Abstract
In the course of determining the sequence of the Dictyostelium discoideum genome we have characterized in detail the quantity and nature of interspersed repetitive elements present in this species. Several of the most abundant small complex repeats and transposons (DIRS-1; TRE3-A,B; TRE5-A; skipper; Tdd-4; H3R) have been described previously. In our analysis we have identified additional elements. Thus, we can now present a complete list of complex repetitive elements in D. discoideum. All elements add up to 10% of the genome. Some of the newly described elements belong to established classes (TRE3-C, D; TRE5-B,C; DGLT-A,P; Tdd-5). However, we have also defined two new classes of DNA transposable elements (DDT and thug) that have not been described thus far. Based on the nucleotide amount, we calculated the least copy number in each family. These vary between <10 up to >200 copies. Unique sequences adjacent to the element ends and truncation points in elements gave a measure for the fragmentation of the elements. Furthermore, we describe the diversity of single elements with regard to polymorphisms and conserved structures. All elements show insertion preference into loci in which other elements of the same family reside. The analysis of the complex repeats is a valuable data resource for the ongoing assembly of whole D. discoideum chromosomes.
[The sequence data described in this paper have been submitted to the GenBank data library under accession nos. AF135841, AF298201, AF298202, AF298203, AF298204, AF298205, AF298206, AF298207, AF298208, AF298209, AF298210 and AF298624.]
Many genomes contain a considerable number of repetitive elements. Repetitive sequences of a genome can be divided into simple repeats, complex repeats, and gene families. Simple repeats consist of tandemly repeated short sequences (mainly mono- to trinucleotides), that can amount to >100 bp. Conversely, complex repeats are much larger. They can reach >5 kb, and they often contain coding sequences. Most of these sequence segments, if complete, are mobile and, therefore, are called transposable elements (TEs). The TEs comprise the major portion of repetitive DNA in many eukaryotes (Smit 1999). Each TE family shows at least one of the following features, which make them distinguishable from gene families: Direct or inverted repeat sequences at the ends, presence of abundant truncated copies, the potential to form secondary structures, and/or lack of coding potential.
Because TEs appear to have no obvious functions for the host cells, they are often considered to be “selfish DNA” (Flavell 1995). Yet, a transposition event can cause spontaneous mutations and may lead to rearrangements in the host genome (Zhang and Peterson 1999; Xiao and Peterson 2000). Thus, these events are considered to be a major source for the variability of genomes. On the other hand, transposition frequently leads to severe genome alterations through deletions, rearrangements, and gene truncations. This, in turn, is an evolutionary pressure against frequent and random transpositions.
TEs are classified according to their intermediate form during transposition as DNA transposons or retroelements. Retroelements are transcribed to RNA for two purposes: Translation of the coding sequences or as intermediates for the transposition, and thus retroelements are amplified via transposition. In contrast, DNA elements are excised and transposed as DNA without amplification. Eukaryotic retroelements can be further divided into long terminal repeat (LTR) elements, non-LTR elements, and retroviruses. Very often, LTRs are found that are not associated with the core segment containing the genes that are needed for transposition (Goodwin and Poulter 2000). In this study, we defined these orphan, or solo, LTRs as elements in their own right.
Repetitive elements are widespread in nature. In yeast, the number of LTR retrotransposons (Ty elements) and the corresponding LTRs reach 3.1% of the genome (Kim et al. 1998). This amount increases considerably in more complex genomes, such as human and maize. Thus, up to 50% of the genomic DNA can be derived from retrotransposons (Smit et al. 1995; SanMiguel et al. 1996). Because of the additional presence of DNA elements, less than half of a genome may be “sense DNA.”
Dictyostelium discoideum, a soil-living amoeba, is an excellent and, therefore, widely used organism for the study of cell motility, signal transduction, cell type differentiation, and developmental processes (Kay and Williams 1999; Noegel and Schleicher 2000). Because of its importance as model organism, D. discoideum was chosen for a genome sequencing project. The genome of D. discoideum is divided into six regular chromosomes ranging in size from 4 Mb to 7 Mb and an additional palindromic multicopy chromosome containing the rRNA genes (Loomis and Kuspa 1997). The six regular chromosomes have a size of 34 Mb. The AT content of the genome is very high (∼78%). In particular, intergenic regions can reach a composition of >95% AT nucleotides. Because of this highly biased nucleotide composition, large bacterial insert clones (>10 kb) are unstable in Eschericia coli host strains. Therefore, we made shotgun libraries from whole genomic or purified chromosomal DNA in pUC plasmids with inserts of moderate size (1 kb–3 kb).
Repeated structures in the genome of D. discoideum have been studied for a long time. Simple repeats have been found to be very abundant in this genome (Firtel and Kindle 1975,1976; Kimmel and Firtel 1985). The first complex transposable repetitive element was described in 1983 (Rosen et al. 1983; Zuker et al. 1983). The quantity of this TE was estimated to be ∼240 full-length and truncated copies. It seems to be the most abundant complex repetitive element in the D. discoideum genome. Several TEs that are associated with tRNAs were also found in D. discoideum (Poole and Firtel 1984; Marschalek et al. 1989). One of these elements was later estimated to reside with ∼30 copies in the genome (Winckler 1998). A further tRNA-associated TE is present in various amounts in different strains (Marschalek et al. 1993). So far, only one DNA transposon has been described (Wells 1999), which is also relatively abundant in the genome. Additional tRNA-associated TEs were detected at the beginning of the genome project by scanning of the produced sequences (Szafranski et al. 1999). Thus, a large-scale sequencing project gives the opportunity to detect even less abundant complex repeats before an assembly.
The length, polymorphism, and abundance of TEs are a barrier for the easy assembly of raw reads into a reliable genomic sequence. Thus, an exhaustive analysis of all repetitive elements is a prerequisite for the assembly.
RESULTS AND DISCUSSION
Definition of Complex Repetitive Elements
The initial analysis of complex repetitive elements was performed on a data set of random raw reads comprising 28 Mb good-quality sequences. This coverage of ≥0.8 × allowed the detection of any genomic feature longer than 1 kb with a reliability of 97% (see Methods). Thus, sequences that are represented several times in the genome could be easily detected using similarity searches comparing each single sequence to the entire data set. Several TEs in the genome of D. discoideum have been described previously (Leng et al. 1998; Winckler 1998; Szafranski et al. 1999; Wells 1999). In addition, one LTR that lacks a core region with coding potential (solo LTR) could be found in previous studies (GenBank accession no. X59570). In a first step, these known elements were reconstructed from our raw reads using their consensus sequences for similarity searches in the data set. The successful reconstruction of the previously described elements was also a test for the feasibility of our approach.
To find additional members belonging to the different classes, similarity searches were performed against the raw reads using the already available sequences. To exclude the possibility that we have missed members in each class, we also searched the database of raw reads with the deduced amino acid sequences of the protein-encoding genes of every transposon using tblastn. In this way, we have identified all members of each previously described class. Even TEs, which are only distantly related to one of the elements, were detected.
TEs are not evenly distributed over the genome (Voytas and Boeke 1993; Voytas 1996). This is caused in part by deleterious mutations that may occur as a consequence of transposition. Thus, TEs tend to accumulate at certain positions in the chromosome and build islands of repetitive elements containing different elements. Taking all these considerations into account, we also searched for parts of new elements at borders and consensus breakpoints of reads from known elements. In this way, we could define additional complex repeats (Tdd-5; DDT class; thug class).
With the progress of the sequencing efforts, we accumulated additional raw reads. Thus, on an extended data set with a genome coverage of 3 ×(100 Mb) we performed an all-reads against all-reads crosscheck. This check revealed the presence of additional sequences that are more abundant than average in the genome, that is, complex repeats and multigene families. These sequences were assembled and further examined. Sequences from multigene families were excluded from the analysis when similarities to known proteins in the GenPept database were found. In addition, sequences without any additional feature of TEs (truncated copies, LTRs, lack of coding potential, clustering in TEs) were also excluded. Using these methods, one additional complex repeat could be found (DGLT-P). In all likelihood, we have indeed identified all complex repeats within the D. discoideum genome.
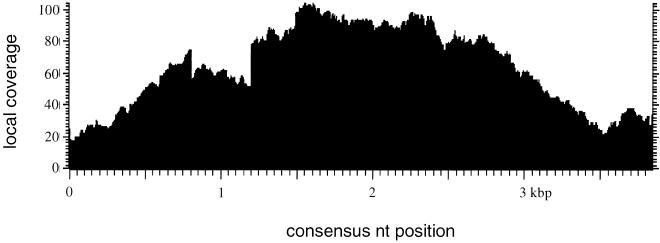
The raw sequences that belong to individual elements were assembled irrespective of polymorphic sites. Consensus breakpoints in single reads that are caused by truncations of a particular element were masked. A typical coverage plot of such an assembly is shown in Figure 1. Interestingly, at the borders of the elements, the sequence coverage is lower than in the core. This feature occurs mostly because of incomplete, truncated, elements, as could be revealed by the analysis of flanking sequences (see also Table 1). Truncation of elements could occur for two reasons: First, retroelements may insert as incomplete copies because of an incomplete reverse transcription; second, the element may be destroyed by the insertion of other DNA segments and rearrangements and deletions.
Figure 1.
Coverage plot for element family Tdd-4 deduced from an alignment of 550 randomly chosen reads using CLUSTALW. Each base of the alignment is represented by a dot.
Table 1.
Insertion Behavior for Element Families Without Insertion Preference for the Vicinity of tRNA Genes
| Transposon | Ends analyzed | Insertion target | |||
|---|---|---|---|---|---|
| Transposon | Genomic ocean | ||||
| No. | % | No. | % | ||
| DIRS-1 | 15 | 13 | 87 | 2 | 13 |
| skipper | 10 | 9 | 90 | 1 | 10 |
| Tdd-4 | 20 | 10 | 50 | 10 | 50 |
| Tdd-5 | 16 | 7 | 44 | 9 | 56 |
| DDT-A | 24 | 20 | 83 | 4 | 18 |
| DDT-B | 29 | 26 | 90 | 3 | 10 |
| DDT-S | 21 | 18 | 86 | 3 | 14 |
| thug-S | 14 | 3 | 21 | 11 | 79 |
| thug-T | 17 | 4 | 24 | 13 | 76 |
| Total | 166 | 110 | 66 | 56 | 34 |
The definition of element borders is difficult because small truncations or target site duplications cannot be detected if a TE is inserted into unknown sequences. Insertions of TEs into known portions of the genome, however, define sharp borders. The insertion of TEs into other TEs has the highest probability because of the genetically neutral nature of this event. Consequently, we found for each TE (except the TRE classes) a location in other TEs. This facilitated the definition of element borders and allowed the determination of target site duplications.
In addition to the underrepresentation of element ends caused by truncations, we also observed a cloning bias against sequences that had very high AT content; that is, AT-rich regions are underrepresented in the clone library. In addition, AT-rich stretches are not as readily sequenced as DNA with equally distributed nucleotides. Overall, these biases led to an additional underrepresentation of the border regions of the complex repetitive elements. Some TE families (TRE3-D; TRE5-C, Tdd-5) represent the least abundant complex repeats in the genome. Consequently, the structure of these elements could not be entirely defined even with the extended data set of 3 × coverage.
To test whether the method described here is exhaustive in finding abundant features in a genome, we analyzed the actin gene family with this approach. The actin genes of D. discoideum have been well studied for a long time (Kindle and Firtel 1978; McKeown et al. 1978). According to hybridization data, the family consists of 17–20 members, of which 15 were sequenced previously (Romans and Firtel 1985). Surprisingly, our statistical analysis of sequence reads suggested that ∼40 copies of that gene family should reside in the genome of D. discoideum (data not shown). Because this quantity differed by a factor of two from the originally estimated number, we decided to assemble the whole actin gene family. This assembly revealed 23 apparently-functional actin genes and two full-length pseudogenes with internal stop codons. In addition, some smaller sequence stretches with homology to actin genes were found. Obviously, our statistical approach led to a slight overestimation of the copy number. Yet we were able to find not only all previously defined actin genes, but also additional members of this gene family. Thus, this analysis should provide a comprehensive view of the complex repeats in the D. discoideum genome.
Classification of the D. discoideum Repeats
Each subgroup of the TEs (LTR retroelements, non-LTR retroelements, and DNA transposons) consists of several classes according to their main features. In all these subgroups of TEs we found new elements that have not been previously described. Table 2 lists all detected elements, and the previously known element families are labeled with an asterisk. Based on the abundance of single reads that belong to a certain element, we give calculations of the nucleotide percentage of each element in the genome (Table 2). Our rounded calculations of the copy number for each element are in good agreement with previous estimations for the known elements based on hybridization techniques (Zuker et al. 1983; Marschalek et al. 1993; Winckler 1998). The fragmentation index is determined from the number of individual ends in each element family divided by the copy number, which is calculated as total quantity of nucleotides of the element divided by the element length. The FI gives a measure for the mean completeness of an element in each TE family. Generally, DNA elements are more fragmented than retroelements. All the elements add up to ∼10% of the whole nuclear genome.
Table 2.
The Transposable Elements of Dictyostelium discoideum
| Subgroup | Class | Transposon | Accession no. | Consensus length (bp) | LTR | TSD (bp) | Genome content (% nt) | Fragment no. | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| type | length (bp) | Nr | ML (N) | FI | |||||||
| LTR | DIRS-1 | DIRS-1* | M11339 | 4826 | IR | 320 | 0 | 3.260 | 235 | 302 | 1.3 |
| transposons | gypsy | skipper* | AF049230 | 6994 | DR | 390 | 5 | 0.997 | 50 | 82 | 1.7 |
| skipper_LTR* | 390 | 0.011 | 10 | n.d. | n.d. | ||||||
| DGLT-A | AF298204 | 5054 | DR | 268 | 4/5 | 0.067 | 5 | 7 | 1.5 | ||
| H3R* | X59570 | 268 | 0.013 | 15 | n.d. | n.d. | |||||
| DGLT-P | AF298205 | (6017) | n.d. | n.d. | n.d. | 0.047 | 10 | 17 | 1.5 | ||
| Non-LTR | TRE3 | TRE3-A* | AF134169 | 5243 | — | — | n.d. | 0.960 | 60 | 82 | 1.3 |
| transposons | TRE3-B* | AF134170 | 5292 | — | — | n.d. | 0.770 | 50 | 68 | 1.3 | |
| TRE3-C* | AF134171 | 4751 | — | — | n.d. | 0.450 | 30 | 36 | 1.1 | ||
| TRE3-D | AF135841 | (2816) | — | — | n.d. | 0.051 | 5 | 16 | 2.7 | ||
| TRE5 | TRE5-A* | X57034 | ∼6200 | — | — | n.d. | 1.220 | 70 | 92 | 1.3 | |
| TRE5-B | AF298209 | ∼5700 | — | — | n.d. | 0.200 | 15 | 36 | 2.1 | ||
| TRE5-C | AF298210 | (890) | — | — | n.d. | 0.012 | 5 | 12 | 3.0 | ||
| DNA | Tdd-4 | Tdd-4* | U57081 | 3843 | IR | 146 | 5 | 0.425 | 40 | 55 | 1.4 |
| transposons | Tdd-5 | AF298206 | 4031 | IR | 183 | 5 | 0.076 | 5 | 20 | 3.3 | |
| DDT | DDT-A | AF298201 | 5169 | IR | 48 | 2 | 0.309 | 20 | 65 | 3.3 | |
| DDT-B | AF298202 | 5471 | IR | 38 | 2 | 0.314 | 20 | 68 | 3.6 | ||
| DDT-S | AF298203 | 758 | IR | 27 | 2 | 0.295 | 130 | 175 | 1.3 | ||
| Unclassified | thug | thug-S | AF298207 | 2192 | IR | 18 | 4 | 0.058 | 10 | 19 | 2.1 |
| thug-T | AF298208 | 1132 | IR | 8 | 4 | 0.038 | 10 | 18 | 1.6 | ||
| Total | 9.573 | 750 | 1195 | ||||||||
DR, direct repeat; IR, inverted repeat; Nr, rounded copy number as estimated from genome content; ML(N), maximum likelihood estimate of fragment number; FI, fragmentation index; TSD, size of target site duplication.
LTR Retrotransposons
In the class of LTR retrotransposons, only skipper (Leng et al. 1998) and DIRS-1 (Cappello et al. 1985) were previously known; a short LTR-like sequence, H3R, had been described (GenBank accession no. X59570). It was speculated that H3R is the LTR of a full-length retrotransposon that had not been found (Winckler 1998). Our analysis confirmed this because we were able to reconstruct a full-length LTR retrotransposon with H3R elements as flanking LTRs. Because of the similarity of the translated ORF to gypsy/Ty3-like elements, we named this transposon Dictyostelium Gypsy-Like Transposon subtype A (DGLT-A). Interestingly, this TE is shorter than previously reported LTR transposons. In contrast to known LTR TEs, it contains only a single open reading frame (ORF), not two. Because most of the DGLT-A copies we examined have this structure, it seems to be evolutionarily conserved. Therefore, DGLT-A may be a functional TE despite its unusual structure.
A second gypsy-like TE, DGLT-P, was discovered, which seems to be nonfunctional. Although there is a slight similarity to proteins of a gypsy-like transposon in certain regions of the element, we were not able to construct a full-length mRNA coding for a reverse transcriptase from the assembled element. Because of its role in both the translation of a mature reverse transcriptase and its use as a template for reverse transcription, the RNA from a retrotransposon is not commonly spliced. This element may not be a complete entity, because we have not found a continuous ORF with features of a reverse transcriptase. Furthermore, it is very unlikely that the mRNA is spliced to result in a functional reverse transcriptase. In additionally, the 5′ sequences adjacent to the TE borders are similar to each other in five out of nine cases. This suggests that these copies are derived from rearrangements and duplications of one single TE, rather than from transposition events. We therefore called this element DGLT-P, in which P stands for pseudotransposon. It is possible that during evolution the balance between the host cell and a functional DGLT-P element could not be maintained. Thus, selection pressure led to D. discoideum cells without a functional DGLT-P.
On the DNA level, the similarity between the elements of the LTR retrotransposon group is very low, but the deduced proteins exhibit similarities of 65% among members of this group. All members of the LTR subgroup in D. discoideum thus belong to the gypsy-like class of LTR retrotransposons. The LTR of DGLT-A (H3R) as well as the LTR of the skipper element family are also present as single short elements in the genome (Table 2). Such solo LTRs can also be found in other genomes (Kim et al. 1998; Goodwin and Poulter 2000).
Non-LTR Retrotransposons
The non-LTR retrotransposons in D. discoideum insert into the genome in a position- and orientation-specific manner. The target sequences for these elements is the vicinity of tRNAs in well-defined distances. The subgroup of non-LTR retrotransposons was divided up into two classes recently, and the families were renamed according to their insertion preference upstream (TRE5) of or downstream (TRE3) from tRNA genes (Szafranski et al. 1999). According to the results from our homology search, four members of the TRE3 class and three members of the TRE5 class exist in D. discoideum. Analysis of the incompletely assembled non-LTR retrotransposon families (TRE3-D and TRE5-C) revealed that all copies of these elements were truncated. This could be caused by incomplete reverse transcription of the RNAs of these elements or the partial deletion of the copied element. Thus, if only one complete copy of each of these elements existed in the genome, the sequences adding up to a complete element may not have been present in our data set. Otherwise, the elements may have been inactivated by truncation during evolution and are no longer functional.
The TRE5-A element shows a modular structure at its distal parts (An-B-core segment-B-C; Winckler 1998). This structure could not be found in the other members of the TRE5 class. Instead, TRE5-C lacks the B module in its module structure (An-core segment-C) and TRE5-B shows only the structure (An-core segment).
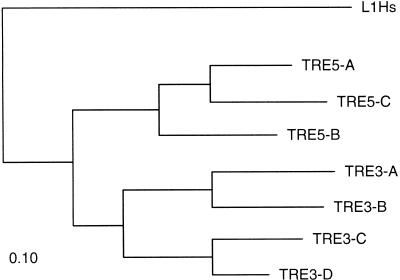
Because there are now several new members of that subgroup available, the similarities among families could be used to construct a phylogenetic tree of the pol proteins (Figure 2). This tree shows that the insertion preferences 3′ or 5′ to tRNA genes of the TRE elements correlates with the tree topology. The two classes presumably diverged from a common ancestor, which may have had no insertion preference, only one of the two observed specificities, or inserted randomly in the 5′- or 3′-vicinity of tRNA genes. Thus, differential insertion preferences were acquired at the same time or shortly after the two evolutionary branches diverged.
Figure 2.
Phylogenetic tree of Dictyostelium discoideum TRE elements calculated with the parsimony method of PHYLIP (see Methods). The tree was rooted with the human L1 element (L1Hs).
DNA Transposons
DNA transposons are not amplified in the process of transposition. They are excised from their genomic sites and inserted as single copies at new locations. Thus, amplification of DNA transposons may occur only when a TE is activated during DNA replication and transposes to a not yet amplified region.
Tdd Elements
Before our analysis, only a single DNA transposon, Tdd-4, was known (Wells 1999). Tdd4 gives rise to two alternatively spliced mRNAs. Both mRNAs code for transposase proteins. We found a related element with a lower copy number in the genome (Table 2). Because we were not able to reconstruct the complete element, we could not analyze the splicing pattern of the corresponding mRNA so far. However, there are indications for the presence of splicing variants. To account for the obvious relationship between the two elements, we called it Tdd-5. However, the similarity between Tdd-4 and Tdd-5 is relatively low. On the protein level it reaches only 50% in a small characteristic region of the transposase protein (data not shown).
DDT Elements
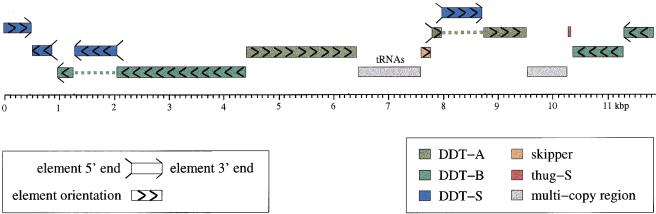
Adjacent to other repetitive elements we found segments of DNA that occur several times in the genome. They are characterized by inverted terminal repeats (ITRs) and a region of tandemly repeated short sequence motifs (TRM). In addition, we found duplicated sequence motifs at the element borders. This is a common feature for TEs because their transposition often leads to target site duplications. Apart from this, no similarities to other TEs neither of D. discoideum or to other organisms were observed. Thus, these segments seem to represent a completely new subclass of potential DNA elements. Accordingly, we named this class Dictyostelium DNA Transposon (DDT; Table 1). This subclass comprises three different elements (Fig. 3). The two larger elements (DDT-A and DDT-B) seem to be full length with 5168 bp and 5521 bp, respectively. Thus, the two large elements are similar in length to other complete DNA transposons or retroelements (Table 2). The third member of this class, DDT-S, is far shorter (758 bp).
Figure 3.
Structures of the members of the Dictyostelium DNA Transposon (DDT) element class. ITR, inverted terminal repeat; TRM, tandemly repeated motif. DDT-A and DDT-B are colinear full length elements, DDT-S consists of ITR and TRM of the full-length elements.
A comparative analysis of DDT-A and DDT-B revealed their coding potential. Each element codes for two proteins. The first ORF on the elements is not interrupted by introns and codes for proteins of 813 (DDT-A) and 815 (DDT-B) amino acids. The second ORF, which codes for proteins of 264 amino acids (DDT-A) and 256 amino acids (DDT-B), respectively, can only be built after splicing three exons from the primary transcript (Fig. 3). Both gene products show no striking similarity to other known proteins.
DDT-S comprises an incomplete element, which mainly consists of ITRs and a TRM region (Fig. 3). We detected no long ORF or similarities to known proteins in this short element. In addition, there is no detectable similarity to the genes of DDT-A and DDT-B. Thus, it is very unlikely that this short member of the DDT class contains coding potential. Nevertheless, the similarity of the ITRs and the existence of similar structural regions adjacent to the ITRs (TRM in Fig. 3) led us to the conclusion, that these three elements build a genuine class. We also detected a subpopulation in the DDT-S family. This subpopulation contains the 5′ end of DDT-S (545 bp). 3′ adjacent to this region there is a stretch of 2630 bp that is nearly identical in all 14 members of this subpopulation. This sequence shows no similarity to other sequences. Thus, this entity may have been created by genomic rearrangements rather than transposition events.
Despite the lack of similarity to other elements, the overall structure of the DDTs suggests that they are DNA transposons. One additional feature of transposable elements is that they are often truncated by insertions of other elements. Thus, we investigated the genome of D. discoideum, asking whether or not we could find a locus where DDTs are truncated. Because, at this time, the coverage of chromosome 2 is the highest, we reconstructed such a locus on this chromosome (Fig. 4). This locus is 12 kb long and consists of different full-length and disrupted elements of the DDT class (nine) and small parts of other TEs (two). Overall, it contains 11 elements of different types. The complete and truncated elements seem to have accumulated step by step. It remains unclear why this locus mainly consists of elements of the same class.
Figure 4.
A locus (GenBank accession no. AF 298624) on chromosome 2 containing multiple DDT class copies. The localization on chromosome 2 was calculated using the contribution of reads from the different chromosome specific libraries to the contig (IMB, unpublished software). Separation of element parts by insertions are marked as dotted lines.
Thug Elements
A second new class of elements is also characterized by its lack of similarity to other known elements. These elements are commonly found in loci that contain other complex repetitive elements (not shown). The thug elements bear some resemblance to so-called miniature inverted-repeat repeat transposable elements (MITEs), which were first detected in plants (Wessler et al. 1995; Surzycki and Belknap 1999). Thug elements also have no coding capacity and have terminal inverted repeats. Furthermore, they have the potential to form stable secondary structures. They share the AT-richness of the MITEs, but this is not a diagnostic feature, given the high AT background of the D. discoideum genome.
Insertion Preferences
The TRE subgroup of TEs integrates highly position-specific upstream of or downstream from tRNA genes, possibly because of an integration mechanism in which the integration complex interacts directly with RNA polymerase III transcription factors, as in yeast Ty3 integration (Connolly and Sandmeyer 1997; Yieh et al. 2000). In addition, DGLT-A also inserts 5′ of tRNA genes. Other elements show no, or only weak, integration site specificity. Whereas no specific sequence pattern is required at the target site of integration, the potential consequences of integration events may prevent TEs from being integrated at random. Thus, during evolution, the TEs accumulate at positions in the genome where the damage to the genome is relatively low. As a consequence thereof, TEs often integrate into other TEs in genomes with densely packed genes, as in lower eukaryotes (Kim et al. 1998).
When looking at the integration preferences for the non-tRNA–associated TEs, we found that they are integrated preferentially into loci where other TEs already reside (Table 1). The coverage plots of each family of elements revealed that there is no preferred truncation or insertion point in any of the TEs. Interestingly, such loci may consist of TEs of mainly one class. This is the case for DDT elements (Fig. 4). It was estimated previously that most of the 240 DIRS-1 elements reside in about seven well-defined loci on the genome (Loomis et al. 1995). Currently, it remains unclear why the same class of elements is preferred as integration sites. No sequence specificity could be detected in the cases examined (DIRS-1 [Cappello et al. 1984]; DDT, this study). Thus, the emergence of TE-class islands may have been caused by the sequential integration of elements after activation of several elements of the same class. Homologous recombination of different loci may also have lead to extension of such islands. Nevertheless, from each family we could detect “orphan” elements, which do not reside in clustered loci (data not shown).
Polymorphisms and Assembly
We analyzed the probability with which each element family can be resolved. Because the different copies of one element family are not identical, particular reads can be collated to a particular copy of a family. Where there are only a few polymorphic sites present in a particular TE family, it may be impossible to resolve loci, especially if they contain full-length or nearly full-length elements. The presence of few polymorphisms is reflected by a low nucleotide diversity value (π) and a high resolution resistance value; that is, the possibility to resolve a particular element copy is low. Because of statistical limitations, the resolution resistance values calculated for Table 3 represent the upper limit of resistance (see Methods).
Table 3.
Nucleotide Diversity and Resolution Resistance of Element Families
| Transposon | Nt diversity (π) | Resolution resistance |
|---|---|---|
| DIRS-1 | 0.0234 | 1537 |
| skipper | 0.0215 | 512 |
| DGLT-A | 0.0042 | 417 |
| DGLT-P | 0.0146 | 59 |
| TRE3-A | 0.0121 | 967 |
| TRE3-B | 0.0143 | 486 |
| TRE3-C | 0.0071 | 774 |
| TRE3-D | 0.0032 | 450 |
| TRE5-A | 0.0104 | 1982 |
| TRE5-B | 0.0079 | 479 |
| TRE5-C | 0.0180 | 51 |
| Tdd-4 | 0.0072 | 667 |
| Tdd-5 | 0.0253 | 76 |
| DDT-A | 0.0211 | 159 |
| DDT-B | 0.0246 | 158 |
| DDT-S | 0.0294 | 842 |
| thug-S | 0.0154 | 161 |
| thug-T | 0.0417 | 55 |
Clearly, the shorter elements (thug, DDT-S) represent no difficulty for the assembly process because sequenced clones can span one entire element. On the other hand, even long elements may be resolved if the number of polymorphic sites is high enough. This is the case for DDT-A and DDT-B.
If some TEs cannot be resolved by ordering the respective reads according to their polymorphic sites, low level sequence coverage from YAC clones will have to be used to determine the proper genomic sequence. This may be required for loci of the TRE classes as well as the DIRS elements.
Conclusion
In this study, we have shown that it is possible to analyze extensively the repetitive element content of a genome at low coverage. All types of TEs could be found, and we calculated their frequency in D. discoideum. Because, for AT-rich genomes, only small bacterial clones are available, at least whole chromosomes (if not the entire genome) have to be assembled. Thus, in one single assembly, much more complex repetitive elements have to be resolved than in assemblies of BACs or PACs. Yet, this study shows that most TEs of D. discoideum can be easily assembled because of the existence of a number of polymorphic sites in each element family. On the other hand, TEs do not contribute significantly to the information content of a genome. Because most of the repetitive elements of this organism seem to be organized in clusters, it might be questionable whether the organization of these clusters has to be resolved to define the genome as completed.
METHODS
Library Construction
Nuclei of D. discoideum were prepared as described (Rogge and Risse 1974) and embedded in LMP agarose (FMC). The DNA was set free from the nuclei by incubating the agarose blocks with SDS-Proteinase K (Roth). To remove the rDNA palindrome (90 kb), the DNA was separated by pulsed field gel electrophoresis. After agarose removal with Agarase (Boehringer) the DNA was treated two times 5 sec each, using a Sonicator (Heat Systems) for fragmentation. The DNA was then separated on an 0.8% agarose gel (6 V/cm for 4 h) and the region that contained fragments in the range of 1 kb–3 kb was cut out. The agarose was removed using the Jetsorb kit (Genomed). The fragments were then ligated into the SmaI site of pUC18 (Craxton 1993).
Sequencing
The pUC18 template DNA for sequencing was purified using the Turbo 96 Kit from Qiagen. Templates were cycle sequenced using Big Dye terminators (PE Biosystems). The sequencing data were collected using ABI377 and ABI3700 sequencers.
Repeat Sequence Clustering
A given seed sequence or the consensus of a repeat sequence alignment was used as a probe to find matching reads in the available genomic shotgun sequences via WU-BLASTN 2.0 (Altschul et al. 1990). Poly(N) stretches in the query sequence having a length >12 bp were masked. BLAST parameters N, M, and W were optimized to maximize both specificity and sensitivity, typically resulting in values of M = 6, N = −18, W = 12. All BLAST matches were filtered against threshold values concerning relative nucleotide identity (∼90%) and BLAST score (∼250). Only the highest-scoring segment pair (HSP) of each matching sequence was included in the growing alignment. Multiple and profile alignments were performed using CLUSTALW V1.8 (Thompson et al. 1994). In an alignment editor, the alignment could be extended in both 5′- and 3′ direction if needed. In this case, the whole procedure was repeated from start until no further readings could be added to the alignment.
Alignment Analysis
All nucleotide polymorphisms in a resulting alignment were verified on trace data level in the GAP4 (Staden et al. 2000) alignment editor. The consensus of these edited alignments are deposited at GenBank (Table 2). Plots of local coverage and a catalogue of polymorphisms were generated using the program AlnK (unpublished software, IMB Jena). To characterize the diverging sequence stretches at truncated positions or the repeat element ends we used WU-BLASTN 2.0 (Altschul et al. 1990) in conjunction with our repeat database. In addition, for the tRNA-associated transposons we used tRNAscan-SE (Lowe and Eddy 1997) or WU-BLASTN 2.0 against a database of known D. discoideum tRNAs.
Consensus Sequence Analysis
Retrotransposon coding sequences could be easily detected as large ORFs in a start/stop codon plot. Sequence similarity search was carried out with WU-BLASTP 2.0, WU-TBLASTN 2.0, or WU-TBLASTX 2.0 (Altschul et al. 1990) against SwissProt database release 39, GenPept database from GenBank release 118, or the local database of complex repeat consensi (http://genome.imb-jena.de/dictyostelium/repeats/index.html).
Phylogenetic Analysis
A TRE pol protein alignment was prepared using CLUSTALW V1.8 (Thompson et al. 1994) with the default alignment parameters except a gap penalty value of 5.0 instead of 10.0. The parsimony method (PHYLIP V3.5) was used to calculate a phylogenetic tree (Fig. 2) from the gapless parts of the alignment.
Probabilistic Model
To calculate the probability of finding a given genomic feature in shotgun sequences, we applied the binomial distribution model. When assuming same lengths every shotgun sequence has the same probability P(H) to hit the feature in an identifiable manner: P(H) = CH/CΩ with CH = number sequence positions resulting in a hit; CΩ = all possible sequence positions, CΩ ≈ lG (size of the genome, ∼34 Mb). Sequence positions resulting in a hit are CH = lR − lO + max (lF −lO, 0) with lF = redundant length of the genomic feature; lR = length of the shotgun sequences (∼300 bp); lO = minimal overlap to allow unambiguous identification of the feature (∼40 bp–50 bp). For a sequence feature that occurs several times nF in the genome, the expected hit number nH for a given number of shotgun sequences nS is E(nH) = nF × nS × P(H) and the maximum-likelihood estimate for the number of features based on the number of observed hits is ML(nF) = nH/(nS × P[H]).
Estimation of Copy Numbers
Two independent methods were used to estimate the distribution of repeat copies in the Dictyostelium genome: (1). The total genomic nucleotide amount of a repeat element was calculated from the sum of nucleotides in the alignment at the current genome coverage value for shotgun reads by extrapolating to a genome coverage value of 1.0. Dividing the nucleotide amount by the consensus sequence length of the repeat element gives the lower limit of copy numbers in each family. (2) An estimation of the fragment number of each element was done by sampling reads comprising element ends and fragmentation events observed in the aligned sequences. A maximum-likelihood estimation of the genomic fragment number was calculated as described above. If then the estimated fragment number was nearly similar to the lower limit of copy numbers calculated as in (1), then the estimate was rather based on the observed truncation events (in the case of DDT-S, DGLT-A, DGLT-P, DIRS-1, skipper, Tdd-4). This excluded the influence of observed cloning and/or sequencing biases that occurred for the distal sequence parts of these TEs.
Nucleotide Diversity
Nucleotide diversity (π) was calculated as described by Nei and Li (1979). To provide a measure of the possibility to resolve repeat copies from shotgun data, we calculated the total expected genomic nucleotide number divided by the number of identified polymorphic sequence features, called “resolution resistance.” Because the number of identified polymorphisms rises with the number of analyzed shotgun sequences, and, therefore, not all polymorphisms were used for the calculation, this measure represents an upper limit for the resolution resistance. However, given that an repeat alignment is covered with shotgun sequences 10 times, ∼95% of all polymophisms having a frequency of ≥0.3 will be uncovered as can be seen from Monte Carlo simulations (results not shown).
Acknowledgments
We thank S. Förste, S. Landmann, R. Müller, S. Rothe, R. Schultz, and N. Zeise for expert technical assistance. This research was supported by the Deutsche Forschungsgemeinschaft (DFG) and the U.S. National Institutes of Health (NIH).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL gernot@imb-jena.de; FAX 49-3641-656255.
Article published on-line before print: Genome Res., 10.1101/gr.162201.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.162201.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Cappello J, Cohen SM, Lodish HF. Dictyostelium transposable element DIRS-1 preferentially inserts into DIRS-1 sequences. Mol Cell Biol. 1984;4:2207–2213. doi: 10.1128/mcb.4.10.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappello J, Handelsman K, Lodish HF. Sequence of Dictyostelium DIRS-1: An apparent retrotransposon with inverted terminal repeats and an internal circle junction sequence. Cell. 1985;43:105–115. doi: 10.1016/0092-8674(85)90016-9. [DOI] [PubMed] [Google Scholar]
- Connolly CM, Sandmeyer SB. RNA polymerase III interferes with Ty3 integration. FEBS Lett. 1997;405:305–311. doi: 10.1016/s0014-5793(97)00200-7. [DOI] [PubMed] [Google Scholar]
- Craxton M. Cosmid sequencing. Methods Mol Biol. 1993;23:149–167. doi: 10.1385/0-89603-248-5:149. [DOI] [PubMed] [Google Scholar]
- Firtel RA, Kindle K. Structural organization of the genome of the cellular slime mold Dictyostelium discoideum: Interspersion of repetitive and single-copy DNA sequences. Cell. 1975;5:401–411. doi: 10.1016/0092-8674(75)90059-8. [DOI] [PubMed] [Google Scholar]
- Firtel RA, Kindle K, Huxley MP. Structural organization and processing of the genetic transcript in the cellular slime mold Dictyostelium discoideum. Fed Proc. 1976;35:13–22. [PubMed] [Google Scholar]
- Flavell AJ. Retroelements, reverse transcriptase and evolution. Comp Biochem Physiol B Biochem Mol Biol. 1995;110:3–15. doi: 10.1016/0305-0491(94)00122-b. [DOI] [PubMed] [Google Scholar]
- Goodwin TJ, Poulter RT. Multiple LTR-retrotransposon families in the asexual yeast Candida albicans. Genome Res. 2000;10:174–191. doi: 10.1101/gr.10.2.174. [DOI] [PubMed] [Google Scholar]
- Kay RR, Williams JG. The Dictyostelium genome project: an invitation to species hopping. Trends Genet. 1999;15:294–297. doi: 10.1016/s0168-9525(99)01770-9. [DOI] [PubMed] [Google Scholar]
- Kim J M, Vanguri S, Boeke JD, Gabriel A, Voytas DF. Transposable elements and genome organization: A comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 1998;8:464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- Kimmel AR, Firtel RA. Sequence organization and developmental expression of an interspersed, repetitive element and associated single-copy DNA sequences in Dictyostelium discoideum. Mol Cell Biol. 1985;5:2123–2130. doi: 10.1128/mcb.5.8.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle KL, Firtel RA. Identification and analysis of Dictyostelium actin genes, a family of moderately repeated genes. Cell. 1978;15:763–778. doi: 10.1016/0092-8674(78)90262-3. [DOI] [PubMed] [Google Scholar]
- Leng P, Klatte DH, Schumann G, Boeke JD, Steck TL. Skipper, an LTR retrotransposon of Dictyostelium. Nucleic Acids Res. 1998;26:2008–2015. doi: 10.1093/nar/26.8.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis WF, Kuspa A. The genome of Dictyostelium discoideum. In: Maeda Y, Inouye K, et al., editors. Dictyostelium-A Model System for Cell and Developmental Biology. Tokyo: Universal Academic Press Inc.; 1997. pp. 15–30. [Google Scholar]
- Loomis WF, Welker D, Hughes J, Maghakian D, Kuspa A. Integrated maps of the chromosomes in Dictyostelium discoideum. Genetics. 1995;141:147–157. doi: 10.1093/genetics/141.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschalek R, Brechner T, Amon-Bohm E, Dingermann T. Transfer RNA genes: Landmarks for integration of mobile genetic elements in Dictyostelium discoideum. Science. 1989;244:1493–1496. doi: 10.1126/science.2567533. [DOI] [PubMed] [Google Scholar]
- Marschalek R, Hofmann J, Schumann G, Bach M, Dingermann T. Different organization of the tRNA-gene-associated repetitive element, DRE, in NC4-derived strains and in other wild-type Dictyostelium discoideum strains. Eur J Biochem. 1993;217:627–631. doi: 10.1111/j.1432-1033.1993.tb18285.x. [DOI] [PubMed] [Google Scholar]
- McKeown M, Taylor WC, Kindle KL, Firtel RA, Bender W, Davidson N. Multiple, heterogeneous actin genes in Dictyostelium. Cell. 1978;15:789–800. doi: 10.1016/0092-8674(78)90264-7. [DOI] [PubMed] [Google Scholar]
- Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noegel AA, Schleicher M. The actin cytoskeleton of Dictyostelium: A story told by mutants. J Cell Sci. 2000;113:759–766. doi: 10.1242/jcs.113.5.759. [DOI] [PubMed] [Google Scholar]
- Poole SJ, Firtel RA. Genomic instability and mobile genetic elements in regions surrounding two discoidin I genes of Dictyostelium discoideum. Mol Cell Biol. 1984;4:671–680. doi: 10.1128/mcb.4.4.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogge H, Risse HJ. A procedure for the isolation of Dictyostelium nuclei. Hoppe Seylers Z Physiol Chem. 1974;355:1467–1470. [PubMed] [Google Scholar]
- Romans P, Firtel RA. Organization of the actin multigene family of Dictyostelium discoideum and analysis of variability in the protein coding regions. J Mol Biol. 1985;186:321–335. doi: 10.1016/0022-2836(85)90108-1. [DOI] [PubMed] [Google Scholar]
- Rosen E, Sivertsen A, Firtel RA. An unusual transposon encoding heat shock inducible and developmentally regulated transcripts. Dictyostelium Cell. 1983;35(1):243–251. doi: 10.1016/0092-8674(83)90227-1. [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z, Bennetzen JL. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- Smit AF. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- Smit AF, Toth G, Riggs AD, Jurka J. Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J Mol Biol. 1995;246:401–417. doi: 10.1006/jmbi.1994.0095. [DOI] [PubMed] [Google Scholar]
- Staden R, Beal KF, Bonfield JK. The Staden package, 1998. Methods Mol Biol. 2000;132:115–130. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- Surzycki SA, Belknap WR. Characterization of repetitive DNA elements in Arabidopsis. J Mol Evol. 1999;48:684–691. doi: 10.1007/pl00006512. [DOI] [PubMed] [Google Scholar]
- Szafranski K, Glockner G, Dingermann T, Dannat K, Noegel AA, Eichinger L, Rosenthal A, Winckler T. Non-LTR retrotransposons with unique integration preferences downstream of Dictyostelium discoideum tRNA genes. Mol Gen Genet. 1999;262:772–780. doi: 10.1007/s004380051140. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas DF. Retroelements in genome organization. Science. 1996;274:737–738. doi: 10.1126/science.274.5288.737. [DOI] [PubMed] [Google Scholar]
- Voytas DF, Boeke JD. Yeast retrotransposons and tRNAs. Trends Genet. 1993;9:421–427. doi: 10.1016/0168-9525(93)90105-q. [DOI] [PubMed] [Google Scholar]
- Wells DJ. Tdd-4, a DNA transposon of Dictyostelium that encodes proteins similar to LTR retroelement integrases. Nucleic Acids Res. 1999;27:2408–2415. doi: 10.1093/nar/27.11.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler SR, Bureau TE, White SE. LTR-retrotransposons and MITEs: Important players in the evolution of plant genomes. Curr Opin Genet Dev. 1995;5:814–821. doi: 10.1016/0959-437x(95)80016-x. [DOI] [PubMed] [Google Scholar]
- Winckler T. Retrotransposable elements in the Dictyostelium discoideum genome. Cell Mol Life Sci. 1998;54:383–393. doi: 10.1007/s000180050168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao YL, Peterson T. Intrachromosomal homologous recombination in Arabidopsis induced by a maize transposon. Mol Gen Genet. 2000;263:22–29. doi: 10.1007/pl00008672. [DOI] [PubMed] [Google Scholar]
- Yieh, L., Kassavetis, G., Geiduschek, E.P., and Sandmeyer, S.B. 2000. The Brf and TBP Subunits of the RNA Polymerase III Transcription Factor IIIB Mediate Position-specific Integration of the Gypsy-like Element, Ty3. J. Biol. Chem. [DOI] [PubMed]
- Zhang J, Peterson T. Genome rearrangements by nonlinear transposons in maize. Genetics. 1999;153:1403–1410. doi: 10.1093/genetics/153.3.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker C, Cappello J, Chisholm RL, Lodish HF. A repetitive Dictyostelium gene family that is induced during differentiation and by heat shock. Cell. 1983;34:997–1005. doi: 10.1016/0092-8674(83)90557-3. [DOI] [PubMed] [Google Scholar]