Abstract
Purpose
The primary aim of this study was to characterize the 6-month overall survival and toxicity associated with second-line capecitabine treatment of advanced pancreatic cancer patients harboring the TYMS *2/*2 allele. The secondary aim was to analyze the response rate and pharmacokinetics of capecitabine-based therapy in this patient population. Lastly, TYMS, ATM and RecQ1 single nucleotide polymorphism were analyzed relative to overall survival in patients screened for study participation.
Methods
Eighty-two patients with stage IV pancreatic cancer were screened for the *2/*2 TYMS allele. Patients with the *2/*2 TYMS polymorphism were treated with capecitabine, 1000 mg/m2 twice daily for 14 consecutive days of a 21 day cycle. Screened patients not possessing TYMS *2/*2 were monitored for survival. Pharmacokinetic analysis was done during Cycle 1 of the therapy.
Results
Sixteen of the 80 screened patients tested positive for *2/*2 TYMS variant. Four out of the 16 eligible patients were treated on study. The study was terminated early due to poor accrual and increased toxicity. Three patients experienced grade 3 non-hematologic toxicities of palmer-plantar erythrodysesthesia, diarrhea, nausea and vomiting. Grade 2 toxicities were similar and occurred in all patients. Only 1 patient was evaluable for response after completion of 3 cycles of therapy. The presence of the *2/*2 TYMS genotype in all of the screened patients trended toward a decreased overall survival.
Conclusion
To our knowledge, this study represents the first genotype-directed clinical trial for patients with pancreatic adenocarcinoma. Although the study was closed early, it appears capecitabine therapy in pancreatic cancer patients harboring the TYMS *2/*2 variant may be associated with increased non-hematologic toxicity. This study also demonstrates the challenges performing a genotype-directed study in the second-line setting for patients with advanced pancreatic cancer.
Keywords: Pancreatic Cancer, Thymidine synthase enhancer region (TYMS), Capecitabine, Gemcitabine, RecQ1, ATM
Introduction
Adenocarcinoma of the pancreas remains a devastating disease. Pancreatic cancer is the 4th leading cause of cancer-related mortality in U.S. annually. The estimated incidence for 2009 was 42,470 new cases and 35,240 anticipated deaths [1]. At the time of diagnosis, 80% of patients have locally advanced or advanced disease for which no curative therapy exist. Seventy to eighty percent of patients treated with curative intent pancreaticoduodenectomy will recur within the first two years after surgical resection and will ultimately succumb to their disease [2–4]. As a result, the overall 5-year survival rate for all patients diagnosed with adenocarcinoma of the pancreas is less than 5%. Standard therapy for patients with advanced disease or for those who recur after surgical resection has been limited to gemcitabine, 5-fluorouracil (5-FU) and most recently erlotinib in combination with gemcitabine [5–7]. Unfortunately, the use of these drugs as single agents or in combination results in a median survival of less than 7 months. The increased understanding of pancreatic ductal adenocarcinoma biology has led number of phase III clinical trials that have combined gemcitabine with either biologic or modern cytotoxic agents but have not shown a survival benefit when compared to gemcitabine except for the modest survival benefit with the addition of capecitabine or erlotinib to gemcitabine [6–11]. These disappointing results demonstrate the need for alternative approaches to the development of pancreatic cancer therapy.
Pharmacogenomics represents a novel approach to drug development that it takes into account the pharmacokinetic variability in drug metabolism amongst individuals as a function of genetic polymorphisms in key drug metabolism genes, such as drug-metabolism enzymes and transport proteins [12]. An initial example of the successful application of pharmacogenomics to augment cancer therapy was demonstrated when this approach was applied to 5-FU dosing in patients with colorectal cancer. Thymidine synthase (TS) is one of the primary targets of the antimetabolite, 5-FU. It is a folate dependent enzyme that catalyzes the conversion of doxyuridine-5′-monophosphate to deoxythymidine-5′-monophosphate, representing the only de novo source of thymidine for DNA synthesis [13–15]. TS regulation is quite complex, but appears to be partially controlled by a polymorphism in the human TS gene enhancer region (TYMS). TYMS consists of variable number of copies of a 28-bp tandem-repeat sequence [16]. Alleles with up to nine copies of this 28-bp repeats have been described, with TYMS*2 [double tandem repeats (S/S)] and TYMS*3 [triple tandem repeats (L/L)] alleles being the most common alleles in the patient populations. The approximate frequency of the TYMS alleles are 20% (*2/*2), 30–55% (*2/*3) and 20–40% (*3/*3). Recent reports identify a G → C single nucleotide polymorphism (SNP) present within the upstream stimulatory factor (USF) consensus element present in repeat 2 of TYMS*3 as highly prevalent in several ethnic groups (non-Hispanic whites 56%, Hispanic whites 47%, African Americans 28%, and Singapore Chinese 37%) [17–19]. This would lead to a decrease in TS transcription by altering USF protein binding to the USF consensus element, functionally converting the *3 allele to a *2 allele. Genotyping of this single nucleotide polymorphism, in addition to the TYMS 28-bp tandem-repeat polymorphism, possess the potential to be a predictive marker of response and toxicity associated with TS-targeted therapy [20, 21].
5-Fluorouracil forms the backbone of chemotherapy for colorectal cancer and is also approved for use in pancreatic cancer. Overexpression of TS in colon cancer patients correlates with primary resistance to 5-FU, poor prognosis and low response to therapy. Colorectal cancer patients who are homozygous for the triple tandem repeats TYMS *3 have significantly higher TS expression than patients who are homozygous for TYMS *2. Patients carrying the *3/*3 polymorphism demonstrate a decreased response rate to 5-FU therapy and have worse outcomes than those patients possessing TYMS *2/*2 alleles [16, 20]. More recently, TS polymorphisms have also been demonstrated to predict response to capecitabine in colorectal cancer [21, 22]. In this small retrospective analysis, Park et al. reported a response rate in patients with *2/*2 alleles of 75% as compared to those with *2/*3 or *3/*3 alleles (25% and 8%, respectively) [22]. However, patients with *2/*2 genotype were also more likely to develop grade 3 toxicity after capecitabine administration than patients with the other variants (100% versus 30–40%). Consistent with these finding, homozygous TYMS*2 expression in patients undergoing neoadjuvant 5-FU based chemoradiation is associated with 60% downstaging of tumors after neoadjuvant therapy compared to TYMS*3 (22%) [21]. Based on these findings, a phase II clinical trial of neoadjuvant chemoradiotherapy in rectal cancer patients was conducted in which neoadjuvant therapy was stratified based upon the pharmacogenomics analysis of TYMS with a primary endpoint of downstaging. Patients expressing either TYMS *2/*2 or *2/*3 ‘Good risk’ were treated with infusional 5-FU plus 45 Gy radiation compared to those patients expressing *3/*3 or *3/*4 ‘Poor risk’ who were treated with 5′FU, irinotecan and 45 Gy radiation. This study indicated that genotype-guided neoadjuvant therapy could improve down-staging rate in both the ‘good risk’ and ‘poor risk’ patient populations. TYMS *3/*3 expression was associated with increased distant recurrence, demonstrating the innate resistance of these cancer cells to 5-FU therapy [23, 24]. This study demonstrated that phase II genotype-directed clinical trials are feasible and may improve patient outcomes.
Capecitabine (Xeloda®) is an oral pro-drug that is absorbed unchanged in the GI tract and subsequently undergoes a series of enzymatic conversions into 5-FU in tumor cells. It has been approved for use in patients with metastatic breast, colon cancer, and pancreatic cancer and is being investigated in numerous other malignant histologic subtypes. In addition to simulating continuous infusion 5-FU, which has been shown to be superior to bolus dosing in the post-operative setting, capecitabine has several properties that may enhance its selectivity for tumor cells. Capecitabine has been shown to selectively upregulate thymidine phosphorylase (TP), which in turn results in the final enzymatic conversion of capecitabine to 5-FU [25]. Following administration of Capecitabine, 5-FU levels are on average 3 times higher in tumor tissue than in surrounding healthy tissue [26]. In pancreatic cancer, single agent capecitabine resulted in 7–8 % objective response rate and a 24 % clinical benefit response [27]. A phase III study has been conducted comparing capecitabine plus gemcitabine to gemcitabine alone and demonstrated a statistically significant survival advantage for the combination with a median overall survival of 8.4 versus 7.2 months [6]. These data demonstrate that capecitabine does possess antitumor activity in pancreatic cancer. Therefore we hypothesized that capecitabine treatment of TYMS genotype-selected patients would improve the 6-month survival of patients with previously treated metastatic pancreatic cancer. Gemcitabine is a prodrug that requires cellular uptake and intracellular phosphorylation in order to exert its mechanism of action [28, 29].
Gemcitabine is an antimetabolite and exerts its cytotoxic effect by incorporating into the DNA strand as the active dFdCTP; thereby, inhibiting DNA synthesis. DNA repair plays an important role in gemcitabine mediated cell death as it acts by inhibiting DNA synthesis. Therefore single nucleotide polymorphisms in gene involved in the DNA repair mechanisms have been evaluated as potential biomarkers of response to gemcitabine. Single nucleotide polymorphisms in the DNA repair genes ATM, RecQ1 and Rad54L have been shown to be associated with improved overall survival in pancreatic cancer patients treated with neoadjuvant gemcitabine based chemoradiation [30, 31]. The ATM gene encodes a protein kinase that plays a key role in the detection and repair of DNA double strand breaks and thus maintenance of genome integrity. This gene is known to be activated by DNA damage and involved in cell cycle arrest, apoptosis and DNA repair [32]. Polymorphisms in ATM gene promote the development of lung cancer, breast cancer and radiosensitivity to radiation [33, 34]. Meanwhile, RecQ1 gene is a member of a family DNA helicases including RecQ5, RecQ4, Bloom (BLM) and Werner (WRN) genes [35]. This family of tumor suppressor genes is involved in DNA repair, s-phase checkpoint and telomere maintenance. The RecQ family helicases interact with cellular topoisomerases, and together these heteromeric complexes manipulate DNA structure to effect efficient DNA replication and genetic recombination and function at the interface between DNA replication and DNA repair [36,37]. Similarly, polymorphisms in the XRCC3 DNA repair gene have been demonstrated to be associated with overall survival in non-small cell lung cancer patients treated with gemcitabine and cisplatin [38]. Many groups have investigated the relationship between DNA repair gene ERCC1 expression in tumors and survival and showed that tumors with low expression are associated with improved survival in NSCLC patients treated with cisplatin and gemcitabine [39]. Polymorphism is ATM G60A and RecQ1 A159C were analyzed in a population of metastatic pancreatic cancer patients treated with gemcitabine in attempt to confirm the previously reported association of ATM and RecQ1 to overall survival in gemcitabine treated patients.
To our knowledge, this clinical trial represents the first genotype-directed clinical trial in the advanced pancreatic adenocarcinoma patient population.
Methods
Patient and Study Design
Patient Selection
Patients were considered eligible for genetic TYMS screening if they met all of the following criteria: Age 18 or older; histologically or cytologically confirmed advanced or metastatic adenocarcinoma of the pancreas: no prior capecitabine therapy; measurable disease; ability to give informed consent; no known second malignancies (other than carcinoma-in-situ of the cervix, superficial skin cancer or superficial bladder cancer); Eastern Cooperative Oncology Group (ECOG) performance status of 0, 1 or 2; adequate hematologic (absolute neutrophil count (ANC) ≥ 1,500/mm3 and platelets ≥100,000/mm3), hepatic (AST/ALT ≤ 2.5 × ULN [or ≤ 5 × ULN if attributable to liver metastases] and total bilirubin ≤ 1.5 ULN) and renal function (creatinine within institutional limits of normal or creatinine clearance [CrCl] > 50 mL/min calculated by the Cockcroft-Gault equation). Screened patients possessing TYMS *2/*2 allele were consider eligible for the treatment portion of the study with capecitabine if they had received at least one gemcitabine containing regimen for metastatic disease; alternatively, patients relapsing within 6 months of completion of gemcitabine-based adjuvant therapy were also eligible. Patients could not have received prior 5-FU or capecitabine-based metastatic disease therapy. Patients must have had adequate end organ function as noted above and have an ECOG performance status of ≤ 2.
Study Design
This was designed as an open-label, non-randomized phase II study. The study protocol was approved by the Johns Hopkins Hospital Institutional Review Board, and all patients gave written consent before any study-specific procedures were preformed. There were two different consent forms for screening and treatment. To avoid delays between first- and second-line treatment, and allow for delays in screening results due to technical and other factors, the screening population included both first- and second-line patients. Genetic screening was performed by testing DNA obtained from peripheral blood mononuclear cells for the presence of the *2/*2 allele of the thymidine synthase enhancer region. Those patients previously treated with gemcitabine who possessed the *2/*2 allele were then eligible to be treated with capecitabine in 2nd line setting. Within 2 weeks prior to capecitabine treatment initiation the patients were reconsented for treatment and underwent a clinical screening assessments, including a medical history and physical examination, ECG, chest x-ray and tumor measurements. Patients who underwent genetic screening and were not homozygous for the TYMS *2/*2 allele were followed for survival, but received treatment according to the management of their primary medical oncologist.
Patients were treated with oral capecitabine 1,000 mg/m2 administered twice daily (2000 mg/m2/d) on days 1 to 14 of a 21 day treatment cycle. Patients received a total of 3 cycles prior to measuring response to therapy as outlined by RECIST criteria. A physical examination, including an assessment of vital signs, physical measurements, and clinical laboratory tests (hematology and blood chemistry), were also performed at all study visits. Adverse events were monitored throughout the study and for 28 days after the last study treatment.
Toxicity Assessment
Adverse events were graded according to the National Cancer Institute’s Common Terminology Criteria for Adverse Events version 3.0. Toxicities were identified by medical history, physical examination, and review of the laboratory studies performed. Capecitabine administration was adjusted or interrupted for treatment related adverse events of grade 2 or greater according to the capecitabine dose modification algorithm in the package insert.
SNP Analysis
Genomic DNA was extracted from peripheral blood samples using the EZ1 BioRobot (Qiagen, Valencia, CA). Amplification of the thymidine synthase polymorphic region was carried out in 20 μl reactions using 1 μl DMSO and 1.5 U Taq Gold polymerase (Applied Biosystems Inc., Foster City, CA). Primers were 5′ FAM-GTGGCTCCTGCGTTTCCCCC 3′ (TS-F) and 5′ GAGCCGGCCACAGGCATG 3′ (TS-R2). One microliter of amplification product was added to a mixture of diformamide and ROX GS500 size standard (Perkin Elmer, Wellesley, MA) and denatured at 95o C for 2 minutes. Samples were then analyzed on an ABI 3100 Genetic Analyzer (Applied Biosystems Inc, Foster City, CA,).
The ATM G60A and RecQ1 A159C polymorphisms were detected using TaqMan genotyping assays (Applied Biosystems Inc, Foster City, CA). ATM (rs664143) reaction consisted of 1X TaqMan Universal PCR Mast Mix (Applied BioSystems Inc, Foster City, CA) and 1X ATM primer/probe mix (Applied Biosystems). RecQ1 (rs13035) reaction consisted of 1X TaqMan Universal PCR Mast Mix, 0.9 μM primers (Forward: CAATCTGGTTCTAAGAATACAGGAGCTA, Reverse: GGCATATACATGCATAAACCATCTTT), 0.25 μM probes (C-Probe: FAM-AGTAACAGTCATATCAGG-MGB, T-Probe: VIC-AGTAACATTCATATCAGGC-MGB). Reactions were set up in a final volume of 25μl with 30 ng DNA in each reaction. Thermal cycling consisted of an initial denaturation of 95°C for 10 minutes, followed by 35 cycles of 92°C for 15 seconds and 60°C for 1 minute, on TaqMan 7900 instrument (Applied BioSystems Inc, Foster City, CA). Directly after amplification, plates were analyzed using ABI Allelic Discrimination Plate Read and Analysis Software (Applied BioSystems Inc, Foster City, CA). Five percent of the samples were subjected to cycle sequencing, and all of the sequencing results were concordant with the TaqMan genotyping results. All specimens were processed and analyzed in the CLIA approved pathology laboratory of Kathy Murphy at Johns Hopkins University.
Pharmacokinetic Analysis
Capecitabine pharmacokinetics studies were performed during cycle 1. Blood samples were taken pre-treatment and post-treatment at 0.25, 0.5, 1, 2, 3, 4, 5, 6 and 8 hours. Trough levels were evaluated by obtaining samples within 30 minutes prior to the dose administration on days 2, 8 and 15 of cycle 1. Blood samples were processed by centrifugation at 1,000 g at 4°C for 10 minutes. For capecitabine samples, tetrahydrouridine, a cytidine deaminase inhibitor, was added at a final concentration of 400 nM to increase the stability of capecitabine and metabolites in plasma during storage in the freezer [40–42]. Analytical analysis of capecitabine and metabolites (5-fluoro-5′-deoxycytodine (5′DFCR), 5-fluoro-5′-dexoxyuridines (5′-DFUR) and 5-FU) were quantitated by LC/MS/MS over the range of 50 to 10,000 ng/mL of plasma [40]. Individual pharmacokinetic parameters were estimated by standard non-compartmental analysis using the WinNonlin version 5.0 (Pharsight Corporation, Mountain View, CA) (Gibaldi M, Perrier D: Pharmacokinetics (ed 2nd). New York, NY, Dekker, 1982).
Statistics
Originally the study was designed to test the primary endpoint of increased 6-month survival rate, compared to historical controls, for patients treated with capecitabine in the second line setting. The planned sample size was 65 patients. With 65 patients, there was a 96% chance of observing at least one occurrence of an adverse event if the true risk of that event was 5% or greater. Assuming a frequency of the S/S variant of 15–20%, we expected to screen approximately 500 patients in order to obtain 65 eligible patients for the treatment portion of the study. Sixty-five treated patients were estimated to be sufficient to demonstrate a 15% increase in 6-month survival rate in patients receiving second line therapy for metastatic pancreatic cancer. The estimate was based on testing a null hypothesis that the true survival rate at 6 months is 15% with a power of 90%, assuming a one-sided exact test for a single proportion; this assumes that all patients will be followed for at least 6 months. Kaplan-Meier methods were planned to estimate the survival data of the patients treated on the study. Unfortunately, due to small sample size the estimates of the survival data were not appropriate. Kaplan-Meier methods were used to estimate the survival data in the screened patient population. The association TYMS, ATM and RecQ1 polymorphism relative to survival in the screened patient population was analyzed using Mantel-Cox log-rank test. Ultimately, this study was closed early due to poor accrual and severe toxicity.
Results
Screened Patients Demographics
Between 1/1/05 and 1/1/08, a total of 80 patients with adenocarcinoma of the pancreas underwent genetic screening to assess the presence of TYMS *2/*2 allele. All tests were performed in a rapid manner with less than 2-week time period from obtaining sample to completion of analysis. The screened patient characteristics can be found in table 1A. The age of screened patients ranged from 42 to 81 years old (median 65) with an equivalent male/female ratio, and performance status was ECOG ≤ 1 in 94% of patients. Sixteen patients (20%) of the total 80 patients screened were found to possess the homozygous S/S allele. Only 17% of screened patients had undergone a previous pancreaticoduodenectomy. Thirty-seven patients (46%) and twenty-two patients (28%) possessed either the heterozygous or homozygous TYMS*3 allele, respectively, resulting in the generation of a triple 28-base pair repeat sequence (Table 2).
Table 1.
Patient Characteristic of all screened patients (A) and those treated in clinical trial (B).
| A. Screened Patient Characteristic | Number |
|---|---|
| Age | |
| Range | 42–81 |
| Median | 65 |
| Sex | |
| Male | 39 |
| Female | 41 |
| Race | |
| White | 74 |
| African American | 3 |
| Asian | 2 |
| Hispanic | 1 |
| Stage at Diagnosis | |
| Stage I | 1 |
| Stage II | 5 |
| Stage III | 8 |
| Stage IV | 63 |
| Performance Status (ECOG) | |
| 0 | 32 |
| 1 | 40 |
| 2 | 5 |
| B. Treated Patient Characteristic | Number |
| Age (Yrs.) | |
| Range | 55–78 |
| Median | 76 |
| Sex | |
| Male | 2 |
| Female | 2 |
| Race | |
| Caucasian | 4 |
| African American | 0 |
| Asian | 0 |
| Stage at Diagnosis | |
| Stage I | 0 |
| Stage II | 0 |
| Stage III | 3 |
| Stage IV | 1 |
| Performance Status (ECOG) | |
| 0 | 1 |
| 1 | 3 |
| Adjuvant Chemotherapy | 3 |
| Gemcitabine | 2 |
| 5-FU | 1 |
| Radiation | 3 |
| Surgery | 3 |
| Chemotherapy Regimens (Metastatic) | |
| Gemcitabine | 3 |
Table 2.
TSER genotype distribution
| TSER Indeterminate | TSER *2/*2 | TSER *2/*3 | TSER *3/*3 |
|---|---|---|---|
| 5(6%) | 16 (20%) | 37 (46%) | 22 (28%) |
Capecitabine treatment of homozygous TYMS*2 patients
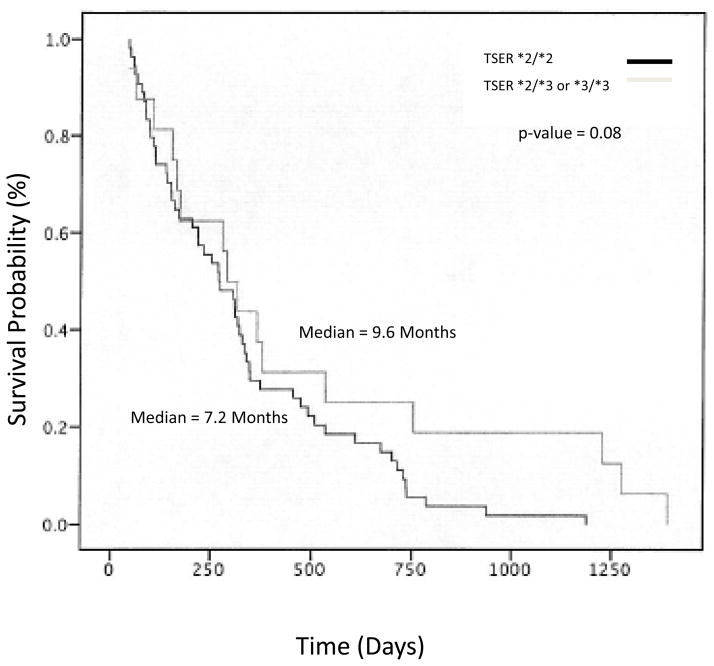
Four (25%) of the 16 patients expressing the homozygous *2/*2 TYMS allele were treated with capecitabine in the second line setting. Twelve of the 16 eligible patients possessed inadequate performance status to receive second line therapy, were treated on other protocols available at the institution, or were treated by their primary oncologist off study. The four patients treated on study possessed good baseline performance status (Table 1B). Three of the four patients had stage III disease at diagnosis. All of these patients had received prior therapy with either radiation or surgery. Three of the four patients had received prior adjuvant chemotherapy. One patient received adjuvant 5-FU and two were treated with gemcitabine in this setting. All patients received an initial capecitabine dose of 1,000 mg/m2 twice daily for days 1 to 14 of a 21 day cycle. However, dose modification was frequent due to toxicity (Table 3). The number of cycles on treatment varied from 1 cycle to a maximum of 3 cycles in one patient. The median number of cycles administered to these patients was 1.5. The daily dose administered ranged from 1,300 to 3,600 mg/day. Pharmacokinetic exposure was analyzed for capecitabine and 5-FU in 2 of the 4 treated patients. High inter-patient variability was observed for capecitabine and metabolite pharmacokinetic parameters, which is consistent with previous literature (Table 4) (43–45). Unfortunately, only one of the four treated patients were evaluable to assess the primary endpoint of 6 month overall survival; thereby, an accurate estimate of the survival data was untenable. However, there was a trend toward decreased median survival in patients with homozygous TYMS*2 alleles when compared to patients with either TYMS*2/*3 or homozygous TYMS*3 in the screened patient population of 82 7.2 months vs 9.6 months (p 0.08), respectively, figure 2.
Table 3.
Capecitabine induced hematologic toxicity (A) and non-hematologic toxicity (B)
| A. Hematologic Toxicity | |||
|---|---|---|---|
| Grade 1 | Grade 2 | Grade 3 | |
| Anemia | 3 | 0 | 0 |
| Neutropenia | 2 | 10 | 0 |
| Thrombocytopenia | 1 | 0 | 0 |
| B. Non-hematologic Toxicity | |||
| Grade 1 | Grade 2 | Grade 3 | |
| Nausea/vomiting | 0 | 1 | 1 |
| Hand Foot syndrome | 0 | 0 | 2 |
| Hypoalbuminemia | 1 | 0 | 0 |
| LFTs | 1 | 0 | 0 |
| Fatigue | 2 | 0 | 0 |
| Diarrhea | 0 | 0 | 2 |
| Mucositis | 0 | 2 | 0 |
| Rash | 1 | 0 | 0 |
Table 4.
Summary of pharmacokinetics parameters of capecitabine, ′DFCR, 5′DFUR, and 5FU.
| Cmax (ng/mL) | Tmax (hr) | AUC (ng*hr/mL) | T1/2 (hr) | |
|---|---|---|---|---|
| Capecitabine | ||||
| Current study | 2467, 11261 | 1.00, 1.05 | 5390, 13767 | 0.43, 0.71 |
| Historical Controls | ||||
| Laheru et al.43 | 3808 ± 1896 (13) | 1.00 (0.50–3.00;13) | 5296 ± 1015 (13) | 0.52 ± 0.17 (13) |
| Soepenberg et al.44 | 5651 ± 5360 (13) | 1.00 (13) | 6810±3904 (13) | 0.98 ± 0.61 (11) |
| Pronk et al.45 | N.R. | N.R. | 5660 ± 2380 (10) | 0.70 ± 0.32 (10) |
| 5′DFCR | ||||
| Current study | 4344, 5596 | 1.00, 1.05 | 10892, 17187 | 0.88, 1.30 |
| Historical Controls | ||||
| Laheru et al.43 | N.R. | N.R. | N.R. | N.R. |
| Soepenberg et al.44 | 4578 ± 2090 (13) | 1.00 (13) | 10299 ± 3663 (13) | 0.99 ± 0.17 (13) |
| Pronk et al.45 | N.R. | N.R. | 8230 ± 3950 (10) | 0.87 ± 0.20 (10) |
| 5′DFUR | ||||
| Current study | 2274, 9660 | 1.00, 1.05 | 8211, 16036 | 0.70, 1.16 |
| Historical Controls | ||||
| Laheru et al.43 | 5787 ± 3996 (13) | 2.00 (0.50–4.00;13) | 10228 ± 4263 (12) | 0.72 ± 0.14 (13) |
| Soepenberg et al.44 | 4906 ± 2928 (13) | 2.00 (13) | 10519 ± 3533 (13) | 0.81 ± 0.10 (10) |
| Pronk et al.45 | N.R. | N.R. | 11800 ± 3780 (10) | 0.71 ± 0.18 (10) |
| 5-FU | ||||
| Current study | 156, 714 | 1.00, 1.05 | 1144 | 0.71 |
| Historical Controls | ||||
| Laheru et al.43 | 102 ± 96 (13) | 2.00 (0.50–4.00; 13) | 176 ± 62 (13) | 0.73 ± 0.17 (13) |
| Soepenberg et al.44 | 211 ± 130 (13) | 2.00 (13) | 434 ± 161 (13) | 1.04 ± 0.57 (10) |
| Pronk et al.45 | N.R. | N.R. | 610 ± 310 (10) | 0.72 ± 0.18 (10) |
Abbreviations: AUC, area under the concentration–time curve; Cmax, maximal plasma concentration; N.R., not reported; Tmax, time of the maximal plasma concentration; T1/2, terminal half-life. For the current study, the individual values are listed. For the historical controls, values are reported as the arithmetic mean ± standard deviation (n). The Tmax is reported as the median (range; n) or as the mean (n) for the Soepenberg and Pronk studies.
The following AUCs were reported: AUC0-inf, AUC0-inf, AUC0-10h, and AUC0-inf in the current, Laheru, Soepenberg, and Pronk studies.
Capecitabine-Induced Toxicity
The results of current clinical study are consistent with previous reports of increased risk of toxicity associated with 5-FU therapy in patients possessing the homozygous TYMS*2 allele. The predominant grade 3 toxicities were non-hematologic (Table 3). Two of the four patients treated developed grade 3 hand and foot syndrome. The other 2 patients developed grade 3 diarrhea despite adequate supportive care. Three of the four patients developed grade 2 stomatitis and nausea vomiting. Only one patient was treated beyond cycle 2. Two patients were treated with only 1 cycle of chemotherapy due the development of grade 2 and 3 mucositis, vomiting and hand and foot syndrome resulting in hospitalization of one of these patients for supportive care. Neither of these patients remained on study beyond day 15 of therapy. One patient was taken off study in cycle 1 due to a combination of disease progression and toxicity, whilst the second patient withdrew consent to participate in the study because of the development non-hematologic toxicity within a single cycle of capecitabine dosing with dose modification. No grade 4 toxicities were observed. Hematologic toxicity in study consisted primarily of grade 1 anemia occurring in 3 of 4 treated patients. One patient experienced grade 2 neutropenia (Table 3). In summary, capecitabine dosing in this patient population produced more non-hematologic severe toxicity than has previously been appreciated when dosing capecitabine in the unselected pancreatic adenocarcinoma population.
Gemcitabine Response Associated SNPs
There was no significant difference in overall survival with respect to either G/G, G/A or A/A genotypes of ATM G60A. Accordingly, the median survival 9.1, 9.6 and 9.0 months was not significantly different amongst the three groups (p=0.8), respectively for the G/G, G/A or A/A genotypes (data not shown). Next, RecQ1 A159C genotype was analyzed with respect to patient overall survival. The results of this analysis were not dissimilar to those obtained with the ATM G60A analysis. There was no association with RecQ1 A159C genotype and survival in this patient population who were gemcitabine refractory. Accordingly, the median survivals of 8.3, 9.0 and 10.1 months for the respective RecQ1genotypes of A/A, A/C and C/C was not significantly different with a p-value of 0.832 (data not shown).
Discussion
Pancreatic cancer remains a devastating disease despite advances in the genetic characterization of the disease. 5-FU and gemcitabine remain the standard of care therapy for this disease. The addition of both cytotoxic and molecular based chemotherapies to these agents has resulted in minimal improvement in the clinical outcomes. These results suggest that alternative strategies must be taken to improve upon the effectiveness of these therapies. This clinical study represents the first attempt to use a genotype-directed strategy to improve the clinical outcome of 5-FU based therapy for pancreatic cancer patients. This study used the thymidylate synthase promoter TYMS*2 homozygous genotype to pre-select patients to receive capecitabine. This strategy was adopted from the results of Park et al demonstrating that the *2/*2 TYMS genotype was associated with both increased efficacy and toxicity of 5-FU therapy in colon cancer patients [22]. Further support for this strategy was demonstrated by Parikh et al who used TYMS genotype-directed therapy to improve pathologic downstaging of rectal cancer by stratifying the neoadjuvant therapy administered based upon TYMS genotypes [24]. Despite the potential benefits demonstrated in colorectal cancer patients, the use of this strategy in second-line pancreatic adenocarcinoma patients proved to be very difficult. One of the primary problems associated with this approach was obtaining appropriate patient enrollment to adequately perform the treatment portion of the study. Unfortunately, of the 16 patients that possessed this genotype, only 4 patients were enrolled in the study. The predominant difficulty associated with enrollment to the treatment portion of the study was the extremely poor prognosis of pancreatic cancer patients requiring second-line therapy. Many of the patients who progressed on gemcitabine were too ill to be enrolled to the treatment portion of the study. Another issue was the number of additional second-line clinical trials open at the institution during the period of patient enrollment onto this study. Lastly, many patients were treated with capecitabine by their local oncologist instead of enrolling on the clinical trial, since they could obtain the drug at their local pharmacy without having to travel to a tertiary care center. While genotype-directed therapy represents a novel approach to the treatment of pancreatic cancer patients, it is difficult to conduct such a study in the second line setting for this patient population, especially with a drug than can easily be used off-label. This strategy may prove to be a more useful treatment strategy either in the adjuvant setting or for first-line metastatic disease.
Despite the small sample size of the treatment portion, the observed toxicity was impressive. All subjects experienced significant grade 2 and 3 non-hematologic toxicity with exposure to capecitabine. The predominant grade 2 and 3 toxicities were diarrhea, nausea/vomiting, hand and foot syndrome and mucositis. One patient was hospitalized due to grade 3 nausea/vomiting, hand and foot syndrome and diarrhea. This is not explained with patient fitness, since all patients had a performance status of ECOG < 1 at time of study entry. In addition, all patients except the 1 patient who died while on study were treated on subsequent clinical trials. The limited data gathered in this study would suggest that pharmacokinetic variability in the studied patient population compared to an unselected patient population did not explain the increase observed toxicity. Exposure levels of capecitabine or 5-FU have not been reported in previous studies evaluating the pharmacogenetics of TYMS as it applies to various forms of 5-FU based chemotherapy. The findings of this study would suggest that other factors such as additional polymorphisms involving other genes important for 5-FU metabolism or pharmacodynamics may be interacting with the TYMS*2 to drive the profound level of toxicity.
Gemcitabine represents the standard of care for patients with advanced pancreatic cancer. The single nucleotide polymorphisms ATM G60A and RecQ1 A159C have previously been demonstrated to be associated with poor overall survival in those patients receiving neoadjuvant gemcitabine-based therapy for resectable early stage pancreatic adenocarcinoma [30,31]. Our finding attempted to extend these findings to the metastatic disease setting, but no statistically significant relationship between ATM, RecQ1 and TYMS genotypes and overall survival were observed. However, despite the limited number of subjects, there did appear to be a trend toward poorer overall survival in patients with metastatic pancreatic cancer possessing the TYMS *2/*2 genotype treated with gemcitabine. This may be more reflective of TYMS genotype association with specific stages of malignancy as has been shown for squamous cell carcinoma of head and neck, colon cancer and non-small cell lung cancer than a direct impact on pathways responsible for gemcitabine induced cell death [46–48]. Recently, SNPs involving a combination of genes involved in gemcitabine transport and metabolism have been associated with survival in patients treated with neoadjuvant gemcitabine-based chemoradiotherapy [49]. These results have not been extended to the metastatic disease setting. These findings suggest that genotype-directed therapy may have a role in determining which patients should receive gemcitabine neoadjuvant therapy but cannot be extended to guide the use of gemcitabine in the metastatic disease setting.
In summary, large studies of genotype-directed therapy for second-line pancreatic adenocarcinoma therapy are difficult to perform successfully, particularly when the investigational agent is available off-study. The poor natural history of this patient population also presents a significant challenge. Therefore application of this strategy may be better utilized in the adjuvant or first-line metastatic disease setting, or with an agent not yet FDA-approved. Another significant limitation to this study was that it was performed as a single institution study. A multi-institutional study would allow for the screening of a larger number of patients, but obtaining funding for such studies can be challenging. Overcoming these obstacles will be important for implementation of genotype-directed therapy and other designs that ultimately may result in the realization of personalized therapy for pancreatic cancer patients.
Fig. 1.
Demonstrates the overall survival probability of all patients screened for the TYMS*2/*2 genotype. The overall survival probability of patients with the homozygous TYMS*2 genotype was analyzed in comparison to those with either the TYMS*2/*3 or homozygous TYMS*3 genotype.
Acknowledgments
This study was approved and funded by the National Comprehensive Cancer Network (NCCN) from general research support provided by Roche Laboratories, Inc. This research was supported by the Analytical Pharmacology Core of the Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins (NIH P30 CA069773). Colin Weekes was supported by a United Negro College Fund UNCF-Merck Science Initiative Postdoctoral Research Fellowship.
We would like to thank Ming Zhao and Ping He for their technical support and Susan Davidson for her quality assurance of the data.
References
- 1.American Cancer Society. Cancer Facts and Figures. 2009. [Google Scholar]
- 2.Yeo TP, Hruban RH, Leach SD, Wilentz RE, Sohn TA, Kern SE, et al. Pancreatic cancer. Curr Probl Cancer. 2002;26(4):176–275. doi: 10.1067/mcn.2002.129579. [DOI] [PubMed] [Google Scholar]
- 3.Abrams RA, Grochow LB, Chakravarthy A, Sohn TA, Zahurak ML, Haulk TL, et al. Intensified adjuvant therapy for pancreatic and periampullary adenocarcinoma: survival results and observations regarding patterns of failure, radiotherapy dose and CA19-9 levels. Int J Radiat Oncol Biol Phys. 1999;44(5):1039–1046. doi: 10.1016/s0360-3016(99)00107-8. [DOI] [PubMed] [Google Scholar]
- 4.Sohn TA, Yeo CJ, Cameron JL, Koniaris L, Kaushal S, Abrams RA, et al. Resected adenocarcinoma of the pancreas-616 patients: results, outcomes, and prognostic indications. J Gastrointest Surg. 2000;4(6):567–579. doi: 10.1016/s1091-255x(00)80105-5. [DOI] [PubMed] [Google Scholar]
- 5.Burris HA, Moore MJ, Andersen J, Green MR, Rothenberg ML, Modiano MR, et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol. 1997;15(6):2403–2413. doi: 10.1200/JCO.1997.15.6.2403. [DOI] [PubMed] [Google Scholar]
- 6.Herrmann R, Bodoky G, Ruhstaller T, Glimelius B, Bajetta E, Schuller J, et al. Gemcitabine plus capecitabine compared with gemcitabine alone in advanced pancreatic cancer: A randomized, multicenter, phase III trial of the swiss group for clinical cancer research and the central European cooperative oncology group. J Clin Oncol. 2007;25(16):2212–2217. doi: 10.1200/JCO.2006.09.0886. [DOI] [PubMed] [Google Scholar]
- 7.Moore MJ, Goldstein D, Hamm J, Figer A, Hecht JR, Gallinger S, et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: A phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Onco. 2007;25(15):1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 8.Volker H, Quietzcsch D, Gieseler F, Gonnermann M, Schonekas H, Rost A, et al. Randomized phase III trial of gemcitabine plus cisplatin compared with gemcitabine alone in advanced pancreatic cancer. J Clin Oncol. 2006;24(24):3946–3952. doi: 10.1200/JCO.2005.05.1490. [DOI] [PubMed] [Google Scholar]
- 9.Louvet C, Labianca R, Hammel P, Lledo G, Zampino MG, Andre T, et al. Gemcitabine in combination with oxaliplatin compared with gemcitabine alone in locally advanced or metastatic pancreatic cancer: Results of a GERCOR and GISCAD phase III trial. J Clin Oncol. 2005;23(15):3509–3516. doi: 10.1200/JCO.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 10.Kindler HL, Friberg G, Singh DA, Locker G, Nattam S, Kozloff M, et al. A phase II trial of bevacizumab plus gemcitabine in patients with advanced pancreatic cancer. J Clin Oncol. 2005;23(31):8033–8040. doi: 10.1200/JCO.2005.01.9661. [DOI] [PubMed] [Google Scholar]
- 11.Van Cutsem E, Vervenne WL, Bennouna J, Humblet Y, Gill S, Van Laethem J-L, et al. Phase III trial of bevacizumab in combination with gemcitabine and erlotinib in patients with metastatic pancreatic cancer. J Clin Oncol. 2009;27(13):2231–2237. doi: 10.1200/JCO.2008.20.0238. [DOI] [PubMed] [Google Scholar]
- 12.Baker SD, Verweij J, Rowinsky EK, Donehower RC, Schellens JHM, Grochow LB, et al. Role of Body Surface Area in Dosing of Investigational Anticancer Agents in Adults, 1991–2001. JNCI Cancer Spectrum. 2002;94:1883–1888. doi: 10.1093/jnci/94.24.1883. [DOI] [PubMed] [Google Scholar]
- 13.Johnston PG, Drake JC, Trepel J, Allegra C. Immunological quantitation of Thymidylate synthase using the monoclonal antibody TS 106 in 5-fluorouracil-sensitive and – resistant human cancer cell lines. Cancer Res. 1992;52:4306–4312. [PubMed] [Google Scholar]
- 14.Johnston PG, Lenz HJ, Leichman CG, Danenberg K, Allegra CJ, Danenberg PV, et al. Thymidylate synthase gene and protein expression correlate and are associated with response to 5-fluorouracil in human colorectal and gastric tumors. Cancer Res. 1995;55:1407–1412. [PubMed] [Google Scholar]
- 15.Leichman CG, Lenz HJ, Leichman L, Danenberg K, Baranda J, Groshen S, et al. Quantitation of intratumoral Thymidylate synthase expression predicts for disseminated colorectal cancer response and resistance to protracted-infusion fluorouracil and weekly leucovorin. J Clin Oncol. 1997;15:3223–3229. doi: 10.1200/JCO.1997.15.10.3223. [DOI] [PubMed] [Google Scholar]
- 16.Horie N, Aiba H, Katsuhiko O, Hojo H, Takeishi K. Functional analysis of DNA polymorphism of the tandemly repeated sequences in the 5′-terminal regulatory region of the human gene for thymidylate synthase. Cell Sturct Funct. 1995;20:191–197. doi: 10.1247/csf.20.191. [DOI] [PubMed] [Google Scholar]
- 17.Pullarkat ST, Stoehlmacher J, Ghaderi V, Xiong Y-P, Ingles SA, Sherrod A, et al. Thymidylate synthase gene polymorphism determines response and toxicity of 5-FU chemotherapy. Pharmacogenomics J. 2001;1:65–70. doi: 10.1038/sj.tpj.6500012. [DOI] [PubMed] [Google Scholar]
- 18.Marsh S, Collie-Duguid ES, Li T, Xiehe L, Mcleod HL. Ethnic variation in the thymidylate synthase enhancer region polymorphism among Caucasian and Asian populations. Genomics. 1999;58:310–312. doi: 10.1006/geno.1999.5833. [DOI] [PubMed] [Google Scholar]
- 19.Marsh S, Ameyaw MM, Githang’a J, Indalo A, Ofori-Adjei D, McLeod HL. Novel thymidylate synthase enhancer region alleles in African populations. Hum Mutat. 2000;16:528–533. doi: 10.1002/1098-1004(200012)16:6<528::AID-HUMU11>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 20.Mandola MV, Stoehlmacher J, Muller-Weeks S, Cesarone G, Yu MC, Lenz H-J, et al. A novel single nucleotide polymorphism within the 5′ tandem repeat polymorphism of the Thymidylate synthase gene abolishes USF-1 binding and alters transcriptional activity. Cancer Res. 2003;63:2898–2904. [PubMed] [Google Scholar]
- 21.Villafranca E, Okruzhnov Y, Dominguez MA, Garcia-Foncillas J, Azinovic I, Martinez E, et al. Polymorphisms of the repeated sequences in the enhancer region of the thymidylate synthase gene promoter may predict downstaging after preoperative chemoradiation in rectal cancer. J Clin Oncol. 2001;19:1779–1786. doi: 10.1200/JCO.2001.19.6.1779. [DOI] [PubMed] [Google Scholar]
- 22.Park DJ, Stoehlmacher J, Zhang W, Tsao-Wei D, Groshen S, Lenz H-J. Thymidylate synthase gene polymorphism predicts response to capecitabine in advanced colorectal cancer. Int J Colorectal Dis. 2002;17:46–49. doi: 10.1007/s003840100358. [DOI] [PubMed] [Google Scholar]
- 23.McLeod HL, Tan B, Malyapa R, Abbey E, Picus J, Myerson R, et al. Genotype-guided neoadjuvant therapy for rectal cancer. J Clin Oncol. 2005;23(16S):3024. [Google Scholar]
- 24.Parikh PJ, Mahasittiwat P, Flexhman JW, Tan B, Mutch MG, Myerson RJ, et al. Predictors of complete pathologic response on a prospective locally advanced rectal cancer trial. J Clin Oncol; Proceedings of ASCO Gastrointestinal Cancer Symposium.2009. Abst 453. [Google Scholar]
- 25.Schuller J, Cassidy J, Dumont E, Roos B, Durston S, Banken L, et al. Preferential activation of capecitabine in tumor following oral administration to colorectal cancer patients. Cancer Chemother Pharmacol. 2000;45(4):291–297. doi: 10.1007/s002800050043. [DOI] [PubMed] [Google Scholar]
- 26.Ishikawa T, Fumiko S, Fukase Y, Sawada N, Ishitsuka H. Positive correlation between the efficacy of capecitabine and doxifluridine and the ratio of thymidine phosphorylase to dihydropyrimidine dehydrogenase activities in tumors in human cancer xenografts. Cancer Res. 1998;58(4):685–690. [PubMed] [Google Scholar]
- 27.Cartwright TH, Cohn A, Varkey JA, Chen YM, Szatrowski TP, Cox JV, et al. Phase II study of oral capecitabine in patients with advanced or metastatic pancreatic cancer. J Clin Oncol. 2002;20(1):160–164. doi: 10.1200/JCO.2002.20.1.160. [DOI] [PubMed] [Google Scholar]
- 28.Fukunaga AK, Marsh S, Murry DJ, Hurley TD, McLeod HL. Identification and analysis of single-nucleotide polymorphisms in the gemcitabine pharmacologic pathway. Pharmacogenomics J. 2004;4(5):307–314. doi: 10.1038/sj.tpj.6500259. [DOI] [PubMed] [Google Scholar]
- 29.Mini E, Nobili S, Caciagli B, Landini I, Mazzei T. Cellular pharmacology of gemcitabine. Ann Oncol. 2006;17(Suppl 5):7–12. doi: 10.1093/annonc/mdj941. [DOI] [PubMed] [Google Scholar]
- 30.Li D, Frazier M, Evans DB, Hess KR, Crane CH, Jiao L, et al. Single nucleotide polymorphisms of RecQ1, RAD54L, and ATM genes are associated with reduced survival of pancreatic cancer. J Clin Oncol. 2006;24(11):1720–1728. doi: 10.1200/JCO.2005.04.4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okazaki T, Jiao L, Chang P, Evans DB, Abbruzzese JL, Li D. Single-nucleotide polymorphisms of DNA damage response genes are associated with overall survival in patients with pancreatic cancer. Clin Cancer Res. 2008;14(7):2042–2048. doi: 10.1158/1078-0432.CCR-07-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3(3):155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 33.Angele S, Romestaing P, Moullan N, Vuillaume M, Chapot B, Friesen M, et al. ATM haplotypes and cellular response to DNA damage: association with breast cancer risk and clinical radiosensitivity. Cancer Res. 2003;63(24):8717–8725. [PubMed] [Google Scholar]
- 34.Kim JH, Kim H, Lee KY, Choe KH, Ryu JS, Yoon HI, et al. Genetic polymorphisms of ataxia telangiectasia mutated affect lung cancer risk. Hum Mol Genet. 2006;15(7):1181–1186. doi: 10.1093/hmg/ddl033. [DOI] [PubMed] [Google Scholar]
- 35.Chakraverty RK, Hickson ID. Defending genome integrity during DNA replication: a proposed role for RecQ family helicases. Bioessays. 1999;21(4):286–294. doi: 10.1002/(SICI)1521-1878(199904)21:4<286::AID-BIES4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 36.Hickson ID. RecQ helicases: caretakers of the genome. Nat Rev Cancer. 2003;3(3):169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 37.Khakhar RR, Cobb JA, Bjerqbaek L, Hickson ID, Gasser SM. RecQ helicases: multiple roles in genome maintenance. Trends Cell Biol. 2003;13(9):493–501. doi: 10.1016/s0962-8924(03)00171-5. [DOI] [PubMed] [Google Scholar]
- 38.de las Penas R, Sanchez-Ronco M, Alberola V, Taron M, Camps C, Garcia-Carbonero R, et al. Polymorphisms in DNA repair genes modulate survival in cisplatin/gemcitabine-treated non-small-cell lung cancer patients. Ann Oncol. 2006;17(4):668–675. doi: 10.1093/annonc/mdj135. [DOI] [PubMed] [Google Scholar]
- 39.Bepler G, Zheng Z, Gautam A, Sharma S, Cantor A, Cress WE, et al. Ribonucleotide reductase M1 gene promoter activity, polymorphisms, population frequencies, and clinical relevance. Lung Cancer. 2005;47(2):183–192. doi: 10.1016/j.lungcan.2004.07.043. [DOI] [PubMed] [Google Scholar]
- 40.Xu Y, Grem JL. Liquid chromatography-mass spectrometry method for the analysis of the anti-cancer agent capecitabine and its nucleoside metabolites in human plasma. J Chromatogr B Analyt Technol Biomed Life Sci. 2003;783(1):273–285. doi: 10.1016/s1570-0232(02)00674-8. [DOI] [PubMed] [Google Scholar]
- 41.Besnard T, Renée N, Etienne-Grimaldi MC, François E, Milano G. Optimized blood sampling with cytidine deaminase inhibitor for improved analysis of capecitabine metabolites. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;870(1):117–120. doi: 10.1016/j.jchromb.2008.05.040. [DOI] [PubMed] [Google Scholar]
- 42.Rudek MA, Zhao M, He P, Hartke C, Gilbert J, Gore SD, et al. Pharmacokinetics of 5-azacitidine administered with phenylbutyrate in patients with refractory solid tumors or hematologic malignancies. J Clin Oncol. 2005;23(17):3906–3911. doi: 10.1200/JCO.2005.07.450. [DOI] [PubMed] [Google Scholar]
- 43.Laheru D, Croghan G, Bukowski R, Rudek M, Messersmith W, Erlichman C, et al. A phase I study of EKB-569 in combination with capecitabine in patients with advanced colorectal cancer. Clin Cancer Res. 2008;14(17):5602–5609. doi: 10.1158/1078-0432.CCR-08-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soepenberg O, Dumez H, Verweij J, Semiond D, de Jonge MJ, Eskens FA, et al. Phase I and pharmacokinetic study of oral irinotecan given once daily for 5 days every 3 weeks in combination with capecitabine in patients with solid tumors. J Clin Oncol. 2005;23(4):889–898. doi: 10.1200/JCO.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 45.Pronk LC, Vasey P, Sparreboom A, Reigner B, Planting AS, Gordon RJ, et al. A phase I and pharmacokinetic study of the combination of capecitabine and docetaxel in patients with advanced solid tumours. Br J Cancer. 2000;83:22–29. doi: 10.1054/bjoc.2000.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Z, Shi Q, Sturgis EM, Spitz MR, Hong WK, Wei Q. Thymidylate synthase 5′ and 3′-untranslated region polymorphisms associated with risk and progression of squamous cell carcinoma of the head and neck. Clin Cancer Res. 2004;10:7903–7910. doi: 10.1158/1078-0432.CCR-04-0923. [DOI] [PubMed] [Google Scholar]
- 47.Ulrich CM, Curtin K, Potter JD, Bigler J, Caan B, Slattery ML. Polymorphisms in the reduced folate carrier, thymidylate synthase, or methionine synthase and risk of colon cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:2509–2516. doi: 10.1158/1055-9965.EPI-05-0261. [DOI] [PubMed] [Google Scholar]
- 48.Shi Q, Zhang A, Neumann AS, Li G, Spitz MR, Wei Q. Case-control analysis of thymidylate synthase polymorphisms and risk of lung cancer. Carcinogenesis. 2005;26:649–656. doi: 10.1093/carcin/bgh351. [DOI] [PubMed] [Google Scholar]
- 49.Javle MM, Okazaki T, Wolff RA, Varadhachary G, Ho L, Crane CH, et al. Combined effect of single nucleotide polymorphisms (SNPs) of gemcitabine metabolic genes on pancreatic cancer survival and drug toxicity. J Clin Oncol; Proceedings ASCO Gastrointestinal Cancers Symposium.2008. Abst 126. [Google Scholar]