Abstract
Evolutionary centromere repositioning is a paradox we have recently discovered while studying the conservation of the phylogenetic chromosome IX in primates. Two explanations were proposed: a conservative hypothesis assuming sequential pericentric inversions, and a more challenging assumption involving centromere emergence during evolution. The complex evolutionary history showed by chromosome IX did not allow us to clearly distinguish between these two hypotheses. Here we report comparative studies of chromosome X in two lemur species: the black lemur and the ringtailed lemur. The X chromosome is telocentric in the black lemur and almost metacentric in the ringtailed lemur. The marker order along these chromosomes, however, was found to be perfectly colinear with humans. Our data unequivocally point to centromere emergence as the most likely explanation of centromere repositioning.
Human centromeric and pericentromeric regions have been shown to be highly plastic (Archidiacono et al. 1995; Eichler et al. 1999; Jackson et al. 1999; Horvath et al. 2000), in sharp contrast to the relative stability of the rest of the genome (Kaessmann et al. 1999). We have recently reported an additional puzzling feature of centromeric regions, the centromere repositioning paradox, discovered while studying the cytogenetic conservation of phylogenetic chromosome IX in primates (Montefalcone et al. 1999). In some species the centromere shows an evolutionary history independent from the surrounding markers. Its position can be reconciled with these markers only by assuming additional, peculiar pericentric inversions (which we named flip-flop inversions) or by hypothesizing evolutionary emergence of centromeres. Distinction between these two alternatives is critical to our understanding of the processes of chromosomal evolution. The flip-flop hypothesis does not imply any new biological mechanism and would just add further support to the opinion that intrachromosomal rearrangements are very frequent in evolution (Muller et al. 2000). Evolutionary appearance of centromeres, on the contrary, would represent a novel, unpredicted biological property.
The X chromosome is considered the most evolutionary conserved chromosome among mammals (Ohno 1973; Chowdhary et al. 1998). Also, its submetacentric shape, as it appears in humans, is highly conserved and very likely represents the mammalian ancestral form. This assumption is strongly supported by the perfect conservation of marker order, including the centromere, between cat (Felix catus) and human X chromosomes (Murphy et al. 1999). We reasoned that the striking X chromosome evolutive conservation was a unique opportunity that would facilitate the testing of centromere repositioning mechanisms. Comparative studies on marker order conservation among the X chromosome of humans (Homo sapiens, HSA) and two Lemuridae species: Eulemur macaco (EMA; black lemur) and Lemur catta (LCA; ringtailed lemur) were performed. These two primates were selected for the study because the morphology of the X chromosome in these two species is quite different from humans. Despite these differences, no marker order discrepancy was observed. Our results strongly indicate, therefore, that the repositioning took place via centromere emergence.
RESULTS
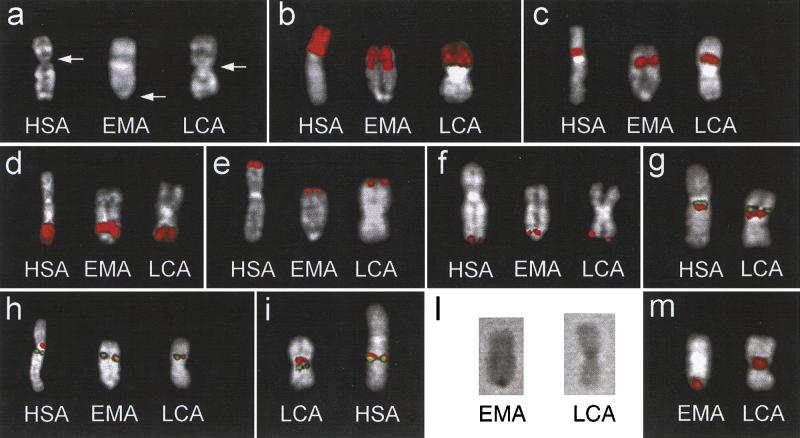
Chromosome X appears telocentric in EMA and almost metacentric in LCA (Fig. 1a). To investigate their evolutionary conservation, we performed fluorescence in situ hybridization (FISH) analysis using different kinds of probes. Human Partial Chromosome Paints (PCP) specific for Xp (#364) and Xp11.2→11.3 (#377) were initially used (Fig. 1b,c). Despite the striking difference in centromere location, no pericentric inversion was detected. This conclusion was also supported by the use of PCPs #102 (Xq; data not shown) and #118 (Xq24→28; Fig. 1d). To investigate marker order conservation in more detail, a panel of 19 human probes spanning the chromosome X from the tip of Xp to the tip of Xq was then used (Table 1). A probe containing the PGPL gene, located inside the PseudoAutosomal Region 1 (PAR1) and mapping 80 to 110 kb from Xpter (Gianfrancesco et al. 1998), was used as a telomeric Xp marker. A cosmid clone (U130F6) containing the HSPRY3 gene was used as a marker of the Xqter region. HSPRY3 maps inside the PAR2, approximately 250 kb from the Xq telomere (Ciccodicola et al. 2000). All the probes were found to hybridize in EMA and LCA as if their X chromosomes were perfectly colinear with the human X. Examples of these experiments are reported in Figure 1e and 1f. The centromere position in EMA was found to be telomeric to all the studied markers, including the HSPRY3 gene (Fig. 1f).
Figure 1.
(a) Q-banded chromosome X from Homo sapiens (HSA), Eulemur macaco (EMA; black lemur), and Lemur catta (LCA, ringtailed lemur). The arrows indicate the centromere. EMA and LCA chromosomes are oriented, in all images, upside-down to match the orientation of human X. (b–f) Examples of fluorescence in situ hybridization (FISH) experiments using PCP #364 (Xp; b), PCP #377 (Xp11.2–Xp11.3; c), PCP #118 (Xq24–28; d), PCP #PGPL probe (PAR1; (e), and U130F6 cosmid clone (HSPRY3 gene, PAR2; f). (g) Cohybridization experiment using probes bA235K20 (green signal) and dJ1015G2 (red signal). Their orientation is identical in both species. (h) Probes dJ598A24 (red) and bA235K20 (green) flank the human centromere. In EMA and LCA, they appear to be almost completely overlapping. (i) Probes dJ1015G2 (red) and dJ715D6 (green) surround the LCA centromere and are well separated by the centromeric heterochromatin. In HSA, they appear much closer. (l) The C-banded chromosome X of EMA and LCA. (m) The signal of DOP-PCR amplified products from microdissected material of the EMA (left) and LCA (right) centromeric regions hybridized to EMA and LCA metaphases, respectively. Only chromosome X is shown (see text for details).
Table 1.
Probes Used in the Study
| Probe | STS | cM | cR | Mapping | |
|---|---|---|---|---|---|
| COS | PGPL | 80-110 kb from Xpter | Xp22.3 | ||
| YAC | 749G10 | DXS1043 | 17.7 | 7.65 | Xp22.2 |
| YAC | 681F6 | DXS987 | 25.5 | Xp22.1 | |
| YAC | 789C8 | DXS989 | 40.6 | Xp22.1 | |
| YAC | 965C6 | DXS997 | 47.0 | 25.91 | Xp21 |
| YAC | 649F6 | DXS991 | 86.9 | Xp11.2 | |
| PAC | dJ598A24 | DXS6967 | Xp11 | ||
| HSA centromere | |||||
| BAC | bA235K20 | WI-14304 | 90.8 | 169.61 | Xq11.2 |
| PAC | dJ1015G24 | DXS8109 | 97.9 | Xq13 | |
| YAC | 933D12 | DXS6809 | 245.07 | Xq21 | |
| YAC | 748C4 | DXS8077 | 108.1 | Xq21 | |
| DXS6740 | 245.17 | ||||
| LCA centromere | |||||
| PAC | dJ715D6 | Xq21.3 | |||
| YAC | 963G9 | DXS1072 | 122.8 | Xq23 | |
| AFMA275ZC9 | 266.47 | ||||
| YAC | 923D3 | DXS1001 | 139.4 | 275.97 | Xq24 |
| YAC | 719H8 | DXS984 | 159.5 | Xq26/2q32 | |
| WI-6425 | 299.53 | ||||
| YAC | 960C9 | DXS1205 | 163.7 | Xq26 | |
| DXS6798 | 302.92 | ||||
| YAC | 878A5 | DXS1215 | 183.8 | 312.58 | Xq27 |
| YAC | 849E12 | DXS8103 | 192.5 | Xq27-28/13q14 | |
| COS | HSPRY3 | 250kb from Xqter | Xq28 (PAR2) | ||
| EMA centromere | |||||
Most of the probes are YACs of the CEPH megalibrary, obtained from the YAC Screening Center (Milan). They were selected after a careful evaluation of their position on the genetic (cM) and radiation hybrid (cR) map (MIT database).
The centromere location in LCA was further investigated using cohybridization experiments, because its different position in respect to HSA could be easily explained by a pericentric inversion of a relatively small chromosomal region. The experiments reported in Figure 1g are crucial in this respect. BAC bA235K20 maps at Xq11.2, below the human centromere; PAC dJ1015G2B, at Xq13 in HSA, is located above the LCA centromere. Both probes are within the hypothetical inverted region; therefore, they would necessarily detect the inversion. Their FISH signals clearly show that their orientation is maintained in both HSA and LCA, thus proving that the difference in centromere position was not caused by a pericentric inversion.
Additional human PAC or BAC probes close to the HSA and LCA centromeres were identified by querying the Sanger Centre database. FISH analysis indicated that dJ598A24 and bA235K20 were very close to the human centromere on the Xp and Xq side, respectively (Fig. 1h). They were cohybridized on EMA and LCA metaphases. The two signals were found to nearly overlap in both species (Fig. 1h). An additional experiment was performed using PACs dJ1015G24 and dJ715D6. These two probes are located at the opposite sides of the LCA centromere, well separated by the centromeric heterochromatin (Fig. 1i, left). In HSA, they appear much closer (Fig. 1i, right).
C-banding technique was used to investigate the nature of the cytogenetic material located at the centromere of EMA and LCA chromosome X. The results are shown in Figure 1l. The two centromeres appear to contain a heterochromatic block comparable in size to the average of the autosomal chromosomes. To test the possibility that this region was the result of a transposition from the ancestral centromeric area or from an autosomal region, we microdissected both centromeres. About 20 samples for each species were collected, DOP-PCR amplified, biotin-labeled, and hybridized on metaphase preparations of EMA, LCA, and HSA. Labeled DOP-PCR material from EMA gave a strong signal on all EMA chromosomes, with the exception of chromosome 1. The signal on chromosome X is shown in Figure 1m. No signal was detected on HSA and LCA chromosomes. The labeled DOP-PCR products from LCA centromere lit up the centromere of the X chromosome of LCA (Fig. 1m) and the DAPI-positive interstitial heterochromatic blocks present on some LCA chromosomes (data not shown). In EMA, a clear FISH signal was detected on the pericentromeric heterochromatic block of chromosomes 6 and 3 (data not shown). No signal was found in HSA. An hypothetical transposition of autosomal material into the X chromosome of LCA was also searched for by hybridizing a complete panel of human WCPs on LCA metaphases. Painting signal of LCA chromosome X was obtained only by the WCP derived from human chromosome X (data not shown). Reciprocal chromosome painting analysis of EMA and HSA chromosomes has been performed by Muller et al. (1997). They report a perfect equivalence of the euchromatic content of human and EMA X chromosomes.
Remains of alphoid sequences at the ancestral X centromere of EMA and LCA were searched for by using p82H α satellite plasmid and a pool of human alphoid clones as probes, hybridized at low stringency. The pool included the alphoid probe specific for chromosome X. Probe p82H recognizes, at low stringency, all the human centromeres (Aleixandre et al. 1987). Both experiments failed to detect any signal on EMA and LCA chromosomes.
DISCUSSION
We have studied the evolutionary conservation of chromosome X in HSA and two Lemuridae species, EMA and LCA, using a panel of appropriate FISH probes. The chromosome X is telocentric in EMA and almost metacentric in LCA. Despite the very apparent morphological differences, our results indicate that marker order is perfectly conserved. Our data, therefore, are an additional example of centromere repositioning in primates. Thanks to the striking evolutive conservation of chromosome X in mammals, the present study represented a unique opportunity of testing the different hypotheses that could be formulated to explain the centromere repositioning phenomenon. The perfect marker conservation in both EMA and LCA does not fit with the flip-flop hypothesis. Indeed, no intermediate form (caused by a single inversion) of the X chromosome has been documented in primates.
Centromere repositioning not mediated by chromosome rearrangement can be hypothesized to occur either through a transposition event that would insert centromeric sequences into an euchromatic region or through neocentromere emergence. FISH experiments using DOP-amplified products of the microdissected centromeric regions of EMA and LCA X chromosomes were used to substantiate a hypothetical insertional transposition from a different centromere. No hints in favor of a transposition event including euchromatic DNA were found. This conclusion is also supported by the systematic use of human chromosome paints on LCA chromosomes (present data) and by reverse painting data reported by Muller et al. (1997) on LCA. A transposition event restricted to heterochromatic material, however, could not be discarded with certainty in EMA. Transposition of centromeric sequences into a distinct centromere has been documented in a prenatal diagnosis case (Verlinsky et al. 1995). As far as we know, however, no examples are available, in mammals, of neocentromere seeding caused by insertional transposition of a centromeric block into a noncentromeric region. On the contrary, since the first convincing report of neocentromere occurrence in humans (du Sart et al. 1997; Barry et al. 2000), several additional cases have been described (Warburton et al. 2000). In one instance, the neocentromere showed a normal familial segregation (Tyler-Smith et al. 1999). Neocentromeres also have been occasionally documented in Drosophila (Williams et al. 1998). Neocentromere emergence, therefore, seems to be the most likely explanation of the centromere repositioning we have documented.
An intriguing issue arising from this conclusion is the loss or gain of heterochromatic material shown, respectively, by the ancestral centromere locus and by the newly formed centromeres. The interpretation of the data is that the heterochromatic material, usually associated with higher eukaryote centromeric regions, has disappeared in both EMA and LCA from the site where the ancestral centromere was located, as a consequence of the deep remodeling of the region following centromere repositioning. Indeed, C-banded material was not detected at the ancestral centromere loci, whereas it was evident at the active centromere of both EMA and LCA. We can hypothesize that heterochromatic material gradually accumulates at the newly formed centromere following neocentromere seeding. Evolution of neocentromeres toward complexity is probably driven by a specific selective pressure (Eichler 1999). Unfortunately, the intermediate evolutionary events of such heterochromatization processes are not tractable. A clear example, however, of the degradation process affecting an inactivated centromere is provided by the ancestral centromere at 2q21 in HSA, which became inactive following the telomere-telomere fusion of the two ancestral phylogenetic chromosomes IIp and IIq (Ijdo et al. 1991). Despite its recent origin (at most 3–5 million years ago), relics of alphoid sequences are barely detectable at 2q21 (Avarello et al. 1992; Baldini et al. 1993), and there is no evidence of C-banded material.
Studies supporting the view that centromere repositioning is a relatively widespread phenomenon in mammalian evolution are starting to emerge (Montefalcone et al. 1999; Band et al. 2000; Iannuzzi et al. 2000; Muller et al. 2000). Altogether, these data indicate that centromere repositioning played a role in mammalian genome evolution.
METHODS
Metaphase spreads were obtained from PHA-stimulated peripheral blood lymphocytes from a normal human donor and from fibroblast cell lines of EMA and LCA. PCPs were generated in our laboratory (see our Web site, http://www.biologia.uniba.it/rmc/1_hy-tutti/1-2_hy_diagrams/X.html ). WCPs, derived from flow-sorted chromosomes, were a gift from Dr. N. Carter (Sanger Centre). WCP specific for chromosome 22 was obtained by Alu-PCR amplification of DNA from a somatic cell hybrid retaining chromosome 22 as the only human chromosome (see our Web site). YAC probes of CEPH megalibrary were obtained from the YAC Screening Centre (Milan). PAC and BAC probes were from the Sanger Centre (http://webace.sanger.ac.uk). The “bA” (RPCI BAC library) or “dJ” (RPCI PAC library) prefix, used by the Sanger Centre to identify the source library, was maintained. Probes PGPL and HSPRY3 were a generous gift of A. Ciccodicola (IIGB, Naples). FISH experiments have been described in detail elsewhere (Montefalcone et al. 1999). Microdissection and DOP-PCR amplification procedures were performed according to Meltzer et al. (1992).
Acknowledgments
The financial support of cofin2000-MURST, AIRC, and Telethon (Grant E.672 to M.R. and E.962 to N.A.) is gratefully acknowledged.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL rocchi@biologia.uniba.it; FAX 39-080-544-3386.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.152101.
REFERENCES
- Archidiacono N, Antonacci R, Marzella R, Finelli P, Lonoce A, Rocchi M. Comparative mapping of human alphoid sequences in great apes using fluorescence in situ hybridization. Genomics. 1995;25:477–484. doi: 10.1016/0888-7543(95)80048-q. [DOI] [PubMed] [Google Scholar]
- Aleixandre C, Miller DA, Mitchell AR, Warburton DA, Gersen SL, Disteche C, Miller OJ. p82H identifies sequences at every human centromere. Hum Genet. 1987;77:46–50. doi: 10.1007/BF00284712. [DOI] [PubMed] [Google Scholar]
- Avarello R, Pedicini A, Caiulo A, Zuffardi O, Fraccaro M. Evidence for an ancestral alphoid domain on the long arm of human chromosome-2. Hum Genet. 1992;89:247–249. doi: 10.1007/BF00217134. [DOI] [PubMed] [Google Scholar]
- Baldini A, Ried T, Shridhar V, Ogura K, D'Aiuto L, Rocchi M, Ward DC. An alphoid DNA sequence conserved in all human and great ape chromosomes: Evidence for ancient centromeric sequences at human chromosomal regions 2q21 and 9q13. Hum Genet. 1993;90:577–583. doi: 10.1007/BF00202474. [DOI] [PubMed] [Google Scholar]
- Band MR, Larson JH, Rebeiz M, Green CA, Heyen DW, Donovan J, Windish R, Steining C, Mahyuddin P, Womack JE, et al. An ordered comparative map of the cattle and human genomes. Genome Res. 2000;10:1359–1368. doi: 10.1101/gr.145900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry AE, Bateman M, Howman EV, Cancilla MR, Tainton KM, Irvine DV, Saffery R, Choo KH. The 10q25 Neocentromere and its inactive progenitor have identical primary nucleotide sequence: Further evidence for epigenetic modification. Genome Res. 2000;10:832–838. doi: 10.1101/gr.10.6.832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccodicola A, D'Esposito M, Esposito T, Gianfrancesco F, Migliaccio C, Miano MG, Matarazzo MR, Vacca M, Franz A, Cuccurese M, et al. Differentially regulated and evolved genes in the fully sequenced Xq/Yq pseudoautosomal region. Hum Mol Genet. 2000;9:395–401. doi: 10.1093/hmg/9.3.395. [DOI] [PubMed] [Google Scholar]
- Chowdhary BP, Raudsepp T, Fronicke L, Scherthan H. Emerging patterns of comparative genome organization in some mammalian species as revealed by Zoo-FISH. Genome Res. 1998;8:577–589. doi: 10.1101/gr.8.6.577. [DOI] [PubMed] [Google Scholar]
- du Sart D, Cancilla MR, Earle E, Mao J, Saffery R, Tainton KM, Kalitsis P, Martin J, Barry AE, Choo KHA. A functional neocentromere formed through activation of a latent human centromere and consisting of non-α-satellite DNA. Nat Genet. 1997;16:144–153. doi: 10.1038/ng0697-144. [DOI] [PubMed] [Google Scholar]
- Eichler EE. Repetitive conundrums of centromere structure and function. Hum Mol Genet. 1999;8:151–155. doi: 10.1093/hmg/8.2.151. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Archidiacono N, Rocchi M. CAGGG repeats and the pericentromeric duplication of the hominoid genome. Genome Res. 1999;9:1048–1058. doi: 10.1101/gr.9.11.1048. [DOI] [PubMed] [Google Scholar]
- Gianfrancesco F, Esposito T, Montanini L, Ciccodicola A, Mumm S, Mazzarella R, Rao E, Giglio S, Rappold G, Forabosco A. A novel pseudoautosomal gene encoding a putative GTP-binding protein resides in the vicinity of the Xp/Yp telomere. Hum Mol Genet. 1998;7:407–414. doi: 10.1093/hmg/7.3.407. [DOI] [PubMed] [Google Scholar]
- Horvath JE, Viggiano L, Loftus BJ, Adams MD, Archidiacono N, Rocchi M, Eichler EE. Molecular structure and evolution of an α satellite/non-α satellite junction at 16p11. Hum Mol Genet. 2000;9:113–123. doi: 10.1093/hmg/9.1.113. [DOI] [PubMed] [Google Scholar]
- Iannuzzi L, Di Meo GP, Perucatti A, Incarnato D, Schibler L, Cribiu EP. Comparative FISH mapping of bovid X chromosomes reveals homologies and divergences between the subfamilies Bovinae and Caprinae. Cytogenet Cell Genet. 2000;89:171–176. doi: 10.1159/000015607. [DOI] [PubMed] [Google Scholar]
- Ijdo JW, Baldini A, Ward DC, Reeders ST, Wells RA. Origin of human chromosome 2: An ancestral telomere-telomere fusion. Proc Natl Acad Sci. 1991;88:9051–9055. doi: 10.1073/pnas.88.20.9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MS, Rocchi M, Thompson G, Hearn T, Crosier M, Guy J, Kirk D, Mulligan L, Ricco A, Piccininni S, et al. Sequences flanking the centromere of human chromosome 10 are a complex patchwork of arm-specific sequences, stable duplications and unstable sequences with homologies to telomeric and other centromeric locations. Hum Mol Genet. 1999;8:205–215. doi: 10.1093/hmg/8.2.205. [DOI] [PubMed] [Google Scholar]
- Kaessmann H, Heissig F, von Haeseler A, Paabo S. DNA sequence variation in a non-coding region of low recombination on the human X chromosome. Nat Genet. 1999;22:78–81. doi: 10.1038/8785. [DOI] [PubMed] [Google Scholar]
- Meltzer PS, Guan XY, Burgess A, Trent JM. Rapid generation of region specific probes by chromosome microdissection and their application. Nat Genet. 1992;1:24–28. doi: 10.1038/ng0492-24. [DOI] [PubMed] [Google Scholar]
- Montefalcone G, Tempesta S, Rocchi M, Archidiacono N. Centromere repositioning. Genome Res. 1999;9:1184–1188. doi: 10.1101/gr.9.12.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, O'Brien PCM, Ferguson-Smith MA, Wienberg J. Reciprocal chromosome painting between human prosimisans (Eulemur macaco macaco and E.fulvus mayottensis) Cytogenet Cell Genet. 1997;78:260–271. doi: 10.1159/000134669. [DOI] [PubMed] [Google Scholar]
- Muller S, Stanyon R, Finelli P, Archidiacono N, Wienberg J. Molecular cytogenetic dissection of human chromosomes 3 and 21 evolution. Proc Natl Acad Sci. 2000;97:206–211. doi: 10.1073/pnas.97.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy WJ, Sun S, Chen ZQ, Pecon-Slattery J, O'Brien SJ. Extensive conservation of sex chromosome organization between cat and human revealed by parallel radiation hybrid mapping. Genome Res. 1999;9:1223–1230. doi: 10.1101/gr.9.12.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Ancient linkage groups and frozen accidents. Nature. 1973;244:259–262. doi: 10.1038/244259a0. [DOI] [PubMed] [Google Scholar]
- Tyler-Smith C, Gimelli G, Giglio S, Floridia G, Pandya A, Terzoli G, Warburton PE, Earnshaw WC, Zuffardi O. Transmission of a fully functional human neocentromere through three generations. Am J Hum Genet. 1999;64:1440–1444. doi: 10.1086/302380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlinsky Y, Ginsberg N, Chmura M, Freidine M, White M, Strom C, Kuliev A. Cross-hybridization of the chromosome 13/21 α satellite DNA probe to chromosome 22 in the prenatal screening of common chromosomal aneuploidies by FISH. Prenat Diagn. 1995;15:831–834. doi: 10.1002/pd.1970150907. [DOI] [PubMed] [Google Scholar]
- Warburton PE, Dolled M, Mahmood R, Alonso A, Li S, Naritomi K, Tohma T, Nagai T, Hasegawa T, Ohashi H, et al. Molecular cytogenetic analysis of eight inversion duplications of human chromosome 13q that each contain a neocentromere. Am J Hum Genet. 2000;66:1794–1806. doi: 10.1086/302924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BC, Murphy TD, Goldberg ML, Karpen GH. Neocentromere activity of structurally acentric mini-chromosomes in Drosophila. Nat Genet. 1998;18:30–37. doi: 10.1038/ng0198-30. [DOI] [PubMed] [Google Scholar]