Abstract
Cancer cells consume large quantities of glucose and primarily use glycolysis for ATP production, even in the presence of adequate oxygen 1,2. This metabolic signature (aerobic glycolysis or the Warburg effect) enables cancer cells to direct glucose to biosynthesis, supporting their rapid growth and proliferation 3,4. However, both causes of the Warburg effect and its connection to biosynthesis are not well understood. Here we show that the tumor suppressor p53, the most frequently mutated gene in human tumors, inhibits the pentose phosphate pathway (PPP) 5. Via the PPP, p53 suppresses glucose consumption, NADPH production, and biosynthesis. The p53 protein binds to glucose-6-phosphate dehydrogenase (G6PD), the first and rate-limiting enzyme of the PPP, and prevents the formation of the active dimer. Tumor-associated p53 mutants lack the G6PD-inhibitory activity. Therefore, enhanced PPP glucose flux due to p53 inactivation may increase glucose consumption and direct glucose toward biosynthesis in tumor cells.
The tumor suppressor p53 invokes anti-proliferative processes, of which the best understood include cell cycle arrest, DNA repair, and apoptosis 6,7. Recent studies suggested that p53 also has a role in modulating metabolism including glycolysis and oxidative phosphorylation 8,9,10. However, the role of p53 in regulating biosynthesis is less understood. The PPP is a main pathway for glucose catabolism and biosynthesis 5. In an oxidative phase, the PPP generates NADPH (nicotinamide adenine dinucleotide phosphate), the principal intracellular reductant required for reductive biosynthesis such as the synthesis of lipid, and an essential precursor for biosynthesis of nucleotides. This is followed by a nonoxidative inter-conversion of ribose 5-phosphate to the intermediates in the glycolytic pathways. Despite the vital role of the PPP in biosynthesis and its close link to glycolysis, the regulation of the PPP in tumor cells remains unclear.
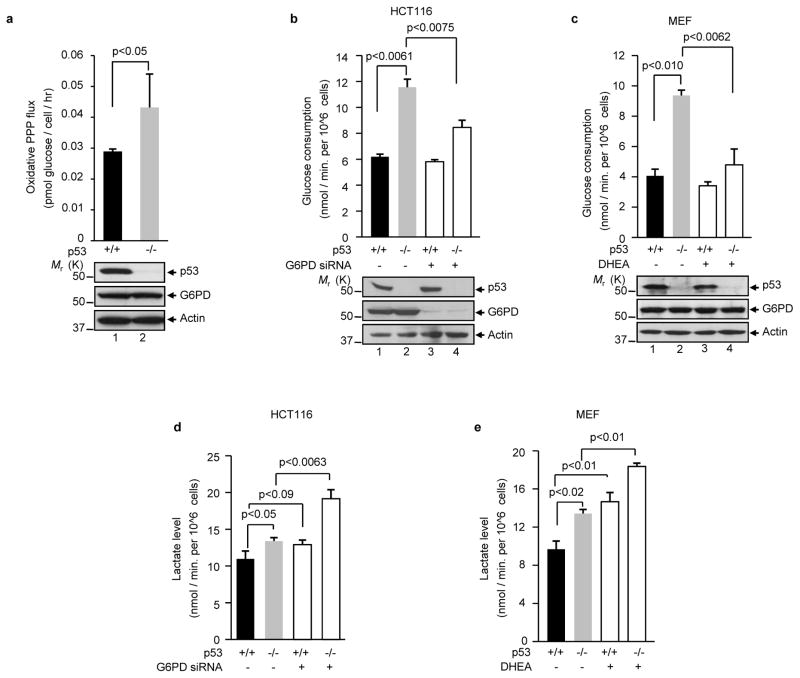
To investigate whether p53 modulates the PPP, we compared the oxidative PPP flux in isogenic p53+/+ and p53−/− human colon cancer HCT116 cells 11. Cells were cultured in medium containing [2-13C]glucose, and the glucose metabolites were measured by nuclear magnetic resonance (NMR) spectroscopy. As shown in Fig. 1a, the absence of p53 resulted in a strong enhancement (~50%) in oxidative PPP flux, suggesting that p53 suppresses the PPP. The absence of p53 concomitantly led to a strong increase in glucose consumption, and this was observed in both HCT116 cells and mouse embryonic fibroblast (MEF) (Figs. 1b, c). Inhibition of G6PD using either small interfering RNA (siRNA) or dehydroepiandrosterone (DHEA) revered the increase in glucose consumption caused by p53 deficiency, while having little effect on glucose consumption in p53+/+ cells (Figs. 1b, c). These results suggest that p53 deficiency increases glucose consumption mainly through an enhanced PPP flux.
Figure 1. p53 deficiency correlates with increases in PPP flux, glucose consumption, and lactate production.
(a) p53+/+ and p53−/− HCT116 cells were cultured in medium containing [2-13C]glucose. Oxidative PPP flux (top) was measured based on the rate of glucose consumption and the ratio of 13C incorporated in to carbon 2 (indicating glycolysis) and carbon 3 (indicating PPP) of lactate by NMR spectroscopy. Molecular weight standards (Mr, in kDa) are indicated on the left. Data shown are means ± S.D. (n=3).
(b, d) p53+/+ and p53−/− HCT116 cells were treated with G6PD siRNA and control siRNA (−). Glucose consumption (b, top) and lactate levels (d) are expressed as mean ± standard deviation (SD) of three independent experiments. The expression of p53, G6PD, and actin (a loading control) is shown at the bottom of (b).
(c, e) p53+/+ and p53−/− MEF cells were treated with 1 mM DHEA or vehicle (−) for 24 h. Glucose consumption (c, top) and lactate levels (e) are measured. Protein expression is shown at the bottom of (c). Data are means ± S.D. (n=3).
The lack of p53 also correlated with elevated lactate production (Figs. 1d, e). However, inhibition of G6PD in these cells increased, rather than decreased, in lactate production regardless of p53 status. Therefore, glucose flux through the PPP may in itself lower lactate production. The suppression of lactate production may be related to the ability of p53 to decrease glycolysis 8 or increase oxidative phosphorylation 9.
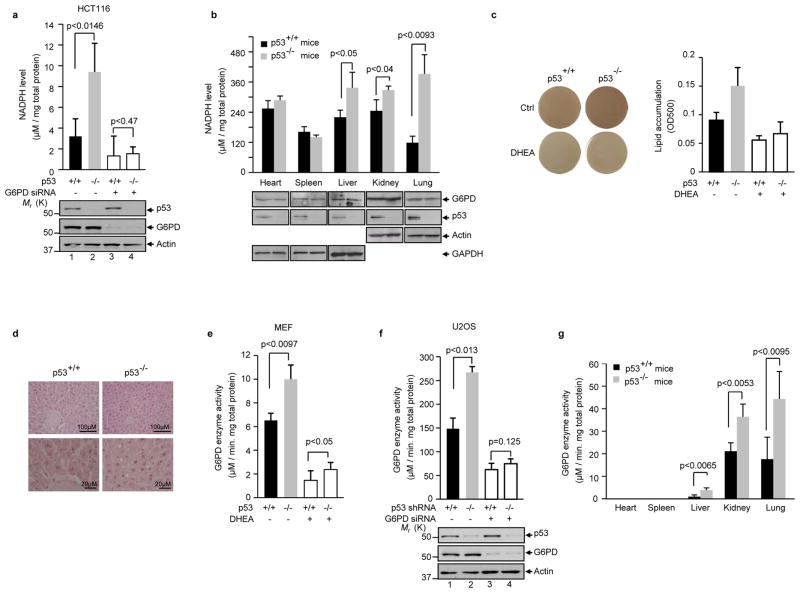
The PPP plays a major role in the production of cellular NADPH. The lack of p53 led to a strong increase in the NADPH level in HCT116 cells (~ 2 folds, Fig. 2a). Likewise, knocking down of p53 in U2OS cells with small hairpin RNA (shRNA) strongly increased NADPH levels (Supplementary Information, Fig. S1a). Treatment with G6PD siRNA minimized the difference in NADPH levels between p53 proficient and deficient cells. To verify the cell culture findings in animals, we compared the NADPH levels in various tissues from p53−/− and p53+/+ mice. The tissues from p53−/− mice – including heart, liver, kidney, and lung – exhibited substantially elevated NADPH levels compared to those in the corresponding tissues from p53+/+ mice (Fig. 2b). The exception was found in the spleen. In this tissue, the activity of G6PD was very low (Fig. 2g), and the PPP might not contribute substantially to the overall NADPH production. Converse to p53 down-regulation, over-expression of p53 led to a strong decrease in NADPH levels (Supplementary Information, Fig. S1b).
Figure 2. p53 regulates NADPH levels, lipid accumulation, and G6PD activity.
(a) NADPH levels (means ± S.D., n=3) in p53+/+ and p53−/− HCT116 cells treated with G6PD siRNA or a control siRNA. Protein expression is shown below.
(b) NADPH levels (means ± S.D., n=3) in tissues from p53+/+ and p53−/− mice maintained on a normal diet. Protein expression is shown below.
(c) p53+/+ and p53−/− MEF cells treated with or without DHEA were cultured in the presence of insulin, rosiglitazone, isobutylmethylxanthine (IBMX), and dexamethasone. Lipid contents were analyzed by Oil Red O staining. Left: Oil Red O-stained dishes. Right: staining was quantified by OD500nm. Data are means ± S.D. (n=3).
(d) Histological sections of liver tissue from p53+/+ and p53−/− mice were stained with hematoxylin and eosin. Arrows indicate fat droplets.
(e) G6PD activity (means ± S.D., n=3) in p53+/+ and p53−/− MEFs treated with or without DHEA.
(f) U2OS cells stably expressed a p53 shRNA or a control shRNA (−) were transfected with a G6PD siRNA or a control siRNA (−). G6PD activity (top) and protein expression (bottom) were analyzed. Data are means ± S.D. (n=3).
(g) G6PD activity in tissues from p53−/− and p53+/+ mice maintained on a normal diet. G6PD activity is the mean ± SD of three p53+/+ or four p53−/− mice.
NADPH is required for the biosynthesis of lipid. To assess the effect of p53 on lipid accumulation, we treated p53+/+ and p53−/− MEF cells with a combination of insulin, rosiglitazone, dexamethasone, and isobutylmethylxanthine, which stimulates lipogenesis 12. The p53−/− MEFs showed enhanced lipid levels compared to p53+/+ MEFs as evaluated by Oil Red O staining (Fig. 2c). The lack of p53 also resulted in higher levels of lipid In HCT116 cells (Supplementary Information, Fig. S1c). The difference in lipid accumulation between p53+/+ and p53−/− cells diminished upon treatment with G6PD siRNA or DHEA. We also evaluated the effect of p53 on the formation of fat droplets in the liver. The liver of p53−/− mice had a larger amount of bigger fat droplets compared to the liver of p53+/+ mice (Fig. 2d). Together, these results suggest that p53 inhibits NADPH production and lipid accumulation by lowering the glucose flux through the PPP.
To investigate the mechanism by which p53 regulates the PPP, we assayed the activity of G6PD, a key regulatory point of the PPP. The lack of p53 correlated with a strong elevation in G6PD activity in both MEF and HCT116 cells (Fig. 2e and Supplementary Information, Figs. S1d, e). Similarly, when p53 was knocked down in U2OS cells with shRNA, G6PD activity nearly doubled (Fig. 2f). Furthermore, in mice tissues where G6PD activity could be adequately detected (e.g. liver, lung, and kidney), the lack of p53 was associated with highly elevated G6PD activity (Fig. 2g). Conversely, over-expression of wild type p53 in the p53-deficient cell lines (H1299 and p53−/−Mdm2−/− MEF) caused a noticeable decrease in G6PD activity (Supplementary Information, Fig. S1f, g). These results show that p53 suppresses G6PD activity.
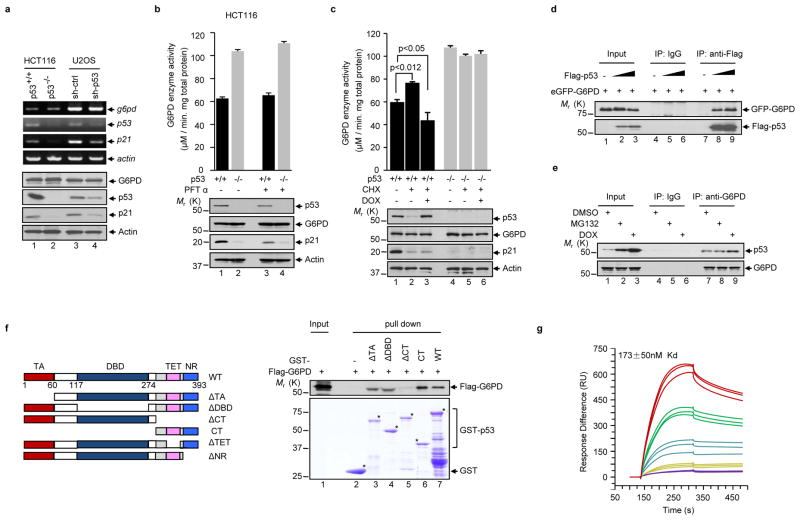
In each of the cell lines and tissues that were examined, the levels of the G6PD protein remained unchanged when p53 was down regulated or overexpressed (Fig. 2a,b,f and Supplementary Information, Fig. S1). Moreover, p53 did not change the level of G6PD transcript (Fig. 3a). To rule out the involvement of other p53 target genes in the inhibition of G6PD, we used an inhibitor of p53 transcriptional activity, pifithrin-α (PFTα)13. PFTα impeded p53-inuced expression of p21, but did not restore p53-inhibited G6PD activity (Fig. 3b and Supplementary Information, Fig. S2a). We also used the protein synthesis inhibitor cycloheximide (CHX), alone or together with the DNA damage agent doxorubicin (DOX). Treatment of p53+/+ HCT116 cells with CHX alone resulted in a lower level of p53, which was accompanied by a higher activity of G6PD (Fig. 3c). Simultaneous treatment with CHX and DOX led to a stabilization of p53 above the basal level seen in unstressed cells, and a concurrent drop of G6PD activity below its basal level (Fig. 3c). As controls, none of these treatments altered G6PD activity in p53−/− HCT116. In addition, the p53 mutant V122A, which has a transactivation activity comparable to or even higher than wild type p53 dependent on the target gene 14, failed to inhibit G6PD (Supplementary Information, Fig. S2b). Moreover, we treated cells with the nuclear export inhibitor leptomycin B to prevent cytoplasmic accumulation of p53 15,16. Leptomycin B reversed p53-mediated inhibition of G6PD (Supplementary Information, Fig. S2c-e). Altogether, these results show that inhibition of G6PD by p53 is independent of transcription or translation and is a cytoplasmic, not nuclear, function of p53.
Figure 3. p53 interacts with G6PD and inhibits its activity independently of transcription.
(a) p53+/+ and p53−/− HCT116 cells, and U2OS cells transfected with p53 shRNA and control (ctrl) shRNA, were analyzed by RT-PCR with reverse transcription (top) and western blot (bottom).
(b, c) p53+/+ and p53−/− HCT116 cells were treated with or without 20 μM of PFTα for 24 h (b), or treated with or without Dox (2 μM) for 1 h, and then with or without cycloheximide (CHX, 20 μM) for 2 h (c). Cells were analyzed for G6PD activity (top) and protein expression (bottom). Data are means ± S.D. (n=3).
(d) H1299 cells were transfected with enhanced GFP-G6PD alone or together with increasing amounts of Flag-p53. Cell lysates were immunoprecipitated with an anti-Flag antibody and an isotype-matching control antibody (IgG). Immunoprecipitated proteins (IP) and 5% input were analyzed by western blot.
(e) p53+/+ HCT116 cells were treated with MG132 (20 μM), doxorubicin (DOX, 2 μM), or vehicle (DMSO). Cell lysates were incubated with anti-G6PD antibody or a control antibody (IgG). IP and input were analyzed by western blot.
(f) Left: Schematic representation of p53 and its deletion mutants. WT, wild-type; TA, transactivation domain; DBD, DNA-binding domain; CT, C-terminal region; TET, tetramerization domain; NR, negative regulation domain. The amino acids at the domain boundaries are indicated. Right: purified GST fusions of wild type and mutant p53 proteins were incubated separately with recombinant Flag-G6PD protein conjugated to beads. Beads-bound and input proteins were analyzed by western blot using anti-Flag (top) and Coomassie blue staining (bottom).
(g) Dissociation constant (KD) for p53 from immobilized G6PD was determined by surface plasmon resonance (BIAcore). A real-time graph of response units (RU) against time is shown. The on rate was 7.53 ± 1.59 × 103 M−1s−1, and the off rate was 1.30 ± 0.03 × 10−3 s−1.
We next investigated whether p53 interacts with G6PD. Flag-tagged p53 specifically associated with enhanced green fluorescent protein (eGFP)-G6PD in vivo (Fig. 3d). Likewise, endogenous p53 interacted with endogenous G6PD (Fig. 3e). G6PD is a cytoplasmic protein while p53 is present in both the cytoplasm and the nucleus, and consistently, the p53-G6PD interaction occurred in the cytoplasm (Supplementary Information, Fig. S2f-h). This interaction was enhanced when cells were treated with the proteasome inhibitor MG132 or the DNA damaging agent DOX, both of which stabilized p53 (Fig. 3d, e and Supplementary Information, Fig. S2h). The binding between p53 and G6PD is direct as shown by a pull-down assay with purified recombinant proteins (Fig. 3f). Analysis of the p53-G6PD binding in real time using surface plasmon resonance (BIAcore) showed that the dissociation constant (KD) of p53 from G6PD was 173 ± 50 nM (Fig. 3g). G6PD is a highly conserved protein, and human p53 interacted with G6PD proteins from both the bacterium L. mesenteroides and the yeast S. cerevisiae (Supplementary Information, Figs. S3a–c).
Consistent with their direct interaction, purified wild type p53 could inhibit purified G6PD in vitro (Figs. 4a, b). Additionally, introducing p53 into two S. cerevisiae strains, AH109 and EGY48, led to more than 60% inhibition of G6PD activity (Supplementary Information, Figs. S3d, e). Expression of p53 also strongly reduced G6PD activity in E. coli (Supplementary Information, Figs. S3f, g).
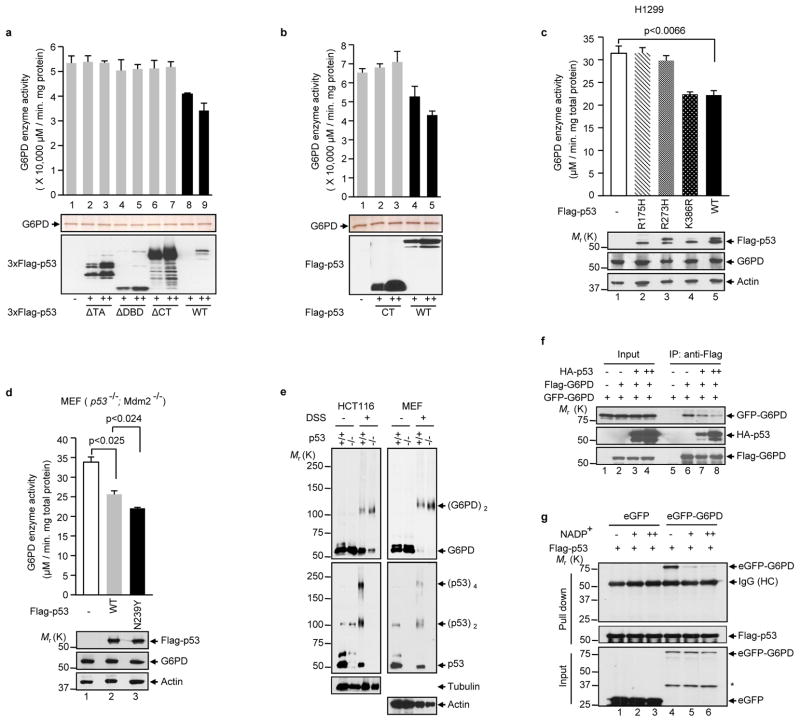
Figure 4. p53 inhibits the formation of dimeric G6PD holoenzyme.
(a, b) Activity of the G6PD protein after being incubated with purified wild type or mutant p53 proteins (top). Proteins were analyzed by silver staining (middle) and anti-Flag western blot (bottom). The p53 proteins were tagged with either three copies (a) or one copy (b) of the Flag epitope. Data are means ± S.D. (n=3).
(c, d) H1299 cells (c) and p53−/−Mdm2−/− MEFs (d) were transfected separately with wild type and mutant p53 proteins as indicated. G6PD activity (top) and protein expression (bottom) were analyzed. The K386R mutation blocks p53 SUMOylation. Data are means ± S.D. (n=3).
(e) Extracts of p53+/+ and p53−/− HCT116 and MEF were treated with and without 5 mM disuccinimidyl suberate (DSS) and analyzed by western blot with antibodies against G6PD, p53, and as controls, tubulin and actin. The positions of various forms of G6PD and p53 are indicated.
(f) H1299 cells were transfected with Flag-G6PD, eGFP-G6PD and different amounts of HA-p53. Cell lysates were incubated with anti-Flag antibody. Input and IP were analyzed by western blot.
(g) Lysates from p53−/− Mdm2−/− MEFs expressing GFP or GFP-G6PD were incubated with Flag-p53 immobilized on M2 beads or control beads in presence of increasing amounts of NADP+ (0, 0.1, and 1 mM). Input and beads-bound (pull down) proteins were analyzed by western blot.
To delineate the structural determinants for p53’s inhibitory activity towards G6PD, we used a panel of p53 deletion mutants. The G6PD interaction domain was mapped to the C-terminal (CT) region of p53 (Fig. 3f and Supplementary Information, Figs. S3b, c). However, the CT region alone was not sufficient for G6PD inhibition; the transactivation (TA) and DNA-binding domain (DBD) were also required (Figs. 4a, b and Supplementary Information, Fig. S1f). A further deletion analysis showed that within the CT region, the negative regulatory (NR) domain, but not the tetramerization (TET) domain, was required for interaction with and inhibition of G6PD (Supplementary Information, Figs. S4a, b).
The involvement of multiple domains of p53 in G6PD inhibition led us to test tumor-associated p53 mutants, the majority of which harbor missense mutations. Unlike wild type p53, three p53 mutants (R175H, R273H, and G279E) showed minimal or no activity in inhibiting G6PD (Fig. 4c and Supplementary Information, Fig. S1g, S2b, S3g), even though at least two of them (R175H and R273H) retained the ability to bind to G6PD (Supplementary Information, Fig. S4c). Another tumor-associated, temperature sensitive mutant (A138V) inhibited G6PD activity at the permissive temperature 32 °C, but not at the non-permissive temperature 37 °C. Moreover, the inhibitory effect at 32 °C was abolished with the introduction of R273H (Supplementary Information, Fig. S4d). To examine the effect of p53 mutants that are expressed at endogenous levels, we used a panel of SW480 cells, in which the endogenous p53 mutant (R273H/P309S) can be inducibly knocked down and, at the same time, exogenous p53 mutants containing either G245S or R248W, alone or in combination with alterations in the activation domain (AD) 1 or 2 17, were inducibly expressed at levels comparable to the endogenous p53 mutant (Ref. 17). Replacement of the endogenous mutant p53 with exogenous mutants caused little or no changes in G6PD activity (Supplementary Information, Fig. S4e). By contrast, a p53 mutant with increased thermodynamic stability (N239Y) 18, exhibited enhanced ability to inhibit G6PD (Fig. 4d). Because tumor-associated mutations impair the native conformation of p53 and the N239Y mutation stabilizes it, the inhibition on G6PD is likely attributed to the native conformation of p53.
How may p53 inhibit G6PD activity? G6PD is in an equilibrium of inactive monomer and active dimer 19,20. In both HCT116 and MEF cells, the lack of p53 led to a strong increase in G6PD dimer and a corresponding decrease in G6PD monomer (Fig. 4e). In a transfection assay, p53 reduced the interaction of two differentially tagged G6PD (Flag-G6PD and eGFP-G6PD) in a dose-dependent manner (Fig. 4f). G6PD requires its substrate NADP+ as the cofactor for the formation of holoenzyme, a property that ensures higher G6PD activity and thus more NADPH production when NADPH/NADP+ ratio drops 5. NADP+ diminished the interaction between G6PD and p53 in a dose-dependent manner (Fig. 4g), suggesting that the binding of p53 to G6PD is incompatible with the binding of NADP+ to G6PD. These results suggest that p53 may disrupt the formation of the dimeric G6PD holoenzyme.
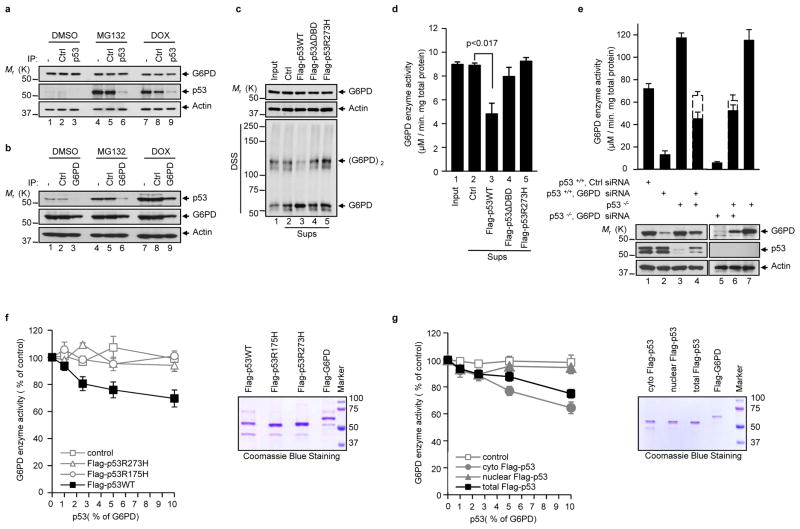
The levels of p53 are kept low in unstressed cells due to its rapid degradation in the proteasome. A semi-quantitative western assay showed the levels of cytoplasmic p53 were approximately 3% that of G6PD. The levels of p53 increased to ~10% that of G6PD when p53 was stabilized by either MG132 or doxorubicin (Supplementary Fig. S5a). Accordingly, the majority of p53 molecules bound to G6PD in unstressed cells, but a small percentage of G6PD bound to p53, as shown by immunodepletion assays (Figs. 5a, b). Only when p53 was stabilized by MG132 and DOX did a significant portion of p53 molecules become separated from G6PD (Fig. 5b).
Figure 5. p53 suppresses G6PD through transient interaction and at substoichiometric ratios.
(a, b) p53+/+ HCT116 cells were treated with DMSO, MG132, and DOX. The cytosolic fraction was immunoprecipitated separately with a control antibody (Ctrl) and anti-p53 (a) or G6PD (b) antibody, plus protein A/G beads. The lysates before (−) and after immunoprecipitation were analyzed by western blot.
(c, d) Lysates from p53−/− MEF cells were first incubated with Flag-tagged p53 proteins immobilized on beads and control beads, and then were separated from the beads. (c) Levels of proteins (top and middle) and G6PD dimerization (bottom) in the input lysates (Input) and supernatant after being incubated with the beads (Sups). (d) G6PD activity in the input lysates and sups. Data in d are means ± S.D. (n=3).
(e) p53+/+ and p53−/− HCT116 cells were either treated separately with G6PD siRNA and a control (Ctr) siRNA, or untreated. Filled columns: actual G6PD activity in the 1:1 mixtures of the indicated lysates (lanes 4 and 6) and in individual lysates (the other lanes). Dashed columns: expected G6PD activity in mixtures based on the averages of the individual lysates. Protein expression is shown at the bottom. The samples were derived from the same conditions. Data are means ± S.D. (n=3).
(f, g) Left: Activity of G6PD after being incubated with total wild type and mutant p53 proteins (f) or cytosolic, nuclear, and total wild type p53 (g). p53 proteins were used at molar ratios of 1, 2.5, 5, and 10% that of G6PD. Right: Coomassie blue staining of purified proteins. Data are means ± S.D. (n=3).
The discrepancy between the amounts of p53 that stably bind to G6PD and the strong effect of p53 on overall G6PD activity raises the possibility that p53 is capable of inhibiting G6PD activity through transient interaction and at sub-stoichiometric ratios. To test whether p53 can inhibit G6PD via transient interaction, we incubated lysates of p53−/− MEF cells with recombinant p53 immobilized on beads and then separated the lysates from the beads. The treated lysates contained virtually all of the G6PD protein and un-detectable amounts of p53 (Fig. 5c, and Supplementary Information, Fig. S5b), but showed much reduced G6PD activity (~50%) and low levels of G6PD dimer (Fig. 5c, d). In the same experiment, un-conjugated control beads and beads conjugated with p53ΔDBD- or p53 R273H protein all failed to inhibit G6PD activity and G6PD dimerization.
To assess whether one p53 molecule can inhibit multiple G6PD molecules, we first mixed extracts of p53+/+ HCT116 cells treated with G6PD siRNA (with low G6PD, but normal p53 level) with extracts of p53−/− HCT116 cells (with normal G6PD level, but no p53). The G6PD activity in the mix was substantially lower than the calculated average of these two extracts (Fig. 5e, column 4), suggesting that the small amounts of p53 protein in the former extracts could significantly inhibit the activity of a much larger amount of G6PD in the latter extracts. In a control experiment, when p53+/+ HCT116 extracts were replaced with p53−/− HCT116 extracts, the G6PD activity in the mix was approximately the average of the two extracts that were used, as expected (Fig. 5e, column 6).
We next mixed purified recombinant proteins at low molar ratios of p53 versus G6PD. At a ratio of 2.5% (approximately the ratio of these proteins in unstressed cells, Supplementary Fig. S5a), p53 decreased G6PD activity by 20% (Fig. 5f), suggesting that one p53 molecule could inactivate up to eight G6PD molecules. This was likely an underestimate because of the lability of p53 protein. The inhibition efficiency decreased at higher ratios of p53 versus G6PD, but increased at a lower molar ratio (Fig. 5f). In contrast, R273H and R175H mutants failed to inhibit G6PD at any ratio examined (Fig. 5f). Altogether, these results suggest that wild type p53 may act as a catalyst to inactive G6PD.
To evaluate cytoplasmic and nuclear pools of p53 in the inhibition of G6PD, we purified p53 proteins from these cellular compartments. Cytoplasmic p53 exhibited robust inhibitory activity towards G6PD. However, nuclear p53 protein showed minimal activity (Fig. 5g). In the cell lysates, cytoplasmic p53 was mainly in monomeric form, while nuclear p53 was mostly in tetrameric form (Supplementary Information, Fig. S5c). This difference remained even after these proteins were purified and adjusted to the same concentration (Supplementary Information, Fig. S5d). Therefore, the cytoplasmic and nuclear p53 proteins appear to be intrinsically different in their oligomerization status and their ability to inhibit G6PD.
The current study identifies an important role for p53 in regulating G6PD. Through this regulation, p53 exerts a powerful surveillance on the metabolic pathways that are critical for both glucose catabolism and biosynthesis. The suppression of G6PD by p53 is evident in unstressed cells and is independent of transcription. Because the majority of cytoplasmic p53 associates with G6PD in unstressed cells (Figs. 5a, b), inhibiting G6PD is likely a main function of cytoplasmic p53 in these cells. In stressed cells where p53 is stabilized, a portion of cytoplasmic p53 becomes free of G6PD and may perform other functions previously attributed to cytoplasmic p53, such as the interaction with Bcl-2 family proteins 21. Notably, p53 can inhibit G6PD through transient interactions and at levels much lower than that of G6PD, suggesting that p53 may act as a catalyst to induce conformational changes in G6PD. This function is attributed to the entire p53 protein, perhaps with the exception of the tetramerization domain, and appears to be specific to the p53 proteins originated from the cytoplasm, but not the nucleus. p53-mediated inhibition of the PPP likely dominates the effect of TIGAR, a previously identified p53 target gene that stimulates the PPP 22; this is shown by the enhanced PPP flux and NADPH production in p53 deficient cells. Given the importance of p53 in suppressing the PPP, the prevalent inactivation of p53 in tumor cells likely accelerates glucose consumption, and, at the same time, directs glucose for rapid production of macromolecules via an increase in the PPP flux. Therefore, p53 inactivation not only contributes to the Warburg effect but also links it to enhanced biosynthesis.
Methods
Antibodies and reagents
Antibodies against the following proteins/epitopes were used with the sources and dilution ratios indicated in parentheses: GFP (Clontech, Palo Alto, CA; 1:4000); p53 (DO-1, Oncogene, Manhasset, NY and Santa Cruz Biotechnology, Santa Cruz, CA; 1:1000); S. cerevisae G6PD and Flag (M2) (Sigma, St Louis, MO); human G6PD (Abcam, Cambridge, UK; 1:4000 and Sigma, St Louis, MO; 1:2000). The following reagents were purchased from Sigma: Flag peptide, glutathione reductase, L. mesenteroides and S. cerevisiae G6PD proteins, dehydroepiandrosterone (DHEA), glucose-6-phosphate, 6-phosphogluconate, NADP+, insulin, rosiglitazone, dexamethasone, isobutylmethylxanthine, doxorubicin and [2-13C]glucose.
Plasmids
The DNA fragments corresponding to the full-length G6PD and p53 were amplified by polymerase chain reaction (PCR) from Marathon-Ready HeLa cDNA (Clontech), and cloned into p3xFlag-myc-CMV-24 (Sigma) and pEGFP-C1 (Clontech). The pGEX-2TK-p53 plasmid was kindly provided by Jörg Kobarg. (Centro de Biologia Molecular Estrutural, Laboratório Nacional de Luz Síncrotron, Rua Giuseppe Máximo Scolfaro 10.000, CP 6192, Campinas, SP, CEP 13084-971, Brazil.) p53 proteins tagged with a single Flag epitope were constructed in pRK5 (Ref. 23). For expressing p53 in yeast, p53 was fused to the Gal4 DNA-binding domain in pGBKT7 and to the E. coli DNA binding protein LexA in pGlida (Clontech). For expressing p53 in bacteria, p53 was N-terminally tagged with a Flag epitope in pET28(a). All deletion and point mutations were made by PCR and confirmed by DNA sequencing.
Analysis of the PPP and glycolysis in cells
The flux through the oxidative branch of the PPP and glycolysis was assessed with carbon-13 nuclear magnetic resonance spectrometry as described previously 24. p53+/+ and p53−/− HCT116 were cultured in triplicate 10 cm plates for three days to approximately 60-70% confluence. After being washed with medium containing no glucose, the cells were incubated with medium containing 10 mM [2-13C]glucose for 9 h. The rates of total glucose consumption and lactate formation were determined with a bench top analyzer (Nova Biomedical, Waltham, MA). Incorporation of 13C in carbon 2 and 3 of lactate is resulted from glucose metabolism via glycolysis and oxidative PPP, respectively. To determine the incorporation of 13C into lactate these carbon positions, the final medium from each culture was analyzed in a 20-mm NMR tube with a 9.4 Tesla spectrometer (Varian, Palo Alto, CA) at 100.66 MHz. A 90° excitation pulse was applied every 6 seconds (fully relaxed), with broadband decoupling used only during 13C data acquisition (no NOE enhancement). Spectra were acquired with 32768 points, a spectral width of 25000 Hz and 3000 excitations. Free induction decays were apodized with exponential multiplication (1 Hz line broadening). The ratio of 13C in carbons 2 (reflecting glycolytic flux) and 3 (reflecting oxidative PPP flux) of lactate and the rate of glucose consumption were used to estimate the oxidative PPP flux.
Glucose consumption and lactate production
Cells were seeded in culture plates and cultured for 6 h. The culture medium was then changed and cells were incubated for an additional 15 h. Glucose levels in the culture medium were measured using the Glucose (GO) assay kit (Sigma). Lactate levels were determined using a Lactate Assay kit (Eton Bioscience Inc).
Cell culture and gene knockdown with shRNA and siRNA
Cells were maintained in standard culture conditions. p53−/− and p53+/+ MEFs were established from embryos of the corresponding mice generated by crossing p53 heterozygote mice (B6.129S2-Trp53tm1Tyj/J, The Jackson Laboratory, Bar Harbor, ME). G6PD siRNAs were purchased from Invitrogen (Catalog No. HSS103891 and HSS103892). G6PD shRNAs were purchased from Open Biosystem. The target sequence for shG6PD #1 (Catalog No. RHS3979-9595543) is 5′-GTCGTCCTCTATGTGGAGAAT-3′, and the target sequence for shG6PD #4 (Catalog No. RHS3979-9595544) is 5′-CAACAGATACAAGAACGTGAA-3′. Expression plasmid for p53 shRNA was made in a vector that co-expresses GFP (kindly provided by David C. Chan, California Institute of Technology, Pasadena, CA, USA.). The targeted sequence for p53 is: 5′-GACTCCAGTGGTAATCTAC-3′ (Ref. 25). siRNAs and shRNA plasmids were transfected into cells using Lipofectamine 2000 (Invitrogen). Stable shRNA transfectants were selected in medium containing puromycin (1 μg/ml) and pooled after culturing for 3–4 weeks.
G6PD enzyme activity
G6PD enzyme activity was determined as described 26. First, the combined activity of G6PD and 6-phosphogluconate dehydrogenase (6PGD), the second enzyme of the PPP that also produces NADPH, was measured by the rate of conversion of NADP+ to NADPH in the presence of glucose-6-phosphate (G6P). The activity of 6PGD alone was then measured by the conversion of NADP+ to NADPH in the presence of 6-phosphogluconate (6PG). G6PD activity was calculated as the difference of these two activities. Cell lysates were added to the reaction buffer containing 50 mM Tris, 1 mM MgCl2, pH 8.1. Substrate concentrations used were: G6P and 6PG (200 μM), and NADP+ (100 μM). Enzyme activities were normalized based on protein concentration, which was determined by a BCA assay kit (Pierce). The data is expressed in arbitrary absorption units.
Immunoprecipitation and indirect immunofluorescence
Immunoprecipitation was performed as described previously 27. Briefly, cell lysates were made by sonication in buffer containing 20 mM HEPES (pH 7.8), 400 mM KCl, 5% Glycerol, 5 mM EDTA, 0.4% NP40, and protease inhibitors, and pre-cleared by centrifugation. The supernatants were then diluted in 20 mM Hepes (pH 7.8), 50 mM KCl, 5% glycerol, 2.5 mM MgCl2, 0.05% NP40 and incubated with indicated protein A/G-Sepharose bound antibodies for 4 h at 4°C. Beads were pre-blocked with 2% BSA in PBS and 0.05% 3-[(3-Cholamidopropyl)-dimethylammonio]-1-propane sulfonate (CHAPS) before immunoprecipitation. After incubation, beads were washed twice with lysis buffer, four times with ice-cold PBS and boiled in 2×loading buffer. Protein samples were resolved by SDS-PAGE.
The indirect immunofluorescence was performed as described previously 23,28. Briefly, cells cultured on coverslips were treated separately with DMSO and etoposide (20 μM) for 24 h. Cells were fixed in 4% paraformaldehyde for 10 min, permeabilized with 0.1% Triton X-100 for 5 min, blocked with 5% BSA and incubated with antibodies as indicated, followed by a Texas-red conjugated anti-rabbit IgG and a FITC-conjugated anti-mouse IgG antibody. The cells were mounted with DAPI-containing medium (Vector Laboratories, Burlingame, CA) and the images were acquired with a confocal microscope.
Protein purification
Flag-tagged G6PD and p53 were expressed in H1299 cells or 293T cells. When indicated, cell lysates were separated into cytosolic and nuclear fractions. Proteins were purified using M2 beads as previously described 23,29. For purifying Flag-G6PD, 10 μM NADP+ was included in the lysis buffer. GST-p53 were expressed in DH5α cells after induction with 2mM IPTG at 37 °C, and purified by glutathione beads (Amersham Biosciences, Piscataway, NJ).
Surface plasmon resonance analysis of p53-G6PD interaction
Surface plasmon resonance analysis was performed at 25 °C with a Pharmacia Biosensor BiAcore 3000 instrument (BiAcore, Uppsala, Sweden) according to the manufacturer’s instruction. S. cerevisae G6PD was immobilized on the surface of a CM5 sensorchip in 10mM sodium acetate buffer (pH4.5) and resulted in 8000 response units. A reference surface was used as a blank to correct for instrumental and buffer effects without protein injection. The amount of protein bound to the sensor chip was monitored by the change in refractive. Purified Flag-p53 was diluted, and ran across each sensor surface at five different concentrations in a running buffer of PBS at a flow-rate of 30μl/min. Flag-p53 protein concentrations were 0.03–0.9 μM. Dissociation constants from five serial dilutions of each protein and other kinetic parameters were calculated by using the BIA Evaluation software.
NADPH level
NADPH levels were determined as described previously 30. Cell lysates were prepared in buffer containing 0.1 M Tris-HCl, pH 8.0, 0.01 M EDTA, and 0.05% (v/v) Triton X-100, sonicated and centrifuged at (2,4000g) at 4 °C. The total levels of NADPH plus NADH in the lysates were assayed by spectrometry (Beckman DU 640 spectrophotometer, Beckman Coulter, Inc., Fullerton, CA) at a wavelength of 341 nm. The levels of NADPH in the lysates were determined based on the decrease in absorbance at 341 nm after NADH was converted to NAD by glutathione reductase.
Lipid synthesis
Lipid accumulation was measured using Oil Red O staining as described 12. Briefly, cells were grown in medium supplemented with insulin (2 μM), dexamethasone (1 μM), and isobutylmethylxanthine (0.25 mM) for 2 days, and in medium supplemented with insulin and rosiglitazone (10−7 M) for an additional 5 days. The medium was changed every other day. Afterwards, cells were fixed with 10% buffered formalin for 16 h at 4 °C and were stained with a filtered Oil Red O solution (0.5% Oil red O in isopropyl alcohol) for 2h at room temperature.
Analysis of p53+/+ and p53−/− mice
Four-week-old p53+/+ and p53−/− mice (B6.129S2-Trp53tm1Tyj/J) were obtained from the Model Animal Research Center (Nanjing University, China) and were maintained on a regular diet. Tissues were excised for the measurement of the G6PD activity and NADPH level. Liver tissues were fixed in 10% formalin and paraffin-embedded. Sections were prepared and stained with haematoxylin and eosin. All animal experiments were performed in accordance with the local Animal Care and Use Committee.
Statistical Analysis
Statistical significance was analyzed by Student’s test and expressed as p value.
Supplementary Material
Acknowledgments
We thank W. Xie for isolating p53+/+ and p53−/− MEFs; X. Chen for SW480 cells; B. Vogelstein and W. El-Deiry for HCT116 cells; J. Cross, N. Li, J. Wu, Y. Mei, A. Stonestrom, W. Tan, H. Liu, Y. Hao, X. Zhao, and Z. Lou for technical assistance; C.B. Thompson and J. Delikatny for helpful comments; and A. Stonestrom and E. Thompson for help with manuscript preparation.
Supported by the China National Natural Science Foundation (31030046), the Ministry of Science and Technology (2010CB912804 and 2011CB966302), and the Chinese Academy of Sciences (KSCX1-YW-R-57) to M.W, and the U.S. National Institutes of Health (CA088868 and GM060911) and the Department of Defense (W81XWH-07-1-0336 and W81XWH-10-1-0468) to X.Y.
Footnotes
Author Contributions: P.J., W.D., M.W., and X.Y. designed the experiments and interpreted results. P.J. and W.D. performed all the experiments except those mentioned below. X.W. performed the experiments on G6PD activity in yeast, the surface plasmon resonance, and lipid droplets in mouse liver. A.M. and P.J. analyzed the oxidative PPP flux. X.G. supplied the p53 wild type and knockout mice. X.Y. wrote the manuscript with the help of P.J. and W.D.
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Warburg O, Posener K, Negelein E. Ueber den Stoffwechsel der Tumoren. Biochem Z. 1924;152:319–344. [Google Scholar]
- 2.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 3.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berg JM, Tymoczko JL, Stryer L. Biochemistry. 2007:577–589. [Google Scholar]
- 6.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 7.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 8.Kondoh H, et al. Glycolytic enzymes can modulate cellular life span. Cancer Res. 2005;65:177–185. [PubMed] [Google Scholar]
- 9.Matoba S, et al. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 10.Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 11.Bunz F, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 12.Tseng YH, et al. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature. 2008;454:1000–1004. doi: 10.1038/nature07221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komarov PG, et al. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999;285:1733–1737. doi: 10.1126/science.285.5434.1733. [DOI] [PubMed] [Google Scholar]
- 14.Inga A, Resnick MA. Novel human p53 mutations that are toxic to yeast can enhance transactivation of specific promoters and reactivate tumor p53 mutants. Oncogene. 2001;20:3409–3419. doi: 10.1038/sj.onc.1204457. [DOI] [PubMed] [Google Scholar]
- 15.Freedman DA, Levine AJ. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol Cell Biol. 1998;18:7288–7293. doi: 10.1128/mcb.18.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stommel JM, et al. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. Embo J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan W, Chen X. Characterization of functional domains necessary for mutant p53 gain of function. J Biol Chem. 2010;285:14229–14238. doi: 10.1074/jbc.M109.097253. M109.097253 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nikolova PV, Henckel J, Lane DP, Fersht AR. Semirational design of active tumor suppressor p53 DNA binding domain with enhanced stability. Proc Natl Acad Sci U S A. 1998;95:14675–14680. doi: 10.1073/pnas.95.25.14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Au SW, Gover S, Lam VM, Adams MJ. Human glucose-6-phosphate dehydrogenase: the crystal structure reveals a structural NADP(+) molecule and provides insights into enzyme deficiency. Structure. 2000;8:293–303. doi: 10.1016/s0969-2126(00)00104-0. [DOI] [PubMed] [Google Scholar]
- 20.Roos D, et al. Molecular basis and enzymatic properties of glucose 6-phosphate dehydrogenase volendam, leading to chronic nonspherocytic anemia, granulocyte dysfunction, and increased susceptibility to infections. Blood. 1999;94:2955–2962. [PubMed] [Google Scholar]
- 21.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. nature07986 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bensaad K, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 23.Tang J, et al. Critical role for Daxx in regulating Mdm2. Nat Cell Biol. 2006;8:855–862. doi: 10.1038/ncb1442. [DOI] [PubMed] [Google Scholar]
- 24.Mancuso A, Sharfstein ST, Tucker SN, Clark DS, Blanch HW. Examination of primary metabolic pathways in a murine hybridoma with carbon-13 nuclear magnetic resonance spectroscopy. Biotechnol Bioeng. 1994;44:563–585. doi: 10.1002/bit.260440504. [DOI] [PubMed] [Google Scholar]
- 25.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 26.Tian WN, et al. Importance of glucose-6-phosphate dehydrogenase activity for cell growth. J Biol Chem. 1998;273:10609–10617. doi: 10.1074/jbc.273.17.10609. [DOI] [PubMed] [Google Scholar]
- 27.Adorno M, et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 28.Du W, et al. Suppression of p53 activity by Siva1. Cell Death Differ. 2009;16:1493–1504. doi: 10.1038/cdd.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang J, et al. A novel transcription regulatory complex containing death domain-associated protein and the ATR-X syndrome protein. J Biol Chem. 2004;279:20369–20377. doi: 10.1074/jbc.M401321200. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Z, Yu J, Stanton RC. A method for determination of pyridine nucleotides using a single extract. Anal Biochem. 2000;285:163–167. doi: 10.1006/abio.2000.4701. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.