Abstract
The presence of additional hox clusters in the zebrafish has led to the hypothesis that there was a whole genome duplication at the origin of modern fish. To investigate the generality of this assumption, we analyzed all available actinopterygian fish gene families, and sequenced nuclear receptors from diverse teleost fish. The origin and timing of duplications was systematically determined by phylogenetic analysis. More genes are indeed found in zebrafish than in mouse. This abundance is shared by all major groups of euteleost fish, but not by eels. Phylogenetic analysis shows that it may result from frequent independent duplications, rather than from an ancestral genome duplication. We predict two zebrafish paralogs for most mouse or human genes, thus expressing a note of caution in functional comparison of fish and mammalian genomes. Redundancy appears to be the rule in fish developmental genetics. Finally, our results imply that the outcome of genome projects cannot be extrapolated easily between fish species.
It has long been supposed that gene duplication plays an essential role in the evolution of genomes and organisms (Ohno 1970). By increasing the number of available protein coding sequences, gene duplication may be an important precursor of evolutionary novelty (Ohno 1970; Holland et al. 1994). One of the best-studied cases of gene duplication probably took place during the early evolution of vertebrates (Ohno 1970): Multiple rounds of whole genome duplications (tetraploidization) could be involved in the morphological diversification of vertebrates. The hox gene complex, present in one copy in amphioxus, the sister group of vertebrates (Spruyt 1998), and four paralogous copies in human and mouse, provides the most spectacular support for this model (Holland et al. 1994; Holland 1999).
The recent increase in sequence data from ray-finned bony fish (Actinopterygii), and especially from model organisms such as the zebrafish, Danio rerio, and the fugu, Takifugu rubripes, has led to several observations of genes for which there were more copies in fish than in mammals. Here again the study of the hox gene complex has provided spectacular examples of gene duplication (Aparicio et al. 1997; Amores et al. 1998; Meyer et al. 1998; Prince et al. 1998a,b; Wittbrodt et al. 1998), with the zebrafish containing seven hox gene complexes. Following phylogenetic analysis of hox gene sequences and genetic mapping, a “chromosome duplication (probably whole genome duplication) in ray-finned fish before the teleost radiation” has been suggested (Amores et al. 1998). The ancestral genome duplication model predicts that more genes should be duplicated in fish than in mammals, that these duplications should predate the divergence of fish lineages, and thus that all fish should share a similar abundance of duplicated genes. Unfortunately, this “more genes in fish” hypothesis (Wittbrodt et al. 1998) is based on few gene families, besides hox complexes, and mainly two species (zebrafish and fugu), although hox complexes from medaka and stripped bass have been characterized.
Our aim in this work is to investigate whether there are indeed more duplicated genes in fish than in mammals, and which species or lineages are concerned. For this, we systematically investigate gene duplications in fish by three complementary approaches: (1) comparison of all available homologous genes between mouse and zebrafish, (2) investigation of the duplication patterns of all genes known in at least three orders of bony fish, and (3) characterization of nuclear hormone-receptor genes, using this superfamily as an alternative marker of genome evolution, in addition to hox genes. Because of lack of sequences in the databases, we essentially sampled teleost fish (Teleostei).
Nuclear receptors were chosen because their strong conservation allows amplification of transcripts by RT-PCR even in divergent species (Marchand et al. 2001), they are dispersed throughout the genome, thus allowing discrimination between gene and genome duplication, and they yield a clear phylogenetic signal allowing construction of robust phylogenies (Laudet et al. 1992). The latter two properties distinguish nuclear receptors from hox genes, although these have other advantages, such as their well documented roles in development.
RESULTS
Twice as Many Duplicated Genes in Zebrafish Than in Mouse
We did the first systematic comparison of gene families between these two major models of vertebrate genetics, checking the phylogenetic tree for each gene family. This allows us to select fish-specific duplications (Fig. 1). For most families investigated (80%), there is one gene in zebrafish and one in mouse, but more than twice as many duplications are detected in the zebrafish than in the mouse lineage (Table 1). The difference is significant by a paired signs test: P = 0.034. One would expect the number of duplicates detected to be directly correlated to the amount of sequencing done, which is 25 times higher for mouse (22,517 coding sequences in Hovergen release 38) than for zebrafish (910 coding sequences in Hovergen). Considering this, many more duplicate genes remain to be discovered in zebrafish than in mouse, in agreement with the experience of fish geneticists on isolate genes. Although we used the mouse because of its preeminence as a genetic and developmental model, results are totally identical with human sequences.
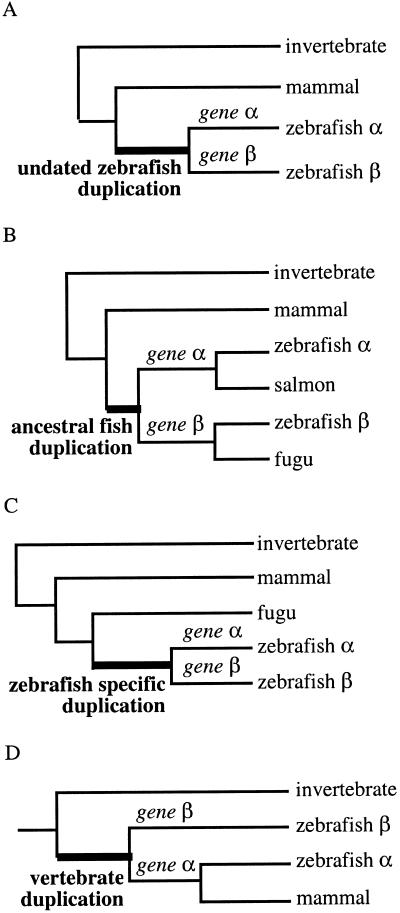
Figure 1.
Determining the origin of duplicated genes by phylogenetic analysis. Branch lengths are arbitrary and do not reflect evolutionary distance. The invertebrate sequence can be replaced by a paralog to root the tree, as described in Methods. Species represented here are arbitrary and depend on sequences available for each gene family. (A) The duplication happened somewhere in the lineage leading to zebrafish. As no other fish sequence is characterized, it may be specific of zebrafish (as in C), or ancestral to fish (as in B). No prediction of the number of genes in other fish is possible. (B) The duplication is shared by several major euteleost fish lineages, proving that the duplication happened in the common ancestor of these fish. The salmon gene should in fact be annotated α, and the fugu gene β. We predict that a salmon β and a fugu α gene should exist, as well as α and β genes in all other euteleost fish, except for secondary losses. (C) The duplication is specific of the zebrafish lineage, and the gene is not duplicated in other major fish lineages. There may be independent duplications in other lineages, but we cannot predict them. (D) The duplication is shared by all vertebrates, even though one of the paralogs was only found in zebrafish. The mammal gene should be annotated α. We predict that a β as well as an α gene should exist in all vertebrates, including mammals, except for secondary losses. Such genes were not used in our analysis.
Table 1.
Distribution of Orthologous Genes between Zebrafish and Mouse
| No. of copies in Zebrafish | No. of Copies in Mouse | No. of Gene Families |
|---|---|---|
| 1 | 1 | 153 |
| 1 | 2 or more | 11 |
| 2 or more | 1 | 26 |
| 2 | 2 | 1 |
| Total | 191 |
Only copies identified as lineage-specific gene duplications by phylogenetic analysis are counted.
Gene Duplications Characterize All Euteleost Fish
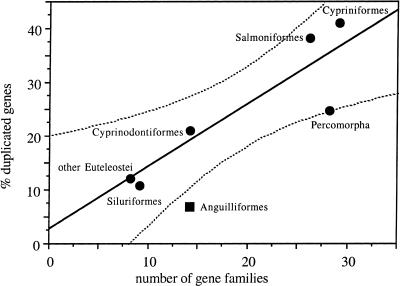
When we systematically sample a wider range of ray-finned fish (Actinopterygii), through phylogenetic analysis of 33 gene families, gene duplications are observed in all lineages (Table 2), and the lineage of zebrafish (Cypriniformes) does not stand out as particularly gene-rich. In fact, the more genes are characterized in a group, the higher the proportion of families with at least one duplication observed, with a significant correlation (R = 0.86; P = 0.014). The correlation increases when only euteleost fish are used (R = 0.89; P = 0.018; Fig. 2). Thus the number of duplicate genes known in an euteleost lineage is mostly dependent on the amount of sequencing done, implying similar frequencies of gene duplication among them. We expect the highest figure observed (41%) to be an underestimate, because sequencing effort is low even in cypriniformes, compared to mammals.
Table 2.
Distribution of Duplicated Genes in Fish Lineages
| Gene family | Actinopterygian lineage | Hovergen family no. | ||||||
|---|---|---|---|---|---|---|---|---|
| Cypriniformes | Percomorpha | Salmoniformes | Cyprinodontiformes | Siluriformes | Other Euteleostei | Anguilliformes | ||
| α globina | specific: 97 | specific: 70 | specific: 100 & 94 | no | no | FAM000215 | ||
| activin β b | specific: 82 | no | no | FAM000307 | ||||
| apo AI | no | no | specific: 100 | FAM001258 | ||||
| androgen receptorb | shared: 83 | specific: 100 | shared: 83 | FAM001375 | ||||
| shared: 83 | ||||||||
| aromatase | shared: 89 | shared: 89 | shared: 89 | shared: 89 | FAM000502 | |||
| CADc | no | no | no | FAM000951 | ||||
| cholecystokininb | shared: 94 | shared: 94 | specific: 100 | FAM003983 | ||||
| shared: 94 | ||||||||
| complement C3 | specific: 100 | no | no | specific: 100 | FAM000664 | |||
| EF1-α | no | no | no | FAM000591 | ||||
| ependymin | specific: 100 | no | specificd | no | no | FAM000398 | ||
| factor B | specific: 100 & 100 | specific: 100 | no | FAM001595 | ||||
| gonadotropin α | specific: 71 | no | specific: 83 | no | no | no | FAM000012 | |
| gonadotropin β | no | no | no | no | no | no | FAM000013 | |
| GnRHe | no | no | no | no | FMA002205 | |||
| GnRH IIe | no | no | no | no | no | FAM002205 | ||
| growth hormone | specific: 100 | no | specific: 100 | no | no | no | FAM000014 | |
| HNF forkhean domain | no | no | no | FAM001266 | ||||
| HSC70/HSP70 | specific: 100 | no | no | no | FAM000300 | |||
| ins-like growth factor II | no | no | no | FAM000006 | ||||
| lactate dehydrogenase A | no | no | no | no | FAM000364 | |||
| lactate dehydrogenase B | no | no | no | FAM000364 | ||||
| Na/H exchange | no | no | no | no | FAM000486 | |||
| OTX–Pit | specific: 100 | no | no | FAM001264 | ||||
| p53 | no | no | no | no | FAM001642 | |||
| prolactin | no | no | no | no | FAM000016 | |||
| Rag-1 | no | no | no | no | no | FAM000556 | ||
| somatolactin | no | specific: 100 | no | no | no | FAM000015 | ||
| TGF β2 | no | no | no | FAM000027 | ||||
| TSH β | no | no | no | FAM000013 | ||||
| trypsinogene | specific: 99 | specific: 100 | no | specific: 100f | FAM001232 | |||
| tyrosinase | no | no | no | FAM000871 | ||||
| tyrosine hydroxylase | no | no | no | FAM000388 | ||||
| zona pelucida ZP2 | specific: 98 | shared: 77 | shared: 77 | shared: 77 | no | FAM001134 | ||
| Number of gene families | 29 | 28 | 26 | 14 | 9 | 8 | 14 | |
| Specific duplicationsg (%) | 34.5 | 10.7 | 34.6 | 7.1 | 0.0 | 12.5 | 0.0 | |
| Total duplicationsh (%) | 41.4 | 25.0 | 38.5 | 21.4 | 11.1 | 12.5 | 7.1 | |
For each lineage in which a gene family has been characterized, the evidence for duplications is shown as follows: (Specific) evidence for a gene duplication specifically in this lineage, followed by bootstrap support; (shared) evidence for a gene duplication shared with at least one other lineage, followed by bootstrap support; (no) that there is no evidence for a gene duplication. When there are two independent gene duplications for the same gene family and the same lineage, both bootstrap supports are reported. Bootstrap support is the proportion of 2000 bootstrap replicates recovering the branch, using Neighbor-Joining with Poisson corrected distances, although other methods were also used (see text).
There may also be a duplication of a globin ancestral to all fish lineages sampled, but phylogenetic evidence is not conclusive.
For these gene families, there is both a duplication shared between lineages, and more recently a specific duplication in some of those lineages.
Carbamyl phosphate synthase.
Neighbor-Joining is not conclusive whether the duplication is shared with esociformes, but Maximum Likelihood supports a salmoniforme-specific duplication.
Gonadotropin-releasing hormone.
Specific to Gadiformes.
Number of gene families with at least one duplication specific of the lineage, divided by the number of gene families sampled for the lineage.
Number of gene families with at least one duplication, specific or shared, divided by the number of gene families sampled for the lineage.
Figure 2.
Correlation between evidence for duplicate genes and number of gene families characterized. (Vertical axis) proportion with a detected duplication among gene families characterized for each actinopterygian lineage. (Horizontal axis) number of gene families characterized for each actinopterygian lineage. The round points represent data for the euteleost lineages listed in Table 2. The square point represents data for Anguilliformes (eels). The straight line is the linear correlation for euteleost lineages (R = 0.89; P = 0.018). The curved dotted lines are 95% confidence interval of the correlation; notice that the point for eels is not included in this interval.
On the other hand, only one duplication is observed in anguilliformes, whereas more than three would be expected considering the number of sequences characterized and the correlation obtained in euteleost fish. In fact, they are the only sampled lineage that falls outside of the 95% confidence interval of the correlation (Fig. 2); this remains true when anguilliformes are used to compute the correlation. Data are scarce for other lineages, but for the three relevant gene families of our dataset, there are zero duplications in noneuteleost fish (data not shown). All this suggests that high levels of gene duplication are characteristic of euteleost fish only.
Several authors have suggested a genome duplication at the origin of fish (Holland et al. 1994; Amores et al. 1998; Postlethwait et al. 1998; Prince et al. 1998a; Meyer et al. 1999). Yet we notice many duplications specific of an order in Table 2, which even constitute a majority of duplications for the well-sampled cypriniformes and salmoniformes. On the other hand, very few duplications are shared by all sampled orders, even excluding anguilliformes to define “fish” as Euteleostei. This is consistent with our observation that most gene phylogenies are at odds with a unique whole-genome duplication at the origin of modern fish (M. Robinson-Rechavi et al., in prep).
Search for Duplicated Nuclear Receptor Genes
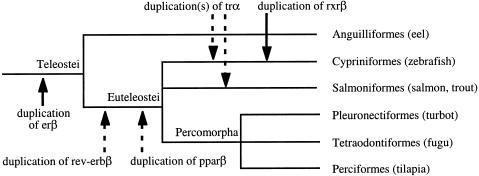
Out of concern that publicly available sequence data may be biased by the sequencing strategies that yielded them, we sequenced nuclear hormone receptors from seven species of fish, searching for duplicate genes (Fig. 3). There are ancient duplications for three nuclear receptors: pparβ (peroxysome proliferators activated receptor; NR1C2 [Nuclear Receptor Nomenclature Committee 1999]) and rev-erbβ (NR1D2) before the divergence of euteleost fish, and erβ (estrogen receptor β NR3A2) before the divergence of all teleosts, including the eel (Anguilliformes). The retinoic X receptor β (rxrβ NR2B2) gene is duplicated specifically in the zebrafish lineage (Cypriniformes). For trα (thyroid hormone receptor α NR1A1) we have identified at least two paralogs in the zebrafish, and two in the Atlantic salmon, but phylogenetic resolution is very poor, and we cannot identify the origin of the duplication(s). On the other hand, despite a total of 10 fish sampled from four major lineages for erα (estrogen receptor α NR3A1), and eight fish sampled from four lineages for trβ (thyroid hormone receptor β NR1A2), we identified no duplication for these genes.
Figure 3.
Duplications detected in a search for fish nuclear receptor genes. The tree represents a simplified phylogeny of the fish in which we characterized new sequences, although more sequences were used when available. Branch lengths are arbitrary, and do not reflect evolutionary distance. Solid arrows indicate origin of duplications, as determined by phylogenetic analysis. Broken arrows indicate possible origin of duplications; we cannot exclude that these are more ancient, but we can exclude that they are more recent. All duplications indicated are specific to actinopterygian fish.
There are fish-specific duplications for five out of seven of these receptors, whereas none is duplicated specifically in mammals. Moreover, all studied lineages are concerned by at least one duplication. This confirms that duplicated genes are abundantly present in all major fish lineages, and are as yet mostly undetected for lack of sequencing compared to mammals. When these nuclear receptor data are added to the previous analyses (data not shown), conclusions remain unchanged, notably the correlation between genes sampled and detection of duplications for euteleosts (R = 0.96), but not for anguilliformes.
Testing the Specificity of Previously Reported Gene Duplications
Several gene families have been cited previously as evidence for a genome duplication ancestral to bony fishes, because more copies were known in a fish than in mammals, but without any phylogenetic analysis. Phylogenetic analysis is necessary to determine whether genes indeed duplicated specifically in fish (Fig. 1A–C), or if the duplication is more ancient, and the copies are lost or undetected in mammals (Fig. 1D). We did a phylogenetic reconstruction of all available genes for each of these families.
Phylogeny significantly supports an ancestral bony fish duplication only for pax6 and sonic hedgehog. Duplications of genes from the TGF-β superfamily (bmp-2, bmp-4, lefty) and of msx homeobox genes are characterized only in the zebrafish, which does not allow to test the origin of the duplication, and hampers robust phylogenetic reconstruction. But they do appear to have duplicated after the divergence between the zebrafish and the mammalian lineages. In the case of msx, this is confirmed by synteny data (Barbazuk et al. 2000). Additional otx (Mori et al. 1994) and engrailed (Ekker et al. 1992) genes have been reported in the zebrafish, but phylogenetic resolution is too low to decide when the duplications took place.
In contradiction with the original report of additional notch genes in the zebrafish (Westin et al. 1997), we find that the notch1b (accession number Y10352) DNA sequence is 100% identical to previously reported notch1a (U57973). Zebrafish notch5 (Y10353) is identical to notch3 (U57975), and indeed groups with mammalian notch3 genes in a phylogenetic tree. As for the sequence reported as zebrafish notch6 (Y10354), it groups with mammalian and fugu notch2 genes, and is most probably zebrafish notch2. These sequences thus do not seem to represent gene duplications. The first reported zebrafish notch sequence (X69088) may represent a separate family of vertebrate notch genes, closest to notch-1. There is thus no evidence for fish specific duplications of notch genes.
DISCUSSION
A High Duplication Rate in Euteleost Fish
Following the discovery of extra hox gene complexes in the zebrafish (Amores et al. 1998; Prince et al. 1998a), the hypothesis of an ancestral genome duplication in bony fishes gained rapid popularity (Postlethwait et al. 1998; Holland 1999; Meyer et al. 1999). If this “more genes in fish” theory (Wittbrodt et al. 1998) fits with the empirical experience of zebrafish geneticists who often notice two copies of their favorite gene where only one was described in mice, it lacks solid backing. Indeed, hox gene complexes only give information on four loci in vertebrates, and cannot be assumed to represent the whole genome. Moreover, to attribute a duplication to an evolutionary lineage, such as bony fish, a phylogenetic analysis specifying the order of events (speciations and duplications) is necessary (Fig. 1). hox genes are not very good phylogenetic markers, and if the specificity of their duplication to bony fishes seems clear, data are not conclusive as to the age of this duplication: before the divergence of bony fishes (Amores et al. 1998), or specifically in the zebrafish lineage (Stellwag 1999). For other genes, reports consist mostly of gene counting (Ekker et al. 1992, 1997; Mori et al. 1994; Westin et al. 1997; Nornes et al. 1998), which is sometimes relevant, but does not allow any inference on the mechanisms or the age of gene or genome duplication. Our phylogenetic analysis of these genes shows the limits of such anecdotal data, because the origin of some duplications is unresolved, and some are resolved for the zebrafish but without information for other fish (Fig. 1A), while a shared fish-specific duplication is supported for only two genes (pax6 and sonic hedgehog). To overcome these difficulties, we have done a systematic study of all available genes in all well-studied bony fish.
It is clear that more duplicate genes will be detected for the most studied gene families and for the most studied species: they have been subjected to different “sequencing pressure.” This bias may be corrected mostly in the future by complete genomes. Meanwhile, the phylogenetic definition of gene duplication (Fig. 1), coupled to the systematic comparison of two species (Table 2), allows a rigorous test of a difference in duplicated genes between zebrafish and mouse. We thus prove for the first time that there are significantly more genes in the new model of vertebrate developmental genetics than in the most studied laboratory mammal. Our conclusions are unchanged, moreover, if human is used instead of mouse (data not shown).
We note that our data do not allow distinguishing between higher rates of gene duplication or lower rates of secondary gene loss. In both cases, the result is the same: the difference between duplication and loss results in an “efficient rate” of gene duplication, which is all we can measure. It is in any case the relevant factor for the number of paralog genes present in genomes. Thus we will not distinguish between these alternative evolutionary mechanisms.
To widen our taxonomic sampling, while still avoiding biases linked to sequencing pressure, we did a statistical correction on all available genes (Fig. 2), but also a systematic search of duplication for several members of a superfamily of genes dispersed through the genome. This allowed us to show that the abundance of gene duplications is specific neither to the zebrafish nor to its order, Cypriniformes. To the contrary, all major euteleost fish groups share similar numbers of duplicate genes. Yet a large number of these duplications occurred after the divergence of fish lineages. Synteny data do not appear conclusive on this question, because both linked and unlinked duplicate genes are found in the zebrafish (Barbazuk et al. 2000; Woods et al. 2000). Thus we show that (1) the zebrafish has more duplicated genes than the mouse, (2) other euteleost fish share similar numbers of duplicate genes, and (3) these gene duplications are often recent, not ancestral. Our conclusion is that, although there may not have been an ancestral genome duplication in fish evolution, independent gene or chromosome duplications are significantly more frequent in each euteleost lineage than in mammals, or are lost less frequently.
Specificity and Role of Gene Duplications
Is this high efficient duplication rate specific to euteleost fish? Other lineages are less well sampled, but the number of duplications detected in eels (Anguilliformes) is significantly lower than expected from the number of gene families sampled (Fig. 2). This remains true adding our new nuclear receptor sequences to the database sequences (not shown). Moreover, we never detected any duplication specific to the eel lineage. As eels are teleosts but not euteleosts, this suggests that the mechanism responsible for high rates of gene duplication was established after the divergence between euteleosts and other fish, but before the diversification of euteleosts.
The picture we obtain of a high frequency of gene duplications in all euteleost fish lineages is consistent with a proposed model of frequent single hox cluster duplications and losses, including specific hox chromosome duplications in cypriniformes (Stellwag 1999), although other explanations specific of the hox cluster are possible. This picture also is reminiscent of the demonstration that spliceosomal introns were gained many times independently in different fish lineages (Venkatesh et al. 1999). Moreover, it seems that genes accumulate substitutions significantly faster in fish than in mammals, independently of duplication (Robinson-Rechavi and Laudet 2001). Fish genomes thus appear very dynamic. What is the role of this dynamism? The additional genes supposed to have resulted from a genome duplication have been correlated with the diversification of ∼25,000 fish species (Vogel 1998), but the sister group of actinopterygians, sarcopterygians, also includes more than 21,000 known species (http://phylogeny.arizona.edu/tree), as diverse as snakes, birds, and whales. This does not suggest any relation between gene diversity and species diversity. We believe that the systematic study of expression patterns and protein specificity of the duplicated copies for many genes holds the key to understanding this major evolutionary thrust in the largest group of vertebrates (for an example in invertebrates, see Christophides et al. 2000).
Our results have three major consequences for the use of euteleost fish as model organisms. (1) We should expect on average to find two genes in fish for each gene identified in human or mouse. Of the 38 gene families with at least one duplication in mouse or zebrafish (43 including the nuclear receptors), more than two thirds have a duplication specific of the zebrafish lineage (72% including the nuclear receptors). Functional characterization of the fish ortholog of a mouse gene thus cannot be complete without a thorough search for a possible duplicate copy, and its eventual functional characterization, too. (2) The information about gene duplication obtained in one fish lineage cannot be extended systematically to another. There may be two copies in the zebrafish, yet only one in the turbot, as for rxrβ for example. Indeed, specific duplications appear abundant in all euteleost lineages well sampled so far. (3) In using data of fish genome projects to detect human genes, less redundancy of paralog genes should be expected in the human genome than in fish.
METHODS
New Nuclear Hormone Receptor Sequences
We searched for duplicate genes of nuclear hormone receptors in seven species of fish (Fig. 3). For this, total RNA was extracted by the guanidinium thiocyanate method, and purified with phenol-chloroform (Chomczynski et al. 1987) from frozen tissues of salmon (Salmo salar), rainbow trout (Oncorhynchus mykiss), zebrafish (D. rerio), tilapia (Oreochromis niloticus), turbot (Scophthalmus maximus) and eel (Anguilla anguilla). RNA from fugu (T. rubripes) was a generous gift from John Wentworth.
Five μg of total RNA were reverse transcribed using random primers or specific primers and Moloney Murine leukemia virus reverse transcriptase (MMLV-RT) in 20 μL of reaction mixture, according to the manufacturer's instructions (GIBCO-BRL, MMLV-RT kit). The resulting cDNA was amplified by PCR in 100 μL volume with 10 mM Tris-HCl pH 8.3, 50 mM KCl, 1.5 mM MgCl2 (Perkin-Elmer), 0.25 mM of each dXTP, 2.5 U Taq Gold DNA polymerase (Perkin-Elmer) and 300 ng of each primer. Degenerate PCR primers designed according to Escriva et al. (Escriva et al. 1999) were used in a “touch-down” PCR assay (Don et al. 1991). The complete cycle is: 94°C, 1 min; hybridization from 55 to 37°C, 1 min; 72°C, 1 min during 40 cycles. PCR products then were cloned into the PCR II vector (Invitrogen), and sequenced. For each amplified receptor, two independent clones were fully sequenced; in case of mismatches a third clone was sequenced for confirmation of the correct sequence.
New sequences were deposited in GenBank under accession nos. AF342936–AF342950. These data were completed by extraction of all available homologous sequences from fishes and other vertebrates from public databases.
Bioinformatic Datasets of Homologous Genes
We characterized 258 protein-coding gene families with at least one copy in zebrafish (D. rerio) and at least one in mouse (Mus musculus) in release 38 (mars 2000) of Hovergen (Duret et al. 1990). By using orthologous genes from species who branched out prior to the zebrafish/mouse divergence (e.g., shark or Drosophila) we ascertained whether the duplications in mouse and zebrafish occurred independently within each lineage. Seven uncertain gene groups were removed, leaving us with a sample of 251 families. Outgroup sequences were either determined in Hovergen, or through a BLAST search (Altschul et al. 1990) against SWISS-PROT (Bairoch et al. 1999). In each case, validity of the outgroup was checked by phylogenetic reconstruction and reference to the literature. When a gene duplication was older than the zebrafish/mouse split, the paralogs were considered as two different families for our study. In some cases, we used paralogous genes whose duplication was older than the zebrafish/mouse split as outgroups. For example, the bmp2 (Bone morphogenic protein 2) tree was rooted with bmp4, and the bmp4 tree rooted with bmp2. The least divergent outgroup sequences were used preferentially in all cases. For 60 families, no outgroup was found, leading to a first dataset of 191 gene families. The complete list of these gene families is available upon request (marc.robinson@ens-lyon.fr).
All protein-coding gene families with at least three orthologs from Actinopterygii were selected from release 38 (mars 2000) of Hovergen (Duret et al. 1994). Families for which bootstrap support (see “Tree-Building Methods”) of all nodes relevant to our study were under 50% were excluded as nonreliable. This led to a second dataset of 33 gene families (Table 2).
We also extracted and aligned all members of each gene family previously cited in the literature in support of a genome duplication in fish. This constituted a third dataset.
Each alignment, corresponding to a gene family, was used for phylogenetic reconstruction. To avoid confusion with gene duplication, we checked for alternative splicing by comparison of sequences at the DNA level outside of the alternatively spliced region, as well as reference to the literature. All alignments and phylogenies are available upon request (marc.robinson@ens-lyon.fr).
Tree-Building Methods
The Hovergen interface includes a phylogenetic tree for each gene family (Duret 1994). For all datasets above, whenever there was any doubt on the quality of this tree, or conclusions were not clear, protein sequences were extracted. This was systematically done for nuclear receptors, and for gene families previously cited in support of the genome duplication hypothesis. Protein sequences were aligned automatically by CLUSTALW (Thomson et al. 1994), with manual correction in Seaview (Galtier et al. 1996). All analyses were done excluding all sites with at least one gap in the alignment.
Trees were reconstructed systematically by Neighbor-Joining (Saitou et al. 1987) with Poisson-corrected distances on amino acids, implemented in Phylo_win (Galtier et al. 1996). Support for branches in the tree was investigated by bootstrap (Felsenstein 1985) with 2000 replicates. If any doubt remained, other methods were used: quartet-puzzling Maximum Likelihood as implemented in Puzzle (Strimmer et al. 1996) with the JonesTaylorThornton (JTT) substitution model (Jones et al. 1992) and gamma distributed rate heterogeneity; Maximum Likelihood as implemented in ProtML (Kishino et al. 1990) if computationally feasible; and Neighbor-Joining with percent accepted mutation (PAM) matrix distances (Dayhoff et al. 1978), as implemented in Phylo_win [32].
Acknowledgments
We thank Franck Delaunay, Laurent Duret, Yann Guiguen, Dan Graur, Anna-Pavlina Haramis, Ioan Negrutiu, and Guy Perrière for critical reading of the manuscript. We thank Association de Recherche sur le Cancer, Centre National pour la Recherche Scientifique, European Molecular Biology Organisation, Fondation pour la Recherche Médicale, Ligue Nationale contre le Cancer, and Ministère de l'Education Nationale for financial support.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL marc.robinson@ens-lyon.fr; FAX 33 4 72 72 80 80.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.165601.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1994;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Aparicio S, Hawker K, Cottage A, Mikawa Y, Zou L, Venkatesh B, Chen E, Krumlauf R, Brenner S. Nat Genet. 1997;16:79–83. doi: 10.1038/ng0597-79. [DOI] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R. The SWISS-PROT protein sequence data bank and its supplement TrEMBL in 1999. Nucleic Acids Res. 1999;27:49–54. doi: 10.1093/nar/27.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbazuk WB, Korf I, Kadavi C, Heyen J, Tate S, Wun E, Bede JA, McPherson JD, Johnson SL. The syntenic relationship of the zebrafish and human genomes. Genome Res. 2000;10:1351–1358. doi: 10.1101/gr.144700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Christophides GK, Livadaras I, Savakis C, Komitopoulou K. Two medfly promoters that have originated by recent gene duplication drive distinct sex, tissue and temporal expression patterns. 2000. Genetics. 2000;156:173–182. doi: 10.1093/genetics/156.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayhoff MO, Schwartz R, Orcutt BC. In: Atlas of Protein Sequence and Structure. Dayhoff MO, editor. 5, Suppl. 3. Washington, DC.: Natl. Biomed. Res. Found.; 1978. pp. 345–352. [Google Scholar]
- Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS. Touchdown PCR to circumvent spurious priming gene duplication. Nucleic Acids Res. 1991;19:81–86. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D, Gouy M. HOVERGEN: a database of homologous vertebrate genes. Nucleic Acids Res. 1994;22:2360–2365. doi: 10.1093/nar/22.12.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekker M, Wegner J, Akimenko MA, Westerfield M. Coordinate embryonic expression of three zebrafish engrailed genes. Development. 1992;116:1001–1010. doi: 10.1242/dev.116.4.1001. [DOI] [PubMed] [Google Scholar]
- Ekker M, Akimenko MA, Allende ML, Smith R, Drouin G, LLangille RM, Weinberg ES, Westerfield M. Relationships among msx gene structure and function in Zebrafish and other vertebrates. Mol Biol Evol. 1997;14:1008–1022. doi: 10.1093/oxfordjournals.molbev.a025707. [DOI] [PubMed] [Google Scholar]
- Escriva H, Robinson M, Laudet V. Evolutionary biology of the nuclear receptor superfamily. In: Picard D, editor. Nuclear receptors. A practical approach. Oxford: Oxford University Press; 1999. pp. 1–28. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: Two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- Holland, P.W., Garcia-Fernandez, J., Williams, N.A., Sidow, A. 1994. Gene duplications and the origins of vertebrate development. Development (Suppl.) 125–133. [PubMed]
- Holland PW. Gene duplication: past, present and future. Cell Dev Biol. 1999;10:541–547. doi: 10.1006/scdb.1999.0335. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kishino H, Miyata T, Hasegawa M. Maximum likelihood inference of protein phylogeny and the origin of chloroplasts. J Mol Evol. 1990;30:151–160. [Google Scholar]
- Laudet V, Hänni C, Coll J, Catzeflis C, Stéhelin D. Evolution of the nuclear receptor gene family. EMBO J. 1992;11:1003–1013. doi: 10.1002/j.1460-2075.1992.tb05139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand O, Safi R, Escriva H, Van Rompaey E, Prunet P, Laudet V. Molecular cloning and characterization of thyroid hormone receptors in teleost fish. J Mol Endocrin. 2001;21:51–65. doi: 10.1677/jme.0.0260051. [DOI] [PubMed] [Google Scholar]
- Meyer A, Schartl M. Gene and genome duplications in vertebrates: The one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr Opin Cell Biol. 1999;1:699–704. doi: 10.1016/s0955-0674(99)00039-3. [DOI] [PubMed] [Google Scholar]
- Mori H, Miyazaki Y, Morita T, Nitta H, Mishina M. Different spatio-temporal expression of three otx homeoprotein transcripts during zebrafish embryogenesis. Brain Res Mol Brain Res. 1994;27:221–231. doi: 10.1016/0169-328x(94)90004-3. [DOI] [PubMed] [Google Scholar]
- Nornesk S, Clarkson M, Mikkola I, Pedersen M, Bardsley A, Martinez JP, Krauss S, Johansen T. Zebrafish contains two Pax6 genes involved in eye development. Mech Dev. 1998;77:185–196. doi: 10.1016/s0925-4773(98)00156-7. [DOI] [PubMed] [Google Scholar]
- Nuclear Receptor Nomenclature Committee. A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97:1–3. doi: 10.1016/s0092-8674(00)80726-6. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. Heidelberg, Germany: Springer-Verlang; 1970. [Google Scholar]
- Postlethwait JH, Yan YL, Gates MA, Horne S, Amores A, Brownlie A, Donovan A, Egan ES, Force A, Gong Z, et al. Vertebrate genome evolution and the zebrafish gene map. Nat Genet. 1998;18:345–349. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- Prince VE, Joly L, Ekker M, Ho RK. Zebrafish hox genes: Genomic organization and modified colinear expression patterns in the trunk. Development. 1998a;125:407–420. doi: 10.1242/dev.125.3.407. [DOI] [PubMed] [Google Scholar]
- Prince VE, Moens CB, Kimmel CB, Ho RK. Zebrafish hox genes: expression in the hindbrain region of wild-type and mutants of the segmentation gene, valentino. Development. 1998b;125:393–406. doi: 10.1242/dev.125.3.393. [DOI] [PubMed] [Google Scholar]
- Robinson-Rechavi M, Laudet V. Evolutionary rates of duplicate genes in fish and mammals. Mol Biol Evol. 2001;181:681–683. doi: 10.1093/oxfordjournals.molbev.a003849. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Spruyt N, Delarbre C, Gachelin G, Laudet V. Complete sequence of the amphioxus (Branchiostoma lanceolatum) mitochondrial genome: Relations to vertebrates. Nucleic Acids Res. 1998;26:3279–3285. doi: 10.1093/nar/26.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwag EJ. Hox gene duplication in fish. Semin Cell Dev Biol. 1999;10:531–540. doi: 10.1006/scdb.1999.0334. [DOI] [PubMed] [Google Scholar]
- Strimmer K, Von Haeseler A. Quartet puzzling: A quartet maximum likelihood method for reconstructing tree topologies. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- Thomson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh B, Ning Y, Brenner S. Late changes in spliceosomal introns define clades in vertebrate evolution. Proc Natl Acad Sci. 1999;96:10267–10271. doi: 10.1073/pnas.96.18.10267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel G. Doubled genes may explain fish diversity. Science. 1998;281:1119–1121. doi: 10.1126/science.281.5380.1119. [DOI] [PubMed] [Google Scholar]
- Westin J, Lardelli M. Three novel Notch genes in zebrafish: Implications for vertebrate Notch gene evolution and function. Dev Genes Evol. 1997;207:51–63. doi: 10.1007/s004270050091. [DOI] [PubMed] [Google Scholar]
- Wittbrodt J, Meyer A, Schartl M. More genes in fish? Bioessays. 1998;20:511–515. [Google Scholar]
- Woods IG, Kelly PD, Chu F, Ngo-Hazelett P, Yan YL, Huang H, Postlethwait JH, Talbot WS. A comparative map of the zebrafish genome. Genome Res. 2000;10:1903–1914. doi: 10.1101/gr.10.12.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]