Abstract
With the human genome sequence approaching completion, a major challenge is to identify the locations and encoded protein sequences of all human genes. To address this problem we have developed a new gene identification algorithm, GenomeScan, which combines exon–intron and splice signal models with similarity to known protein sequences in an integrated model. Extensive testing shows that GenomeScan can accurately identify the exon–intron structures of genes in finished or draft human genome sequence with a low rate of false-positives. Application of GenomeScan to 2.7 billion bases of human genomic DNA identified at least 20,000–25,000 human genes out of an estimated 30,000–40,000 present in the genome. The results show an accurate and efficient automated approach for identifying genes in higher eukaryotic genomes and provide a first-level annotation of the draft human genome.
A first draft of the human genomic sequence has been completed (International Human Genome Sequencing Consortium 2001; Venter et al. 2001). To make most effective use of these data for evolutionary and functional studies, one must first identify the locations, exon–intron structures, and encoded proteins of the thousands of genes that this sequence contains. For example, human genetic studies relying on polymorphic markers such as SNPs will benefit from knowledge of gene structures in the genomic neighborhood of the polymorphism. Furthermore, microarray studies using human expressed sequence tags (ESTs) require gene structure information to help in identification of putative regulatory regions. In addition, inferences about the probable presence or absence of particular genes or gene families in the human genome depend on reliable gene annotation. Full-length cDNA sequencing is the most definitive way to characterize human gene structure. However, full-length cDNA sequence data are presently available for only 10,000 human genes (Maglott et al. 2000), less than one-third of the total using conservative recent estimates of human gene numbers (Ewing and Green 2000; Roest Crollius et al. 2000). To identify the remaining genes that lack available cDNA sequence will require other methods.
Two classes of computational approaches are commonly used to detect genes in genomic sequences: (1) statistically based ab initio gene-finding algorithms (for reviews, Claverie 1997; Burge and Karlin 1998) such as GENSCAN (Burge and Karlin 1997), HMMGene (Krogh 2000), Fgenes (Salamov and Solovyev 2000), GRAIL (Xu and Uberbacher 1997), and Genie (Reese et al. 2000), which use compositional properties of exons, introns, and other gene features to predict gene locations; (2) local alignment methods such as the BLAST family of programs (Gish and States 1993; Altschul et al. 1997), which detect sequence similarity to known genes, proteins, or ESTs. Each of these approaches has particular strengths and limitations. For example, the ab initio gene-finding program GENSCAN (generally considered to be among the most accurate programs of its type) can predict the precise locations of 70%–80% of coding exons in sequences containing single genes using compositional properties of the genomic sequence alone with only a few percent missed or wrong exons (Burge and Karlin 1997). However, the accuracy of such programs on large genomic sequences containing multiple genes appears to be significantly lower, with a higher rate of apparent false-positive predictions (Dunham et al. 1999). On the other hand, local sequence alignment algorithms such as BLASTX (Gish and States 1993) detect similarities between open reading frames (ORFs) in a genomic sequence and known proteins. BLASTX hits can often indicate the approximate locations of many coding exons in a genomic sequence but cannot identify every exon and do not accurately delineate exon boundaries. In addition to local alignment methods such as BLASTX, two more recently developed algorithms, Procrustes (Gelfand et al. 1996) and GeneWise (Birney and Durbin 2000) use global alignment of a homologous protein to translated ORFs in a genomic sequence for gene prediction. Although these methods can be highly accurate, they predict exactly one gene per genomic sequence, require close similarity to identify complete genes (Guigó et al. 2000), and are so computationally intensive as to be impractical for many genomic-scale applications.
Therefore, we sought to develop a method that would effectively combine the distinct types of evidence used by these two classes of methods, sequence similarity and exon–intron composition, into one integrated computer algorithm. In devising such an algorithm, we had three principal goals: (1) to build on the strengths of BLAST and GENSCAN and to incorporate aspects of the probabilistic models underlying these two methods into a coherent whole; (2) to develop a method efficient and reliable enough to be run without human supervision on an entire vertebrate genome; (3) to focus on predicting gene structure as accurately as possible in the typical case, when one or more homologous but not identical proteins are available. These goals have largely been achieved in the GenomeScan algorithm described below. Applying this method to the human genome gives a large set of reliably inferred exon–intron structures of genes with sequence similarity to known proteins from human or other organisms and provides a first layer of annotation on the draft human genome.
RESULTS
GenomeScan Model and Algorithm
The basic idea of our approach is to combine sequence similarity information, which can indicate the rough locations of many coding exons, with modeling of exon–intron and splice signal composition to aid in identification of additional exons and for determination of precise exon–intron boundaries. Although this general idea has been explored by other investigators (Birney and Durbin 2000; Reese et al. 2000), the approach introduced here is different and in some ways more general than those used previously and gives superior results across a broad range of conditions, as discussed below. Our new method derives from the probabilistic model of the exon–intron structure and compositional features of human genes used by GENSCAN, which has been described in detail previously (Burge 1997; Burge and Karlin 1997). This is a semi-Markov or generalized Hidden Markov Model (HMM; Rabiner 1989; Kulp et al. 1996) in which components of a gene such as exons, introns, 5′ and 3′ untranslated regions (UTRs), are modeled as abstract states corresponding to variably sized stretches of DNA sequence. The HMM architecture allows enforcement of the natural grammatical order of a gene—promoter precedes 5′ UTR precedes initial coding exon, etc. Each state (e.g., internal coding exon) has an associated length and is thought of as generating a DNA sequence of this length according to a probabilistic model of the sequence composition of that state (e.g., a model of coding region composition). In this model, each possible gene structure or set of gene structures that may be present in the sequence corresponds to an ordered list of states with associated lengths and is referred to as a parse of the sequence. For a given genomic sequence, the gene structure predicted by GENSCAN corresponds to the parse that has maximum probability under its HMM model of gene structure/sequence composition.
The crucial difference between GENSCAN and GenomeScan is that in the latter algorithm the predicted gene structure corresponds to the parse that has maximum probability conditional on available similarity information. Here, the similarity information will be the results of a BLASTX search of the input genomic sequence against an appropriate protein database, but the algorithm could be adapted to use other sorts of information such as the results of comparisons of homologous genomic regions (Batzoglou et al. 2000; Roest Crollius et al. 2000). The first step in our method is to convert the information present in a set of BLASTX hits into a corresponding set of probabilistic statements about the likelihood that coding exons occur at particular places in the query genomic sequence. Each BLASTX hit alters the probabilities of the various parses of the genomic sequence in the GenomeScan model, increasing the likelihood of parses that are consistent with the BLASTX information and reducing the likelihood of those that are not, as described in Methods. Because not every BLAST hit represents true homology between the query genomic sequence and the subject protein, the possibility that the hit may be artifactual (e.g., a BLAST false-positive or pseudogene) is explicitly considered and assigned an appropriate probability in the GenomeScan model. Therefore, the final prediction is generally compatible with most, but not necessarily all, BLASTX hits that have been provided.
Three modifications of this basic framework have been made that improve the accuracy significantly: (1) BLASTX hits that fall very near the N- or C-terminus of a subject protein are used to aid in identification of initiation or termination codons, respectively; (2) pairs of BLASTX hits which are adjacent in the same subject protein and have proper separation in the query genomic sequence (≥60 bases, the minimum length of a human intron) are used to identify putative intronic regions; and (3) multiple overlapping BLASTX hits, which generally provide redundant information, are pruned in a preprocessing step, keeping only the strongest (lowest P-value) hit in each cluster of overlapping hits. (These and other aspects of the algorithm are described in more detail in Methods.) By default, the GenomeScan program prints only those predicted genes that have one or more BLASTX hits overlapping predicted exons, so all GenomeScan-predicted genes have at least modest similarity to a known protein as assessed by BLAST. Because of this, it is perhaps more accurate to refer to the output of this program as gene inferences rather than gene predictions (we used both).
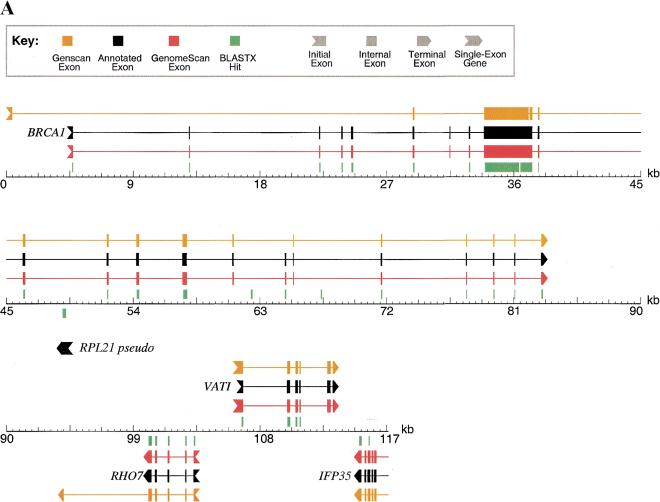
Sample GenomeScan Predictions in Human BAC-Sized Genomic Regions
As part of our testing, GenomeScan was run using GenomeScript (see below) on several large, well-annotated human genomic sequences using only available mouse proteins from GenPept Release 118 (June 2000) to aid in predictions. The results of GENSCAN, BLASTX, and GenomeScan were then compared to the annotated gene structures to identify strengths and weaknesses of these methods for exon and gene identification—two representative examples are shown in Figure 1. The first example (Fig. 1A) shows a 117-kbp genomic contig from human chromosome 17q21 containing four annotated genes and one pseudogene (Smith et al. 1996). In this example, GENSCAN predicts the exon–intron structures of three of the four genes quite accurately (RHO7, VATI, IFP35) but has great difficulty with the fourth gene (BRCA1), missing seven of the first eight exons. As homologs of most of these genes have been sequenced in mouse, a majority of the exons give BLASTX hits using the two-stage BLAST protocol that is part of GenomeScript (see below), showing the power of this simple method when mammalian homologs are available. Nevertheless, a total of eight annotated exons in this sequence do not have a corresponding BLASTX hit even at the extremely reduced significance threshold used in this procedure and there are three apparent false-positive BLASTX hits (at ∼63 kbp, ∼67 kbp, and overlapping the RPL21 pseudogene at ∼49 kbp). Comparing the GenomeScan output to that of BLASTX and GENSCAN, in many cases it is clear that BLASTX is helping GenomeScan to identify exons missed by GENSCAN in the expected way (e.g., the first five exons of BRCA1). However, because of the probabilistic way in which similarity information is treated in the GenomeScan algorithm, not all of the predictions agree completely with BLASTX. For example, the program does not predict exons overlapping any of the three false-positive BLASTX hits in this sequence. Furthermore, in this sequence, GenomeScan correctly predicts all eight of the exons that lack BLASTX hits, one of which was also missed by GENSCAN (exon 7 of BRCA1, at ∼31.5 kbp). This example shows clearly that GenomeScan is not simply the additive combination of GENSCAN and BLASTX but effectively integrates these two imperfect sources of information and occasionally even makes simple inferences on its own (e.g., using the fact that exon 8 of BRCA1 is incompatible in terms of intron phase with exon 6 to aid in identification of exon 7, see Fig. 1A).
Figure 1.
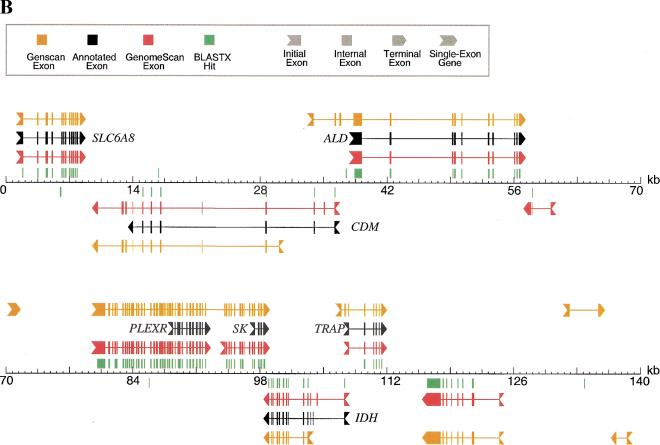
Examples of GenomeScan predictions. GenomeScan was run with GenomeScript, using similarity to available mouse proteins from GenPept Release 118 (June 2000). Two examples are shown. Exons and genes on the forward strand are shown above the sequence line; reverse strand exons and genes are shown below the sequence line. BLASTX hits with P < 0.05 are shown as green blocks above or below the sequence line, according to the reading frame/strand indicated by BLAST. (A) GenBank locus HUMBRCA1 (accession no. L78833). (B) GenBank locus HSU52111 (accession no. U52111). Only the first 140 kbp (of 153 kbp) of the sequence is shown for clarity. The extra predicted exons upstream of PLEXR and SK and the extra predicted gene at ∼118 kb are supported by several human ESTs (accession nos. AW663636, AA514687, AW071821, and others).
A genomic region from human chromosome Xq28 containing seven annotated genes (Brenner et al. 1997) is illustrated in Figure 1B. In this example, GENSCAN predicts gene boundaries very poorly and produces three apparent false-positive gene predictions (at ∼71 kbp, ∼133 kbp, and ∼137 kbp). As in the previous example, mouse homologs of most of the genes present are available and the two-step BLAST procedure is able to identify the majority of exons, but there are several apparent false-positive BLASTX hits. GenomeScan is again able to identify the gene structures present far more accurately than either GENSCAN or BLASTX and correctly ignores all but one of the incorrect BLASTX hits, producing only one apparent false positive predicted gene (at ∼57 kbp). GenomeScan also identifies putative additional exons of two genes (PLEXR and SK) and predicts a novel gene at ∼119 kbp which is supported by BLASTX (and GENSCAN) but not annotated in the GenBank record. These extra predicted exons/genes are supported by other evidence (see Fig. 1 legend).
In applications, the GenomeScan algorithm is integrated with database searches using a procedure called GenomeScript, which is described in Methods. In brief, this script does the following to an input genomic sequence: (1) mask repetitive elements; (2) identify peptides with significant similarity to regions of the genomic sequence using BLASTX and/or BLASTP; (3) compare all ORFs in the masked genomic sequence to these peptides using a more sensitive BLASTX search; (4) run GenomeScan on the masked genomic sequence using the BLASTX results as input.
Comparison of Gene Identification Algorithms
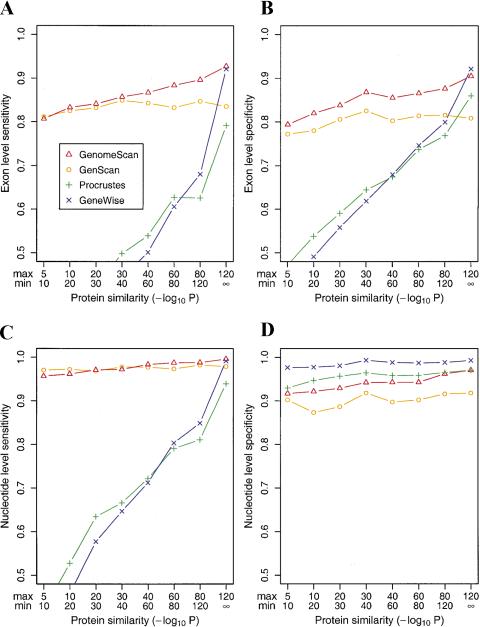
The accuracy of GenomeScan was tested by running the program on sets of genomic sequences with known gene locations, using proteins with various levels of similarity as input and comparing the predicted genes to the known genes in these sequences. Accuracy was measured primarily in terms of the fraction of known exons and genes identified (sensitivity) and in terms of the proportion of predicted exons that correspond to known exons/genes (specificity). GenomeScan was first run on the SingleGene dataset (see Methods), consisting of 175 human genomic sequences each containing a single gene that was used in the testing of other similarity-based gene finding programs by Guigó et al. (2000). The results (Fig. 2) show a steady increase in accuracy as similarity to the subject protein increases, as expected. When only a very weakly similar protein is available (BLASTP P-value between 10−5 and 10−10), GenomeScan has only a slight advantage over GENSCAN, predicting ∼80% of annotated exons correctly in this set, with similar levels of specificity. In this dataset, the accuracy of GENSCAN is somewhat higher than for the gene finders HMMGene (Krogh 2000) and GRAIL (Xu and Uberbacher 1996) by most measures (accuracy statistics are listed in the Fig. 2 legend). The gap between GENSCAN and GenomeScan increases steadily with increasing similarity, and >90% of exons are exactly predicted (with comparable specificity) when a very strongly similar protein is available (BLASTP P-value <10−120). Other measures of sequence similarity besides BLAST P-value, such as bit score and percent identity, give qualitatively similar results (data not shown). Importantly, the accuracy of GenomeScan is significantly higher than either GENSCAN or the similarity-based gene finders Procrustes and GeneWise, which in turn are generally more accurate than BLASTX itself (Guigó et al. 2000), across a broad range of similarity levels. Only when a very strongly similar protein is available (P<10−120) does another method (GeneWise) achieve comparable exon-level accuracy. Nucleotide level sensitivity (fraction of coding nucleotides predicted correctly) in this dataset is qualitatively similar (Fig. 2C) to exon level sensitivity (Fig. 2A). However, nucleotide level specificity (fraction of predicted coding nucleotides that are truly coding) is much less variable across similarity levels and between methods (Fig. 2D), with GeneWise performing consistently slightly better than Procrustes or GenomeScan. The discrepancy between the nucleotide and exon-level specificity values observed for Procrustes and GeneWise appears to result from the inherent conservative bias of these methods and their tendency to end predicted exons close to the end of the aligned region, irrespective of the locations of splice sites or initiation/termination signals.
Figure 2.
Exon- and nucleotide-level accuracy of similarity-based gene-prediction programs as a function of protein similarity. (A) Exon-level sensitivity (ESn: percent of exons predicted exactly) and (B) exon-level specificity (ESp: percent of predicted exons exactly correct) were calculated for subsets of the SingleGene dataset and grouped according to the level of BLASTP similarity (in the context of a database search) between the encoded protein and the protein used in the prediction for GenomeScan, Procrustes, and GeneWise as described by Guigó et al. 2000). The definitions of the subsets and number of genes per subset were as follows: 10−5 > P >10−10 (90); 10−10 > P > 10−20 (103); 10−20 > P >10−30 (102); 10−30 > P > 10−40 (97); 10−40 > P >10−60 (114); 10−60 > P > 10−80 (97); 10−80 > P > 10−120 (97); and P < 10−120 (72). For example, 114 of the 175 sequences in the SingleGene dataset had a homolog with BLAST P-value in the range 10−60< P < 10−40. For sequences in this subset, GenomeScan was run using the results of a BLASTX run of the genomic sequence against the top hit in the nonredundant protein database that had sequence similarity in the desired range (10−40 > P > 10−60). GeneWise and Procrustes data, run using the same peptides as input, are from Guigó et al. (2000). (C) Nucleotide-level sensitivity (NSn: percent of coding nucleotides predicted correctly) and (D) nucleotide-level specificity (NSp: percent of predicted coding nucleotides that are correct). Accuracy statistics on the SingleGene dataset as a whole for the ab initio gene-prediction methods GENSCAN, HMMGene 1.1, and GRAIL 3.1, respectively, were as follows: ESn (0.79, 0.75, 0.47); ESp (0.77, 0.68, 0.61); NSn (0.93, 0.86, 0.68): NSp (0.91, 0.74, 0.94).
An advantage of the SingleGene dataset is that accuracy statistics have been meticulously calculated for a variety of similarity-based gene-finding algorithms, allowing direct comparison between methods. However, the sequences in this dataset are relatively small finished genomic sequences containing single genes, which are not representative of the human genome as a whole. In its present state of sequencing, the publicly available human genome is represented both by very long finished sequences and smaller rough draft contigs often containing no genes or partial genes. Therefore, the accuracy measured in the SingleGene set is likely to represent upper limits rather than typical values for these methods.
Representative Sets of Finished and Draft Human Genome Sequences
To obtain measures of the accuracy of GenomeScan that would be more representative of its probable performance on the bulk of available human genomic sequences, we constructed two new datasets, DraftGene and FinishGene, as described in Methods. These two datasets represent the same 206 human genes sequenced in draft and finished form, respectively. Properties of the SingleGene, DraftGene, and FinishGene datasets are summarized in Table 1, together with corresponding data for the September 2000 freeze of the GoldenPath assembled human genome sequence (http://genome.ucsc.edu), which represents an assembly of finished and draft human genomic sequences. Comparing the data on sequence size, gene density, and C + G-percent content between the DraftGene and GoldenPath datasets reveals some biases in the DraftGene set. Probably because of the gene-centric way it was constructed, the DraftGene set has somewhat higher gene density and C + G-percent content than the GoldenPath and is probably also biased toward shorter genes because of the requirement that the whole gene fit into a single BAC clone (mostly <200 kbp). Nevertheless, these data show that the DraftGene and FinishGene sets are much more similar to the GoldenPath in all respects than is the small, gene-dense SingleGene dataset.
Table 1.
Summary of Sequence Sets Used in This Study
| Variable | Dataset | |||
|---|---|---|---|---|
| SingleGene | FinishGene | DraftGene | GoldenPath | |
| No. of sequences | 175 | 194 | 1038 | 156500 |
| No. of complete genes (partial) | 175 | 206 | 116 (256) | — |
| Mean sequence lengths (kbp) | 7 | 96 | 14 | 17 |
| No. of genes/Mbp (estimated) | 144 | 17 | 14 | (10) |
| No. of exons/complete gene (partial) | 5.0 | 7.0 | 5.7 (3.0) | — |
| Mean C + G% | 49.6 | 45.1 | 45.2 | 39.9 |
| No. of aa/complete protein (partial) | 324 | 404 | 321 (170) | — |
Datasets are described in Methods. Some genes in the DraftGene set are represented by multiple partial genes in different draft contigs, data for these genes are listed in parentheses. Gene density in the GoldenPath set assumes 30,000 human genes in a 3000-Mbp genome.
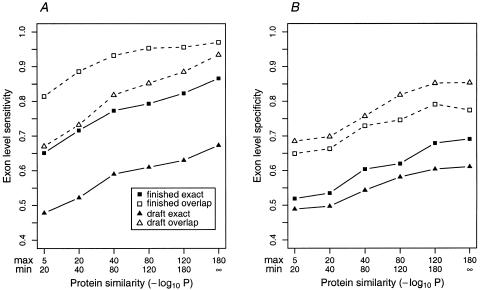
Prediction accuracy was measured by running GenomeScan on both sets using proteins with various levels of similarity as input and comparing the predicted gene structures to the cDNA-derived annotations. Results are shown in Figure 3. The results for GENSCAN + BLASTP were as follows: GENSCAN-predicted genes that have a P<10−5 BLASTP hit to the September 2000 nonredundant protein database (GenPept + PDB + SwissProt + PIR) are listed in the Fig. 3 legend. GENSCAN + BLASTP represents a possible alternative gene-annotation strategy that has not been extensively tested previously. As for the SingleGene set, both sensitivity and specificity increase steadily as a function of protein similarity and GenomeScan has a significant advantage over GENSCAN when a protein with at least moderate similarity is available. Most significantly, the specificity (proportion of predicted exons that are correct) is far higher for GenomeScan than for GENSCAN + BLASTP in these datasets. As expected, accuracy is a bit lower overall in draft sequences than finished sequences, but this is reflected primarily in the prediction of exact exon boundaries (Fig. 3, solid squares and triangles). In terms of overlap between predicted and annotated exons (unfilled squares and triangles) as opposed to exact exon-boundary prediction, the accuracy of GenomeScan is similar in finished and draft sequences, with slightly lower sensitivity but higher specificity in draft sequences. Both of these differences are attributable to the fragmentation of genes into multiple contigs that results from draft sequencing: Small contigs containing only one or a few exons are more likely to be missed, whereas BLASTX searches against smaller genomic regions result in fewer false-positive hits. The specificity values listed are likely to represent lower bounds for these methods since the datasets consist of long human genomic sequences that almost certainly contain additional exons/genes not yet sequenced at the cDNA level and predictions matching these exons/genes are counted as wrong. Nevertheless, when a protein with at least moderate similarity is available (P < 10−40), >80% of annotated exons are overlapped by GenomeScan-predicted exons and >70% of predicted exons overlap an annotated exon, even in draft sequences.
Figure 3.
Exon-level accuracy of GenomeScan as a function of protein similarity in draft and finished sequences. GenomeScan was run on subsets of the FinishGene and DraftGene datasets, grouped according to the level of similarity to the nearest proteins used in the predictions. (A) Exon-level sensitivity (percent of annotated exons predicted exactly) is displayed with solid squares/triangles and solid lines; overlap sensitivity (percent of annotated exons overlapped by a predicted exon) by open squares/triangles and dashed lines. (B) Exon-level specificity (percent of predicted exons exactly correct) is displayed with solid squares/triangles and solid lines. Overlap specificity (percent of predicted exons overlapped by an annotated exon) is displayed by open squares/triangles and broken lines. For comparison, overlap exon-level sensitivity and specificity values for GENSCAN + BLASTP (GENSCAN predictions that have a BLASTP hit with P < 10−5 against the nonredundant protein database) were 0.90 and 0.48, respectively, in the FinishGene dataset and 0.87 and 0.47, respectively, in the DraftGene dataset.
Accuracy in these datasets was also measured at the gene level (Table 2). Because GenomeScan, like GENSCAN, is able to predict partial, as well as complete genes, the fragmentation of DraftGene genes into multiple contigs presents no fundamental obstacle to these algorithms. Gene level accuracy was measured by counting the proportion of the exons of a gene that were covered (overlapped) by GenomeScan predicted exons: A gene is completely covered if all of its exons are covered by predicted exons, partially covered if some but not all exons are covered, or missed if no exon was covered. The results (Table 2) show that when a protein with at least moderate similarity or stronger (P < 10−40) is available, only a negligible fraction of genes are missed (≤1%) in both finished and draft sequences and >70% of genes are completely covered in finished sequences, compared to ∼50% in draft data. Only when the level of similarity is very weak (P > 10−20) does the algorithm miss a significant proportion of genes: ∼1 in 10 in finished sequences and ∼1 in 7 in draft data. Significantly, the number of extra predicted genes (those not overlapping annotated genes) remains relatively low, at ∼5%–10% of the total number of predicted genes in both finished and draft sequences, at all levels of protein similarity. These data suggest a relatively low rate of false-positives for GenomeScan, especially considering that some fraction of these predictions likely represent additional unannotated genes. Another important property of a gene-prediction program is the ratio between the number of complete and partial genes predicted and the number of real genes present. Table 2 shows that this ratio is close to 1:1 in finished data at all levels of similarity but varies between 1.1:1 and 1.6:1 in draft data, with higher ratios occurring at higher levels of protein similarity. This reflects the intrinsic fragmentation of the genes into multiple contigs in draft sequences (1.8 contigs per gene on average) in the DraftGene set and the higher proportion of exon-containing contigs predicted correctly by GenomeScan at higher levels of protein similarity.
Table 2.
Gene Level Accuracy of GenomeScan as a Function of Protein Similarity in DraftGene and FinishGene Datasets
| Variable | Similarity category/dataset | |||||
|---|---|---|---|---|---|---|
| 10−5 > P > 10−20 | 10−40 > P > 10−80 | 10−120 > P > 10−180 | ||||
| Draft | Finish | Draft | Finish | Draft | Finish | |
| No. of genes in dataset | 174 | 174 | 151 | 151 | 93 | 93 |
| % of fragmented genes | 42 | 0 | 43 | 0 | 55 | 0 |
| No. of predicted genes* | 186 | 172 | 205 | 159 | 152 | 104 |
| Genes completely covered (%) | 38 | 58 | 48 | 71 | 57 | 73 |
| Genes partially covered (%) | 49 | 32 | 51 | 28 | 42 | 27 |
| Genes missed (%) | 13 | 10 | 1 | 1 | 1 | 0 |
| No. of “extra” predicted genes* | 18 | 14 | 19 | 10 | 8 | 11 |
Sequences were grouped according to the level of similarity between the encoded protein and the available database proteins used in the predictions as described in the legend to Fig. 3. All known genes in the FinishGene set are complete (all coding exons present in a single sequence). Some genes in the DraftGene set represented by multiple “partial genes” in different draft contigs; these are listed as fragmented genes. Known genes were classified as completely covered if all exons were covered by GenomeScan predicted exons; partially covered, if some exons (but not all) were covered by GenomeScan predicted exons; and missed, if no exon was covered by a GenomeScan-predicted exon. GenomeScan predicted genes which did not overlap any known gene are listed as “extra” predicted genes.
Includes predicted partial genes as well as complete genes.
To explore how well the accuracy results obtained on the FinishGene dataset would extrapolate to larger finished regions of the human genome, we compared the GenomeScan predicted genes in the May 19, 2000, version of the finished human chromosome 22 (Chr22) sequence (Dunham et al. 1999) with the annotation provided by the Sanger Centre. The results of this comparison are summarized in Table 3. Notably, >90% of the exons in known and related genes were covered by GenomeScan-predicted exons, confirming the high exon level sensitivity observed in Figure 2A. At the gene level, 95% of known genes and 88% of related genes were covered, roughly comparable to the gene level accuracy reported in Table 2. Table 3 also shows a low rate of gene splitting by GenomeScan with <10% of genes overlapping multiple predicted genes in all categories. In addition, <10% of known or related genes are predicted as parts of chimeric genes by GenomeScan, compared to 27% by GENSCAN (data not shown). This shows a substantial improvement by GenomeScan in terms of defining gene boundaries, a known weakness of GENSCAN. Overall, approximately two-thirds of the 648 genes predicted by GenomeScan on Chr22 overlapped known or related genes. An additional 12% matched annotated predicted genes or immunoglobulin gene segments (listed as “Other” in Table 3), whereas 11% matched annotated pseudogenes and 11% did not match any annotated gene (“extra predicted genes”). Because all of these extra genes have at least moderate BLAST similarity to known proteins, most are likely to represent additional genes or pseudogenes not yet annotated by the Chr22 team. Therefore, we conclude that the rate of false-positive GenomeScan predictions in Chr22 is at the most 11% (probably far lower), and that an additional ∼11% of predictions represent probable pseudogenes. The rate of false-positive predictions including pseudogenes, therefore, is between 11% and 22%. From Figure 3 and Table 2, the specificity of GenomeScan in draft sequence is comparable to that in finished sequence, suggesting that similar rates of false-positives can be extrapolated to the GoldenPath human genome sequence, which comprises an assembly of all publicly available finished and draft sequences.
Table 3.
Comparison of GenomeScan-Predicted Genes on Human Chromosome 22 with Annotated Genes
| Variable | Category of gene | |||
|---|---|---|---|---|
| Known | Related | Pseudo | Other | |
| Total no. of genes annotated in Chr22 | 307 | 120 | 132 | 245 |
| Percent of annotated genes in category covered by GenomeScan-predicted genes | 95 | 88 | 67 | 67 |
| Percent of annotated genes in category overlapping multiple GenomeScan-predicted genes | 9 | 6 | 1 | 1 |
| Percent of annotated exons in category covered by GenomeScan-predicted exons | 94 | 92 | 74 | 74 |
| Percent of all GenomeScan-predicted genes which match annotated genes in category | 51 | 15 | 11 | 12 |
Genes were predicted with GenomeScan in the masked May 19, 2000 version of the Chr22 sequence, and compared to the Sanger Centre annotation (http://www.sanger.ac.uk/HGP/Chr22/cwa_archive/Release_2_19-05-2000.shtml). Known genes, related genes, predicted genes, immunoglobulin “gene segments”, and pseudogenes are distinguished in the annotation; the “Other” category includes annotated predicted genes and gene segments. GenomeScan predicted a total of 648 genes in the Chr22 sequence, of which 11% did not overlap any annotated gene; thus, the entries in the last row of the table total 89% rather than 100%.
Application to the Human Genome
In a large-scale computational analysis, genes were identified with GenomeScan in the entire September 2000 GoldenPath human genome sequence as described in Methods. A total of 38,647 complete and partial human genes were predicted in this dataset using similarity to proteins from the nonredundant protein database (September 2000 version). In light of the results obtained above in the DraftGene dataset (Table 2), which is similar in many respects to the GoldenPath (Table 1), we estimate that each gene detected by GenomeScan in the GoldenPath sequence is likely to be represented by ∼1.4–1.5 predicted (complete and partial) genes on average (C.B. Burge and R.-F. Yeh, data not shown). Therefore, the total number of distinct genes represented by this set is ∼38,647/1.5 = ∼26,000 to 38,647/1.4 = ∼28,000. Correcting for the estimated rate of false-positives and pseudogenes derived from Chr 22 (11%–22%), this set represents 20,000–25,000 distinct human genes.
DISCUSSION
The process of identifying genes in higher eukaryotic genomes is complicated by several factors, including complex gene organization, the presence of large numbers of introns and repetitive elements, and the sheer size of the genomic sequence. These issues are particularly acute for the human genome, which totals over 3 billion base pairs and contains far more intronic and repetitive sequences than any previously sequenced eukaryotic genome. To aid in the annotation of gene locations in the human genome, we have developed a novel algorithm, GenomeScan, which combines sequence similarity information with models of exon–intron and splice signal composition to identify genes. Systematic tests of the accuracy of GenomeScan showed that it is more accurate than existing ab initio and similarity-based algorithms across a broad range of similarity levels (Fig. 2) and is able to detect all but a few percent of genes in both draft and finished genomic sequence, provided only that a moderately similar homologous protein is available (Table 2). Approximately 80% of exons are identified in draft sequence and ∼90% in finished sequence at moderate levels of protein sequence similarity (Fig. 3) with relatively low rates of false-positive predictions.
Application of this method to the available human genome sequence produced a set of 38,647 putative complete and partial genes, which we designate the GenomeScan2000 gene set. Correcting for the estimated rate of false-positives and pseudogenes (11%–22%; Table 3), between 78% and 89% of predictions in this set are likely to represent functional human genes. To investigate the properties of this gene set in more detail, the coding regions of all predicted complete and partial genes were compared to available cDNA and EST databases using BLASTN (Altschul et al. 1997). The number of predicted complete genes with >2 exons, partial genes, and all genes (including also one- and two-exon genes) that matched complete cDNAs from the RefSeq database (September 2000 version) are listed in Table 4. The large number of predicted partial genes reflects the fragmentation of genes in the underlying GoldenPath genomic sequence rather than a property of GenomeScan per se, as only a few percent of partial genes are predicted in finished sequences (data not shown). Therefore, this fraction will decline as sequencing of the human genome is completed. Of all predicted complete and partial genes, 41.5% had a BLASTN hit with ≥98% identity over 100 bp or more to a RefSeq cDNA. Predicted gene structures that differ from corresponding RefSeq cDNA alignments can be used to assess the accuracy of exon–intron prediction and may suggest alternatively spliced isoforms. An additional 32.5% of predicted genes had a BLASTN hit with ≥97% identity over 100 bp to a publicly available human EST sequence (dbEST September 2000). These GenomeScan genes provide a link between ESTs and corresponding putative peptide sequences that can aid in assigning function to genes represented only by fragmentary EST data, including many ESTs on current human gene microarrays (e.g., Iyer et al. 1999). The remaining 26% of predicted genes did not match any RefSeq cDNA or human EST using these criteria. These predicted genes likely contain a higher proportion of pseudogenes than the other subsets, but they also probably contain the highest proportion of interesting novel genes that perhaps are expressed at too low a level or in too restricted a set of tissues to be efficiently sampled by EST sequencing. It has been estimated that ∼20% of human genes are not represented in current EST databases (Ewing and Green 2000).
Table 4.
Summary of GenomeScan-predicted Genes and Partial Genes in the Human Genome
| Similarity category | Type of predicted gene | ||||||
|---|---|---|---|---|---|---|---|
| Complete genes (>2 exons) | Partial genes | All genes (partial + complete) | |||||
| No. of genes | No. of exons/gene | No. of aa/gene | No. of genes | No. of exons/gene | No. of genes | % of all predicted genes | |
| Known (cDNA) | 5698 | 9.6 | 496 | 8901 | 4.9 | 16040 | 41.5 |
| Protein + EST | 4502 | 8.8 | 510 | 6537 | 5.5 | 12546 | 32.5 |
| Proteins only | 2767 | 5.2 | 303 | 4600 | 3.1 | 10061 | 26.0 |
| All | 12967 | 8.4 | 460 | 20038 | 4.7 | 38647 | 100.0 |
Genes were predicted in the September 2000 GoldenPath human genome sequence as described in Methods. Predicted coding sequences (CDS) were first compared to cDNAs in the RefSeq cDNA database (September 2000) using BLASTN; those which had a hit at least 100 bp long with at least 98% identity are listed as “known”. The remaining predicted coding sequences were searched against dbEST (September 2000 release) using BLASTN; those which had a hit at least 100 bp long with at least 97% identity are listed as “Protein + EST”. All other predicted genes are categorized as “Protein only” because all GenomeScan-predicted genes have at least modest similarity to a known protein. Statistics are listed separately for predicted partial genes and predicted complete genes with at least three exons; the category “all genes” includes these two groups as well as predicted 1- and 2-exon genes.
The average sizes of the encoded proteins and number of exons per gene for GenomeScan predicted genes are also listed in Table 4. These data show that predicted complete genes that have EST or cDNA hits are comparable in size to the average human gene—at least 450 amino acids, distributed across ∼9 exons (International Human Genome Sequencing Consortium 2001)—and that predicted partial genes are on average about half the size of a typical gene. Of predicted complete genes with >2 exons, ∼76% had a hit to a human EST (as defined above), consistent with the estimated coverage of human genes by available ESTs (80%; Ewing and Green 2000). Interestingly, the fraction of predicted partial genes that had hits to human ESTs (71%) was close to that seen for complete genes, suggesting that this subset contains a comparably high fraction of functional genes. However, only ∼47% of predicted one- and two-exon genes had EST hits, suggesting that this subset of predictions may be enriched for nonexpressed pseudogenes or other false-positives. Comparing across human chromosomes, the fraction of predicted complete genes with >2 exons that had EST hits was roughly constant, between 62% and 84% (data not shown), with one striking exception: Only 27% of predicted multi-exon genes on the Y chromosome had EST hits. This suggests that either (1) most Y genes are very poorly expressed or expressed only in tissues not well-sampled by EST databases, or (2) that the vast majority of predicted genes on the Y chromosome represent pseudogenes. The latter explanation is consistent with previous studies indicating that the Y chromosome contains a higher than usual proportion of pseudogenes (Lahn and Page 1999).
Based on comparisons of the human genome to one-third of the genome of the pufferfish Tetraodon nigroviridis, the human genome was estimated to contain ∼28,000–34,000 genes (Roest Crollius et al. 2000). These estimates, based on genomic sequence conservation, are comparable to some estimates based on EST sequences (Ewing and Green 2000) but much lower than others (Liang et al. 2000), underscoring the difficulty of EST clustering and of accounting for artifacts such as unprocessed mRNA or contaminating genomic DNA in cDNA libraries. Updated human–pufferfish comparisons have refined the human gene number estimate to ∼30,000 (H. Roest Crollius et al., in prep.) and other recent analyses have placed human gene number estimates in the 30,000–40,000 range (International Human Genome Consortium 2001; Venter et al. 2001). Based on our estimates that the GenomeScan2000 set represents 20,000–25,000 expressed genes, we conclude that we have identified approximately two-thirds of all human genes. Consistent with this estimate, 65% of Exofish ecores (genomic regions conserved between human and Tetraodon comprising almost exclusively coding DNA) fall inside GenomeScan-predicted exons (H. Roest Crollius et al., in prep.). This fraction is significantly higher in finished sequences (data not shown), suggesting that finishing of the draft human genome sequence will enable a significantly larger fraction of human genes to be identified using automated approaches. Ecore analysis also provides a way to assess and compare the completeness of different human gene annotations. For example, 46% of ecores fall within genes annotated by the Ensembl project (Hubbard and Birney 2000; http://www.ensembl.org), implying that the GenomeScan2000 gene set contains a significantly larger number of human genes. Some of the extra ecores that fall outside of genes annotated in the current GenomeScan and Ensembl datasets may represent additional exons of genes identified by these methods. Others will undoubtedly represent genes missed by these approaches, and some might represent false-positives of the Exofish approach.
How to identify the remaining human genes? One promising general approach is to test for expression of those GENSCAN-predicted genes that fall outside of the boundaries of genes annotated by other methods (e.g., GenomeScan, Ensembl) using sensitive experimental techniques. Recently, a microarray-based method has been applied to identify expressed genes based on coexpression of sets of adjacent exons predicted by GENSCAN (Shoemaker et al. 2001). Using this approach, evidence was found supporting the expression of the vast majority of genes predicted by GENSCAN on Chr22, including more than half of the predictions that lacked similarity to any known gene, protein, or EST as of the time of completion of the Chr22 sequence. A major strength of the microarray method is that it can be scaled up to test hundreds of thousands of predicted exons (Shoemaker et al. 2001). However, such microarrays cannot directly determine whether two exons form part of the same transcript, relying on correlation of expression patterns to make such inferences (Burge 2001). An alternative approach that can directly detect splicing of exons in the same message is to use RT-PCR with radio-labeled primers targeted to pairs of adjacent predicted exons, followed by sequencing of the amplified product. Applying this strategy to predicted novel genes on human Chr22 suggests that a significant number of human genes not similar to known proteins or ESTs can be identified with this approach (C.B. Burge et al., in prep.).
Both the microarray and RT-PCR data confirm the presence of a significant fraction of human genes that are not similar to any previously identified in lower organisms and will very likely lead to increases in estimates of human gene numbers. Exon locations identified by these approaches or by alignment of available EST or cDNA sequences to the human genome could potentially be integrated into the GenomeScan algorithm. EST sequences in particular represent a rich source of information about gene expression because millions are available in public databases. However, a small but significant fraction of EST sequences appear to derive from unprocessed RNA or contaminating genomic DNA (Wolfsberg and Landsman 1997), causing problems for automated gene prediction methods. For example, Krogh (2000) found that the incorporation of EST matches into the sophisticated HMMGene algorithm resulted in lower overall accuracy, with increases in sensitivity more than offset by decreases in specificity, and we have observed similar results in our preliminary efforts to incorporate EST similarity into GenomeScan (data not shown). For this reason, we have chosen not to include EST information in constructing the GenomeScan2000 gene set. However, we expect that improved methods for filtering and assembly of EST sequences should address these problems in the near future.
Alternative splicing is increasingly recognized as an important and widespread form of gene regulation that is thought to affect more than half of human genes (International Human Genome Sequencing Consortium 2001). However, the mechanisms underlying alternative splicing are not well understood and computational analysis of this phenomenon will require more specialized tools than those described here. When GenomeScan is run on a genomic sequence containing a known alternatively spliced gene, the predicted gene generally includes most or all of the alternatively spliced exons, often corresponding to the longest ‘possible’ alternative product (‘possible’ in quotes because in some cases the exons are mutually exclusive, so this longest product is never observed). For example, running GenomeScan on the massively alternatively spliced Drosophila Dscam locus (GenBank accession no. AF260530; Schmucker et al. 2000) using BLASTX results against the human DSCAM protein (accession no. AAF27525) produces a long predicted gene that includes many of the 95 known alternative exons present in this locus. Similar results are obtained for other known alternatively spliced genes (data not shown). In terms of annotating novel putative alternatively spliced genes, GenomeScan predictions, therefore, may be most useful in indicating a plausible set of exons, but detailed prediction of exactly which spliced isoforms occur in the cell is beyond the scope of this method. Other methods based on EST alignment may help in this regard. Full-length cDNA sequencing still provides the gold standard for determining exon–intron structures and alternative splicing of genes (Kawai et al. 2001).
Comparative genomics should also prove to be a powerful approach for identifying and annotating gene locations as additional vertebrate genomes are sequenced (Batzoglou et al. 2000; Roest Crollius et al. 2000). Given the generality of the model used by GenomeScan, it should be relatively straightforward to integrate comparative genomic information, such as TBLASTX alignments of homologous human and Tetraodon genomic regions into the algorithm.
The work described here represents one of the first reliable, fully automated approaches for annotating gene locations in a higher eukaryotic genome. The GenomeScan2000 gene set is freely available for downloading and analysis at http://genes.mit.edu/genomescan. Integration of this set with other automated human gene annotations, such as those produced by the Ensembl project (http://www.ensembl.org), should be particularly useful for future experimental and computational analyses of the human proteome. The GenomeScan algorithm can be accessed at http://genes.mit.edu/genomescan.html. Because the accuracy of the underlying gene model does not vary significantly between different groups of vertebrates (Burge and Karlin 1997), the method described here should work equally well for gene identification in other vertebrate genomes, such as zebrafish, mouse, and rat, as these sequences are determined.
METHODS
Information in BLASTX Hits
In an HMM model like that used by GENSCAN, each possible gene structure or set of gene structures that may be present in the sequence corresponds to an ordered list of states with associated lengths; such a list is referred to as a parse and designated with the Greek letter φ. Because the model determines a probability for generating each possible parse φi and sequence S, the predicted gene structure for an HMM model like GENSCAN is taken to be the parse that maximizes the joint probability P(φi, S) over all possible parses of the given input genomic sequence S. The crucial difference here is that we wish instead to maximize the joint conditional probability P (φi, S|Γ), conditional on similarity information Γ, such as the results of a BLASTX search. The first step in our method is to convert the information present in a set of BLASTX hits to a given human genomic sequence into a corresponding set of probabilistic statements about the likelihood that coding exons occur at particular places in the sequence. Each BLASTX hit alters the probabilities of the various parses of the genomic sequence in the GenomeScan model, increasing the likelihoods of parses that are consistent with the BLASTX information and reducing the likelihoods of those that are not, as described below. Because the boundaries of BLASTX hits correspond only roughly to the boundaries of coding exons, parses are only required to have an exon (of the appropriate reading frame) that overlaps the central, highest scoring region of the BLASTX hit, termed the centroid of the hit to be considered consistent. Intuitively, the portion of a BLAST alignment that has the most strongly positive BLOSUM62 score is most likely to be internal to a coding exon. Therefore, the centroid of a BLASTX hit is defined using a steepest-slope heuristic as the position C in the genomic sequence with steepest slope over a window of 15 codons centered at C in the cumulative BLASTX score plot. Cases in which the cumulative score plot is significantly multimodal are handled by breaking the BLASTX hit into multiple single-mode segments with different centroids. BLASTX hits that extend over a distance of ≥100 codons in the genomic sequence are converted to a series of segments, with centroids equally spaced along the length of the hit at ∼75-bp intervals.
Each BLASTX hit B to a genomic sequence is converted to an equivalent Genoa hit G (genome annotation) that summarizes the information present in the hit, including the genomic coordinates, centroid, reading frame, and P-value. Any parse of the sequence containing an exon that overlaps the centroid of the Genoa hit in the appropriate reading frame is said to be consistent with G, and the set of all such parses is designated ΦG; all other parses are said to be inconsistent with G . Of course, not every BLAST hit represents true homology between the query genomic sequence and the subject protein. This issue is made explicit by formally distinguishing the event HG, that the region of the genomic sequence corresponding to the BLASTX hit is functional and homologous to the target protein, from the event AG, that the similarity represented is artifactual. The phrase functional and homologous is meant to include only expressed, translated genes (i.e., excluding pseudogenes). Intuitively, the probability ΡA = P(AG) that a Genoa hit is artifactual should be related to the BLASTX P-value ΡB. However, the proportion of P < 10−10 BLASTX hits in the genome that represent pseudogenes is likely to be orders of magnitude higher than one in 1010, for example. Thus, ΡA is likely to be far higher than ΡB in general but is difficult to estimate precisely. In practice, we use a root-r heuristic, setting ΡA = (ΡB)1/r, where r is a small integer. For example, with r = 5 a ΡB = 10−5 BLASTX hit would be treated as having a ΡA = 10−1 = 10% chance of representing an artifact. The default value of r applied to results of the second BLAST in GenomeScript (against a restricted peptide database) is 10, and this value was used in all of the analyses described here. This heuristic works well in practice, but other ways of estimating ΡA may be worth investigating.
Consider the special case when the similarity information consists of a single Genoa hit, G. Letting ΡG = P(HG) —so that P(AG) = 1 − ΡG —the joint conditional probability P(φi, S|Γ) is defined as:
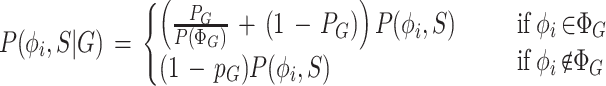 |
1 |
where P(φi, S) is the GENSCAN joint probability and P(ΦG) is the unconditional probability that one of the parses in ΦG is correct. The term 1 − ΡG (always ≤1) in the φi ∉ΦG case can be thought of as the penalty that is applied to parses that are inconsistent with the Genoa hit G. The more complicated term
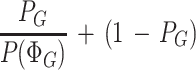 |
(always ≥1) can be thought of as the bonus that is applied to parses which are consistent with G. In this way, gene structures that are consistent with the similarity information in a Genoa hit are favored by the GenomeScan model, but inconsistent parses are not completely ruled out since the possibility that the Genoa hit may be artifactual is explicitly considered in the model and assigned an appropriate probability.
Equation 1 can be derived as follows. Observing that the information in a Genoa hit G affects the probabilities of parses and sequences only through the mutually exclusive events HG, AG, we have P(φi, S|G) = ΡGP(φi, S|HG) + (1 − ΡG)P(φi S|AG). By definition of HG, P(φi, S|HG) = 0 for any parse that is inconsistent with G. For any parse that is consistent with G, we have P(S|φi, HG) = P(S|φi, ΦG) = P(S|φi) under the assumption that HG influences the model only through the event ΦG and that one of the parses consistent with G is correct. Because HG implies ΦG, but does not otherwise affect the relative likelihood of any particular parse, P(φi, S|HG) = P(φi, S|ΦG) = P(φi,S)/P(ΦG), for any parse φiεΦG and P(φi, S|HG) = 0 for any parse φi∉ΦG. The quantity P(ΦG) may be calculated using a modification of the forward–backward algorithm described previously (Rabiner 1989; Burge 1997). In the event AG, we have no additional information about the likelihood of any particular parse or sequence, so P(φi, S|AG) = P(φi, S) for any parse φi.
Multiple nonredundant Genoa hits are handled similarly, making an assumption that is essentially equivalent to independence between distinct Genoa hits. This quasi-independence assumption provides a good approximation to P(φi, S|Γ) and allows straightforward applications of the Viterbi, forward and backward algorithms, which is one of the principal virtues of HMM models.
Initiation Codons, Termination Codons, and Introns
Suppose that residues 6–50 of a protein match genomic coordinates 116–250. It then stands to reason that an ATG located five codons upstream at position 101 in the genomic sequence (if one occurs there) has higher likelihood of representing an initiation codon than some other randomly chosen ATG in the sequence. In this case, the probability of any parse that involves an initiation codon at position 101 is increased by the factor Cstart (the default value of this parameter is 1e6, determined empirically). An analogous argument and treatment applies to termination codons. In addition, suppose that BLASTX hit B1 matches residues 101–150 of protein P to nucleotides 850–999 of the genomic sequence with P-value ΡB1 and BLASTX hit B2 matches residues 151–200 of protein P to nucleotides 2001–2150 of the genomic sequence with P-value PB2. Assuming that both BLASTX hits represent functional homology, the presence of an intron extending roughly from coordinates 1000–2000 in the genomic sequence is strongly implied; this situation is handled in the model by generating a special type of Genoa intron hit that spans 1030–1970 with P-value ΡI = 1 − (1 −ΡB1)(1 −ΡB2), assuming independence between hits. The 30-bp offset (1030 rather than 1000, 1970 rather than 2000) is used to ensure that the specified intronic region is very unlikely to overlap with either of the flanking exons. Internally, the GenomeScan program reduces the probabilities of parses that involve an exon in the region specified by a Genoa intron hit in a manner analogous to the way that regular Genoa hits reduce the probabilities of parses that do not contain overlapping exons. Additional technical details of the GenomeScan method are described in the GenomeScan documentation at http://genes.mit.edu/genomescan.
GenomeScript
GenomeScan is integrated with other analysis tools in a procedure called GenomeScript. GenomeScript performs the following series of analyses on an input genomic sequence: (1) mask the repetitive elements using RepeatMasker (http://www.genome.washington.edu/UWGC/analysistools/repeatmask.html); (2) run GENSCAN on masked genomic sequence, search predicted peptides against an appropriate protein database, and retrieve protein hits that achieve a desired level of significance (default: E < 10−5); (3) run a second BLASTX search of the masked genomic sequence against this subset of peptides with increased gap penalties (−G 20, −E 3) and relaxed E-value cutoff (default: E < 0.05) and convert the output to Genoa format (see above); and (4) run GenomeScan on the masked genomic sequence using the Genoa hits from the previous step as input. The second BLASTX search with relaxed E-value cutoff allows very sensitive detection of coding exons (as seen in Fig. 1) but produces a certain level of false-positive hits, most of which GenomeScan is able to filter out (as in Fig. 1). A variant of this procedure replaces step 2 above (GENSCAN + BLASTP) by BLASTX of the masked genomic sequence against the protein database. This variant procedure increases the run time of the BLAST database search (typically rate-limiting) by one or two orders of magnitude but has little effect on accuracy (data not shown), and so was not used in the applications described here.
Datasets
The SingleGene dataset of 175 human genes was constructed by deleting three sequences (accession nos. X63578, U34879, and Y07661) from the h178 dataset constructed by Guigó et al. (2000). These three sequences have apparent annotation errors—unrealistically short introns or evidence for additional unannotated genes. The FinishGene and DraftGene datasets were constructed as follows. First, all available full-length human cDNAs were aligned to available draft (htgs) and finished human genomic sequences from GenBank release 118 using the spliced alignment algorithm mRNAvsGen (L.L. Lim and C.B. Burge, unpubl.), which is similar in concept to the sim4 program (Florea et al. 1998) but is tailored specifically for aligning cDNA sequences rather than ESTs. Next, pairs of finished/draft genomic sequences were identified that contained alignments to the same full-length cDNA. A total of 194 such pairs were identified for which at least one cDNA was aligned across its entire length to the finished genomic sequence and at least the first and last exons of the gene were found in contigs from the same draft BAC, implying that the BAC covers the genomic locus encoding the cDNA. These 194 finished sequences, containing 206 genes also sequenced in draft form, comprise the FinishGene dataset. Those draft contigs containing exons or introns from these same 206 genes (as determined by BLASTN) form the DraftGene dataset. On average, exons from each gene in the DraftGene set were represented by 1.8 different draft contigs, reflecting the modest level of fragmentation resulting from the rough draft sequencing strategy. The median number of contigs per DraftGene BAC was 18; the median sequence coverage was 4.3-fold (one quarter of draft BACs did not have coverage information). For both datasets, the exon–intron structures derived by mRNAvsGen alignments of the corresponding full-length cDNAs were treated as sequence annotation for the purposes of calibrating gene prediction accuracy. The FinishGene and DraftGene datasets are at http://genes.mit.edu/genomescan/datasets.
Implementation
The GenomeScan program was written in the C programming language and has been compiled and run on a variety of Unix/Linux platforms. Run time for GenomeScan grows roughly linearly with sequence length in the typical case; typical run time for a 100-kb genomic sequence on a Pentium III 500 MHz Linux workstation is ∼10 sec. The program requires ∼0.5 MB of RAM per kbp of sequence so that 1–2 Mbp genomic sequences can be analyzed on a computer with 1 GB or more of RAM. GenomeScan was run using GenomeScript with default parameters on contigs from the GoldenPath human genome on the BioCluster at the Compaq Enterprise System Lab. The BioCluster comprises 25 ES40 nodes with four processors (667 MHz Alpha EV67) and 4-GB memory each. Before running GenomeScript, the GoldenPath was masked with RepeatMasker (http://www.genome.washington.edu/UWGC/analysistools/repeatmask.html; June 19, 2000, version) and broken into individual contigs, breaking at gaps represented by 100 or more unknown nucleotides (Ns) or when necessary to produce a maximum practical contig size of 500 kbp. Total run time for the September 2000 GoldenPath was approximately 48 h.
Acknowledgments
We thank Roderic Guigó, Eric Lander, David Lipman, and David Page for helpful discussions; Phillip Sharp for comments on the manuscript; Liana Lareau for expert technical assistance; Anders Krogh for providing the HMMGene software; Doug Hyatt for providing GRAIL, Compaq for computer resources; and Merck and MIT for support. C.B.B. is a recipient of a Burroughs Wellcome Fund Innovation Award in Functional Genomics. L.P.L. is supported by United States Public Health Service MERIT award R37-GM34277 to Phillip A. Sharp. This work was made possible by the public availability of the human genome sequence and annotation, and we thank all contributing sequencing centers and scientists.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL cburge@mit.edu; FAX 617-253-3128.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.175701.
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzoglou S, Pachter L, Mesirov JP, Berger B, Lander ES. Human and mouse gene structure: Comparative analysis and application to exon prediction. Genome Res. 2000;10:950–958. doi: 10.1101/gr.10.7.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Durbin R. Using GeneWise in the Drosophilaannotation experiment. Genome Res. 2000;10:547–548. doi: 10.1101/gr.10.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner V, Nyakatura G, Rosenthal A, Platzer M. Genomic organization of two novel genes on human Xq28: Compact head to head arrangement of IDH gamma and TRAP delta is conserved in rat and mouse. Genomics. 1997;44:8–14. doi: 10.1006/geno.1997.4822. [DOI] [PubMed] [Google Scholar]
- Burge CB. “Identification of genes in human genomic DNA.” Ph.D. thesis. California: Stanford University; 1997. [Google Scholar]
- ————— Chipping away at the transcriptome. Nat Genet. 2001;27:232–234. doi: 10.1038/85772. [DOI] [PubMed] [Google Scholar]
- Burge CB, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- ————— Finding the genes in genomic DNA. Curr Opin Struct Biol. 1998;8:346–354. doi: 10.1016/s0959-440x(98)80069-9. [DOI] [PubMed] [Google Scholar]
- Claverie J-M. Computational methods for the identification of genes in vertebrate genomic sequences. Hum Mol Genet. 1997;6:1735–1744. doi: 10.1093/hmg/6.10.1735. [DOI] [PubMed] [Google Scholar]
- Dunham I, et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Analysis of expressed sequence tags indicates 35,000 human genes. Nature Genet. 2000;25:232–234. doi: 10.1038/76115. [DOI] [PubMed] [Google Scholar]
- Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand MS, Mironov AA, Pevzner PA. Gene recognition via spliced sequence alignment. Proc Natl Acad Sci. 1996;93:9061–9066. doi: 10.1073/pnas.93.17.9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gish W, States DJ. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- Guigó R, Agarwal P, Abril JF, Burset M, Fickett JW. An assessment of gene prediction accuracy in large DNA sequences. Genome Res. 2000;10:1631–1642. doi: 10.1101/gr.122800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard T, Birney E. Open annotation offers a democratic solution to genome sequencing. Nature. 2000;403:825. doi: 10.1038/35002770. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JCF, Trent JM, Staudt LM, Hudson J, Jr, Boguski MS, et al. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- Kawai J, et al. Functional annotation of a full-length mouse cDNA collection. Nature. 2001;409:685–690. doi: 10.1038/35055500. [DOI] [PubMed] [Google Scholar]
- Krogh A. Using database matches with HMMGene for automated gene detection in Drosophila. Genome Res. 2000;10:523–528. doi: 10.1101/gr.10.4.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp D, Haussler D, Reese MG, Eeckman FH. A generalized hidden Markov model for the recognition of human genes in DNA. Proc ISMB. 1996;4:134–142. [PubMed] [Google Scholar]
- Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- Liang F, et al. Gene Index analysis of the human genome estimates approximately 120,000 genes. Nat Genet. 2000;25:239–240. doi: 10.1038/76126. [DOI] [PubMed] [Google Scholar]
- Maglott DR, Katz KS, Sicotte H, Pruitt KD. NCBI's LocusLink and RefSeq. Nucleic Acids Res. 2000;28:126–128. doi: 10.1093/nar/28.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabiner L. A tutorial on hidden Markov models and selected applications in speech recognition. Proc IEEE. 1989;77:257–285. [Google Scholar]
- Reese MG, Kulp D, Tammana H, Haussler D. Genie—Gene finding in Drosophila melanogaster. Genome Res. 2000;10:529–538. doi: 10.1101/gr.10.4.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roest Crollius H, Jaillon O, Bernot A, Dasilva C, Bouneau L, Fischer C, Fizames C, Winckes P, Brottier P, Queties F, et al. Estimate of human gene number provided by genome-wide analysis using Tetraodon nigroviridisDNA sequence. Nat Genet. 2000;25:235–238. doi: 10.1038/76118. [DOI] [PubMed] [Google Scholar]
- Salamov AA, Solovyev VV. Ab initio gene finding in Drosophilagenomic DNA. Genome Res. 2000;10:516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker DD, Schadt EE, Armous CD, He YD, Garrett-Engele P, McDonagh PD, Loerch PM, Leonardson A, Lum PY, Cavet G, et al. Experimental annotation of the human genome using microarray technology. Nature. 2001;409:922–927. doi: 10.1038/35057141. [DOI] [PubMed] [Google Scholar]
- Smith TM, Lee MK, Szabo CI, Jerome N, McEwen M, Taylor M, Hood L, King MC. Complete genomic sequence and analysis of 117 kb of human DNA containing the gene BRCA1. Genome Res. 1996;6:1029–1049. doi: 10.1101/gr.6.11.1029. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Xu Y, Uberbacher EC. Automated gene identification in large-scale genomic sequences. J Comp Biol. 1997;4:325–338. doi: 10.1089/cmb.1997.4.325. [DOI] [PubMed] [Google Scholar]
- Wolfsberg TG, Landsman DA. Comparison of expressed sequence tags (ESTs) to human genomic sequences. Nucleic Acids Res. 1997;25:1626–1632. doi: 10.1093/nar/25.8.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]