Abstract
Cobalamin (Cbl, B12) is an essential micronutrient required to fulfill the enzymatic reactions of cytosolic methylcobalamin-dependent methionine synthase and mitochondrial adenosylcobalamin-dependent methylmalonyl-CoA mutase. Mutations in the MMACHC gene (cblC complementation group) disrupt processing of the upper-axial ligand of newly internalized cobalamins, leading to functional deficiency of the vitamin. Patients with cblC disease present with both hyperhomocysteinemia and methylmalonic acidemia, cognitive dysfunction, and megaloblastic anemia. In the present study we show that cultured skin fibroblasts from cblC patients export increased levels of both homocysteine and methylmalonic acid compared to control skin fibroblasts, and that they also have decreased levels of total intracellular folates. This is consistent with the clinical phenotype of functional cobalamin deficiency in vivo. The protein changes that accompany human functional Cbl deficiency are unknown. The proteome of control and cblC fibroblasts was quantitatively examined by two dimensional in-gel electrophoresis (2D-DIGE) and liquid chromatography-electrospray ionization-mass spectrometry (LC/ESI/MS). Major changes were observed in the expression levels of proteins involved in cytoskeleton organization and assembly, the neurological system and cell signaling. Pathway analysis of the differentially expressed proteins demonstrated strong associations with neurological disorders, muscular and skeletal disorders, and cardiovascular diseases in the cblC mutant cell lines. Supplementation of the cell cultures with hydroxocobalamin did not restore the cblC proteome to the patterns of expression observed in control cells. These results concur with the observed phenotype of patients with the cblC disorder and their sometimes poor response to treatment with hydroxocobalamin. Our findings could be valuable for designing alternative therapies to alleviate the clinical manifestation of the cblC disorder, as some of the protein changes detected in our study are common hallmarks of known pathologies such as Alzheimer’s and Parkinson's diseases as well as muscular dystrophies.
INTRODUCTION
Cobalamin (Cbl, B12) is an essential micronutrient required by all cells in the body. Methylcobalamin (MeCbl) and adenosylcobalamin (AdoCbl) are required cofactors in the catalytic reactions of cytosolic methionine synthase [1–2] and mitochondrial methylmalonyl-CoA mutase [1–2], respectively. Synthesis of AdoCbl and MeCbl from other Cbl derivatives is dependent on the MMACHC (for methylmalonic aciduria combined with homocystinuria type C) gene product, which defines the cblC complementation group [3]. The MMACHC protein is responsible for processing the upper-axial ligands of dietary Cbls prior to Cbl coenzyme biosynthesis [4–6]. In cblC patients, who form the largest group of inborn errors of cobalamin metabolism, the inability to synthesize MeCbl results in increased concentrations of homocysteine in blood as well as decreased blood levels of methionine. The inability to synthesize AdoCbl results in increased levels methylmalonic acid in blood, spinal fluid and urine.
It was initially thought that patients in the cblC complementation group were neonates and infants with clinical findings that included failure to thrive, acute neurological deterioration, developmental delay, multisystem organ dysfunction, and hematological abnormalities including megaloblastic anemia [7–8]. However, a late-onset form of the cblC disease was established in 1984 [9]. A previously asymptomatic 14-year old girl presented with acute onset of neurological symptoms including dementia, myelopathy, and motor-neuron disease [9]. The patient was diagnosed with cblC disease, thus establishing the age-related heterogeneity of this condition. In 1997, Rosenblatt et al., reviewed 50 cblC patients, of whom six patients had late-onset disease [10]. The authors noted that late-onset patients had better survival and response to treatment, and less neurological sequelae compared to early-onset patients [10]. Today hundreds of cblC patients have been identified with different mutations in the MMACHC gene and a variety of clinical presentations [11].
Herein, we investigate the proteome of cblC mutant fibroblasts isolated from genetically unrelated and severely ill patients with early-onset disease. We observed that the cblC cell lines produced increased levels of both homocysteine and methylmalonic compared to normal cell lines, and that supplementation of the cell cultures with hydroxocobalamin (HOCbl) did not reduce the export of homocysteine in cblC cell lines to the levels observed in normal fibroblasts. An assessment of the cblC proteome and its comparison with that of normal fibroblasts was conducted using 2D-DIGE and LC/ESI/MS. The identification of changes at the protein level bridges the gap existing between the role(s) of the MMACHC gene product in vitro and the clinical manifestations of functional Cbl deficiency in humans.
EXPERIMENTAL PROCEDURES
Materials
[57Co]-CNCbl was purchased from MP Biomedical (Solon, OH). [57Co]-HOCbl was prepared by decyanation of [57Co]-CNCbl as described previously [12]. Hydroxocobalamin hydrochloride was purchased from Sigma (Saint Louis, MO). Antibodies for CLIC4 and UCHL1 were purchased from Abcam (Cambridge, MA). Antibodies for PDI and GRP94 (anti-KDEL monoclonal antibody) were purchased from Stratagene (Ann Arbor, MI). Antibodies for VIM and human recombinant VIM were purchased from R&D Systems (Minneapolis, MN).
Human fibroblast cell lines
Control and cblC mutant fibroblasts were grown in Advanced Dulbecco’s Modified Eagle Medium (aDMEM) (Gibco, Grand Island, NY) containing 9.07 µM folic acid and supplemented with 10 % fetal bovine serum (FBS) (final Cbl concentration, 66 pM) penicillin (100 units/ml) and streptomycin (0.1 mg/ml). When indicated, cells were supplemented with aDMEM containing 723 nM HOCbl. Fibroblast cell lines from patients with severe cblC disease were obtained from the Repository for Mutant Human Cell Strains, Montreal Children’s Hospital, Montreal, Canada. Culture, handling and disposal of human cell lines were performed according to protocols approved by the Cleveland Clinic’s IRB/IACUC committees. Genetic and clinical information for the cblC patient fibroblasts used in present study can be found in our previous publication [5]. Control and cblC mutant fibroblasts were utilized between passages 7 and 11.
Determination of homocysteine and methylmalonic acid in conditioned culture medium
After culturing cells for 7 days, the conditioned culture medium was then collected, centrifuged at 1,000 rpm for 10 min, filtered through a 0.22 µm filter (Millipore, Billerica, MA) and stored at 4˚C. Total homocysteine in conditioned medium was determined by the method of Jacobsen et al. using monobromobimane and HPLC with fluorescence detection [13]. The concentration of methylmalonic acid in conditioned medium was determined by gas chromatography and mass spectrometry by a method modified from Hoffmann et al [14]. Values were normalized to cellular protein concentration. Total protein concentration was determined by the bicinchoninic acid assay (Thermo Scientific, Rockford, IL) using bovine serum albumin as a standard.
Determination of total intracellular Cbl and folates
Total intracellular Cbl and folate were determined using the SimulTRAC-SNB Radioassay kit vitamin B12[57Co]/Folate [125I], MP Biomedicals (Orangeburg, NY, USA). Cbl and folate values were normalized to cellular protein concentration.
Cobalamin uptake studies
Cells were seeded at an initial density of 25% to 30% and allowed to grow for 24 h. After 24 h, half of the conditioned culture medium was replaced with fresh medium and [57Co]-HOCbl was added to a final concentration of 0.20 nM (specific activity: 379 µCi/µg HOCbl). Uptake was followed by counting the radioactivity in a γ-counter at 6, 12, 24, 48 and 72 h, both in spent medium and in washed cell pellets. Total Cbl values were normalized to cellular protein concentration.
Processing of [57Co]-HOCbl by normal and cblC fibroblasts
Processing experiments were carried out as described in a previous report utilizing [57Co]-HOCbl (final concentration in the culture medium ≈ 0.125 nM) as the sole source of Cbl [5]. Cells were grown for 48 h, harvested and the intracellular Cbl profile was determined by HPLC as described previously [12].
CyDye labeling of the normal and cblC proteomes
To detect and identify differentially expressed proteins between normal and cblC mutant fibroblasts, we used the fluorescence technique 2D-DIGE [15]. In addition, we investigated the effect of HOCbl supplementation on the proteome of normal and cblC fibroblasts. In order to minimize variations associated with growth conditions, cell extracts for 2D-DIGE analysis were prepared from a single batch of normal and cblC fibroblasts (i.e., same passage number), which were split and grown simultaneously for 7 days, either in the presence or absence of 723 nM HOCbl. Cells were harvested by trypsinization, washed 3 times with phosphate buffered saline and stored at −80 °C until ready for workup. The cell pellets were freeze-thawed three times, and lysed with 0.2 ml of homogenization buffer (40 mM Tris, 7 M Urea, 2 M thiourea, 4% CHAPS, pH 8.6 containing the complete mini protease inhibitor cocktail (Roche, Indianapolis, IN) per cell pellet. The concentration of protein was determined on a 5 µl aliquot using the 2-D Quant kit (GE Healthcare, Piscataway, NJ). Five hundred micrograms of total protein per cell line were divided into two aliquots of 250 µg each and subjected to a “clean-up” procedure using the 2-D Clean up kit (GE Healthcare). The final pellet from each tube was resuspended with 50 µl of lysis/homogenization buffer and the 2 aliquots of each sample were pooled prior to protein determination. The samples were stored at −20 ˚C until CyDye labeling. Proteins from normal and the 3 cblC cell lines were individually labeled with the fluorescent dyes Cy2, Cy3, and Cy5 as recommended by the manufacturer (see below), mixed, and co-resolved on 3 different analytical gels (total protein ∼150 µg). A preparative gel containing ∼600 µg of total unlabelled protein (pooled normal and cblC samples) was run simultaneously. Table 1S in the Supplementary data summarizes the experimental set up.
CyDye labeling procedure
CyDye stock solutions were prepared according to the manufacturer’s instructions. Briefly, each CyDye was resuspended to a final concentration of 1 nmol/ml with dimethylformamide, mixed, centrifuged at 12,000 rpm for 30 s, and stored at −20 ˚C until further use. An internal standard was prepared as follows: 250 µg of protein from each sample were pooled and lysis buffer was added to adjust the protein concentration to 2 mg/ml. The internal standard pool is used for two purposes: 1) One aliquot (200 µg) is labeled with Cy2 to be used as the internal standard on each of the 3 analytical gels (Table S 1) and 2) the remainder (800 µg) is used for the preparative gel that will be spot picked for MS analysis. The internal standard (200 µg) was labeled with Cy2 by addition of 1,600 pmol of Cy2 (400 pmol dye/50 µg protein). One hundred fifty micrograms of the normal human foreskin fibroblasts sample was labeled with Cy5 and 50 µg of each cblC mutant sample was labeled with Cy3 under the same conditions, i.e., 400 pmol dye/50 µg protein. The samples were mixed, centrifuged and allowed to react for 30 min in the dark at 4 ˚C. The labeling reaction was stopped by addition of 1 µl of 10 mM lysine. The mixture was vortexed, centrifuged, left on ice for 10 min in the dark and then stored at −80 ˚C.
2-Dimensional difference gel electrophoresis
For the analytical gels, three tubes containing mixtures of 50 µg (25 µl) of one Cy3-labeled sample, 50 µg (25 µl) of one Cy5 labeled sample and 50 µg (25 µl) of the Cy2 labeled internal standard were prepared. Disulfide bonds were reduced by addition of 2% dithiothreitol (DTT) to the pooled samples (final concentration of DTT = 0.2%). Samples were vortexed and 0.37 ml of DeStreak solution (GE Healthcare) was added to each mixture. The resulting protein concentration was ∼ 0.33 mg/ml (150 µg/450 µl). For the preparative gels (P1 and P2), an aliquot of 0.60 mg (in 0.3 ml) of total protein was placed into a microfuge tube, and 2% DTT was added to a final concentration of 0.2%. The samples were vortexed, and 100 µl of DeStreak solution (GE Healthcare) was added. The final concentration of protein was ∼1.33 mg/ml (0.60 mg/450 µl). Samples (0.45 ml) were subjected to isoelectrofocusing using an Amersham IPGphor system (GE Healthcare). The isoelectrofocusing strips were rehydrated for 11 h at 20 ˚C using 50 μA/strip and 30V, using the following conditions: step 1: 0.5 h, 0–250 V; step 2: 0.5 h, 250–6,000 V; step 3: 5 h, 6,000 V; step 4: 10 h, 6,000 V for a total of 78 kV. The strips were then equilibrated with 7 ml of SDS Equilibration Buffer #1 (reducing buffer: 6 M urea, 2% SDS, 30% glycerol, 1.6% DTT, 50 mM Tris and 0.002% bromophenol blue, pH 8.8) and incubated with gentle shaking for 15 min, at room temperature. The solution was carefully removed and 10 ml of SDS equilibration buffer #2 (alkylation buffer: 6 M urea, 2% SDS, 30% glycerol, 2% iodoacetamide, 50 mM Tris and 0.002% bromophenol blue, pH 8.8) were added. The isoelectrofocusing strips were then electrophoresed on a 24 cm 7.0% - 20% gradient pre-cast polyacrylamide SDS-PAGE gel (Jule Inc, Milford, CT) for 4 h at constant power (5 watts/strip for 1 h; 20 watts/strip for 3 h) in an Ettan DALT™ twelve vertical electrophoresis system (GE Healthcare) using the Laemmli buffer system [16]. The preparatory scale gel was fixed and stained with GelCode Blue™ Coomassie stain (Pierce, Rockford, IL).
Decyder® image analysis of differentially expressed proteins
Fluorescence images of the gels were acquired on a Typhoon 9400 scanner (GE Healthcare). Cy2, Cy3, and Cy5 images for each gel were scanned with a resolution of 200 pixels. The image files were then analyzed with DeCyder software using published [15, 17] and manufacturer’s recommendations (GE Healthcare). Only proteins with a fold change equal or greater than ± 2.0 and a statistical significance of at least 95% (p ≤ 0.05) were selected for identification. A representative scheme for 2D-DIGE image analysis using DeCyder® software is given in Fig. S1. Candidate spots were cut manually, trypsinized and analyzed by LC/ESI/MS. All fluorescence gel images (raw data) are provided in the Supplementary Data (Fig. S2–S7).
Protein digestion, LC/ESI/MS and protein identification
For protein digestion, the bands were cut to minimize excess polyacrylamide, divided into a number of smaller pieces, washed and destained. The gel pieces were then washed with water and dehydrated in acetonitrile. The bands were alkylated with iodoacetamide prior to the in-gel digestion step. All of the bands were digested in-gel by adding 5 µl of 20 ng/µl trypsin in 50 mM ammonium bicarbonate and incubating the samples overnight at room temperature. The resulting peptides were extracted from the polyacrylamide gel in two aliquots of 30 µl using 50% acetonitrile with 5% formic acid. These extracts were combined and evaporated to a final volume of less than 10 µl in a Speedvac, and were then resuspended in 1% acetic acid to make up a final volume of ∼30 µl for LC/ESI/MS analysis using a Thermo Scientific linear trap quadrupole linear ion-trap mass spectrometer system. The HPLC column was a self-packed 9 cm x 75 µm id Phenomenex Jupiter C18 reversed-phase capillary chromatographic column. Ten µl volumes of the extract were injected, and the peptides eluted from the column (acetonitrile/0.10 % formic acid gradient, flow rate: 0.3 µl/min) were introduced into the source of the mass spectrometer online. The electrospray ion source was operated at 2.5 kV. The tryptic digest was analyzed using the data dependent multitask capability of the instrument acquiring full scan mass spectra to determine peptide molecular weights and product ion spectra to determine amino acid sequence in successive instrument scans. This mode of analysis produces approximately 2,500 collisionally induced dissociation (CID) spectra of ions ranging in abundance over several orders of magnitude. The data were analyzed by using all CID spectra collected in the experiment to search the National Center for Biotechnology Information reference sequence database with the search program Mascot version 2.0. All matching spectra were verified by manual interpretation. The interpretation process was aided by additional searches using the programs Sequest and Blast as needed.
Validation of 2D-DIGE results by an alternative method
This study was designed to identify statistically significant changes in protein levels of normal versus the MMACHC proteome, rather than looking into changes in individual MMACHC patients. The analysis was designed to be quantitative and comparative in that it contrasts three runs of the normal proteome versus three runs for the MMACHC proteome (one for each of the three patients). Of the proteins identified as differentially expressed by 2D-DIGE and mass spectrometry, some were selected for further validation based on the availability of antibodies, activity assays and reagents. The selection process focused on : a) spots that contained more than one protein, b) proteins which, based on current knowledge, might be involved in the pathobiology of the MMACHC disease (cytoskeleton, nervous system) and c) proteins with known housekeeping functions and whose expression was unaffected by the presence or absence of HOCbl (GST, CLIC4). In all cases, the results agreed well with the expression patterns retrieved from 2D-DIGE and mass spectrometry analysis.
Western blots
Western blots for chloride intracellular channel 4 (CLIC4), vimentin (VIM), protein disulfide isomerase (PDI), glucose regulated protein 94 (GRP94), and ubiquitin C-terminal esterase L1 (UCHL1) were done according to standard procedures. Briefly, total proteins (50 µg per lane) were resolved on a 12.5% SDS-PAGE gel, transferred onto a PVDF immobilon membrane, and blotted with the appropriate antibody. Detection was done by chemiluminescence using a commercially available kit (ECL Western Lightning Chemiluminescent Kit, Perkin-Elmer NEL104, Waltham, MA). Quantization of Western blots was done using ImageJ software (http://rsbweb.nih.gov/ij/). To account for variations on protein loading in SDS-PAGE runs, expression levels of the target protein were expressed as a ratio of its band intensity divided by that of a protein not detected as being differentially expressed by 2D-DIGE (in this case GRP94).
Immunocytochemistry
Immunostaining of human VIM was performed according to a protocol provided by Abcam. Briefly, cells were grown on NUNC 8-well chamber slides (Fisher Scientific, Pittsburgh, PA) to a final density of ∼ 50%, the conditioned culture medium discarded and the cells were rinsed briefly in phosphate buffered saline (PBS). The samples were then fixed with ice-cold acetone for 4 min, which results in a partial destruction of the cell membrane, and washed twice with PBS. Cells were incubated in blocking solution (1% bovine serum albumin in PBS + 0.25% Triton X-100) for 30 min, and then incubated in primary antibody overnight at 4 ˚C. Negative controls were incubated in the same solution without the primary antibody. After three washes in PBS the cells were incubated with secondary antibody for 1 h at room temperature in the dark. Nuclei were stained with 4',6-diamidino-2-phenylindole.
Activity assays and ELISA
Fresh cell lysates for activity assays and/or enzyme-linked immunoassay (ELISA) were prepared according to the manufacturer’s directions. Glutathione-S-transferase (GST) activity was determined using a commercial kit (Cayman Chemical, Ann Arbor, MI). Annexin V (ANXA5) was determined using an ELISA kit (Immunoclone® Annexin V ELISA kit, American Diagnostica Inc, Stanford, CT).
Analysis of pathways and networks
Bioinformatic analysis of the results was conducted using Ingenuity Pathway Analysis® software (www.ingenuity.com). Only proteins with a fold change of equal or greater than ± 2.0 (p< 0.05) were included in the study. In addition, homocysteine, methylmalonic acid, folic acid and Cbl were included, as these metabolites were determined under the same conditions utilized for the 2D-DIGE experiments (7 days in culture, with and without HOCbl supplementation). The activities of the enzymes methionine synthase, methylmalonyl-Co-A mutase, cob(I)alamin adenosyltransferase and methyltetrahydrofolate reductase were included, as a comparison between normal and five other cblC fibroblast cell lines was available from the literature and the activity of these enzymes appears to follow a highly reproducible pattern among patients [18].
Statistics
All results were expressed as mean ± standard deviation of the mean. In all cases non-paired two-tailed Student’s t-tests or 1-way ANOVA were used to determine significance of the DIGE results, and false discovery rates were applied to all comparisons. Fischer’s exact test was used by Ingenuity Pathway Analysis® to determine the p values for all of the network and pathway analyses. In all cases, a p value of p ≤ 0.05 was considered statistically significant.
RESULTS
In this work we compared the proteome of cblC mutant fibroblasts with the proteome of normal fibroblasts, and in so doing, determined if intracellular Cbl coenzyme deficiency results in differential expression of proteins.
Cellular morphology
Mutant fibroblasts from the cblC complementation group, especially WG2176 and WG3354, displayed an altered cell morphology compared to control cell lines when seeded at low density (Fig. S8 in Supplementary Data). Despite the altered morphology observed in non-confluent cultures, all cblC cell lines had similar doubling times (Fig. S9), and after 72–96 h in culture no differences could be seen in morphology, cell number or total protein content between normal and cblC fibroblasts. Nonetheless, late-stage apoptosis and senescence were examined by DNA laddering and in situ measurement of β-galactosidase activity, respectively. No significant differences were found between normal and cblC fibroblasts (Fig. S10 and Fig. S11 in Supplementary Data). All studies described in the following sections were performed with cells that were cultured for a total of 7 days. This was designed to mimic a chronic stage of elevated Hcy and MMA ex vivo, while assuring that at the time of harvesting, all four cell lines were in the stationary growth phase (i.e., fully confluent).
Production of homocysteine and methylmalonic acid
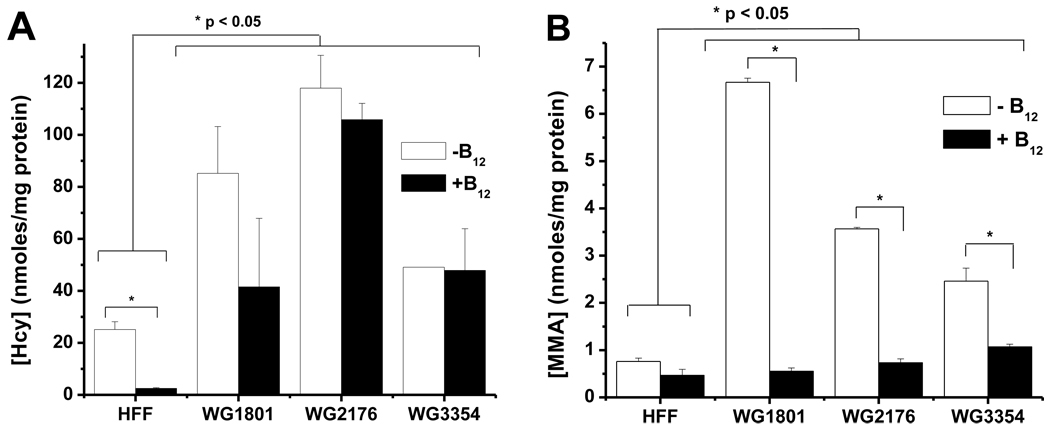
Based on the failure of cblC fibroblasts to synthesize AdoCbl and MeCbl, we hypothesized that cultured cblC cells will export higher levels of homocysteine and methylmalonic acid to the culture medium compared to normal skin fibroblasts. Fig. 1 shows the levels of homocysteine and methylmalonic acid measured in the conditioned culture medium after 7 days. All cblC cell lines produced significantly higher levels of both homocysteine (Fig. 1A) and methylmalonic acid (Fig. 1B) compared to the control cell line. These results suggest that the cblC fibroblast cell lines have a functional Cbl deficiency. Importantly, it was observed that methylmalonic acid levels were maintained close to normal values in cblC mutants supplemented with HOCbl (Fig. 1B), whereas this response was not observed for homocysteine production (Fig. 1A).
Fig. 1.
Determination of homocysteine (Hcy) and methylmalonic acid (MMA) in culture media of normal (HFF) and cblC fibroblast cell lines (WG 1801, WG 2178, WG 3354) after 7 days. All three cblC patients produced significantly higher amounts of Hcy and MMA compared to normal cells (panels A and C). Supplementation of the culture medium with 723 nM hydroxocobalamin (B12) did not cause a significant decrease in the levels of Hcy but proved to be very efficient in normalizing MMA levels (panels B and D, respectively). Statistically significant differences were established by independent Student’s t-test, at α=0.05. N=3, two independent experiments.
Cobalamin uptake and cofactor biosynthesis
Cobalamin uptake and the ability of the cblC cell lines to synthesize AdoCbl and MeCbl were also examined. All cblC cell lines displayed slightly decreased uptake of [57Co]-HOCbl and failure to retain intracellular Cbls after 48 h in culture (Fig. S12 in Supplementary Data). This is consistent with earlier studies by Mellman et al. with other cblC mutant fibroblasts [19]. We had previously shown that cblC fibroblasts were unable to convert [57Co]-CNCbl into [57Co]-labeled coenzymes [5]. The ability of cblC cell lines to utilize [57Co]-HOCbl as a substrate for cofactor biosynthesis was investigated. While the normal cell line utilized [57Co]-HOCbl as substrate for [57Co]-MeCbl and [57Co]-AdoCbl biosynthesis (Table 1), none of the cblC fibroblasts were able to synthesize either [57Co]-AdoCbl or [57Co]-MeCbl, except for patient WG1801 (21.7 % [57Co]-AdoCbl, no [57Co]-MeCbl). Unutilized [57Co]-HOCbl was the principal intracellular form isolated from the cblC mutant cell lines. These data suggest that with the exception of patient cell line WG1801, the cblC mutants cannot utilize [57Co]-HOCbl as a substrate for the biosynthesis of [57Co]-AdoCbl through the putative mitochondrial machinery originally suggested in the literature [20–21].
Table 1.
Utilization of [57Co]-HOCbl by human normal (HFF) and cblC mutant fibroblasts (WG1801, WG2176 and WG3354) for Cbl coenzyme biosynthesis.
| % intracellular Cbla | ||||
|---|---|---|---|---|
| HFF | WG1801 |
WG2176 | WG3354 | |
| HOCbl | 25.9 ± 5.3 | 78.3 ± 18.4 | 100 | 100 |
| AdoCbl | 13.4 ± 6.8 | 21.7 ± 1.7 | ND | ND |
| MeCbl | 60.7 ± 3.0 | ND | ND | ND |
Results are expressed as mean ± standard deviation (n = 3).
ND: less than 1% or undetectable.
Intracellular levels of cobalamin and folate
It was of interest to determine the total levels of intracellular Cbl in normal versus cblC mutant cell lines that were grown with and without supplementation with HOCbl. Surprisingly, the cblC cell lines displayed slightly higher levels of total intracellular Cbl compared to normal fibroblasts when grown in the absence of exogenous HOCbl (Table 2). Supplementation of the cell cultures with 723 nM HOCbl (supraphysiological concentration) increased the levels of intracellular Cbl in all four cell lines, normal and cblC mutants, however the cblC cell lines did not achieve the levels observed in the normal cell line (Table 2).
Table 2.
Intracellular levels of cobalamin and folic acid in normal (HFF) and cblC mutant fibroblasts (WG1801, WG2176 and WG33541
| Cell line | Total intracellular Cbl (pg/mg protein) |
Total intracellular folates (ng/mg protein) |
||
|---|---|---|---|---|
| − OHCbl | +HOCbl | −HOCbl | +HOCbl | |
| Control | 931 | 9,895 | 18.4 | 68.5 |
| WG1801 | 1,287 | 6,803 | 17.1 | 36.0 |
| WG2176 | 1,251 | 4,663 | 10.9 | 22.0 |
| WG3354 | 1,367 | 4,822 | 12.0 | 25.0 |
Total intracellular Cbl and folates were determined on lysates from cells grown in the presence or in the absence of exogenous HOCbl (723 nm), for 7 days. Each value represents total Cbl or total folates for three pooled lysates per cell line.
We examined the total levels of intracellular folate in normal and mutant cblC cell lines, since intracellular folate retention is dependent on the activity of methionine synthase. With the exception of patient cell line WG1801, the cblC cell lines displayed lower levels of intracellular folate compared to normal cells in the absence of exogenous HOCbl (Table 2). Supplementation of the cell cultures with HOCbl resulted in increased levels of total folates in all the cell lines. However, none of the cblC mutant fibroblasts reached the intracellular levels of Cbl or folate observed in the normal cell line.
Identification of differentially expressed proteins in normal and cblC mutant human fibroblasts by 2D-DIGE and LC/ESI/MS
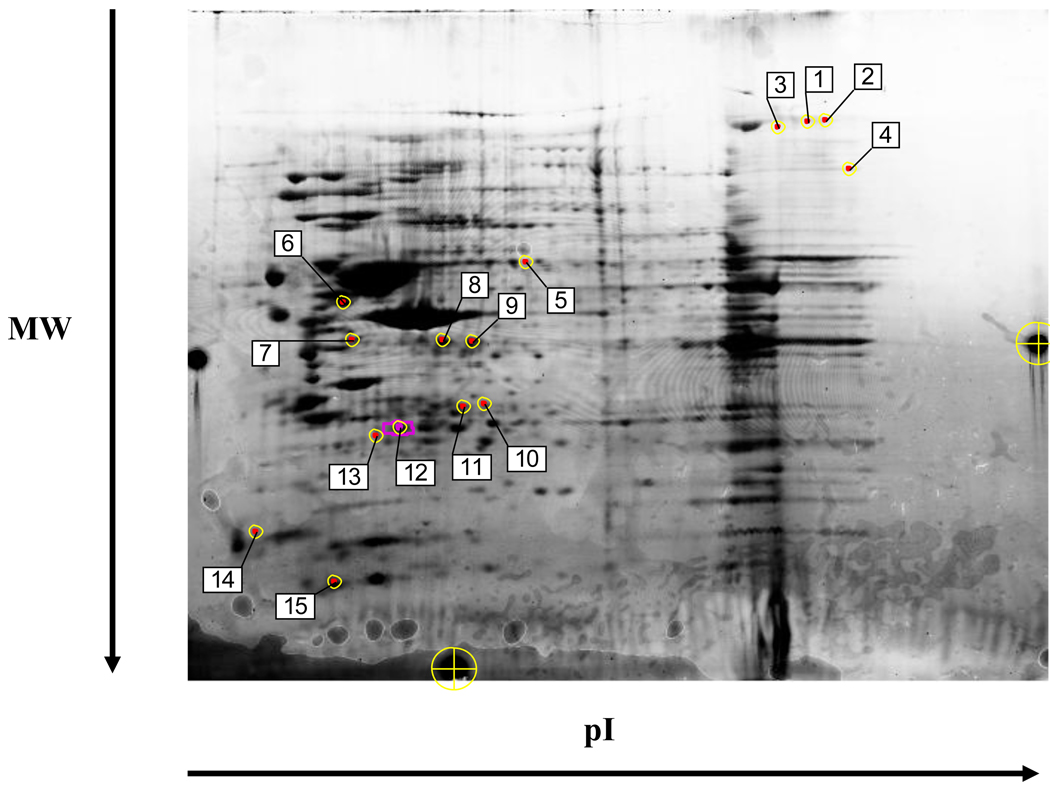
The proteome of normal and cblC fibroblasts grown in the absence of exogenous HOCbl was examined first. A representative set of fluorescent scans from normal versus patient WG2176 is given in Fig. 1. The quantitative analysis was performed such that only proteins down or up-regulated in all three patients versus normal cell lines were taken into account for further analysis (see Experimental Procedures for details, and Fig. S1 in Supplementary Data). Comparison of the normal fibroblast proteome with that from cblC mutant fibroblasts allowed us to select 15 spots with significant variation (Fig. 3). These protein spots exhibited differences in standardized average spot volume ratios ≥ 2.0 and a t-test with p< 0.05 (n=3). Among the 15 spots, 5 were over-expressed in the cblC mutants, whereas 10 were under-expressed in the cblC fibroblasts. These differentially expressed proteins spots were picked manually and analyzed by LC/ESI/MS. All of the spots were found to contain one or more proteins.
Fig. 3.
Differential expression of proteins in three cblC mutants compared to normal fibroblasts. Labels 1–15 indicate the spots selected for identification by mass spectrometry. A statistical significance of 95% confidence and a stringency of ± 2-fold in the expression ratio of control versus cblC mutants was used to analyze the dataset. The preparative gel shown in the picture was loaded with 600 µg of total protein and stained with Coomassie blue overnight. Protein spots are identified in Table 4.
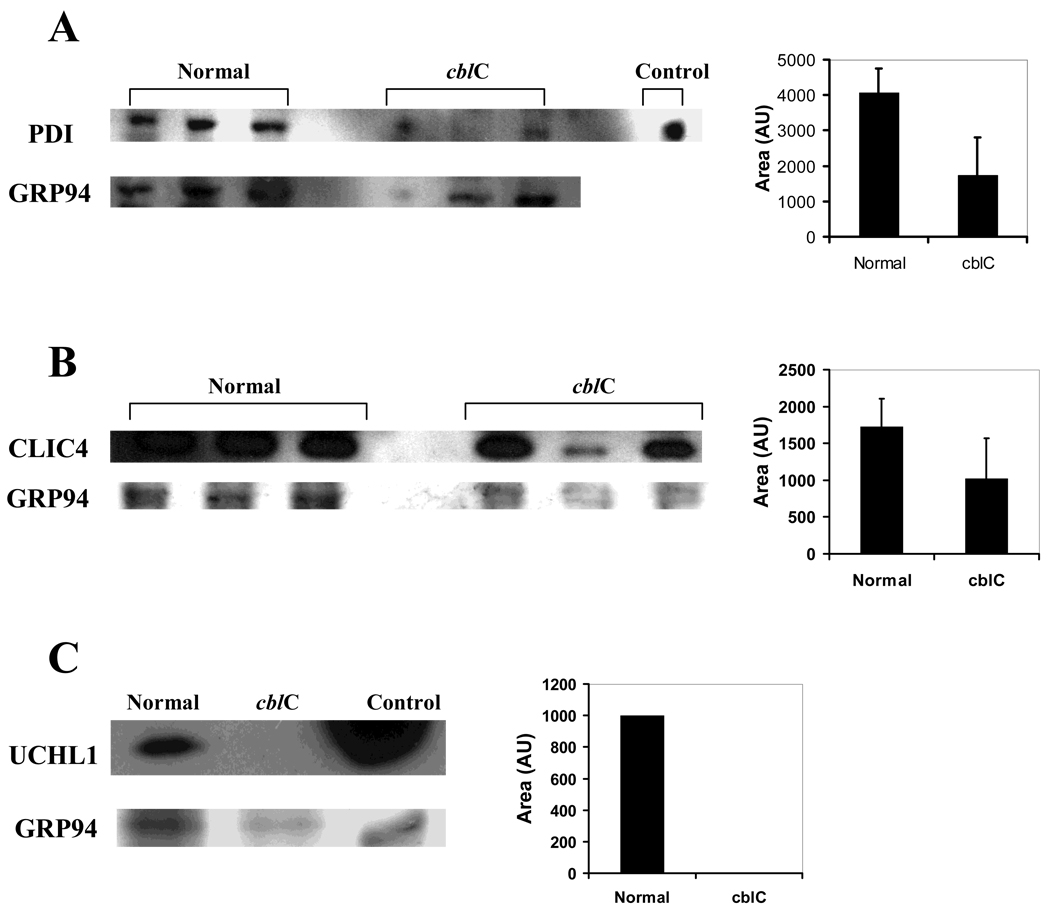
A summary of the proteins that were differentially expressed in normal versus cblC fibroblasts is given in Table 3. Major changes were observed in proteins related to cytoskeleton, nervous system, signaling and cellular detoxification. Further validation of the results using alternative techniques was also conducted, especially in cases where one spot contained more than one protein. Fig. 4 shows the western blot results for protein disulfide isomerase (PDI) (panel A), chloride intracellular ion channel 4 (CLIC4) (panel B), and ubiquitin carboxyl hydrolase L1 (UCHL1) (panel C).
Table 3.
Differentially expressed proteins in cblC mutants compared to normal fibroblasts (N=3, α=0.05) grown in the absence of exogenous HOCbl.
| Spot | Protein Name | Gene | Gene ID | Fold Change |
No peptides identified (% coverage) |
Mascot score |
t-test (p value) |
|---|---|---|---|---|---|---|---|
| 1 | HDL binding protein | HDLBP | 42716280 | 2.87 | 10 (9%) | * | 0.018 |
| 2 | HDL binding protein | HDLBP | 42716280 | 3.09 | 28 (27%) | 807 | 0.029 |
| 4 | Eukaryotic translation elongation factor 2 |
EEF2 | 4503483 | 2.5 | 29 (36 %) | 1234 | 0.042 |
| 7 | Serine (or Cys) proteinase inhibitor (protease inhibitor 6, PI-6, or serpin B6) |
SERPINB6 | 41152086 | −2.09 | 23 (59%) | 2322 | 0.032 |
| 7 | Caldesmon-1, isoform 2, Ct truncated |
CALD1 | 4826657 | −2.09 | 15 (32%) | 956 | 0.032 |
| 8 | β-actin | ACTB | 4501885 | −2.11 | 16 (50%) | 1116 | 0.024 |
| 9 | Tubulin alpha, ubiquitious |
TUBA1B | 57013276 | −2.17 | 8 (24%) | 907 | 0.028 |
| 11 | Chloride intracellular channel 4 |
CLIC4 | 7330335 | −2.04 | 22 (72%) | 1507 | 0.018 |
| 2 | Collagen type VI, alpha 2, isoform 2C2 |
COL6A2 | 115527062 | 2.1 | 8 (17%) | 295 | 0.028 |
| 3 | Collagen type VI, alpha 1 precursor |
COL6A1 | 87196339 | 2.1 | 16 (16%) | 662 | 0.028 |
| 12 | Ubiquitin carboxyl esterase L1 |
UCHL1 | 21361091 | −2.19 | 19 (66%) | 1261 | 0.021 |
| 5 | Plastin 3 | PLS3 | 7549809 | −2.9 | 25 (46%) | 1471 | 0.023 |
| 6 | Vimentin | VIM | 62414289 | −2.44 | 44 (65%) | 3652 | 0.032 |
| 10 | Glutathione transferase omega 1 |
GSTO1 | 4758484 | −2.1 | 9 (39%) | 172 | 0.0096 |
| 13 | Glutathione transferase | GSTP1-1 | 4504183 | −2.84 | 8 (45%) | 821 | 0.016 |
| 12 | Glutathione transferase M3 |
GSTM3 | 23065552 | −2.19 | 13 (52%) | 281 | 0.021 |
| 5 | PDI associated 3 precursor |
ERp57 | 21361657 | −2.9 | 17 (34%) | 529 | 0.023 |
| 15 | SH3 domain binding Glu-rich protein like 3 |
SH3BGRL3 | 13775198 | −2.23 | 4 (43%) | 312 | 0.042 |
| 15 | S100 Ca-binding protein A6 |
S100A6 | 7657532 | −2.23 | 4 (24%) | 121 | 0.042 |
| 14 | ribosomal protein S14 | RPS14 | 5032051 | 2.45 | 4 (27%) | 260 | 0.033 |
| 14 | H2B histone family member A |
HIST1H2BG | 4504257 | 2.45 | 3 (25%) | 90 | 0.033 |
| 8 | Glyceraldehyde-3- phosphate dehydrogenase |
GAPDH | 7669492 | −2.11 | 10 (42%) | 761 | 0.024 |
This protein was identified manually and via Sequest analysis.
Fig. 4.
Western blots of protein disulfide isomerase (PDI panel A), chloride intracellular channel 4 (CLIC4 panel B) and ubiquitin carboxyl esterase L1 (UCHL1 panel C). Cell lysates were obtained from cells grown without HOCbl supplementation. Control lanes contain purified standards (PDI in panel A and UHCLI in panel C). Pooled samples were run for UCHL1 (n=3). Band density was determined using ImageJ software. In all cases, the expression levels of the target protein (bar graphs) was expressed as a ratio of its band intensity divided by that of a protein not detected as differentially expressed by 2D-DIGE (GRP94).
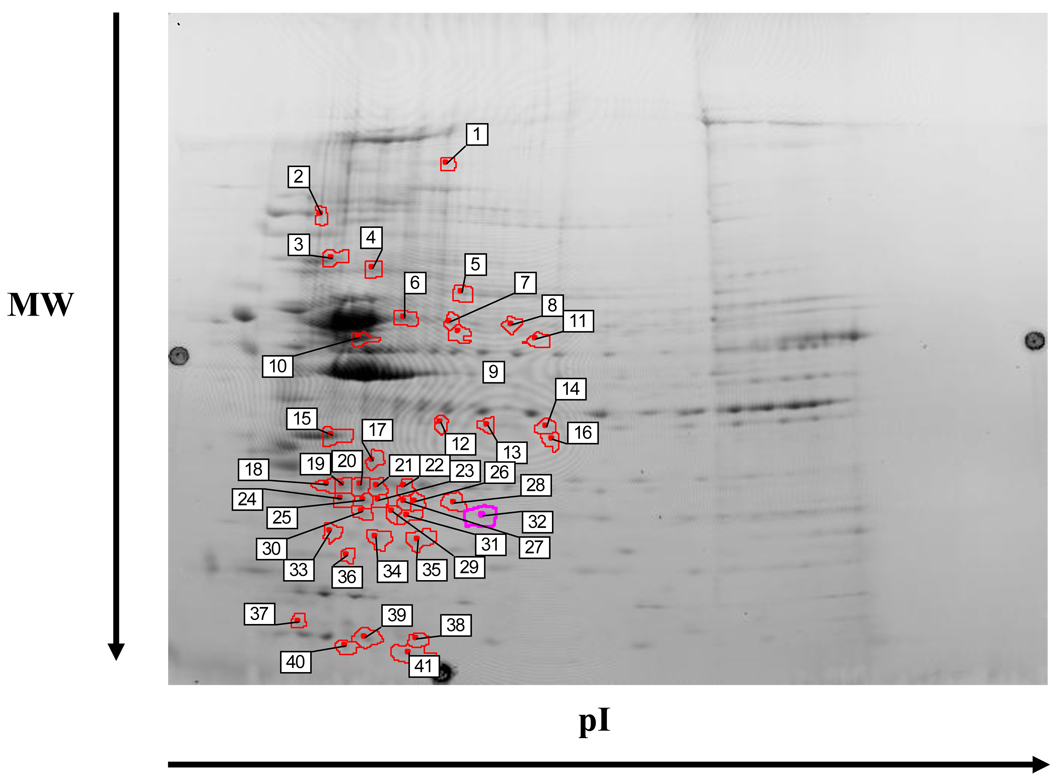
The effects of HOCbl supplementation on the proteome of cblC fibroblasts was then examined. A total of 41 spots displayed significant changes in expression (Fig. 5). LC/ESI/MS analysis of the spots identified 37 proteins. Remarkably, all 37 proteins identified as differentially expressed were down-regulated in the cblC fibroblasts (Table 4). This indicates that supplementation of the cell cultures with a supraphysiological dose of HOCbl induced a global down-regulation of the cblC proteome with respect to the normal cell lines. Furthermore, treatment of cblC fibroblast with HOCbl normalized the expression of high density lipoprotein binding protein (HDLBP), collagen type VI alpha 1 precursor (COL6A1), eukaryotic elongation factor 2 (EEF2), glutathione transferase omega 1 (GSTO1), GSTM3 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). This suggests that supplementation with HOCbl corrected some of the alterations caused by its deficiency or insufficiency.
Fig. 5.
Differential expression of proteins in three cblC mutants compared to normal fibroblasts grown in the presence of 723 nM HOCbl. Labels 1–41 indicate the spots selected for identification by mass spectrometry. A statistical significance of 95% confidence and a stringency of ± 2-fold in the expression ratio of control versus cblC mutants was used to analyze the dataset. The preparative gel shown in the picture was loaded with 600 µg of total protein and stained with Coomassie blue overnight. Protein spots are identified in Table 4.
Table 4.
Differentially expressed proteins in cblC mutants compared to normal fibroblasts (N=3, α=0.05) grown in the presence of 723 nM HOCbl.
| Spot | Protein Name | Gene | Gene ID | Fold Change |
No peptides identified (% coverage) |
Mascot score |
t-test (p value) |
|---|---|---|---|---|---|---|---|
| 1 | Collagen type VI, isoform 2C2 |
COL6A2 | 115527062 | −2.72 | 18 (19%) | 953 | 0.039 |
| 1 | Vinculin isoform VCL | VCL | 4507877 | −2.72 | 21 (27%) | 660 | 0.039 |
| 2 | Hsp 90 protein 1, beta | HSP90AB1 | 20149594 | −2.03 | 33 (49%) | 1758 | 0.042 |
| 2 | Hsp 90 alpha (cytosolic), class A, member 1, isoform 2 |
HSP90AA1 | 154146191 | 27 (40%) | 1393 | 0.042 | |
| 3 | Hsp 70, protein 5, | HSPA5 | 16507237 | −2.28 | 44 (59%) | 3310 | 0.031 |
| 3 | PDI associated 4 | PDIA4 | 4758304 | −2.28 | 14 (21%) | 435 | 0.031 |
| 4 | Hsp70 protein 8, isoform 1 |
HSPA8 | 5729877 | 33 (57%) | 2218 | 0.042 | |
| 4 | Annexin VI isoform 1 | ANXA6 | 71773329 | −2.15 | 43 (64%) | 1678 | 0.042 |
| 4 | ATPase, H+ transporting, lysosomal |
ATP6V1A | 19913424 | −2.15 | 16 (35%) | 701 | 0.042 |
| 5 | Lamin A/C, isoform 2 | LMNA | 5031875 | −2.3 | 46 (61%) | 2672 | 0.049 |
| 5 | Dihydropyrimidase - like 2 |
DPYSL2 | 4503377 | 26 (65%) | 1622 | 0.049 | |
| 5 | chaperonin containing TCP1, sub3 |
CCT3 | 63162572 | −2.3 | 22 (47%) | 829 | 0.049 |
| 15 | Annexin 5 | ANXA5 | 4502107 | −2.08 | 19 (60%) | 1285 | 0.031 |
| 6 | Vimentin | VIM | 62414289 | −5.03 | 52 (71%) | 3908 | 0.031 |
| 6 | PDI associated 3 precursor |
PDIA3 | 21361657 | −2.38 | 21 (56%) | 1140 | 0.042 |
| 6 | Tubulin alpha 6 | TUBA1C | 14389309 | −2.38 | 15 (45%) | 1014 | 0.042 |
| 8 | Phosphoglycerate dehydrogenase |
PHGDH | 23308577 | −2.13 | 22 (49%) | 2316 | 0.033 |
| 7 | tryptophanyl-tRNA synthetase isoform a |
WARS | 47419914 | −2.63 | 27 (67%) | 2129 | 0.033 |
| 8 | inosine monophosphate dehydrogenase 2 |
IMPDH2 | 66933016 | −2.13 | 17 (39%) | 778 | 0.042 |
| 11 | Septin 11 | SEPT11 | 8922712 | −2.25 | 22 (50%) | 1237 | 0.031 |
| 13 | Annexin 5, A2, isoform I |
ANXA2 | 50845388 | −2.90 | 9 (26%) | 355 | 0.039 |
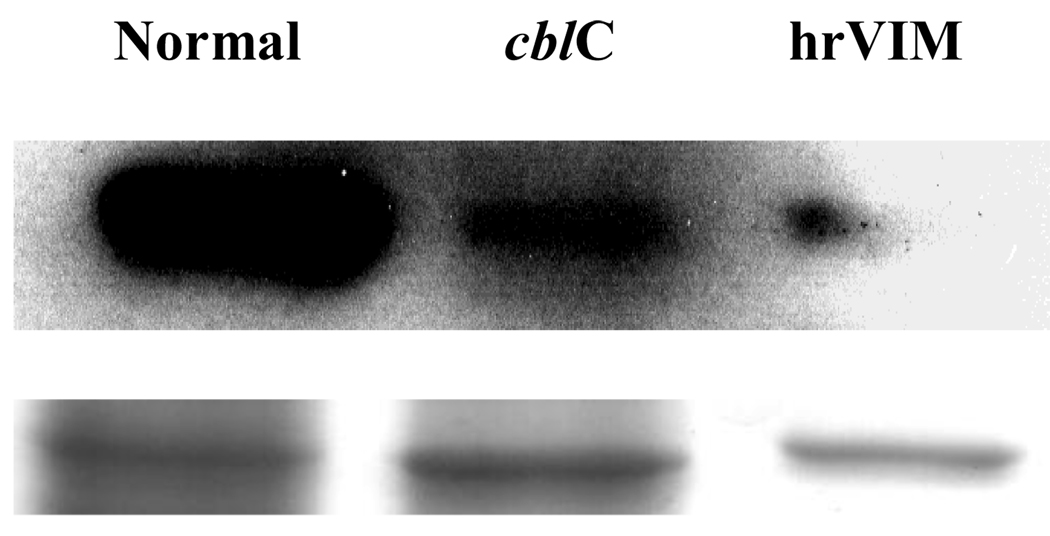
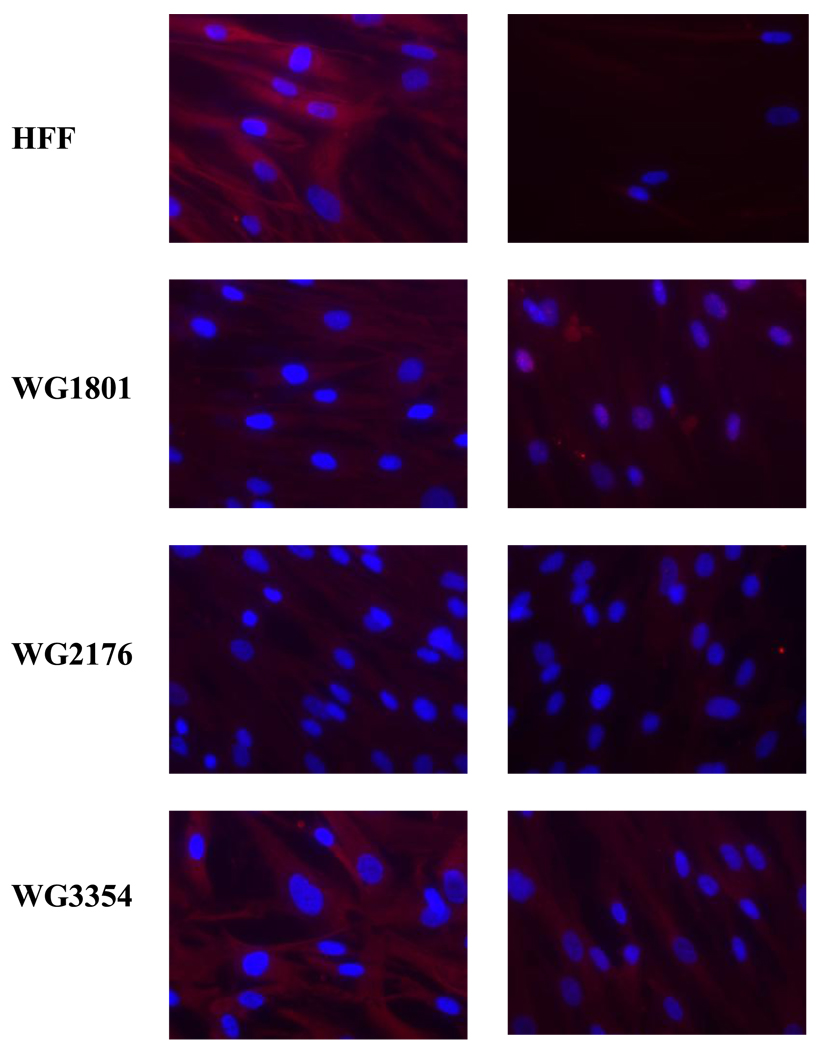
In the absence of exogenous HOCbl, collagen type VI isoform 2C2 (COL6A2) was upregulated in the cblC fibroblasts (COL6A2cblC/COL6A2HFF = 2.10), whereas the opposite result was obtained for cells grown in the presence of HOCbl (COL6A2cblC/COL6A2HFF = – 2.72). This indicates a profound effect of HOCbl supplementation on the expression levels of this important structural protein. In the absence of HOCbl, vimentin expression was decreased in cblC fibroblasts compared to normal fibroblasts (VIMcblC/VIMHFF = −2.44). In the presence of exogenous HOCbl the ratio was decreased to −5.03. This finding was validated by western blot (Fig. 6) and immunocytochemistry (Fig. 7). Immunocytochemistry results for human VIM from cells cultured in the absence of exogenous HOCbl are shown in Fig. 7. The cblC cell lines presented decreased staining compared to the normal cell line. Patient fibroblasts WG3354 exhibited an almost normal VIM phenotype.
Fig. 6.
Western blots for vimentin (VIM). Human normal and cblC fibroblasts grown in the presence of 723 nM HOCbl were stained with anti-hrVIM. Upper row: detection; Bottom row: coomassie staining of the SDS-PAGE. Vimentin was stained with Texas Red whereas nuclei were stained with DAPI. Each lane represents pooled normal and cblC patient cells (n=3). hrVim: human recombinant vimentin.
Fig. 7.
Immunocytochemistry of human normal (HFF) and cblC fibroblasts (WG1801, WG2176 and WG3354) with anti-hrVIM. Cells were grown without HOCbl supplementation. Left column: staining; right column: staining in the absence of primary antibody (negative controls). Vimentin was stained with Texas Red and nuclei were stained with DAPI.
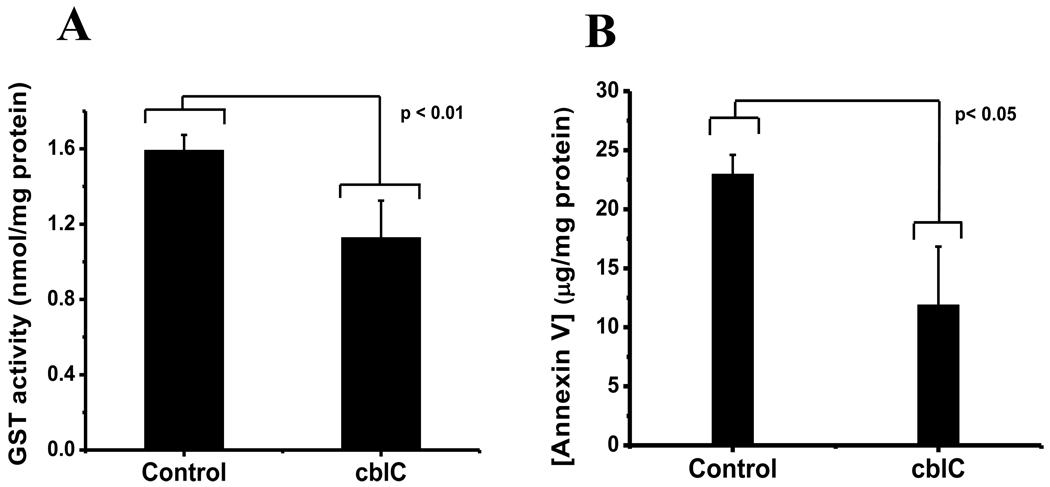
A number of proteins in the cblC proteome remained down-regulated regardless of supplementation with HOCbl. These include PDI associated precursor 3 (PDIA3), CLIC4, UCHL1, GST, SH3 domain binding Glu-rich protein (SH3DBGRP) and S100 Ca-binding protein A6 (S100A6). Supplementation with HOCbl caused the differential expression of a number of proteins that were not identified or differentially expressed in cells that were grown without exogenous HOCbl. These included: vinculin isoform VCL (VCL), heat shock protein 90 (HSP90), heat shock protein 70 (HSP70), annexins VI isoform 1 (ANXA6) and V (ANXA5), septin 11 (SEPT11), peroxiredoxins 1 (PRDX1) and 6 (PRDX6), cofilin 1 (CFL1), Parkinson disease protein 7 (PARK7, also known as DJ-1), transgelin 2 (TAGLN2) and others (see Table 4). Validation assays for GST and annexin V are shown in Fig. 8. Lower GST activity was determined in cblC fibroblasts compared to the normal cell line, which correlates with the lower levels of expression observed in the 2D-DIGE experiments. Lower concentrations of annexin V were found in cblC fibroblasts compared to normal cells, which again confirms our 2D-DIGE results. Overall, these results indicate that: 1) the cblC skin fibroblast proteome differs substantially from that of normal fibroblasts, and 2) supplementation with HOCbl does not completely restore the cblC proteome to that observed in normal fibroblasts.
Fig. 8.
Glutathione transferase (GST) activity (panel A) and concentration of annexin V (panel B) in normal and cblC fibroblasts grown in the presence of 723 nM HOCbl. Fresh cell lysates were prepared and the assays conducted according the manufacturer’s directions (see Experimental Procedures).
Network analysis of the normal and cblC proteomes in the absence of exogenous HOCbl
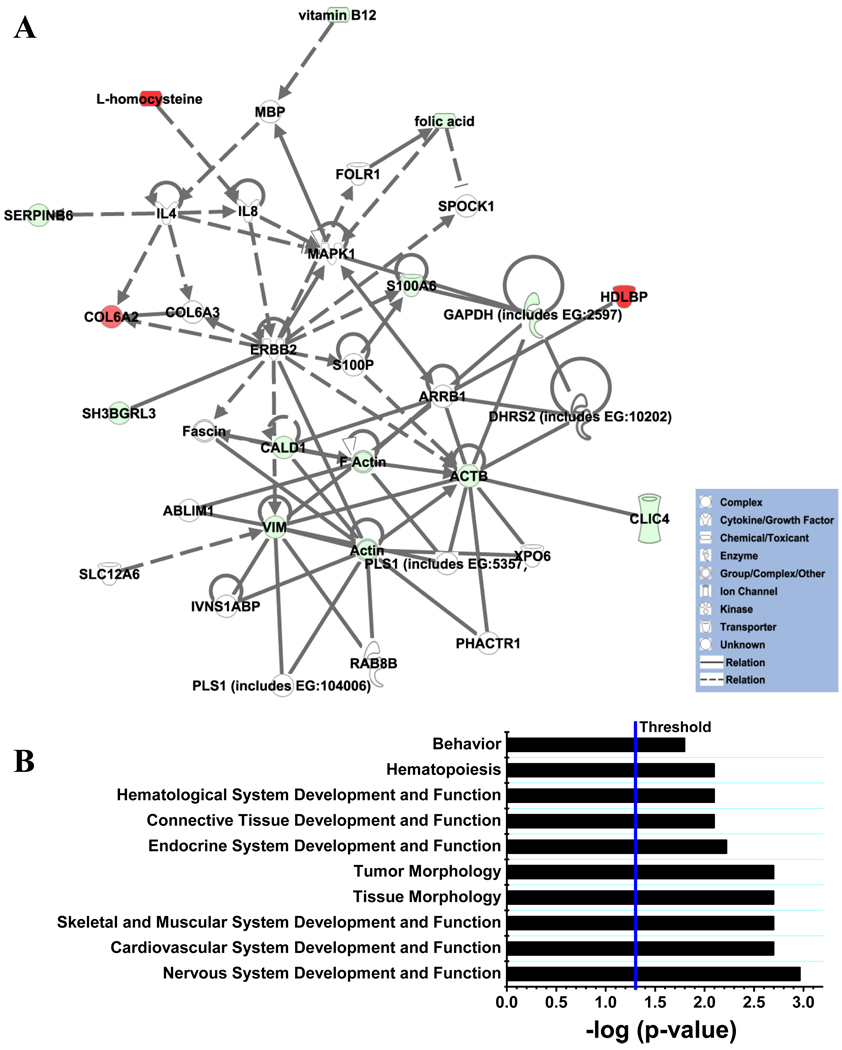
A comparative analysis of the normal and cblC proteomes and relevant metabolites (homocysteine, methylmalonic acid, folate and vitamin B12) generated two statistically significant cellular networks: 1) cell cycle, gene expression and drug metabolism, and 2) genetic disorders, skeletal and muscular disorders and lipid disorders. A list of the proteins involved in these two networks is shown in Table 5. Pathway analysis of these interactions established ranks for diseases and disorders derived from the proteome of patients with the cblC disorder. A graphical view of the second network and the top ranked physiological systems and functions derived from the analysis is shown in Fig. 9. These associations confirm that the cblC mutation leads to broad metabolic dysfunction. The results of the analysis of pathways correlates with the clinical features of the cblC disorder: The cblC complementation group is a genetic and metabolic disease, which presents with neurological, hematological, cardiovascular, skeletal and muscular abnormalities as the principal clinical manifestations. Statistically significant associations with other functions (hepatic system disease, behavioral problems) were also found, some of which have been described less commonly in patients with the cblC disease [11, 22–30]. This confirms that the cblC patient cell lines chosen for this study provide global insights into the general biochemical and functional hallmarks of the cblC disease, rather than focusing in the case-specific metabolic changes.
Table 5.
List of molecules displaying direct or indirect interactions with differentially expressed proteins determined by 2D-DIGE. Each set of molecules (IDs 1 and 2) was found to belong to a common functional network.
| ID | Molecules in Network | Score | Focus Molecules |
Top Functions |
|---|---|---|---|---|
| 1 | ANXA4, ANXA11, CLIC4, coumarin, EEF2, FCGRT, glutamyl-Se- methylselenocysteine, glutathione transferase, GST, GSTA1, GSTA2, GSTA5, GSTM1, GSTM2, GSTM3 (includes EG:14864), GSTM3 (includes EG:2947), GSTO1, GSTP1, HIST1H2BG, HNF4A, IL6, JUN, MTHFR, MTR, MUT, MYC, PDGF BB, PDIA3, PLS3, PNO1, RPS14, TGFB1, TUBA1B, UCHL1, VDAC2 |
34 | 14 | Cell Cycle, Gene Expression, Drug Metabolism |
| 2 | ABLIM1, ACTB, Actin, ARRB1, CALD1, CLIC4, COL6A2, COL6A3, DHRS2 (includes EG:10202), ERBB2, F Actin, fascin, folic acid, FOLR1, GAPDH (includes EG:2597), HDLBP, IL4, IL8, IVNS1ABP, L-homocysteine, MAPK1, MBP, PHACTR1, PLS1 (includes EG:104006), PLS1 (includes EG:5357), RAB8B, S100A6, S100P, SERPINB6, SH3BGRL3, SLC12A6, SPOCK1, VIM, vitamin B12, XPO6 |
31 | 13 | Genetic Disorder, Skeletal and Muscular Disorders, Lipid Metabolism |
Fig. 9.
A. Genetic disorders, skeletal and muscular disorders and lipid disorders network generated from 2D-DIGE and metabolites data through comparative analysis of normal and cblC fibroblasts grown without exogenous HOCbl. Solid lines represent direct interactions between molecules, whereas dashed lines represent indirect interactions between molecules. Color code: red: upregulated proteins or metabolites; green: downregulated proteins or metabolites. The intensity of green and red node colors indicates the degree of down or upregulation, respectively. B. Top ranked associations for physiological systems development and functions predicted by IPA® based on 2D-DIGE and metabolite data for normal and cblC fibroblasts grown without exogenous HOCbl. Threshold: −log p value (p = 0.05).
Network analysis of the normal and cblC proteomes in cells supplemented with HOCbl
The same analysis was conducted utilizing the 2D-DIGE and metabolite data gathered from normal and cblC fibroblasts supplemented with HOCbl. Three relevant networks were detected: 1) cancer, post-translational modification and protein folding, 2) genetic disorder, skeletal and muscular disorder and cancer, and 3) cellular assembly and organization, small molecule biochemistry and molecular transport. A summary of the proteins involved in each network is given in Table 6. Our DIGE results showed that supplementation with HOCbl did not completely reverse the protein expression pattern of the cblC cell lines to that observed in normal fibroblasts. In line with the experimental evidence, network analysis determined strong associations with neurological, hematological and skeletal and muscular dysfunctions. This was not surprising, as the majority of the patients with the cblC inborn error are only partially responsive to therapy with HOCbl [31]. Indeed, severely-ill patients do not show substantial improvement of their cognitive and motor functions upon long-term treatment with HOCbl [10].
Table 6.
Genes directly and indirectly associated with the cblC phenotype supplemented with HOCbl, linked to functions by pathway analysis.
| ID | Molecules in Network | Score | Focus Molecules |
Top Functions |
|---|---|---|---|---|
| 1 | Actin, actin-Actn-Ptk2-Pxn-Vcl, ANXA2, ANXA5, ANXA6, CCT3, CFL1, Ck2, CORO2B, DIS3L2, DPYSL2, ERK, F Actin, GSTP1, HSP, Hsp70, Hsp90, HSP90AA1, HSP90AB1, HSPA5, HSPA8, LMNA, PACRG, PARK7, PDIA3, PDIA4, PLS1 (includes EG:104006), RPS27A, S100A6, TMEM132A, TPI1, UCHL1, VCL, VIM, WARS |
55 | 22 | Cancer, Post- Translational Modification, Protein Folding |
| 2 | AHCY, AKT1, ANGPT1, ATP6V1A, CDKN2A, COL6A1, COL6A2, COL6A3, ERBB2, folic acid, FOLR1, G3BP1, HYOU1, IL8, IMPDH2, L-homocysteine, LPA, MTR, MUT, nitric oxide, NOTCH1, PDIA4, PROS1, S100A6, SEPT9, SEPT11, SH3BGRL3, SPOCK1, TAGLN2, TPD52, TRAF6, TUBA1C, TUBB2C, TUFM, vitamin B12 |
30 | 14 | Genetic Disorder, Skeletal and Muscular Disorders, Cancer |
| 3 | ACTB, ADRM1, CAP1, CCT2, CCT3, CCT4, CFL1, CLIC4, DHRS2 (includes EG:10202), GOT1, HERC5, HK1, peroxidase (miscellaneous), PHGDH, PPIA (includes EG:5478), PRDX1, PRDX2, PRDX4, PRDX6, RAB8B, RPL22, RPLP0 (includes EG:6175), RPS9, RPS18, RPS19, RPS4X, RPSA, SEPT2, SLC25A4, SLC25A5, SLC2A4, SRXN1, VDAC1, VDAC2 |
18 | 9 | Cellular Assembly and Organization, Small Molecule Biochemistry, Molecular Transport |
DISCUSSION
The primary objective of this study was to analyze the cblC proteome in skin fibroblasts from 3 patients with severe cblC disease and to compare it with the proteome in control skin fibroblasts. A general assessment of the metabolic features of the cblC and normal cell lines was also carried out. All cblC cell lines exported increased levels of Hcy and MMA compared to normal cells. Supplementation with HOCbl reduced the levels of MMA in all cblC cell lines (Fig. 1B). However, only one of the three cblC cell lines was able to utilize HOCbl as substrate for AdoCbl biosynthesis (Table 1). Our results do not preclude the possibility that small amounts of AdoCbl (undetectable by HPLC) synthesized by some of the cblC cell lines might have been sufficient to reduce the production and export of MMA. Supplementation with HOCbl was not effective in substantially reducing the levels of Hcy (Fig. 1A). The cblC cell lines employed in our study displayed reduced uptake and a greatly reduced capacity to utilize HOCbl for Cbl coenzyme biosynthesis compared to normal cells. This was in line with the finding that all cblC mutant cell lines take up or retain less amounts of Cbl compared to the normal fibroblasts, and this is also mirrored in the total levels of intracellular folates. One simple explanation for this result is that cblC cell lines do not retain Cbl because they are unable to process it to a form suitable for coenzyme biosynthesis. This suggests that a functional cblC protein is required not only for the decyanation and dealkylation of cobalamins, but also to maintain the homeostasis of related metabolites. Tetrahydrofolate, the biologically active form required for DNA biosynthesis, is regenerated by the enzyme methionine synthase. Presumably, the inability of the cblC cell lines to utilize HOCbl as a substrate for the biosynthesis of MeCbl leads to a blockage in the biosynthesis of tetrahydrofolate by methionine synthase. This could explain the observation that the cblC mutant cell lines had lower levels of folate compared to normal cells under conditions of cobalamin sufficiency. This is in line with the “methyl trap hypothesis” of Herbert and Zalusky [32] in which N5-mehtyltetrahydrofolate is no longer utilized as a substrate for Cbl-dependent methionine synthase and is exported from the cblC fibroblasts.
Proteins involved in muscular and skeletal functions
Substantial changes in protein expression levels between normal and cblC fibroblasts were found for cytoskeletal proteins with structural and regulatory roles. These include COL6A1, COL6A2, VIM, tubulin alpha (TUBA1B), β-actin (ACTB), VCL, plastin 3 (PLS3), lamin A/C isoform 2 (LMNA), chaperonin TCP1 (CCT3), caldesmon 1 (CALD1), CFL1 and TAGLN2. Interestingly, changes in cytoskeletal proteins have been reported for a patient cell line belonging to the complementation group D (cblD) [33] and also, for human fibroblasts grown under conditions of folate deficiency [34].
Our results showed that the production of COL6A2 was upregulated in cblC fibroblasts grown without HOCbl supplementation, and down-regulated in cblC cells grown in the presence of HOCbl. These findings suggest that increased levels of Hcy and/or MMA may be responsible for the up-regulation of COL6A2, isoform 2C2 in cblC fibroblasts and that even a modest decrease in the levels of these metabolites could have a substantial impact in the expression pattern of this protein. Collagen VI is a major structural component of microfibrils. In a 2D-DIGE proteomic study performed with normal and patient fibroblasts belonging to the cblD complementation group, an up-regulation of COL6A2 was also noted [33]. The patient described in the latter study presented with isolated methylmalonic aciduria.
In addition, a study performed in human smooth muscle cells demonstrated that high levels of Hcy cause an up-regulation in the production of collagen, which could be related to the pathogenesis of homocystinuria [35]. In support of this proposal, patients with untreated homocystinuria have widespread premature atherosclerosis with intimal thickening and collagen-rich fibrous plaques [36]. Based on these findings, it is reasonable to hypothesize that muscular dystrophy and cardiomyopathies, two common presentations of the cblC disorder, may be caused by an excessive production of collagens as a result of increased levels of Hcy. This is further supported by the observation that cblC patients show some improvement of their motor functions after prolonged therapy with HOCbl [7, 37].
Human vimentin was also down-regulated in cblC fibroblasts (Table 3). VIM is a cytoskeletal protein whose major role is to stabilize the architecture of the cytoplasm. We found that cblC fibroblasts expressed lower levels of VIM compared to normal cells, and that the difference was exacerbated by supplementation with HOCbl (Table 4). It is important to mention that the expression pattern of VIM is highly tissue-specific [38]. Therefore, correlations between the expression levels of this protein in fibroblasts with the clinical manifestation of the cblC disorder are difficult. Low levels of some major cytoskeleton structural proteins, namely β-actin and tubulin-α, were also found in cblC fibroblasts compared to normal cells grown without HOCbl. Supplementation of the cell cultures with HOCbl corrected for the observed down-regulation of β-actin, but was without effect on the levels of tubulin-α. To our knowledge, there is only one report linking Cbl administration with the expression of actin (the alpha-smooth muscle actin isoform). Isoda et al. reported that the overexpression of actin in liver (a marker of fibrosis) induced by the carcinogenic agent dimethylnitrosamine can be suppressed by administration of CNCbl [39].
Proteins involved in neurological functions
UCHL1 (also known as PGP9.5), a major component of the ubiquitin-proteasome protein degradation system, is down-regulated in cblC fibroblasts (Table 3). UCHL1 is one of the most abundant proteins in the brain (1–2% of the total soluble protein) and was reported to be exclusively localized in neurons [40] and in cells of the diffuse neuroendocrine system and their tumors [41]. Down-regulation and extensive oxidative modification of UCHL1 have been observed in brain tissue of patients with Alzheimer’s, as well as Parkinson's diseases [42–44]. Down-regulation of UCHL1 in cblC fibroblasts was not restored to normal levels upon supplementation with HOCbl. Therefore, down-regulation of UCHL1 could be partially responsible for the documented inability of cblC patients to achieve normal cognitive performance after prolonged treatment with HOCbl [7].
Two key proteins involved in neurological functions also appear to be down-regulated in cblC fibroblasts grown in the presence of exogenous HOCbl: DJ-1 (also known as PARK7) and dihydropyrimidase –like2 (DPYLS2) (Table 3). DJ-1 belongs to a family of peptidases that acts as a positive regulator of androgen receptor-dependent transcription. DJ-1 may also function as a redox-sensitive chaperone, as a sensor for oxidative stress, and it is thought to protect neurons from oxidative damage [45]. Defects in this gene are the cause of early-onset Parkinson disease 7 [45–46]. DPYLS2 is a protein that presents homology to dihydropyrimidase and is expressed mainly in the fetal and neonatal brains of mammals and chickens [47]. Little is known about this family of proteins, however, they are thought to be intracellular transducers in the development of the nervous system [47].
Proteins involved in intracellular trafficking and protein folding
A number of proteins involved in protein folding and intracellular trafficking are down-regulated in cblC fibroblasts compared to the normal cells, and exogenously added HOCbl did not correct this phenotype. These include: PDIA3, PDIA4, heat-shock proteins 90AB1, 90AA1, A5 and A8 (HSP90AB1, HSP90AA1, HSPA5 and HSPA8, respectively), ANXA5, ANXA56, a lysosomal –H+ transporting ATPase (ATP6VIA), and voltage dependent anion channels 1 and 2 (VDAC1 and VDAC2, respectively) (Table 3). Importantly, PDI, HSP70 and HSP90 play important roles in folding of newly synthesized proteins or stabilizing and refolding of denatured proteins after stress [48–49]. Annexins are a family of Ca-dependent and membrane-binding proteins, which are involved in membrane trafficking and various other processes including signaling, proliferation, differentiation, and inflammation [50–52]. Lysosomal ATP6VIA is a vacuolar multi-subunit enzyme that mediates acidification of eukaryotic intracellular organelles, a critical step for processes such as protein sorting, zymogen activation, receptor-mediated endocytosis and synaptic vesicle proton gradient generation [53]. The voltage dependent anion channels (VDACs) are the major channels by which small hydrophilic molecules can pass through the mitochondrial outer membrane. The expression of both VDAC1 and VDAC2 was lower in cblC fibroblasts than in normal cells, a pattern that did not vary when cells were grown in the presence of HOCbl.
Disturbances in protein folding, maturation and trafficking have been strongly associated with the neuropathology of Huntington’s disease (HD), Alzheimer’s disease and Parkinson's disease [54–55]. A number of proteins involved in these pathways are down-regulated in cblC fibroblasts, and their expression levels did not respond to supplementation with HOCbl. These alterations could contribute to the neurological deterioration observed in cblC patients, which in the majority of the cases, can only be partially alleviated by treatment with Cbl.
General metabolism and cellular detoxification
A number of proteins involved in general metabolism and cellular detoxification are down-regulated in the cblC proteome. These include: HDLBP, GAPDH, GST (various isoforms), CLIC4, phosphoglycerate dehydrogenase (PHDGH), tryptophanyl–tRNA synthetase isoform a (WARS), inosine monophosphate dehydrogenase 2 (IMPDH2), triosephosphate isomerase 1 (TPI1), and PRDX 1, 2 and 6. Although HDLBP (also known as vigilin) was upregulated in cblC fibroblasts grown without exogenous HOCbl, no differences in the expression levels of this protein were found for cells grown with HOCbl supplementation. HDLBP specifically binds to HDL and may function in the removal of excess cellular cholesterol [56].
There are two proteins whose expression patterns were unaffected by the presence of added HOCbl: CLIC4 and GST. CLIC4 encompasses a group of proteins that regulate fundamental cellular processes such as stabilization of cell membrane potential, transepithelial transport, maintenance of intracellular pH and regulation of cell volume.
Three isoforms of GST were down-regulated in cblC fibroblasts grown without HOCbl supplementation: GSTO1, GST, and GSTM3. Of these, only GST remained down-regulated under conditions of HOCbl supplementation, whereas the expression levels GSTO1 and GSTM3 did not differ significantly from that of normal cell lines. The GST family of proteins uses glutathione in the process of biotransformation of drugs, xenobiotics and oxidative stress. A recent report showed significant associations between the age of onset of Alzheimer’s and Parkinson's diseases and polymorphisms of GST omega 1 and 2 [57]. Our activity assays confirmed that cblC fibroblasts have reduced total GST activity. This may compromise the detoxification of metabolites that could potential aggravate the manifestation of the cblC disease.
Three members of the peroxiredoxin family were down-regulated in cblC fibroblasts grown with HOCbl supplementation: PRDX1, 2 and 6. Peroxiredoxins are responsible for the detoxification of hydrogen peroxide as well as of organic peroxides, thus exerting a protective role against oxidative damage [58]. Importantly, down-regulation of some of these detoxifying proteins has been reported in a model of folate deficiency [59]. Down-regulation of phase II metabolizing enzymes such as GST and peroxiredoxins is associated with a reduced capacity to eliminate carcinogens and therefore, an increased risk of cancer [59].
In summary, examination of the cblC proteome indicates that protein expression patterns are significantly different in the biochemical background of a defective MMACHC (cblC) gene. We observe that major changes affect various aspects of cellular metabolism and regulation, including cytoskeleton assembly and reorganization, nervous system proteins, signaling and cellular detoxification. Some of the proteins identified by this study have been strongly associated with skeletal and muscular diseases as well as neurological diseases (these include collagen VI, vimentin, actin, and UCHL1, respectively). This finding is consistent with the clinical manifestations of the cblC disorder. We anticipate that the identification of some proteins whose expression was strongly affected by the cblC mutation could be useful for designing alternative therapies to alleviate the symptoms of the cblC disease that do not respond sufficiently to the current approaches (HOCbl supplementation alone or in combination with other drugs).
The analysis of pathways and networks based on 2D-DIGE results and the measurement of relevant metabolites (Hcy, MMA, vitamin B12 and folic acid) shows that the cblC mutation strongly relates to dysfunctions involving the neurological, skeletal and muscular and hematological systems among others. This is in line with the commonly reported manifestations of the cblC mutation. It could be interesting to test whether some of the therapies utilized to treat patients with skeletal and muscular diseases would be also effective for alleviating related symptoms in patients with mutations in the cblC gene; we now know that at least some of the proteins involved in the progression of these diseases appear to have a common functional relationship. Although these notions remain speculative until further research is conducted, our results open new avenues for the investigation and treatment of the most common inborn error in human cobalamin metabolism.
Supplementary Material
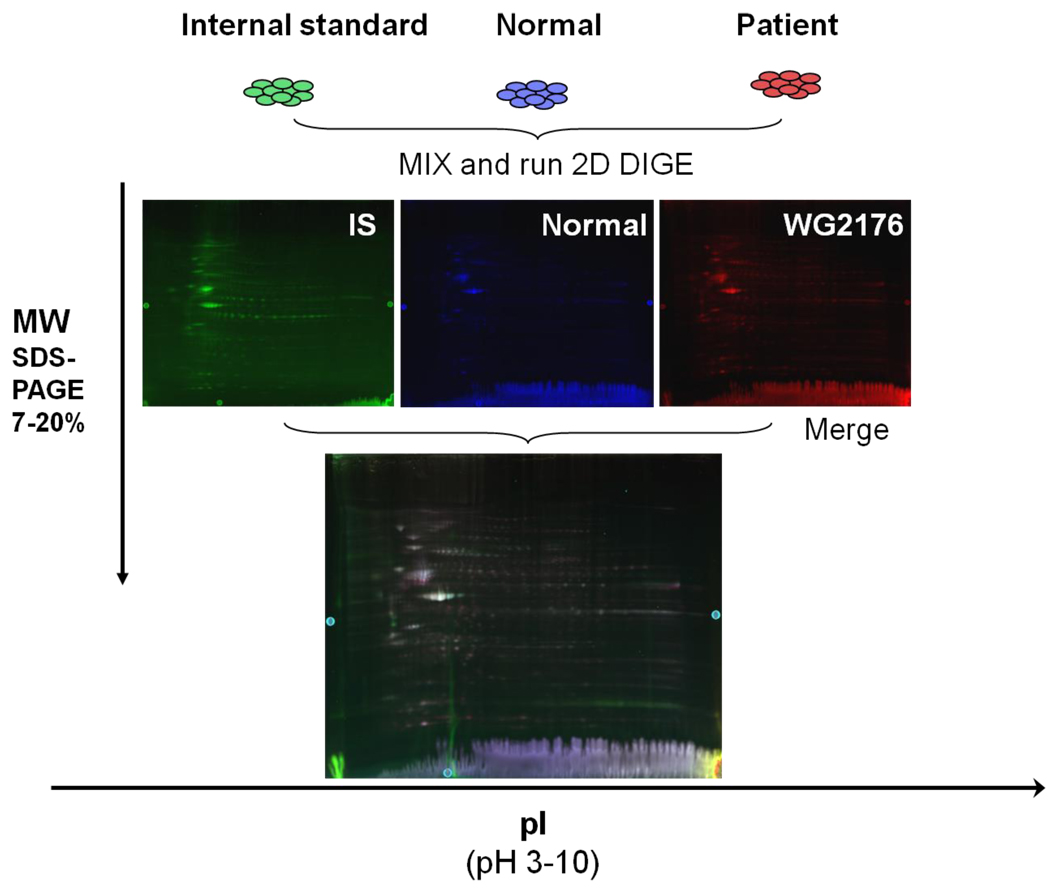
Fig. 2.
Individual and merged fluorescence scan images of Cy2-, Cy3- and Cy5-labeled proteins from normal and cblC fibroblasts. The proteins were labeled with either Cy2 (blue, normal), Cy3 (green, internal standard), and Cy5 (red, cblC patient WG2176). The 3 labeled aliquots were admixed and run on a single 2D-DIGE gel. The Cy2 images were acquired with a 488 nm excitation laser and a 520 nm emission filter; the Cy3 images used a 532 nm excitation laser with a 580 nm emission filter; and the Cy5 images were acquired with a 633 nm excitation laser and a 670 nm emission filter. The merged image shows that the CyDye labeled spots co-migrated to the exact same position and neither color was more prevalent in the merged image. Thus, CyDye labeling does not change the pI or the molecular weight of any given protein.
ACKNOWLEDGEMENTS
This research was supported by grants from the National Heart, Lung and Blood Institute of the National Institutes of Health (HL 71907 and HL 52234 to DWJ).
Abbreviations
- Cbl
cobalamin (B12)
- MeCbl
methylcobalamin
- AdoCbl
adenosylcobalamin
- MMACHC
methylmalonic aciduria combined with homocystinuria type C
- CNCbl
cyanocobalamin
- HOCbl
hydroxocobalamin
- 2-D DIGE
2-dimensional difference gel electrophoresis
- aDMEM
advanced Dulbecco’s modified Eagle medium
- HPLC
high-performance liquid chromatography
- PMSF
phenylmethanesulfonylfluoride
- ELISA
enzyme-linked immunosorbent assay
- DTT
dithiothreitol
- SDS
sodium dodecyl sulfate
- IEF
isoelectric focusing
- LC/ESI/MS
liquid chromatography-electrospray ionization-mass spectrometry
- CID
collisionally induced dissociation
- ANOVA
analysis of variance
- TNF-α
tumor necrosis factor-α
- CLIC4
chloride intracellular channel 4
- VIM
vimentin
- PDI
protein disulfide isomerase
- PDIA3
protein disulfide isomerase associated precursor 3
- PDIA4
protein disulfide isomerase associated precursor 4
- GRP94
glucose-regulated protein 94
- UCHLI
ubiquitin C-terminal esterase L1
- ANXA5
annexin V
- ANXA6
annexin VI isoform 1
- HDLBP
high density lipoprotein binding protein
- COL6A1
collagen type VI alpha 1 precursor
- COL6A2
collagen type VI isoform 2C2
- EEF2
eukaryotic elongation factor 2
- GST
glutathione-S-transferase
- GSTO1
glutathione-S-transferase omega 1
- GSTM3
glutathione-S-transferase mu 3
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- SH3DBGRP
SH3 domain binding Glu-rich protein
- S100A6
S100 Ca-binding protein A6
- VCL
vinculin isoform
- HSP90
heat shock protein 90
- HSP90AB1
heat shock protein 90 beta
- HSP90AA1
heat shock protein 90 class A member 1 isoform 2
- HSP70
heat shock protein 70
- HSPA5
heat shock protein 70 protein 5
- HSP70A8
heat shock protein 70 protein 8 isoform 1
- SEPT11
septin 11
- PRDX1
peroxiredoxin 1
- PRDX6
peroxiredoxin 6
- CFL1
cofilin 1
- PARK7 (also known as DJ-1)
Parkinson disease protein 7
- TAGLN2
transgelin 2
- TUBA1B
tubulin alpha
- ACTB
β-actin
- PLS3
plastin 3
- LMNA
lamin A/C isoform 2
- CCT3
chaperonin TCP1
- CALD1
caldesmon 1
- DPYLS2
dihydropyrimidase-like protein 2
- ATP6VIA
lysosomal H+ transporting ATPase
- VDAC1
voltage dependent anion channel 1
- VDAC2
voltage dependent anion channel 2
- PHDGH
phosphoglycerate dehydrogenase
- WARS
tryptophanyl-tRNA synthetase isoform a
- IMPDH2
inosine monophosphate dehydrogenase 2
- TPI1
triosephosphate isomerase 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Apendix A. Supplementary data. Supplementary data to this article can be found online at doi:
REFERENCES
- 1.Banerjee R, Chowdhury S. Chemistry and Biochemistry of B12. New York: Wiley-IEEE; 1999. Methylmalonyl-CoA mutase. [Google Scholar]
- 2.Matthews RG. Chemistry and Biochemistry of B12. New York: Wiley-IEEE; 1999. Cobalamin-dependent methionine synthase. [Google Scholar]
- 3.Lerner-Ellis JP, Tirone JC, Pawelek PD, Dore C, Atkinson JL, Watkins D, Morel CF, Fujiwara TM, Moras E, Hosack AR, Dunbar GV, Antonicka H, Forgetta V, Dobson CM, Leclerc D, Gravel RA, Shoubridge EA, Coulton JW, Lepage P, Rommens JM, Morgan K, Rosenblatt DS. Identification of the gene responsible for methylmalonic aciduria and homocystinuria, cblC type. Nat Genet. 2006;38:93–100. doi: 10.1038/ng1683. [DOI] [PubMed] [Google Scholar]
- 4.Kim J, Gherasim C, Banerjee R. Decyanation of vitamin B12 by a trafficking chaperone. Proc Natl Acad Sci U S A. 2008;105:14551–14554. doi: 10.1073/pnas.0805989105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hannibal L, Kim J, Brasch NE, Wang S, Rosenblatt DS, Banerjee R, Jacobsen DW. Processing of alkylcobalamins in mammalian cells: A role for the MMACHC (cblC) gene product. Mol Genet Metab. 2009;97:260–266. doi: 10.1016/j.ymgme.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim J, Hannibal L, Gherasim C, Jacobsen DW, Banerjee R. A human vitamin B12 trafficking protein uses glutathione transferase activity for processing alkylcobalamins. J Biol Chem. 2009;284:33418–33424. doi: 10.1074/jbc.M109.057877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersson HC, Marble M, Shapira E. Long-term outcome in treated combined methylmalonic acidemia and homocystinemia. Genet Med. 1999;1:146–150. doi: 10.1097/00125817-199905000-00006. [DOI] [PubMed] [Google Scholar]
- 8.Rosenblatt DS, Fenton WA. Inherited disorders of folate and cobalamin transport and metabolism. In: Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Kinzler KW, Vogelstein B, editors. The Metabolic & Molecular Bases of Inherited Disease. New York: Mc-Graw Hill; 2001. pp. 3897–3933. [Google Scholar]
- 9.Shinnar S, Singer HS. Cobalamin C mutation (methylmalonic aciduria and homocystinuria) in adolescence: a treatable cause of dementia and myelopathy. N Engl J Med. 1984;311:451–454. doi: 10.1056/NEJM198408163110707. [DOI] [PubMed] [Google Scholar]
- 10.Rosenblatt DS, Aspler AL, Shevell MI, Pletcher BA, Fenton WA, Seashore MR. Clinical heterogeneity and prognosis in combined methylmalonic aciduria and homocystinuria (cblC) J Inherit Metab Dis. 1997;20:528–538. doi: 10.1023/a:1005353530303. [DOI] [PubMed] [Google Scholar]
- 11.Lerner-Ellis JP, Anastasio N, Liu J, Coelho D, Suormala T, Stucki M, Loewy AD, Gurd S, Grundberg E, Morel CF, Watkins D, Baumgartner MR, Pastinen T, Rosenblatt DS, Fowler B. Spectrum of mutations in MMACHC, allelic expression, and evidence for genotype-phenotype correlations. Hum Mutat. 2009;30:1072–1081. doi: 10.1002/humu.21001. [DOI] [PubMed] [Google Scholar]
- 12.Hannibal L, Axhemi A, Glushchenko AV, Moreira ES, Brasch NE, Jacobsen DW. Accurate assessment and identification of naturally occurring cellular cobalamins. Clin Chem Lab Med. 2008;46:1739–1746. doi: 10.1515/CCLM.2008.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobsen DW, Gatautis VJ, Green R, Robinson K, Savon SR, Secic M, Ji J, Otto JM, Taylor LM., Jr Rapid HPLC determination of total homocysteine and other thiols in serum and plasma: sex differences and correlation with cobalamin and folate levels in normal subjects. Clin Chem. 1994;40:873–881. [PubMed] [Google Scholar]
- 14.Hoffmann G, Aramaki S, Blum-Hoffmann E, Nyhan WL, Sweetman L. Quantitative analysis for organic acids in biological samples: batch isolation followed by gas chromatographic-mass spectrometric analysis. Clin Chem. 1989;35:587–595. [PubMed] [Google Scholar]
- 15.Marouga R, David S, Hawkins E. The development of the DIGE system: 2D fluorescence difference gel analysis technology. Anal Bioanal Chem. 2005;382:669–678. doi: 10.1007/s00216-005-3126-3. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Karp NA, Kreil DP, Lilley KS. Determining a significant change in protein expression with DeCyder during a pair-wise comparison using two-dimensional difference gel electrophoresis. Proteomics. 2004;4:1421–1432. doi: 10.1002/pmic.200300681. [DOI] [PubMed] [Google Scholar]
- 18.Suormala T, Baumgartner MR, Coelho D, Zavadakova P, Kozich V, Koch HG, Berghauser M, Wraith JE, Burlina A, Sewell A, Herwig J, Fowler B. The cblD defect causes either isolated or combined deficiency of methylcobalamin and adenosylcobalamin synthesis. J Biol Chem. 2004;279:42742–42749. doi: 10.1074/jbc.M407733200. [DOI] [PubMed] [Google Scholar]
- 19.Mellman I, Willard HF, Youngdahl-Turner P, Rosenberg LE. Cobalamin coenzyme synthesis in normal and mutant human fibroblasts. Journal of Biological Chemistry. 1979;254(No.23):11847–11853. [PubMed] [Google Scholar]
- 20.Fenton WA, Ambani LM, Rosenberg LE. Uptake of hydroxocobalamin by rat liver mitochondria. Binding to a mitochondrial protein. J Biol Chem. 1976;251:6616–6623. [PubMed] [Google Scholar]
- 21.Fenton WA, Rosenberg LE. Mitochondrial metabolism of hydroxocobalamin: Synthesis of adenosylcobalamin by intact rat liver mitochondria. Arch Biochem Biophys. 1978;189:441–447. doi: 10.1016/0003-9861(78)90232-1. [DOI] [PubMed] [Google Scholar]
- 22.Loewy AD, Niles KM, Anastasio N, Watkins D, Lavoie J, Lerner-Ellis JP, Pastinen T, Trasler JM, Rosenblatt DS. Epigenetic modification of the gene for the vitamin B(12) chaperone MMACHC can result in increased tumorigenicity and methionine dependence. Mol Genet Metab. 2009;96:261–267. doi: 10.1016/j.ymgme.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 23.Tsai AC, Morel CF, Scharer G, Yang M, Lerner-Ellis JP, Rosenblatt DS, Thomas JA. Late-onset combined homocystinuria and methylmalonic aciduria (cblC) and neuropsychiatric disturbance. Am J Med Genet A. 2007;143A:2430–2434. doi: 10.1002/ajmg.a.31932. [DOI] [PubMed] [Google Scholar]
- 24.Morel CF, Lerner-Ellis JP, Rosenblatt DS. Combined methylmalonic aciduria and homocystinuria (cblC): phenotype-genotype correlations and ethnic-specific observations. Mol Genet Metab. 2006;88:315–321. doi: 10.1016/j.ymgme.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Guigonis V, Fremeaux-Bacchi V, Giraudier S, Favier R, Borderie D, Massy Z, Mougenot B, Rosenblatt DS, Deschenes G. Late-onset thrombocytic microangiopathy caused by cblC disease: association with a factor H mutation. Am J Kidney Dis. 2005;45:588–595. doi: 10.1053/j.ajkd.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 26.Harding CO, Pillers DA, Steiner RD, Bottiglieri T, Rosenblatt DS, Debley J, Michael Gibson K. Potential for misdiagnosis due to lack of metabolic derangement in combined methylmalonic aciduria/hyperhomocysteinemia (cblC) in the neonate. J Perinatol. 2003;23:384–386. doi: 10.1038/sj.jp.7210955. [DOI] [PubMed] [Google Scholar]
- 27.Bodamer OA, Rosenblatt DS, Appel SH, Beaudet AL. Adult-onset combined methylmalonic aciduria and homocystinuria (cblC) Neurology. 2001;56:1113. doi: 10.1212/wnl.56.8.1113. [DOI] [PubMed] [Google Scholar]
- 28.Augoustides-Savvopoulou P, Mylonas I, Sewell AC, Rosenblatt DS. Reversible dementia in an adolescent with cblC disease: clinical heterogeneity within the same family. J Inherit Metab Dis. 1999;22:756–758. doi: 10.1023/a:1005508620919. [DOI] [PubMed] [Google Scholar]
- 29.Enns GM, Barkovich AJ, Rosenblatt DS, Fredrick DR, Weisiger K, Ohnstad C, Packman S. Progressive neurological deterioration and MRI changes in cblC methylmalonic acidaemia treated with hydroxocobalamin. J Inherit Metab Dis. 1999;22:599–607. doi: 10.1023/a:1005517727451. [DOI] [PubMed] [Google Scholar]
- 30.Rosenblatt DS, Aspler AL, Shevell MI, Pletcher BA, Fenton WA, Seashore MR. Clinical heterogeneity and prognosis in combined methylmalonic aciduria and homocystinuria (cblC) J Inherit Metab Dis. 1997;20:528–538. doi: 10.1023/a:1005353530303. [DOI] [PubMed] [Google Scholar]
- 31.Froese DS, Zhang J, Healy S, Gravel RA. Mechanism of vitamin B12-responsiveness in cblC methylmalonic aciduria with homocystinuria. Mol Genet Metab. 2009;98:338–343. doi: 10.1016/j.ymgme.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 32.Herbert V, Zalusky R. Interrelations of vitamin B12 and folic acid metabolism: folic acid clearance studies. Journal of Clinical Investigation. 1962;41:1263–1276. doi: 10.1172/JCI104589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richard E, Monteoliva L, Juarez S, Perez B, Desviat LR, Ugarte M, Albar JP. Quantitative analysis of mitochondrial protein expression in methylmalonic acidemia by two-dimensional difference gel electrophoresis. J Proteome Res. 2006;5:1602–1610. doi: 10.1021/pr050481r. [DOI] [PubMed] [Google Scholar]
- 34.Katula KS, Heinloth AN, Paules RS. Folate deficiency in normal human fibroblasts leads to altered expression of genes primarily linked to cell signaling, the cytoskeleton and extracellular matrix. J Nutr Biochem. 2007;18:541–552. doi: 10.1016/j.jnutbio.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 35.Majors AK, Sengupta S, Jacobsen DW, Pyeritz RE. Upregulation of smooth muscle cell collagen production by homocysteine-insight into the pathogenesis of homocystinuria. Mol Genet Metabol. 2002;76:92–99. doi: 10.1016/s1096-7192(02)00030-6. [DOI] [PubMed] [Google Scholar]
- 36.Majors A, Ehrhart LA, Pezacka EH. Homocysteine as a risk factor for vascular disease. Enhanced collagen production and accumulation by smooth muscle cells. Arterioscler Thromb Vasc Biol. 1997;17:2074–2081. doi: 10.1161/01.atv.17.10.2074. [DOI] [PubMed] [Google Scholar]
- 37.Andersson HC, Shapira E. Biochemical and clinical response to hydroxocobalamin versus cyanocobalamin treatment in patients with methylmalonic acidemia and homocystinuria (cblC) J Pediatr. 1998;132:121–124. doi: 10.1016/s0022-3476(98)70496-2. [DOI] [PubMed] [Google Scholar]
- 38.Perreau J, Lilienbaum A, Vasseur M, Paulin D. Nucleotide sequence of the human vimentin gene and regulation of its transcription in tissues and cultured cells. Gene. 1988;62:7–16. doi: 10.1016/0378-1119(88)90575-6. [DOI] [PubMed] [Google Scholar]
- 39.Isoda K, Kagaya N, Akamatsu S, Hayashi S, Tamesada M, Watanabe A, Kobayashi M, Tagawa Y, Kondoh M, Kawase M, Yagi K. Hepatoprotective effect of vitamin B12 on dimethylnitrosamine-induced liver injury. Biol Pharm Bull. 2008;31:309–311. doi: 10.1248/bpb.31.309. [DOI] [PubMed] [Google Scholar]
- 40.Wilson PO, Barber PC, Hamid QA, Power BF, Dhillon AP, Rode J, Day IN, Thompson RJ, Polak JM. The immunolocalization of protein gene product 9.5 using rabbit polyclonal and mouse monoclonal antibodies. Br J Exp Pathol. 1988;69:91–104. [PMC free article] [PubMed] [Google Scholar]
- 41.Doran JF, Jackson P, Kynoch PA, Thompson RJ. Isolation of PGP 9.5, a new human neurone-specific protein detected by high-resolution two-dimensional electrophoresis. J Neurochem. 1983;40:1542–1547. doi: 10.1111/j.1471-4159.1983.tb08124.x. [DOI] [PubMed] [Google Scholar]
- 42.Castegna A, Aksenov M, Thongboonkerd V, Klein JB, Pierce WM, Booze R, Markesbery WR, Butterfield DA. Proteomic identification of oxidatively modified proteins in Alzheimer's disease brain. Part II: dihydropyrimidinase-related protein 2, alpha-enolase and heat shock cognate 71. J Neurochem. 2002;82:1524–1532. doi: 10.1046/j.1471-4159.2002.01103.x. [DOI] [PubMed] [Google Scholar]
- 43.Choi J, Levey AI, Weintraub ST, Rees HD, Gearing M, Chin LS, Li L. Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson's and Alzheimer’s diseases. J Biol Chem. 2004;279:13256–13264. doi: 10.1074/jbc.M314124200. [DOI] [PubMed] [Google Scholar]
- 44.Butterfield DA, Gnjec A, Poon HF, Castegna A, Pierce WM, Klein JB, Martins RN. Redox proteomics identification of oxidatively modified brain proteins in inherited Alzheimer's disease: an initial assessment. J Alzheimers Dis. 2006;10:391–397. doi: 10.3233/jad-2006-10407. [DOI] [PubMed] [Google Scholar]
- 45.Abou-Sleiman PM, Healy DG, Quinn N, Lees AJ, Wood NW. The role of pathogenic DJ-1 mutations in Parkinson's disease. Ann Neurol. 2003;54:283–286. doi: 10.1002/ana.10675. [DOI] [PubMed] [Google Scholar]
- 46.Annesi G, Savettieri G, Pugliese P, D’Amelio M, Tarantino P, Ragonese P, La Bella V, Piccoli T, Civitelli D, Annesi F, Fierro B, Piccoli F, Arabia G, Caracciolo M, Ciro Candiano IC, Quattrone A. DJ-1 mutations and parkinsonism-dementia-amyotrophic lateral sclerosis complex. Ann Neurol. 2005;58:803–807. doi: 10.1002/ana.20666. [DOI] [PubMed] [Google Scholar]
- 47.Kitamura K, Takayama M, Hamajima N, Nakanishi M, Sasaki M, Endo Y, Takemoto T, Kimura H, Iwaki M, Nonaka M. Characterization of the human dihydropyrimidinase-related protein 2 (DRP-2) gene. DNA Res. 1999;6:291–297. doi: 10.1093/dnares/6.5.291. [DOI] [PubMed] [Google Scholar]
- 48.Appenzeller-Herzog C, Ellgaard L. The human PDI family: versatility packed into a single fold. Biochim Biophys Acta. 2008;1783:535–548. doi: 10.1016/j.bbamcr.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 49.Gregersen N. Protein misfolding disorders: pathogenesis and intervention. J Inherit Metab Dis. 2006;29:456–470. doi: 10.1007/s10545-006-0301-4. [DOI] [PubMed] [Google Scholar]
- 50.Raynal P, Pollard HB. Annexins: the problem of assessing the biological role for a gene family of multifunctional calcium- and phospholipid-binding proteins. Biochim Biophys Acta. 1994;1197:63–93. doi: 10.1016/0304-4157(94)90019-1. [DOI] [PubMed] [Google Scholar]
- 51.Gerke V, Creutz CE, Moss SE. Annexins: linking Ca2+ signalling to membrane dynamics. Nat Rev Mol Cell Biol. 2005;6:449–461. doi: 10.1038/nrm1661. [DOI] [PubMed] [Google Scholar]
- 52.Grewal T, Enrich C. Molecular mechanisms involved in Ras inactivation: the annexin A6-p120GAP complex. Bioessays. 2006;28:1211–1220. doi: 10.1002/bies.20503. [DOI] [PubMed] [Google Scholar]
- 53.Stevens TH, Forgac M. Structure, function and regulation of the vacuolar (H+)-ATPase. Annu Rev Cell Dev Biol. 1997;13:779–808. doi: 10.1146/annurev.cellbio.13.1.779. [DOI] [PubMed] [Google Scholar]
- 54.Matus S, Glimcher LH, Hetz C. Protein folding stress in neurodegenerative diseases: a glimpse into the ER. Curr Opin Cell Biol. 2011 doi: 10.1016/j.ceb.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 55.Vidal R, Caballero B, Couve A, Hetz C. Converging pathways in the occurrence of endoplasmic reticulum (ER) stress in Huntington's disease. Curr Mol Med. 2011;11:1–12. doi: 10.2174/156652411794474419. [DOI] [PubMed] [Google Scholar]
- 56.Plenz G, Kugler S, Schnittger S, Rieder H, Fonatsch C, Muller PK. The human vigilin gene: identification, chromosomal localization and expression pattern. Hum Genet. 1994;93:575–582. doi: 10.1007/BF00202827. [DOI] [PubMed] [Google Scholar]
- 57.Takeshita H, Fujihara J, Takastuka H, Agusa T, Yasuda T, Kunito T. Diversity of glutathione s-transferase omega 1 (a140d) and 2 (n142d) gene polymorphisms in worldwide populations. Clin Exp Pharmacol Physiol. 2009;36:283–286. doi: 10.1111/j.1440-1681.2008.05058.x. [DOI] [PubMed] [Google Scholar]
- 58.Knoops B, Loumaye E, Van Der Eecken V. Evolution of the peroxiredoxins. Subcell Biochem. 2007;44:27–40. doi: 10.1007/978-1-4020-6051-9_2. [DOI] [PubMed] [Google Scholar]
- 59.Duthie SJ, Mavrommatis Y, Rucklidge G, Reid M, Duncan G, Moyer MP, Pirie LP, Bestwick CS. The response of human colonocytes to folate deficiency in vitro: functional and proteomic analyses. J Proteome Res. 2008;7:3254–3266. doi: 10.1021/pr700751y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.