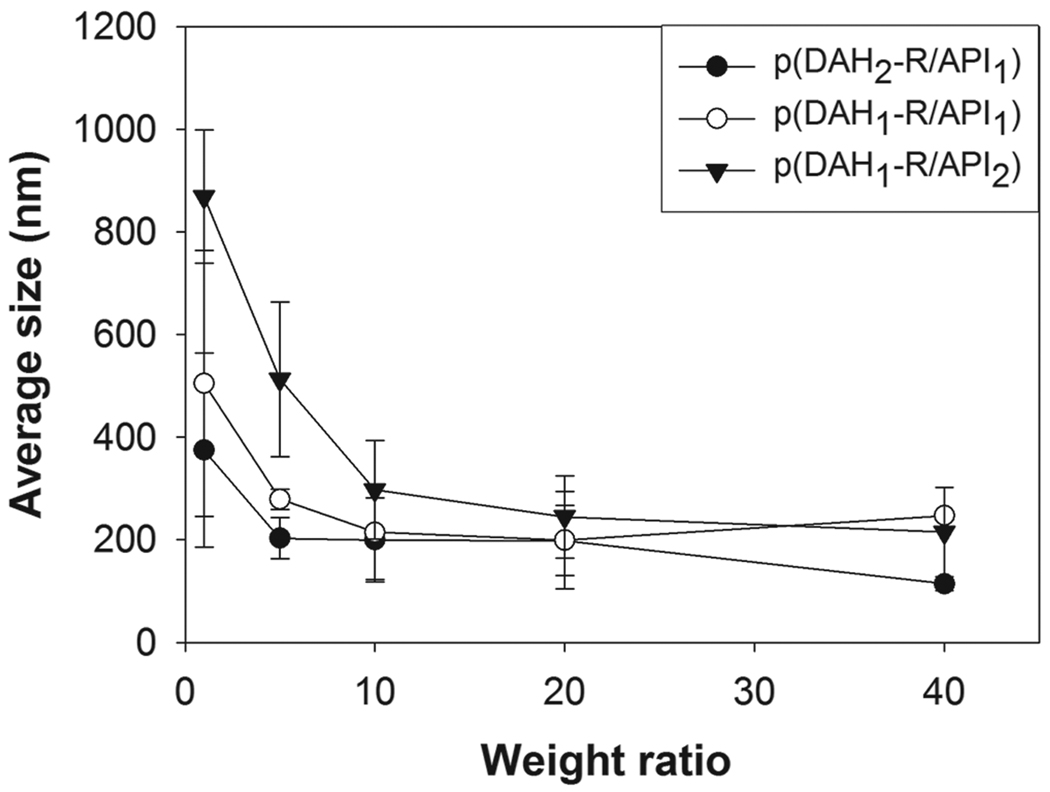
Average sizes of polyplexes were measured by using a Zeta-sizer as shown in Figure 5. All p(DAH-R/API) polyplexes have sizes of about 200 nm or less at above weight ratio of 20, which is a prerequisite for efficient cellular uptake [26]. However, size of each polyplex seemed to be increased according to the following order at low weight ratios less than 20; p(DAH2-R/API1) < p(DAH1-R/API1) < p(DAH1-R/API2). Thus, it was revealed that electrically neutral imidazole groups of API can’t support pDNA condensation in Hepes buffer (pH 7.4) and that primary amines and guanidines of DAHR have beneficial effect on compact condensation of pDNA, which corresponds with agarose gel electrophoresis results.
Figure 5.
Average size measurements of p(DAH-R/API) polyplexes.
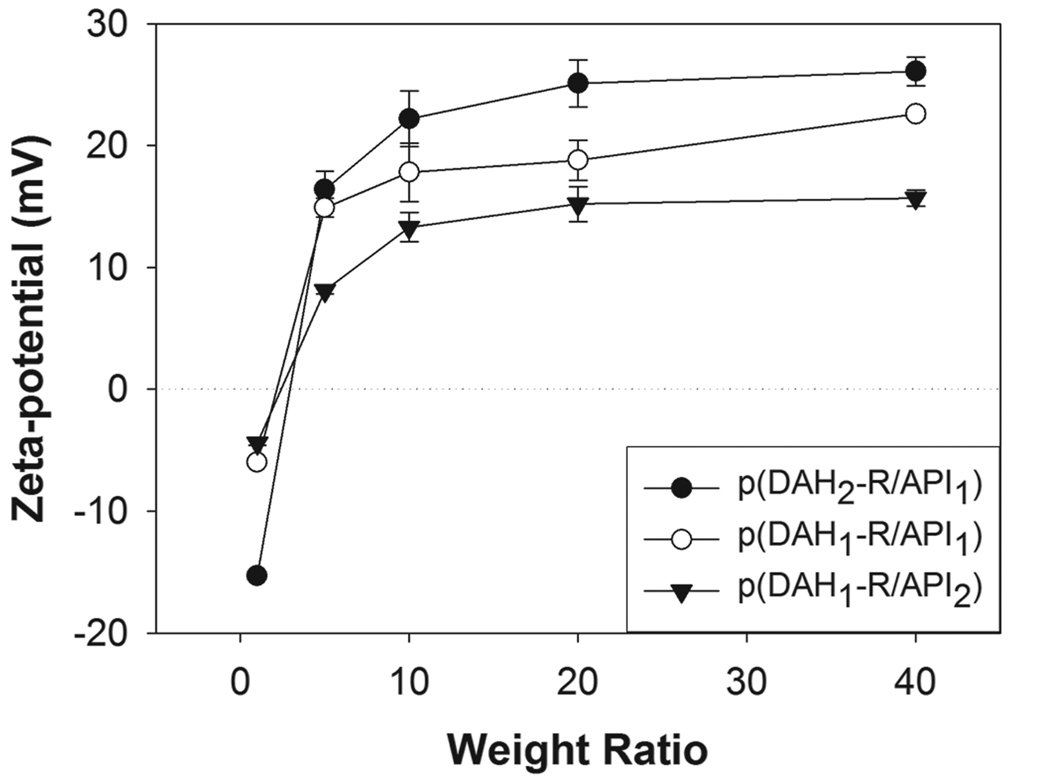
Distinctly different behaviors of polyplex formation of p(DAH-R/API)s were also observed in Zeta-potential measurements (Figure 6). All Zeta-potentials of polyplexes were negative at a weight ratio of 1. However, all polyplexes displayed increasing Zeta-potential profiles with increasing weight ratios over 1 as following order; p(DAH1-R/API2) < p(DAH1-R/API1) < p(DAH2-R/API1), until reaching their maximum values (p(DAH2-R/API1) : 22~26 mV, p(DAH1-R/API1) : 18~23 mV, p(DAH1-R/API2) : 13~16 mV). The inversely proportional relation between API portions of p(DAH-R/API)s and Zeta-potentials shows that higher portions of electrically neutral imidazole groups of API lead to lower Zeta-potentials of the polyplexes at pH 7.4.
Figure 6.
Zeta-potential measurements of p(DAH-R/API) polyplexes.
3.5. Cytotoxicity
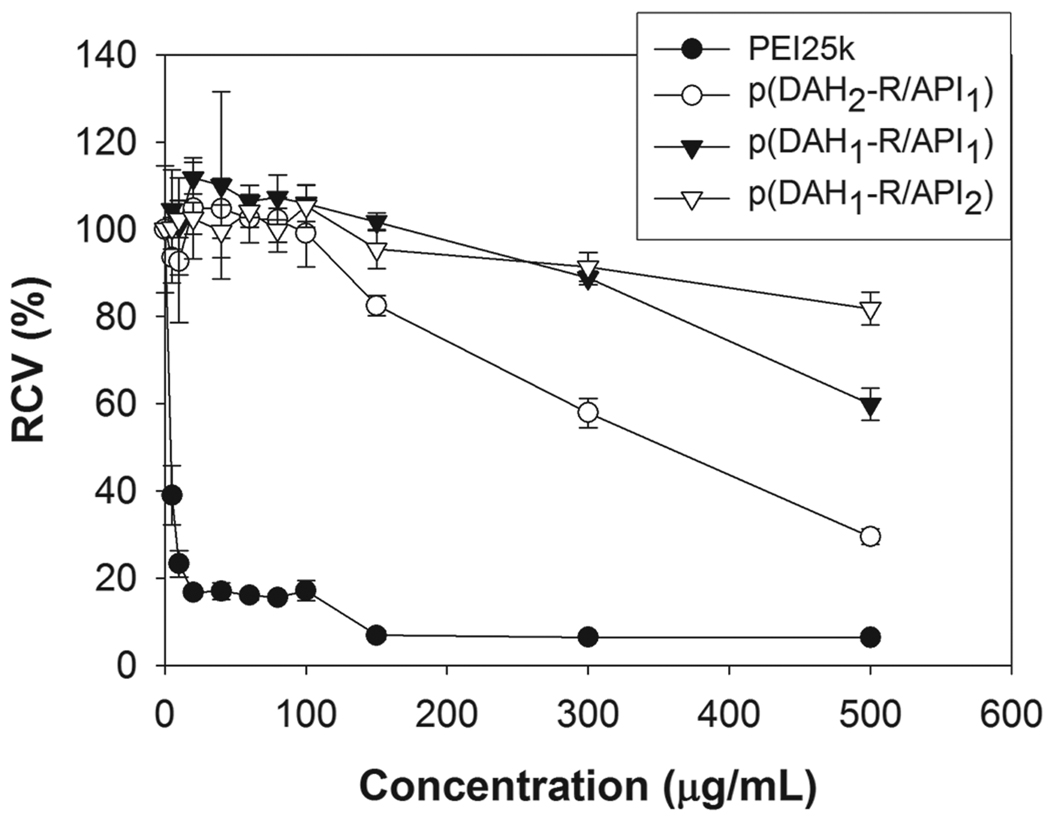
The cytotoxicities of p(DAH-R/API)s were investigated by MTT assay in C2C12 cells as shown in Figure 7. The molecular weights as well as the charge densities of cationic polymers were reported to function as key parameters for the interaction with cell membranes and consequently, for the cell damage [27]. Accordingly, relative cell viability (RCV) for PEI25k, used as a control, was abruptly decreased to 23% even at 10 µg/mL concentration and to less than 6% at over 100 µg/mL, displaying its well-known severe cytotoxicity which arises from its high charge density and molecular weight. RCVs of p(DAH-R/API)s were found to be about 100% or higher even at a concentration of 100 µg/mL, meaning that cytotoxicities of p(DAH-R/API)s were marginal due to their biodegradability.
Figure 7.
MTT assay results in C2C12 cells. PEI25k was used as a control. RCV (relative cell viability) is presented as relative value (%) to untreated cells.
However, decreasing RCVs of p(DAH-R/API)s were evaluated with increasing concentration over 100 µg/mL, meaning the augmenting cytotoxicities of the polymers as following order; p(DAH1-R/API2) < p(DAH1-R/API1) < p(DAH2-R/API1). This cytotoxicity profile shows that high density of arginine graft can cause some cytotoxicity by the interaction of strong positive charges with cell compartments, which was also presented by previous study about cytotoxicity of mono- and di-arginine-conjugated PAMAM dendrimers [28].
3.6. Flow cytometry
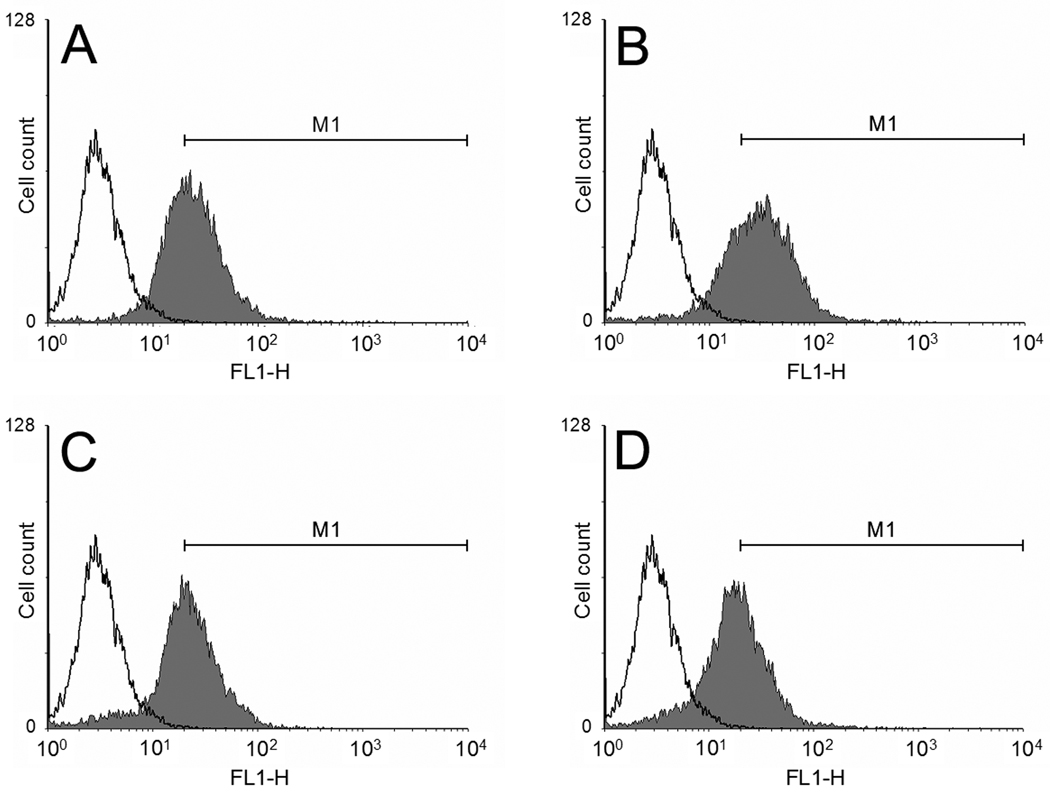
Cellular uptakes of polyplexes were examined in C2C12 cells by flow cytometry with YOYO-1 iodide labeled pDNA (Figure 8). Peak of each polyplex processed by WinMDI software was presented in comparison with pDNA peak. Region of gate M1 was set up where the portion of counted events for control (cell only) was 0.35%. Based on this gate, cellular uptake of each polyplex was quantified as followings; PEI25k : 60.6 %, p(DAH2-R/API1) : 69.6 %, p(DAH1-R/API1) : 52.0 %, p(DAH1-R/API2) : 43.9%.
Figure 8.
Flow cytometry results in C2C12 cells. White peaks: cell only. Gray peaks: polyplexes. (A) PEI25k, (B) p(DAH2-R/API1), (C) p(DAH1-R/API1), (D) p(DAH1-R/API2). YOYO-1 iodide labeled pDNA was used. M1 region was set up as a gate for the estimation of cellular uptake values.
It was clearly demonstrated that cellular uptake of all p(DAH-R/API) polyplexes shows positive correlation with the degree of arginine-graft. Cell penetrating ability of arginine moiety, induced by formation of less polar ion pairs with negatively charged cell membrane via bidentate hydrogen bonding, may be associated with these results, as previously proposed by Wender and colleagues [29]. However, the fact that cellular uptake of PEI25k polyplex is still high despite absence of arginine, shows cellular uptake of polyplexes also can be significantly mediated by their surficial positive charges and Zeta-potentials. This expectation is well supported by our previous work reporting that bioreducible polyplexes having similar Zeta-potentials shows similar cell uptake regardless of their arginine-graft [17].
3.7. Transfection experiments
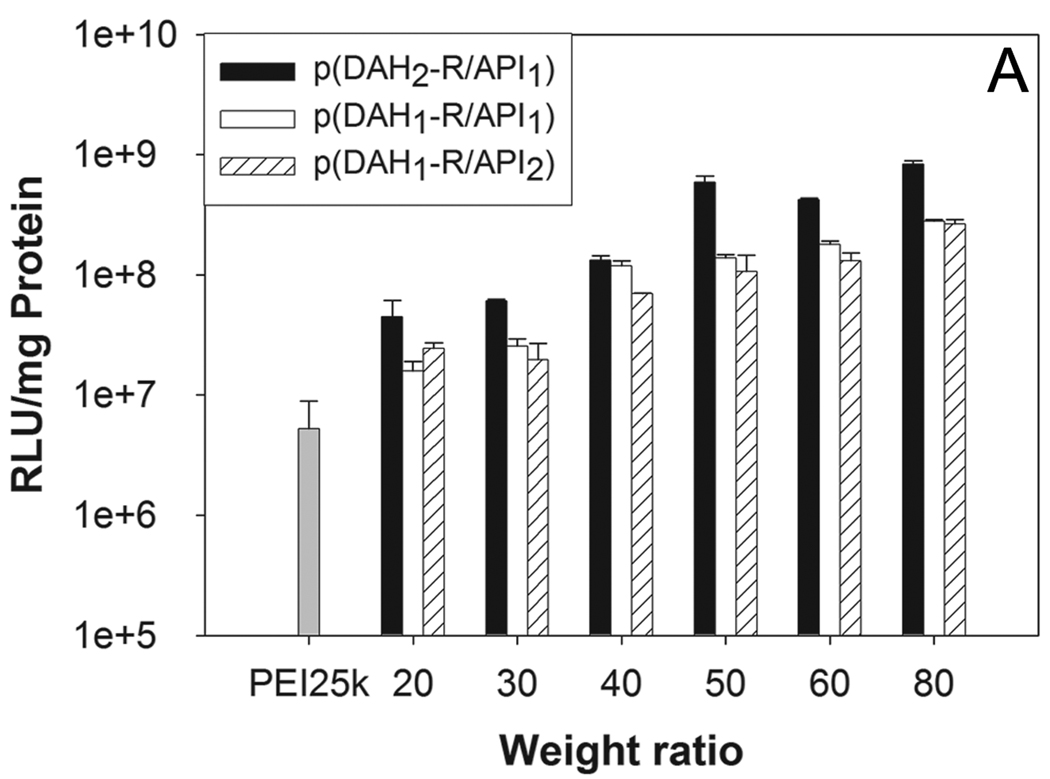
The transfection efficiency (TE) of p(DAH-R/API)s were evaluated by using luciferase reporter gene in C2C12 cells. PEI25k was used as a control. In the absence of serum (Figure 9A), all p(DAH-R/API)s showed augmented TE in accordance with increasing weight ratios. At a weight ratio of 80, p(DAH2-R/API1), p(DAH1-R/API1), and p(DAH1-R/API2) displayed 160, 53, and 50 fold higher TE than PEI25k, respectively. This results indicate that p(DAH-R/API)s possesses high transfection ability due to arginine moieties, as already proposed for ABP.
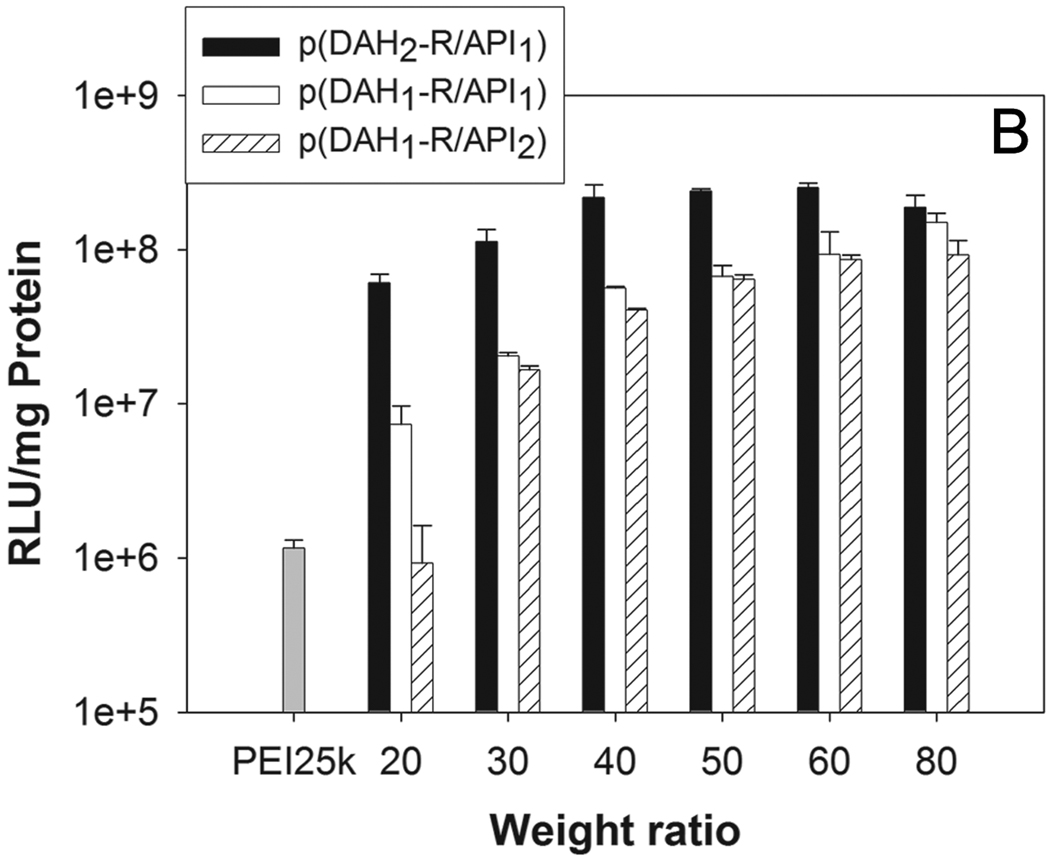
Figure 9.
Transfection results in C2C12 cells. (A) non-serum condition, (B) 10% serum condition. RLU (relative light unit) values from luciferase assay were normalized by total cellular proteins (mg). PEI25k polyplex was used as a control at a weight ratio of 1.
In the presence of serum (Figure 9B), all p(DAH-R/API)s also showed similar proportional correlation between weight ratios and their TEs, except TE of p(DAH2-R/API1) which has its maximum at a weight ratio of 60. In serum condition, the polymers displayed much improved TEs than PEI25k (162, 129, and 79 fold higher at a weight ratio of 80, respectively). TEs in serum condition were found to be reduced to 41 ~ 54% of TEs in serum-free condition over weight ratios of 40, when TE of PEI25k in serum condition were decreased to 22% of TE in serum-free condition. Introduction of API which is hydrophobic at physiological pH to polymers, leading to low Zeta-potential values, may enhance the stability in serum condition due to reduction of nonspecific interactions between polyplexes and serum compartments. However, considering the fact that TE of ABP having only arginines as grfted moieties in serum condition was just decreased to 85% of TE in serum-free condition at a weight ratio of 40 [17], it can be presumed that hindering interaction of polyplexes from serum compartments by introduction of API doesn’t override the benefit loss from the decrease of grafted arginine moieties for transfection ability.
3.8. Transfection experiments with chloroquine or nigericin
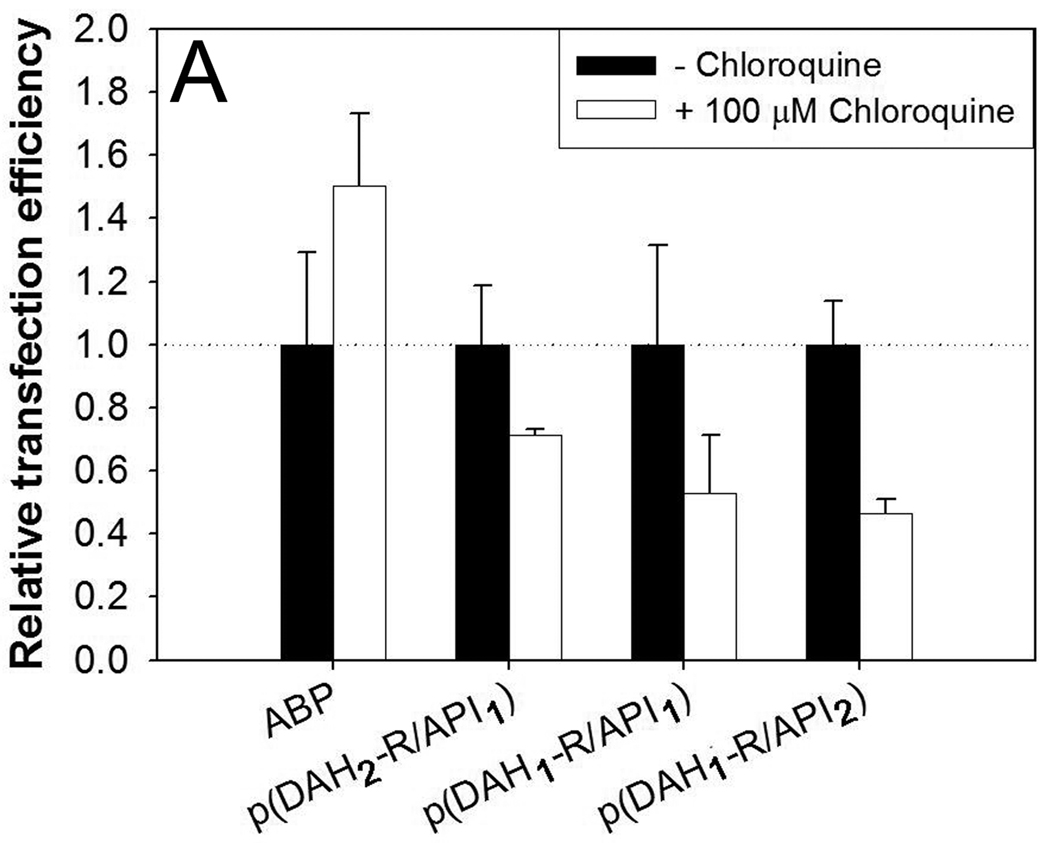
Transfection experiments were also performed with treatment of two chemical reagents, chloroquine or nigericin in order to investigate endosomal escape mechanism of polyplexes. Chloroquine is known as an endosome disruptive agent by pH buffering in endosome [30]. Nigericin is a carboxylic ionophore which inhibits endosomal acidification by monovalent cation exchange [31]. ABP polyplex was used as a control.
As shown in Figure 10A, all TEs of p(DAH-R/API)s were decreased to 46~71% in the presence of chloroquine compared with TEs in the absence of chloroquine. TE of ABP was rather increased to 150% when treated with chloroquine. It was reported that chloroquine can support endosomal escape of polyplexes via its own proton sponge effect and thus enhance their transfection efficiency, proven for polyplexes having no endosome buffering ability, like poly[r-(4-aminobutyl)-l-glycolic acid] (PAGA) [32]. However, chloroquine could not increase TE of p(DAH-R/API)s in this experiment, revealing that these bioreducible polymers may have their own ability for endosomal escape such as endosome buffer, as already shown in Section 3.2.2. Reduction of TE in chloroquine condition may rely on the displacement of polymers by chloroquine from pDNA in polyplexes, which was already suggested by Cheng et al [33]. p(DAH-R/API) polyplexes may be more susceptible to this displacement by chloroquine because p(DAH-R/API)s were formerly shown to have lower pDNA condensing ability than ABP. This may enhance exposure of dissociated pDNA to hostile environment containing DNase, leading to the decrease of transgene expression.
Figure 10.
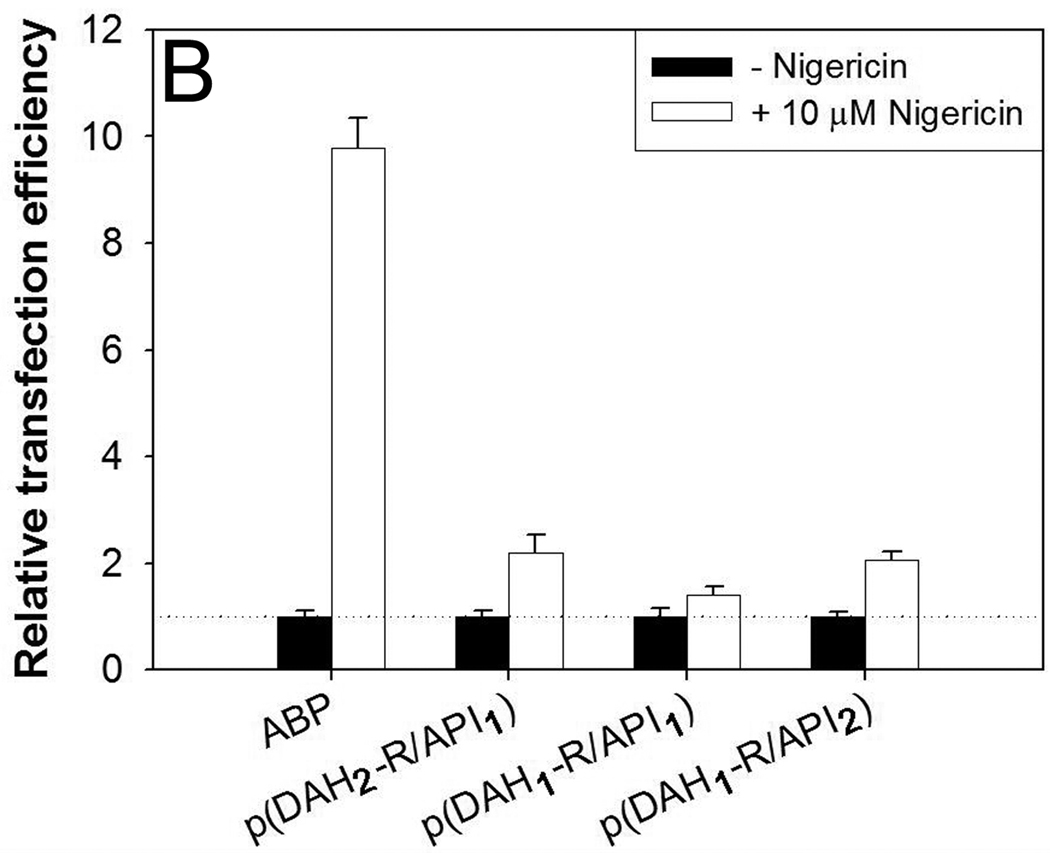
Transfection results in C2C12 cells in the presence of (A) 100 µM chloroquine and (B) 10 µM nigericin. Relative transfection efficiency: experimental values/control values × 100 (%).
Secondly, transfection experiments were performed in the presence of nigericin. On the contrary to our expectation, TEs of p(DAH-R/API)s in the presence of nigericin were increased to 140~220% values in comparison with TEs in the absence of nigericin (Figure 10B). Interestingly, TE of ABP was significantly increased to about 10 fold higher value in the presence of nigericin. Nigericin inhibits endosomal acidification by monovalent cation exchange, finally interrupting the transporting process from endosome to lysosome. It was reported that cationic polymers having endosome buffering capacity such as PEI25k show reduced TE in the presence of nigericin because their endosome buffering moieties can’t function for endosome disruption via proton sponge effect in physiological pH [32].
Therefore, increased TEs of p(DAH-R/API)s demonstrate that these polyplexes can escape from endosome by different pathway in the nigericin condition, which is independent from its endosome buffering capacity. Considering the structure of p(DAH-R/API)s and ABP, it is postulated that arginine moieties with cell penetrating ability may enhance polyplex transport to cytosol through endosomal membrane as well. This hypothesis is supported by the previous study about guanidinium-rich transporters suggesting the possibility of both endosome and plasma membrane penetrating ability of guanidine [34]. Remarkably increased TE of ABP (~ 10 fold) in the nigericin condition can be obviously explained by this hypothesis because ABP has only arginine-grafted moieties.
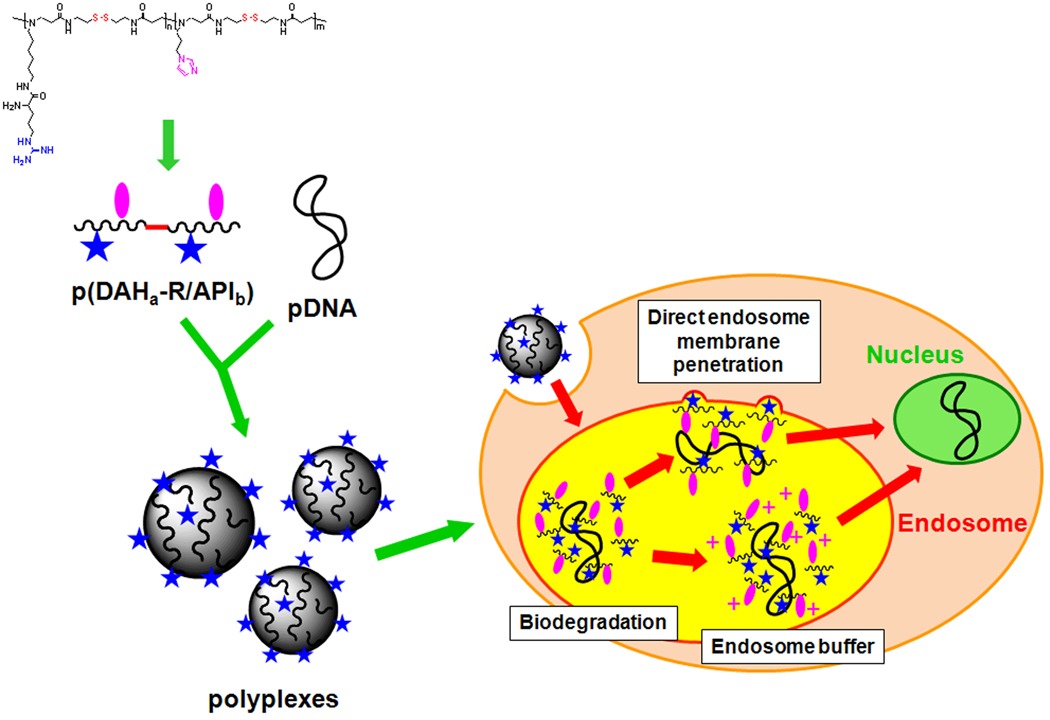
In conclusion, it is thought that p(DAH-R/API) polyplexes internalized by endocytosis may escape from endosome by direct endosome membrane penetration due to its cell penetrating ability as well as endosome disruption due to endosome buffering ability (graphically illustrated in Scheme 2).
Scheme 12.
Schematic diagram of the proposed endosome escape mechanism of p(DAH-R/API) polyplexes.
Regardless of lower cellular uptake efficiency than PEI25k, p(DAH1-R/API1) and p(DAH1-R/API2) showed higher transfection efficiency. As mentioned above, efficient escape mechanism of p(DAH-R/API)s is thought to induce superior transfection efficiency. Additionally, nuclear localization ability of guanidine of p(DAH-R/API) also may mediate high transfection efficiency as proposed in our previous study [35].
4. Conclusion
p(DAH-R/API)s, bioreducible polymeric gene delivery carriers containing DAH-R as cell penetrating moiety and API as endosome buffering moiety were synthesized with three different composition ratios and examined regarding their structural effects on properties for gene delivery. It was elucidated that both endosome buffering capacities and cytotoxicity issues of the polymers were improved but pDNA condesing abilities, Zeta-potential values, cellular uptakes, and transfection efficiencies of the polyplexes were decreased in accordance with increasing API portion in p(DAH-R/API)s. Modulation of endosome buffering capacities of arginine-grafted bioreducible polymers by introduction of API was found to benefit some features of polyplexes but not to improve gene delivery potency, suggesting that grafted arginine moieties play a crucial role for efficient gene delivery instead of endosome buffering moieties. Moreover, transfection results with chloroquine or nigericin proposed the unique transfection mechanism that p(DAH-R/API) polyplexes may escape from endosomes by direct endsome membrane penetration of arginine moieties as well as endosome buffering abilities of the polymers after cellular uptake, which supports and emphasizes the importance of arginine moieties for polymeric gene delivery systems.
Acknowledgments
This work was supported by NIH grants (HL065477-10, CA107070-06) and Research fund (WCU 200900000000024) of the Ministry of Education, Science and Technology, Korea. We thank Monika Sima for FPLC-SEC experiments and thank Prof. You Han Bae for generous permission of Zetasizer usage.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Luo D, Saltzman WM. Synthetic DNA delivery systems. Nat. Biotechnol. 2000;18:33–37. doi: 10.1038/71889. [DOI] [PubMed] [Google Scholar]
- 2.Park TG, Jeong JH, Kim SW. Current status of polymeric gene delivery systems. Adv. Drug. Deliv. Rev. 2006;58:467–486. doi: 10.1016/j.addr.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Jeong JH, Kim SW, Park TG. Molecular design of functional polymers for gene therapy. Prog. Polym. Sci. 2007;32:1239–1274. [Google Scholar]
- 4.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat. Rev. Drug Discov. 2005;4:581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 5.Neu M, Fischer D, Kissel T. Recent advances in rational gene transfer vector design based on poly(ethylene imine) and its derivatives. J. Gene Med. 2005;7:992–1009. doi: 10.1002/jgm.773. [DOI] [PubMed] [Google Scholar]
- 6.Wong SY, Pelet JM, Putnam D. Polymer systems for gene delivery-past, present, and future. Prog. Polym. Sci. 2007;32:799–837. [Google Scholar]
- 7.Lynn DM, Langer R. Degradable poly(β-amino esters): synthesis, characterization, and self-assembly with plasmid DNA. J. Am. Chem. Soc. 2000;122:10761–10768. [Google Scholar]
- 8.Lim Y-b, Kim C-h, Kim K, Kim SW, Park J-s. Development of a safe gene delivery system using biodegradable polymer, poly[α-(4-aminobutyl)-L-glycolic acid] J. Am. Chem. Soc. 2002;122:6524–6525. [Google Scholar]
- 9.Wang J, Mao H, Leong KW. A novel biodegradable gene carrier based on polyphosphoester. J. Am. Chem. Soc. 2001;123:9480–9481. doi: 10.1021/ja016062m. [DOI] [PubMed] [Google Scholar]
- 10.Goh SL, Murthy N, Xu M, Fre´chet JMJ. Cross-linked microparticles as carriers for the delivery of plasmid DNA for vaccine development. Bioconjugate Chem. 2004;15:467–474. doi: 10.1021/bc034159n. [DOI] [PubMed] [Google Scholar]
- 11.Luten J, van Nostrum CF, De Smedt SC, Hennink WE. Biodegradable polymers as non-viral carriers for plasmid DNA delivery. J. Control. Release. 2008;126:97–110. doi: 10.1016/j.jconrel.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 12.Lin C, Engbersen JFJ. Effect of chemical functionalities in poly(amido amine)s for non-viral gene transfection. J. Control. Release. 2008;132:267–272. doi: 10.1016/j.jconrel.2008.06.022. [DOI] [PubMed] [Google Scholar]
- 13.Gosselin MA, Guo W, Lee RJ. Efficient gene transfer using reversibly crosslinked low molecular weight polyethylenimine. Bioconjugate Chem. 2001;12:989–994. doi: 10.1021/bc0100455. [DOI] [PubMed] [Google Scholar]
- 14.Oupicky D, Parker AL, Seymour LW. Laterally stabilized complexes of DNA with linear reducible polycations: Strategy for triggered intracellular activation of DNA delivery vectors. J. Am. Chem. Soc. 2002;124:8–9. doi: 10.1021/ja016440n. [DOI] [PubMed] [Google Scholar]
- 15.Miyata K, Kakizawa Y, Nishiyama N, Harada A, Yamasaki Y, Koyama H, Kataoka K. Block catiomer polyplexes with regulated densities of charge and disulfide cross-linking directed to enhance gene expression. J. Am. Chem. Soc. 2004;126:2355–2361. doi: 10.1021/ja0379666. [DOI] [PubMed] [Google Scholar]
- 16.Christensen LV, Chang CW, Kim WJ, Kim SW, Zhong Z, Lin C, Engbersen JFJ, Feijen J. Reducible poly(amido ethylenimine)s designed for triggered intracellular gene delivery. Bioconjugate Chem. 2006;17:1233–1240. doi: 10.1021/bc0602026. [DOI] [PubMed] [Google Scholar]
- 17.Kim T-i, Ou M, Lee M, Kim SW. Arginine-grafted bioreducible poly(disulfide amine) for gene delivery systems. Biomaterials. 2009;30:658–664. doi: 10.1016/j.biomaterials.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao M, Weissleder R. Intracellular cargo delivery using tat peptide and derivatives. Med. Res. Rev. 2004;24:1–12. doi: 10.1002/med.10056. [DOI] [PubMed] [Google Scholar]
- 19.Futaki S. Membrane-permeable arginine-rich peptides and the translocation mechanisms. Adv. Drug Deliv. Rev. 2005;57:547–558. doi: 10.1016/j.addr.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 20.Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr J. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proc. Natl. Acad. Sci. U.S.A. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Putnam D, Zelikin AN, Izumrudov VA, Langer R. Polyhistidine-PEG: DNA nanocomposites for gene delivery. Biomaterials. 2003;24:4425–4433. doi: 10.1016/s0142-9612(03)00341-7. [DOI] [PubMed] [Google Scholar]
- 22.Cho YW, Kim JD, Park K. Polycation gene delivery systems: escape from endosomes to cytosol. J. Pharm. Pharmacol. 2003;55:721–734. doi: 10.1211/002235703765951311. [DOI] [PubMed] [Google Scholar]
- 23.Ou M, Wang X-L, Xu R, Chang C-W, Bull DA, Kim SW. Novel Biodegradable poly(disulfide amine)s for gene delivery with high efficiency and low cytotoxicity. Bioconjugate Chem. 2008;19:626–633. doi: 10.1021/bc700397x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin C, Zhong Z, Lok MC, Jiang X, Hennink WE, Feijen J, Engbersen JFJ. Random and block copolymers of bioreducible poly(amido amine)s with high- and low-basicity amino groups: Study of DNA condensation and buffer capacity on gene transfection. J. Control. Release. 2007;123:67–75. doi: 10.1016/j.jconrel.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 25.Wyatt PJ. Light scattering and the absolute characterization of macromolecules. Anal. Chim. Acta. 1993;272:1–40. [Google Scholar]
- 26.Zauner W, Ogris M, Wagner E. Polylysine-based transfection systems utilizing receptor-mediated delivery. Adv. Drug Deliv. Rev. 1998;30:97–113. doi: 10.1016/s0169-409x(97)00110-5. [DOI] [PubMed] [Google Scholar]
- 27.Fischer D, Li Y, Ahlemeyer B, Krieglstein J, Kissel T. In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis. Biomaterials. 2003;24:1121–1131. doi: 10.1016/s0142-9612(02)00445-3. [DOI] [PubMed] [Google Scholar]
- 28.Kim T-i, Bai CZ, Nam K, Park J-s. Comparison between arginine conjugated PAMAM dendrimers with structural diversity for gene delivery systems. J. Control. Release. 2009;136:132–139. doi: 10.1016/j.jconrel.2009.01.028. [DOI] [PubMed] [Google Scholar]
- 29.Rothbard JB, Jessop TC, Lewis RS, Murray BA, Wender PA. Role of membrane potential and hydrogen bonding in the mechanism of translocation of guanidinium-rich peptides into cells. J. Am. Chem. Soc. 2004;126:9506–9507. doi: 10.1021/ja0482536. [DOI] [PubMed] [Google Scholar]
- 30.Wolfert MA, Seymour LW. Chloroquine and amphipathic peptide helices show synergistic transfection in vitro. Gene Ther. 1998;5:409–414. doi: 10.1038/sj.gt.3300606. [DOI] [PubMed] [Google Scholar]
- 31.Uherek C, Fominaya J, Wels W. A modular DNA carrier protein based on the structure of diphtheria toxin mediates target cell-specific gene delivery. J. Biol. Chem. 1998;273:8835–8841. doi: 10.1074/jbc.273.15.8835. [DOI] [PubMed] [Google Scholar]
- 32.Lim Y-b, Kim S-m, Suh H, Park J-s. Biodegradable, endosome disruptive, and cationic network-type polymer as a highly efficient and nontoxic gene delivery carrier. Bioconjugate Chem. 2002;13:952–957. doi: 10.1021/bc025541n. [DOI] [PubMed] [Google Scholar]
- 33.Cheng J, Zeidan R, Mishra S, Liu A, Pun SH, Kulkarni RP, Jensen GS, Bellocq NC, Davis ME. Structure-function correlation of chloroquine and analogues as transgene expression enhancers in nonviral gene delivery. J. Med. Chem. 2006;49:6522–6531. doi: 10.1021/jm060736s. [DOI] [PubMed] [Google Scholar]
- 34.Rothbard JB, Jessop TC, Wender PA. Adaptive translocation: the role of hydrogen bonding and membrane potential in the uptake of guanidinium-rich transporters into cells. Adv. Drug Deliv. Rev. 2005;57:495–504. doi: 10.1016/j.addr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 35.Kim T-i, Lee M, Kim SW. A guanidinylated bioreducible polymer with high nuclear localization ability for gene delivery systems. Biomaterials. 2010;31:1798–1804. doi: 10.1016/j.biomaterials.2009.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]