Abstract
Centromere protein A (CENP-A) is an essential histone H3-related protein that constitutes the specialized chromatin of an active centromere. It has been suggested that this protein plays a key role in the epigenetic marking and transformation of noncentromeric genomic DNA into functional neocentromeres. Neocentromeres have been identified on more than two-thirds of the human chromosomes, presumably involving different noncentromeric DNA sequences, but it is unclear whether some generalized sequence properties account for these neocentromeric sites. Using a novel method combining chromatin immunoprecipitation and genomic array hybridization, we have identified a 460-kb CENP-A-binding DNA domain of a neocentromere derived from the 20p12 region of an invdup (20p) human marker chromosome. Detailed sequence analysis indicates that this domain contains no centromeric α-satellite, classical satellites, or other known pericentric repetitive sequence motifs. Putative gene loci are detected, suggesting that their presence does not preclude neocentromere formation. The sequence is not significantly different from surrounding non-CENP-A-binding DNA in terms of the prevalence of various interspersed repeats and binding sites for DNA-interacting proteins (Topoisomerase II and High-Mobility-Group protein I). Notable variations include a higher AT content similar to that seen in human α-satellite DNA and a reduced prevalence of long terminal repeats (LTRs), short interspersed repeats (SINEs), and Alus. The significance of these features in neocentromerization is discussed.
Neocentromeres are fully functional centromeres that originate on some human marker chromosomes from interstitial chromosomal DNA segments (Choo 1997a). Neocentromeres, like the normal centromeres, maintain correct mitotic segregation properties of the marker chromosomes. Today, >40 neocentromeres have been described on more than two-thirds of the human chromosomes (Warburton et al. 2000). Characteristically, neocentromeres are devoid of the highly repetitive sequences found in normal centromeres. In at least two different marker chromosomes examined, the presence of >20 functionally important centromere proteins that are known to be associated with normal active centromeres suggests basic structural and functional similarities between neocentromeres and normal centromeres (Saffery et al. 2000).
Centromere protein A (CENP-A) is among the earliest recognized centromere components from studies using the sera of patients with the autoimmune disease CREST (Calcinosis, Raynaud's phenomenon, Esophageal dysmotility, Sclerodactyly, Telangiectasia; Earnshaw and Rothfield 1985). The protein is a 17-kD histone H3 homolog (Sullivan et al. 1994) and is associated with active centromeres and neocentromeres (Warburton et al. 1997; Voullaire et al. 1999). Chromatin immunoprecipitation in HeLa cells (Vafa and Sullivan 1997) and Indian muntjac cells (Vafa et al. 1999) reveals CENP-A binding to centromere-associated DNA. CENP-A is essential for chromosome segregation, as demonstrated in antibody injection experiments (Figueroa et al. 1998; Valdivia et al. 1998) and gene disruption experiments in mouse and in Caenorhabditis elegans (Buchwitz et al. 2000; Howman et al. 2000). The capacity of CENP-A to organize centromeric DNA has also been demonstrated in vitro, where CENP-A replaces histone H3 for the packaging of a circular plasmid-template DNA containing α-satellite into the familiar “beads-on-a-string” primary chromatin structure (Yoda et al. 2000). Atomic force microscopy of the reconstituted nucleosomes has shown that CENP-A would form nucleosomes that appear to deposit in a regular fashion on the α-satellite DNA template.
To date, only one human neocentromere at chromosomal band 10q25.2 on the marker chromosome mardel(10) has been characterized beyond the molecular cytogenetic level. Initial immunofluorescence/FISH mapping on mechanically stretched metaphase chromosomes identified a genomic YAC clone of ∼640 kb spanning the entire neocentromere domain (du Sart et al. 1997). An 80-kb BAC subclone, E8, was obtained from the CREST centromere protein-binding domain (Cancilla et al. 1998). Further restriction and high-density PCR analyses and, subsequently, direct DNA sequencing of this 80-kb region revealed no difference between the DNA sequence of the neocentromerized genomic segment and the progenitor pre-active genomic segment from the patient's father (Barry et al. 2000). These studies provide evidence for the sequence independence of neocentromere formation.
It is likely that the processes of centromerization and neocentromerization are epigenetically controlled (Choo 2000). A model has been proposed in which different primary sequences of DNA may possess different potential for centromerization (Choo 2000; Maggert and Karpen 2000). In the normal situation on a human chromosome, the site of the highest centromerization potential along the whole chromosome will be the α-satellite-containing DNA. It is only in cases of chromosomal rearrangements that result in the loss of most or all of the α-satellite sequences that the potential of various non-α-satellite genomic DNA to form neocentromeres becomes challenged. The possible existence of hotspots with increased neocentromerization propensity has been investigated by comparing the cytogenetic location of ∼40 reported neocentromeres (Warburton et al. 2000). It is of particular interest that eight neocentromeres were formed on 13q, eight on 15q, and five on 3q. Of the cases associated with 13q, five have demonstrated neocentromerization at the 13q32 region. Further investigations of whether these chromosomal regions contain unusual characteristics predisposing them to neocentromerization will require the identification and detailed sequence analysis of the underlying DNA.
Here, we describe a new procedure combining chromatin immunoprecipitation and genome array screening that allows the rapid identification of the DNA that is directly associated with neocentromere-specific chromatin. Application of this procedure on a previously described neocentromere formed as a result of inverted duplication of the short arm of human chromosome 20 reveals a CENP-A-binding domain of 460 kb. The detailed sequence analysis of this domain is also described.
RESULTS
Preliminary Localization of Neocentromere to a 3–4-Mb Domain by Immunofluorescence/FISH Mapping
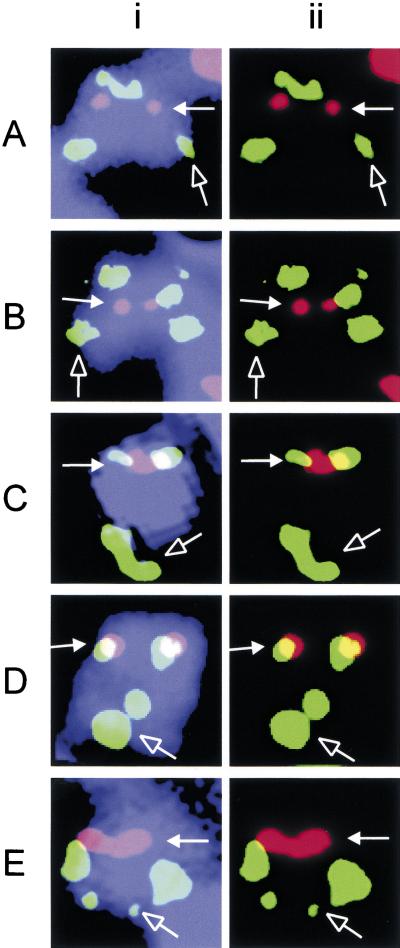
We have described previously the cytogenetic characterization of a neocentromere at 20p12 (Voullaire et al. 1999). Here, the precise location of this neocentromere in the genome was studied by immunofluorescence/FISH analysis on a transformed lymphoblastoid cell line established from the patient. A genomic array spanning ∼18 Mb around the 20p12 neocentromere region was constructed using mapped bacterial artificial chromosomes (BAC) or P1-based artificial chromosomes (PAC) (Fig. 1). When each of the BAC/PAC sequences was used in FISH, two sets of signals were obtained on the marker chromosomes because of the inverted duplication (Fig. 2, green signals). The position of the neocentromere was marked by immunofluorescence using the CREST#6 antiserum (Fig. 2, red signals). As the location of the BAC/PAC probes approached the neocentromere core antigen binding site, the corresponding FISH signals moved from the initial p′ side (Fig. 2A,B) to overlapping signals (Fig. 2C) before emerging on the q′ side (Fig. 2D,E) of the CREST#6 signals. By a preliminary scoring of a limited number of metaphases, the neocentromere could be localized within a 3–4 Mb region within the array. Use of this technique to further narrow the neocentromere position was unreliable because of the highly condensed state of the chromosome. This low-resolution problem probably explained the apparent overlapping of both PACs dJ727I10 and dJ742J24 with the CREST#6 signals (Fig. 2C,D), despite the two PACs being known to be ∼1 Mb apart (Fig. 1B). Attempts to increase the resolution by mechanical stretching of the marker chromosome, for example, through reduction of the cell concentration and/or increasing the speed of cytospinning (du Sart et al. 1992), were unsuccessful, possibly because of the small size of this chromosome. Furthermore, the intense signals that spread over a broad area (see Fig. 2) caused by the relatively large BAC/PAC probes also posed serious limitations to the mapping resolution. A method that overcame these limitations was therefore desirable and was developed.
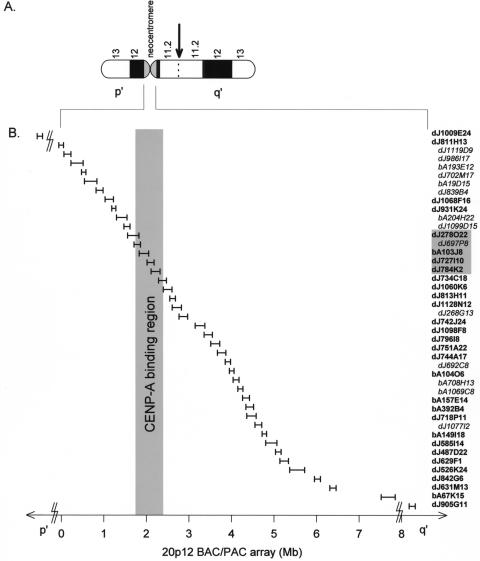
Figure 1.
20p12 BAC/PAC array. (A) Ideogram of the invdup(20p) marker chromosome showing the inverted duplication breakpoint in band 20p11.2 (arrow) and the activation of a neocentromere at 20p12. p′ and q′ indicate the short and long arms of the marker chromosome relative to the neocentromere, respectively. (B) BAC/PAC array constructed from public databases around the neocentromere. Distances (in Mb units) within the array are plotted relative to PAC dJ811H13 on the p′ arm of the marker chromosome. The size of each horizontal bar is proportional to the corresponding insert size of the BAC or PAC. Addresses of the BAC/PACs are listed on the right. The prefix “dJ” denotes a PAC while “bA” indicates a BAC. PACs dJ1009E24 and dJ905G11 map outside the array and are indicated by broken lines. BAC/PACs that were not used in the CIA analysis (see Methods) are shown in italics and recessed. Shaded box represents the CENP-A binding region (see Results).
Figure 2.
Immunofluorescence/FISH analysis of 20p12 neocentromere. Immunofluorescence was performed using CREST#6 (red) to mark the position of the neocentromere (solid arrow). FISH was performed using PACs dJ1009E24 (A), dJ811H13 (B), dJ727I10 (C), dJ742J24 (D), and dJ905G11 (E). Two sets of green FISH signals are observed on the marker chromosome invdup (20p) because of the inverted duplication of the probed segment; the signal set at the distal q′ arm is indicated by an open arrow. The combined image in (i) shows the relative positions of the signals on the marker chromosome (blue); (ii) shows only the images for the green and red signals in (i). Colocalization of the two signals appears as yellow in (ii).
CIA Analysis Defines a 460-kb CENP-A Binding Region
For the rapid, single-step identification of the critical neocentromere DNA sequences, we tested a novel method combining the use of chromatin immunoprecipitation and array (CIA) analysis. Because CENP-A binds specifically to centromere DNA at the nucleosomal level (Vafa and Sullivan 1997; Vafa et al. 1999) and is essential for centromere organization (Howman et al. 2000), we decided to use this protein as a marker for functional neocentromere chromatin. DNA obtained from chromatin immunoprecipitation (Johnson et al. 1998) using an antibody specific to human CENP-A was amplified by DOP–PCR (Telenius et al. 1992) and radioactively labeled by random priming. The immunoprecipitated DNA was used as a probe on a dot blot containing an array of the 20p12 BAC/PACs (Fig. 3). The signal obtained from each spot was compared to that on a duplicate blot hybridized to the total input DNA. As a positive control, cloned α-satellite DNA (αRI) (Jorgensen et al. 1988) was spotted onto the membrane and similarly hybridized. Quantitation by phosphorimaging indicated 200–400 × enhancement of the α-satellite DNA spot signal, showing the fidelity of the immunoprecipitation procedure (data not shown).
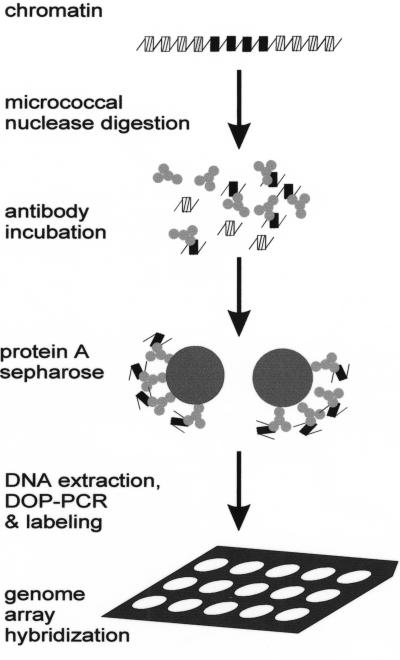
Figure 3.
Schematic representation of the combined chromatin immunoprecipitation and array (CIA) analysis.
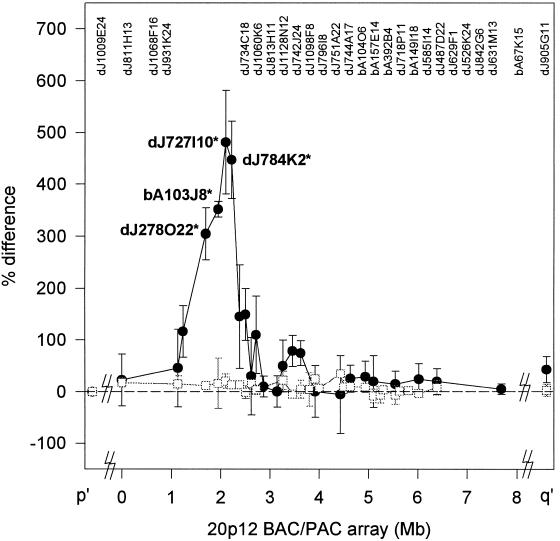
Similar hybridization analysis of the BAC/PAC array gave a distinct signal peak that represented the CENP-A-binding domain of the neocentromere (Fig. 4, filled circles and solid lines). The neocentromere-specific chromatin was contained within the four clones dJ278O22, bA103J8, dJ727I10, and dJ784K2 (P < 0.01 when compared with the baseline values of surrounding BAC/PAC clones), which together defined a genomic region of ∼460 kb. No significant peak was observed when the CIA procedure was repeated on a different control normal human lymphoblast cell line (Fig. 4, open squares and dotted lines) or when preimmune or unrelated antisera were used (data not shown).
Figure 4.
 |
CENP-A-Binding Domain Shows Slightly-Increased AT Content
The AT nucleotide content of the CENP-A-binding region was compared with those of the surrounding region using the completed primary nucleotide sequences of the Celera Discovery Database. To facilitate this analysis, these regions were arbitrarily divided into 37 smaller segments of predominantly 125 kb each (described in Table 1 legend). Analysis of the overall sequence composition of the entire arrayed region revealed an AT content for each of the segments ranging from 62.8% to 56.4%, with an average of 60.8%. This value was slightly higher than the reported average AT content of the genome of 58.0% (Smit 1999). The average AT content of the CENP-A binding region was 61.1% and was not significantly different from the value of 60.7% for the non-CENP-A-binding sequences of the region. Interestingly, when the AT content for human α-satellite DNA was calculated from its consensus sequence (Choo et al. 1991; Choo 1997b), a similar higher-than-genome-average value of 62.6% was obtained (J. Craig, unpubl.). These analyses suggest that the overall chromosomal region containing the CENP-A-binding domain and surrounding sequences has a slightly higher AT content compared to that of the total genomic DNA.
Table 1.
Summary of the in silico Analysis of the CENP-A-Binding Region of the 20p12 Neocentromere
| Sequence motifs | CENP-A- binding region | Nonbinding region | P value | Genome average | AT-rich average (>64%) | |
|---|---|---|---|---|---|---|
| Base composition | AT, % | 61.1 | 60.7 | >0.05 | 58.0a | n/a |
| Repeats | Tandem repeatsb, % | 2.11 | 2.20 | >0.05 | n/a | n/a |
| Satellitescf, % | 0 | 0 | 1 | n/a | n/a | |
| LINEs, % | 15.1 | 17.3 | >0.05 | 16.6a | 23.1a | |
| L1, % | 11.6 | 14.1 | >0.05 | 13.1a | 20.0a | |
| SINEs, % | 5.49 | 7.23 | <0.05 | 14.9a | 6.5a | |
| Alus, % | 3.99 | 5.67 | <0.05 | 12.5a | 6.0a | |
| MIRs, % | 1.50 | 1.47 | >0.05 | n/a | n/a | |
| LTRs, % | 6.94 | 9.13 | <0.05 | 7.4a | 7.4a | |
| DNA elements, % | 2.54 | 3.40 | >0.05 | n/a | n/a | |
| Protein binding motifs | HMG1de | 17.6 | 19.3 | >0.05 | 17.7f | n/a |
| Topo IIde | 0.07 | 0.06 | >0.05 | n/a | n/a | |
| CENP-Beg | 0 | 0 | 1 | n/a | n/a | |
| pJαde | 0.01 | 0.02 | >0.05 | n/a | n/a |
To facilitate statistical analysis of differences between the two regions, the 460-kb CENP-A-binding region was split into three equal segments of 125 kb and one 87 kb segment, which were analyzed separately. Similarly, the nonbinding regions were split into 31 segments of 125 kb and two of 144 kb and 142 kb. Results are averages for each region unless otherwise stated, and P values from t-tests comparing both regions are also given. Where known, values for average genomic sequences and genomic AT-rich sequences are quoted in the final two columns.
Tandem repeats were classed as being ≥2 copies of any repeat of unit length ≥2 bp.
Satellite repeats (with % homology to consensus): α-satellite (80%); β-satellite (100%); γ-satellite (100%); 48-bp repeat (100%); classical satellites Ia and Ib (100%); classical satellite III (100%, ≥2 copies in tandem).
per kb.
100% homology to consensus.
From Barry et al. (1999).
Number of matches.
n/a, not applicable/attempted.
CENP-A-Binding Domain Shows Lower Prevalence of LTRs, SINEs, and Alus
Using BLAST analysis, none of the sequences in the CENP-A binding region showed significant sequence similarity to the consensus-centromeric α-satellite DNA or the pericentric β-satellite, γ-satellite, and classical satellites IA and IIB (Choo 1997b; Table 1). Similarly, sequences related to the 28-bp AT-rich tandem repeat (AT28) previously identified as an ∼600-bp domain in the 10q25.2 neocentromere region (Barry et al. 1999; Koch 2000) were also not found, although various other smaller (10–50-bp) tandem repeats that ranged in copy number from 10 to 28 were detected.
Examination of the patterns of other repetitive sequences including the human transposable elements and other classes of interspersed repeats indicated that these were not significantly different from the surrounding genomic sequences as scanned with RepeatMasker. The only exceptions were the LTRs, SINEs, and Alus. The prevalence of LTRs in the CENP-A-binding domain (mean = 6.94%) was significantly lower than that of the non-CENP-A-binding sequences (mean = 9.13%; P < 0.05; Table 1) and slightly lower than that of the genome average (7.4%). Similarly, the prevalence of SINEs and Alus (mean = 5.49% and 3.99%, respectively) was lower in the CENP-A-binding region than in the non-CENP-A-binding region (mean = 7.23% and 5.67% and P < 0.05, respectively). The prevalence of SINEs and Alus in the CENP-A-binding region was also lower than the average content of these sequence motifs in the genome (14.1% and 11.8%, respectively; Smit 1999).
Genes May Be Present in the CENP-A-Binding Region
When scanned with the CPGPLOT using stringent parameters (length > 400 bp with a GC content of 55%), a highly probable CpG island indicative of the presence of a functional gene was detected within the CENP-A-binding region. In addition, in accordance with the information provided by the NCBI database, at least one other novel uncharacterized gene was present in the CENP-A-binding region. Furthermore, PowerBLAST search for an EST in the CENP-A-binding domain revealed that each of the four segments in the CENP-A-binding region contained at least one EST, suggesting that these sequences might contain expressed genes. Further analysis using the GRAIL gene prediction program also suggests the presence of exons and ESTs in the whole CENP-A-binding region.
Normal Prevalence of Putative DNA-Binding Protein Motifs
The prevalence of putative sequence motifs for several known DNA-binding proteins was determined. Two of these motifs were the degenerate CENP-B box (TTCGNN NNANNCGGG; Kipling et al. 1995) and the pJα sequence (GTGAAAAAG; Gaff et al. 1994). No significant sequence identity was identified for the CENP-B box motif, while a low level of the pJα motif that was not significantly different from that seen in the non-CENP-A binding domain was observed (Table 1). The distribution of Topoisomerase II (TopoII) and high-mobility-group protein I (HMGI) motifs was also determined. The 18-bp TopoII-binding sequence RNYNNC NNCYNGKTYNY (Spitzner and Muller 1988) was also searched for. HMGI is a protein that recognizes stretches of six A or T nucleotides (Solomon et al. 1986). The array WWWWWWS was used to search for this pattern, as it allowed the omission of overlapping matches where there were more than six A or T nucleotides. Both the TopoII and HMGI motifs were found to be distributed throughout the CENP-A binding region at frequencies that were not significantly different from those of other adjacent sequences (P > 0.05; Table 1).
DISCUSSION
CIA Analysis Allows the Rapid Identification of the Functional DNA Domain of a Neocentromere and Other Applications
The immunofluorescence/FISH procedure we used initially to investigate the genomic region corresponding to the 20p12 neocentromere has been previously employed to identify the CREST protein-binding domain of a human 10q25 neocentromere on the mardel(10) chromosome (du Sart et al. 1997). A higher resolution was achieved in that study because we were more successful in mechanically stretching the mardel(10) marker chromosome and because significantly smaller cosmid probes of ∼35-kb insert sizes were used. In contrast, mechanical stretching of the invdup(20p) chromosome was difficult, probably because of its small size, and the significantly larger BAC/PAC probes (average 150 kb) gave intense FISH signals that compromised resolution. The CIA method described in this study overcomes these problems, alleviates the need to subclone the BAC/PACs to provide smaller probes, and eliminates the tedious task of scoring large numbers of metaphases. It offers a general and rapid molecular approach to identify the functional domain of any neocentromere for which a genomic array is available. Our demonstration that the method is discriminatory in a cell line containing three copies of the DNA corresponding to the neocentromere region (two from the 20p12 inverted duplication marker chromosome and one from the normal chromosome 20) indicates that it can be directly applied to patient cell lines without the need to isolate the chromosome of interest in somatic cell hybrids.
In addition to mapping active neocentromere domains, the CIA method should be useful for studying other chromosomal or genomic properties for which specific antibodies or differential physico-chemical characteristics are available or discernible. These characteristics include chemical modifications (e.g., histone acetylation and phosphorylation), differences in detergent or salt extractability (such as scaffold attachment regions), imprinting status, and higher-order chromatin conformational changes. The completion of the sequencing of many different genomes and the implementation of sophisticated automation and microarray formats should allow these studies to be performed at the level of the whole genome.
Localization and Sequence Analysis of 460 kb of CENP-A-Specific Chromatin at the 20p12 Neocentromere
CENP-A is an essential centromere protein that constitutes centromere-specific nucleosomes by replacing histone H3 in the core particle (Wolffe and Pruss 1996; Yoda et al. 2000). Using an antibody to CENP-A in CIA analysis, we have delineated a 460-kb genomic DNA domain that associates specifically with this protein at the 20p12 neocentromere. This domain defines a critical region of centromere-specific chromatin and is the first direct demonstration at the molecular level of the extent to which CENP-A binds to a neocentromere or any higher eukaryotic centromere. Within the 460-kb region, it is at present unclear whether CENP-A constitutes and replaces histone H3 in all the nucleosomes or whether it alternates with histone H3 in some unknown ratios to achieve the most stable centromeric chromatin structure (Choo 2000). Future studies will be required to discriminate between these two possibilities.
Sequence analysis of the CENP-A-binding region reveals the absence of related sequences that are found on normal centromeric and pericentromeric regions, including α-, β-, and γ-satellites, classical satellites, 48-bp repeats, and ATRS. Absence of these sequences conforms to the epigenetic theory that neocentromerization involves activation of underlying normal genomic DNA without the insertion of pre-existing centromeric DNA or other sequence alterations (Choo 2000; Maggert and Karpen 2000).
The CENP-A-binding domain appears to reside in a larger 20p12 genomic region (represented by the different BAC/PACs in our array) of slightly increased AT content compared to that for the general genome. The positioning of nucleosomes is thought to depend partly on the primary DNA sequence (Wolffe 1995). Because AT-rich sequences are more bendable, they may be more suitable for the formation of compact centromere chromatin (Luger et al. 1997). In vitro studies have indeed shown that nucleosomes seem to be more compact and arranged more regularly on α-satellite DNA when they are formed with CENP-A instead of histone H3 (Yoda et al. 2000). It therefore appears that an increased AT composition may provide a more favorable disposition for neocentromere formation in interstitial chromosomal regions.
We have analyzed the distribution of the different classes of interspersed repeats within the CENP-A-binding domain, including SINEs, LINEs, Alus, MIRs, LTRs, and various subclasses of DNA transposable elements (for review, see Smit 1999). The results indicate that except for the retrotransposable LTRs, SINEs, and Alus, the CENP-A-binding sequences show a relatively normal distribution of these interspersed repeats. The prevalence of LTRs in the CENP-A-binding domain is significantly lower than that of the surrounding DNA. The significance of this is unclear. It has been proposed that an abundance of such retrotransposable elements in any particular sequence may be indicative of, or contribute to, its instability (Calabretta et al. 1982), and thus the lower level of LTRs may provide a more stable environment for neocentromere formation. In addition, the SINE and Alu content of the CENP-A-binding region is significantly lower than that of the surrounding region. Indeed, given that the AT content of the CENP-A-binding region is 61.1%, we would have expected a relatively high SINE and Alu content of ∼14.1% and 11.8%, respectively (Smit 1999).
We have analyzed the prevalence of binding sites for DNA-interacting proteins including CENP-B, pJα, HMGI, and TopoII. These proteins have been shown or proposed to be associated with eukaryotic centromeres (Choo 1997b). CENP-B and pJα boxes are abundantly found in subsets of human α-satellite (Muro et al. 1992; Romanova et al. 1996). These sequence motifs are either absent (CENP-B box) or present (pJα) at a level that is not significantly different from that expected randomly.
HMGI proteins bind to DNA stretches containing six A or T nucleotides and have been shown to bind the 172-bp α-satellite repeats of the African green monkey in vitro. It has been proposed to facilitate the formation of higher-order nucleoprotein complexes by interacting with the minor groove of the DNA helix and binding to irregular structures (Johns 1982; Straus and Varshavsky 1984; Grosschedl et al. 1994). TopoII has been shown to alter the topology of DNA and is thought to have a role in a host of cellular processes requiring the modulation of double-stranded DNA, including chromosome condensation and segregation (Earnshaw and Heck 1985; Earnshaw et al. 1985; Gasser and Laemmli 1986; Roca 1995). This protein is evenly distributed on chromosomes and thus has been suggested to play a role in maintaining the integrity of the centromere structure and function (Rattner et al. 1996; Sumner 1996). Our analysis has revealed an abundance of putative HMGI and TopoII sites in the CENP-A binding region. However, these sites are randomly distributed at a frequency similar to those of the adjacent non-CENP-A-binding regions. Thus, it can be concluded that the presence of these sites is unlikely to play a significant role in neocentromerization. It is interesting that each of the BAC/PAC clones in the CENP-A-binding domain contains at least one EST, reflecting the possible presence of genes. Genes are generally not found in normal centromeres, and centromeric heterochromatin is known to silence gene activity (Elgin 1996). The question of whether these genes are expressed can be tested experimentally and will be the subject of future work.
Comparison of the sequences of the CENP-A-binding DNA for the invdup (20p) neocentromere with an 80-kb (E8) DNA previously identified on a mardel(10) neocentromere by CREST antibody using immunofluorescence/FISH technique (du Sart et al. 1997; Cancilla et al. 1998) has revealed that, despite some variations in the AT, LTR, SINE, and Alu contents of the invdup (20p) neocentromere and the presence of an AT-rich AT28 tandem repeat on the E8 DNA, both DNA sequences are not remarkably different from bulk genomic sequences. Neither the increased AT content nor the decreased LTR, SINE, and Alu level detected in the CENP-A-binding domain of invdup(20p) are seen on the E8 sequence (Barry et al. 1999). Conversely, the AT-rich AT28 tandem repeat detected on E8 is absent on the invdup (20p) DNA. However, it is noteworthy that the E8 DNA was identified using a technique that is quite different in sensitivity and resolution compared to the CIA technique and, as such, does not provide an unequivocal comparison. The availability of the CIA procedure should allow the CENP-A-binding DNA of not only mardel(10) but other neocentromeres to be readily identified for detailed sequence comparison and to further investigate if some general characteristics common to all neocentromeres exist.
METHODS
Chemicals and Cell lines
All chemicals used were of molecular biology grade and purchased from Sigma-Aldrich. An Epstein-Barr virus–transformed lymphoblastoid cell line was established from the patient with the marker chromosome invdup (20p). The cell line was cultured in RPMI 1640 supplemented with 20% fetal calf serum. Two lymphoblastoid cell lines from anonymous normal individuals were used as controls.
Antisera
Antisera CREST#6, provided by S. Wittingham and T. Kaye (Walter and Eliza Hall Institute of Medical Research, Melbourne), was from a patient with the autoimmune CREST disease, containing antibodies against centromere components including CENP-A and CENP-B (du Sart et al. 1997). Antihuman CENP-A polyclonal antibody was a whole serum produced by immunizing rabbit with synthetic peptide of the N terminus of human CENP-A amino acid sequences and has been shown to be useful in centromere studies (Howman et al. 2000). Other secondary antibodies were purchased from the Jackson ImmunoResearch Laboratory.
PAC and BAC Array
PAC and BAC clones were obtained from the BACPAC Resources Center. An array map around the cytogenetic location of 20p12 was constructed (Fig. 1). The sequence and mapping data were mainly produced by the Chromosome 20 Sequencing Group at the Sanger Centre and were obtained from the World Wide Web (http://www.sanger.ac.uk/HGP/Chr20). Gaps were filled by screening a human genomic BAC library, RPCI-11, using the end sequences of adjacent BAC/PACs. Addresses with prefixes of “dJ” are PACs and those with “bA” are BACs. The BAC/PAC contig was verified and extended using sequence data from the Celera Discovery System and Celera's associated databases (http://www.celera.com). This resulted in 99.8% complete sequences for the 4.5-Mb contig. DNA was obtained using the MAXI plasmid preparation kit (QIAGEN) following the manufacturer's instructions. Probes for FISH were labeled with digoxigenin by nick translation (Nick Translation Kit, Roche Diagnostics).
Immunofluorescence/FISH Analysis
This was performed as described previously (du Sart et al. 1997). Briefly, metaphase chromosomes were obtained by treating actively replicating lymphoblasts with 10 μM nocodazole at 37°C for 1 h. Then, 200 μL of hypotonically swelled cells (5 × 104 cells/mL) was centrifuged onto clean microscope slides using Cytopsin 2 (Shandon) at 1000 r.p.m. for 5 min. Cells were then incubated with 1 : 100 CREST#6 antisera and detected with 1 : 100 Texas Red conjugated rabbit antihuman antibody. Slides were fixed in 4% formalin and then postfixed in ice-cold methanol/acetic acid before denaturation for 7 min in 70% formamide in 2× SSC (pH 7.0) at 82°C followed by dehydration in ice-cold ethanol. Then, 200 ng of digoxigenin-labeled PAC/BAC DNA preannealed with Cot-1 DNA (Roche) was hybridized to the slides at high stringency (50% formamide at 37°C). Hybridization was detected using 1 : 50 mouse antidigoxigenin antibodies and 1 : 50 goat FITC conjugated antimouse antibodies after three high-stringency washes in 0.1× SSC at 61°C. Chromosomes were counterstained using 4,6-diamindo-2-phenylindole (DAPI; 0.25 μg/mL) in Vectashield antifade mountant (Vector Laboratories).
Epifluorescence microscopy was performed on a Zeiss Axoplan II (Carl Zeiss) mounted with appropriate filter sets. Images were digitally acquired using a cooled charge-coupled device video camera (SenSys 2, Photometrics) connected to a PowerMac G3 personal computer controlled by the software IP Lab version 2.5.5 (Scanalytics).
Chromatin Immunoprecipitation and Array (CIA) Analysis
Chromatin immunoprecipitation was carried out as described (Johnson et al. 1998) with slight modifications. Briefly, cells were grown to 70% confluence and harvested. About 107 cells were incubated in TBS (0.01 M Tris-HCl [pH 7.5], 3 mM CaCl2, 2 mM MgCl2 with 0.1mM phenylmethylsulphonyl fluoride [PMSF] and proteinase inhibitors [Complete, Proteinase Inhibitor Cocktail Tablet, Roche]) with 0.25% Tween 40 at 4°C on a roller stirrer for 2 h before extruding the nuclei using 30 strokes with the “Tight” or “A” prestle on a Dounce homogenizor (Wheaton). Nuclei were separated from cytoplasmic debris by centrifugation at 1500 g for 20 min at 4°C through a 25%/50% discontinuous sucrose gradient. Oligonucleosomes were produced by digesting the nuclei with micrococcal nuclease (USB) in digestion buffer (0.32 M sucrose, 50 mM Tris-HCl at pH 7.5, 4 mM MgCl2, 1 mM CaCl2, 0.1 mM PMSF) at a concentration of 80 U/mg DNA at 37°C for 10 min. The reaction mix was then centrifuged at 15,000 g at 4°C. The supernatant contained mainly mononucleosomes. The pellet fraction was further processed by incubation with lysis buffer (1 mM Tris-HCl at pH 7.5, 0.2 mM EDTA, 0.2 mM PMSF, and proteinase inhibitors) on ice for 1 h. The final supernatant containing oligonucleosomes was then obtained by centrifugation at 15,000 g for 5 min at 4°C. The two supernatant fractions were pooled and precleared by the incubation with 1 : 1000 dilution of the preimmunized rabbit serum and 1% protein A-sepharose (Amerham-Pharmcia) at 4°C. After preclearing, the supernatant was obtained by centrifugation at 250 g for 5 min at 4°C. This fraction was used immediately for immunoprecipitation (input fraction). Equal volumes of the supernatant and incubation buffer (50 mM NaCl, 20 mM Tris-HCl at pH 7.5, 5 mM EDTA, 0.1mM PMSF, and protease inhibitors) were incubated with 1 : 500 anti-CENP-A at 4°C overnight. The immune complexes were then captured by incubating in 12.5% protein A-sepharose at 4°C for 2 h. At the end of the incubation, the protein A-sepharose was washed extensively in a stepwise manner in buffer A (50 mM Tris-HCl at pH 7.5, 10 mM EDTA) containing 50, 100, and 150 mM NaCl. Bounded immune complexes were then eluted with 2 vol of 1% SDS. DNA (bound fraction) was extracted from the eluate by phenol/chloroform/isoamyl alcohol extraction. Probes were then generated by degenerate oligonucleotide primed PCR (DOP–PCR) reaction (Telenius et al. 1992) using 500 ng of immunoprecipitated DNA.
An array of genome DNA was generated by immobilizing 100–200 ng of BAC/PAC DNA on Nylon membrane (Hybond N+, Amersham) in a dot blot format (Minifold SRC-96, Schleicher and Schuell). Between 500 and 600 ng of PCR-amplified DNA was labeled radioactively by random priming and used as probes. About 5 μg of human Cot-1 DNA was used for preannealing the probes. High-stringency hybridization and washes were carried out at 65°C. All blots were analyzed using the phosphorimager system (Storm 860 Gel and Blot Imaging System and the software ImageQuaNT version 4.2, Molecular Dynamics) running on an IBM-compatible personal computer. The ratio of the corresponding spot from hybridization with the bound fraction was compared to that of the input fraction. To correct for the inter- and intraexperimental variations, normalization was performed with the values of the PAC dJ1009E24, which mapped close to but distinct from the neocentromere region (see Fig. 1). The normalized ratio was then compared for the invdup(20p) cell line with a control cell line by expressing the difference of the two as a ratio of the value of the control.
To represent the data graphically, the midpoint of each BAC/PAC was used to plot the data point on the BAC/PAC array. The genomic distance was calculated relative to dJ811H13 on the p′ side of the neocentromere.
In Silico Sequence Analysis
Analysis was performed on 4.5 Mb of DNA. To facilitate handling and comparison of properties within the region, the sequence was divided into smaller regions. The CENP-A binding region was divided into three segments of 125 kb and one segment of 87 kb whereas the surrounding regions were divided into 31 segments of 125 kb and one segment of 145 kb and one of 142 kb.
The AT composition of the entire contig was calculated using the Web-based RepeatMasker (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker). This program was also used to identify simple repetitive regions and human transposable elements. AT-rich islands were determined with BASEPAIRPLOT and CPGPLOT on a Web-based interface (http://www.angis.org.au) at Australian National Institute for Genome Information Service (ANGIS). Tandem repeats were identified with TANDEM at ANGIS. Sequence motifs such as pJα, TopoII, and HMGI were determined by using BLAST at NCBI and FINDPATTERNS at ANGIS. Gene prediction was carried out by searching CpG islands using CPGPLOT (EMBOSS; http://bioweb.pasteur.fr) and by using the Web-based GRAIL version 1.3 (http://compbio.ornl.gov/Grail-1.3/).
Acknowledgments
A.W.I.L. was supported by a Melbourne International Research Scholarship and an International Postgraduate Research Scholarship. This work was supported by National Health and Medical Research Council of Australia.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL choo@cryptic.rch.unimelb.edu.au; FAX 61-3-9348-1391.
Article published on-line before print: Genome Res., 10.1101/gr.167601.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.167601.
REFERENCES
- Barry AE, Howman EV, Cancilla MR, Saffery R, Choo KHA. Sequence analysis of an 80kb human neocentromere DNA. Hum Mol Genet. 1999;8:217–227. doi: 10.1093/hmg/8.2.217. [DOI] [PubMed] [Google Scholar]
- Barry AE, Bateman M, Howman EV, Cancilla MR, Tainton KM, Irvine DV, Saffery R, Choo KHA. The 10q25 neocentromere and its inactive progenitor have identical primary nucleotide sequence: Further evidence for epigenetic modification. Genome Res. 2000;10:832–838. doi: 10.1101/gr.10.6.832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwitz BJ, Ahmad K, Moore LL, Roth MB, Henikoff S. A histone-H3-like protein in C. elegans. Nature. 2000;401:547–548. doi: 10.1038/44062. [DOI] [PubMed] [Google Scholar]
- Calabretta B, Robberson DL, Barera-Saldana HA, Lambrou TP, Saunders GF. Genome instability in a region of human DNA enriched with Alu repeat sequences. Nature. 1982;296:219–225. doi: 10.1038/296219a0. [DOI] [PubMed] [Google Scholar]
- Cancilla MR, Tainton KM, Barry AE, Lorionov V, Kouprina N, Resnick MA, du Sart D, Choo KHA. Direct cloning of human 10q25 neocentromere DNA using transformation-associated recombination (TAR) in yeast. Genomics. 1998;47:399–404. doi: 10.1006/geno.1997.5129. [DOI] [PubMed] [Google Scholar]
- Choo KHA. Centromere DNA Dynamics: Latent centromeres and neocentromere formation. Am J Hum Genet. 1997a;61:1225–1233. doi: 10.1086/301657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— . The centromere. Oxford: Oxford University Press; 1997b. [Google Scholar]
- ————— Centromerisation. Trends Cell Biol. 2000;10:182–188. doi: 10.1016/s0962-8924(00)01739-6. [DOI] [PubMed] [Google Scholar]
- Choo KH, Vissel B, Nagy A, Earle E, Kalitsis P. A survey of the genomic distribution of α-satellite DNA on all human chromosomes, and derivation of a new consensus sequence. Nucleic Acid Res. 1991;19:1170–1182. doi: 10.1093/nar/19.6.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Sart D, Cancilla MR, Earle E, Mao J-I, Saffery R, Tainton KM, Kalitsis P, Martyn J, Barry AE, Choo KHA. A functional neo-centromere formed through activation of a latent human centromere and consisting of non-alpha-satellite DNA. Nat Genet. 1997;16:144–153. doi: 10.1038/ng0697-144. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Heck M. Localization of topoisomerase II in mitotic chromosomes. J Cell Biol. 1985;100:1716–1725. doi: 10.1083/jcb.100.5.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw WC, Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Halligan B, Cooke CA, Heck M, Liu L. Topoisomerase II is a structural component of mitotic chromosome scaffolds. J Cell Biol. 1985;100:1706–1715. doi: 10.1083/jcb.100.5.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin SCR. Heterochromatin and gene regulation in Drosophila. Curr Opin Genet Dev. 1996;6:193–202. doi: 10.1016/s0959-437x(96)80050-5. [DOI] [PubMed] [Google Scholar]
- Figueroa J, Saffrich R, Ansorge W, Valdivia M. Microinjection of antibodies to centromere protein CENP-A arrests cells in interphase but does not prevent mitosis. Chromosoma. 1998;107:397–405. doi: 10.1007/s004120050323. [DOI] [PubMed] [Google Scholar]
- Gaff C, du Sart D, Kalitsis P, Ianello R, Nagy A, Choo KHA. A novel nuclear protein binds centromeric alpha satellite DNA. Hum Mol Genet. 1994;3:711–716. doi: 10.1093/hmg/3.5.711. [DOI] [PubMed] [Google Scholar]
- Gasser S, Laemmli U. The organisation of chromatin loops: Characterisation of a scaffold attachment site. EMBO J. 1986;5:511–518. doi: 10.1002/j.1460-2075.1986.tb04240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosschedl R, Giese K, Pagel J. HMG domain proteins: Architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994;10:94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- Howman EV, Fowler KJ, Newson AJ, Redward S, MacDonald AC, Kalitsis P, Choo KHA. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc Natl Acad Sci. 2000;97:1148–1153. doi: 10.1073/pnas.97.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns EW. The HMG Chromosomal Proteins. New York: Academic Press; 1982. [Google Scholar]
- Johnson CA, O'Neill LP, Mitchell A, Turner BM. Distinctive patterns of histone H4 acetylation are associated with defined sequence elements within both heterochromatic and euchromatic regions of the human genome. Nucleic Acid Res. 1998;26:994–1001. doi: 10.1093/nar/26.4.994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen AL, Kolvraa S, Jones C, Bak AL. A subfamily of alphoid repetitive DNA shared by the NOR-bearing human chromosome 14 and 22. Genomics. 1988;3:100–109. doi: 10.1016/0888-7543(88)90139-5. [DOI] [PubMed] [Google Scholar]
- Kipling D, Mitchell A, Masumoto H, Wilson H, Nicol L, Cooke H. CENP-B binds a novel centromeric sequence in the Asian mouse Mus caroli. Mol Cell Biol. 1995;15:409–4020. doi: 10.1128/mcb.15.8.4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch J. Neocentromeres and α satellite: A proposed structural code for functional human centromere DNA. Hum Mol Genet. 2000;9:149–154. doi: 10.1093/hmg/9.2.149. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Maggert KA, Karpen GH. Acquisition and metastability of centromere identity and function: Sequence analysis of a human neocentromere. Genome Res. 2000;10:725–728. doi: 10.1101/gr.10.6.725. [DOI] [PubMed] [Google Scholar]
- Muro Y, Masumoto H, Yoda K, Nozaki N, Ohashi M, Okazaki T. Centromere protein B assembles human centromeric α-satellite DNA at the 17-bp sequence. J Cell Biol. 1992;116:1081–1093. doi: 10.1083/jcb.116.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner JB, Hendzel MJ, Sommer Furbee C, Muller MT, Bazett-Jones DP. Topoisomerase IIα is associated with the mammalian centromere in a cell cycle– and species-specific manner and is required for proper centromere/kinetochore structure. J Cell Biol. 1996;134:1097–1107. doi: 10.1083/jcb.134.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca J. The mechanism of DNA topoisomerases. Trends Biochem Sci. 1995;20:156–160. doi: 10.1016/s0968-0004(00)88993-8. [DOI] [PubMed] [Google Scholar]
- Romanova LY, Deriagan GV, Mashkova TD, Tumeneva IG, Mushegian AR, Kisselev LL, Alexandrov IA. Evidence for selection in evolution of alpha satellite DNA: The central role of CENP-B/pJα binding region. J Mol Biol. 1996;261:334–340. doi: 10.1006/jmbi.1996.0466. [DOI] [PubMed] [Google Scholar]
- Saffery R, Irvine DV, Griffiths B, Kalitsis P, Wordeman L, Choo KHA. Human centromeres and neocentromeres show identical distribution patterns of >20 functional important kinetochore-associated proteins. Hum Mol Genet. 2000;9:175–185. doi: 10.1093/hmg/9.2.175. [DOI] [PubMed] [Google Scholar]
- Solomon M, Strauss F, Varshavsky A. A mammalian high mobility group protein recognizes any stretch of size A.T base pairs in duplex DNA. Proc Natl Acad Sci. 1986;89:1695–1699. doi: 10.1073/pnas.83.5.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev. 1998;6:743–748. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- Spitzner JR, Muller MT. A consensus sequence for cleavage by vertebrate DNA topoisomerase II. Nucleic Acids Res. 1988;16:5533–5556. doi: 10.1093/nar/16.12.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straus F, Varshavsky A. A protein binds to satellite DNA repeat at three specific sites that would be brought into mutual proximity by DNA folding in the nucleosome. Cell. 1984;37:889–901. doi: 10.1016/0092-8674(84)90424-0. [DOI] [PubMed] [Google Scholar]
- Sullivan KF, Hechenberger M, Masri K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner A. The distribution of topoisomerase II on mammalian chromosomes. Chromo Res. 1996;4:4–5. doi: 10.1007/BF02254938. [DOI] [PubMed] [Google Scholar]
- Telenius H, Carter NP, Bebb CE, Nordenskjold M, Ponder BAJ, Tunnacliffe A. Degenerate oligonucleotide–primed PCR: general amplification of target DNA by a single degenerate primer. Genomics. 1992;13:718–725. doi: 10.1016/0888-7543(92)90147-k. [DOI] [PubMed] [Google Scholar]
- Vafa O, Sullivan KF. Chromatin containing CENP-A and α-satellite DNA is a major component of the inner kinetochore plate. Curr Biol. 1997;7:897–900. doi: 10.1016/s0960-9822(06)00381-2. [DOI] [PubMed] [Google Scholar]
- Vafa O, Shelby RD, Sullivan KF. CENP-A associated complex satellite DNA in the kinetochore of the Indian muntjac. Chromosoma. 1999;108:367–374. doi: 10.1007/s004120050388. [DOI] [PubMed] [Google Scholar]
- Valdivia MM, Figueroa J, Iglesias C, Ortiz M. A novel centromere monospecific serum to a human autoepitope on the histone H3-like protein CENP-A. FEBS Let. 1998;422:5–9. doi: 10.1016/s0014-5793(97)01583-4. [DOI] [PubMed] [Google Scholar]
- Voullaire L, Saffery R, Davies J, Earle E, Kalitsis P, Slater H, Irvine DV, Choo KHA. Trisomy 20p resulting from inverted duplication and neocentromere formation. Am J Med Genet. 1999;85:403–408. [PubMed] [Google Scholar]
- Warburton, P.E., Cooke, C.A., Bourassa, S., Vafa, O., Sullivan, B.A., Stetten, G., Gimelli, G., Warburton, D., Tyler-Smith, C., Sullivan, K.F., et al. Immunolocalisation of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol.7: 901–904. [DOI] [PubMed]
- Warburton PE, Dolled M, Mahmood R, Alonso A, Li S, Naritomi K, Tohma T, Nagai T, Hasegaqa T, Ohashi H, et al. Molecular cytogenetic analysis of eight inversion duplications of human chromosome 13q that each contain a neocentromere. Am J Hum Genet. 2000;66:1794–1806. doi: 10.1086/302924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe AF. Chromatin: structure and function. 2nd ed. New York: Academic Press; 1995. [Google Scholar]
- Wolffe AP, Pruss D. Deviant nucleosomes: The functional specialisation of chromatin. Trends Genet. 1996;12:58–62. doi: 10.1016/0168-9525(96)81401-6. [DOI] [PubMed] [Google Scholar]
- Yoda K, Ando S, Morishita S, Houmura K, Hashimoto K, Takeyasu K, Okazaki T. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc Natl Acad Sci. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]