Abstract
Aspergillus species have the ability to produce a wide range of secondary metabolites including polyketides that are generated by multi-domain polyketide synthases (PKSs). Recent biochemical studies using dissected single or multiple domains from PKSs have provided deep insight into how these PKSs control the structural outcome. Moreover, the recent genome sequencing of several species has greatly facilitated the understanding of the biosynthetic pathways for these secondary metabolites. In this review, we will highlight the current knowledge regarding polyketide biosynthesis in Aspergillus based on the domain architecture of non-reducing, highly reducing, and partially reducing PKSs, and PKS-non-ribosomal peptide synthetases.
Keywords: Secondary metabolites, Fungi, Polyketide, Nonribosomal peptides
Introduction
Polyketides are structurally diverse secondary metabolites including polyphenols, polyenes, and macrolides. These natural products are produced by polyketide synthases (PKSs) from a variety of organisms including bacteria, fungi, and plants (Fischbach and Walsh 2006; Hertweck 2009). PKSs are categorized into three subtypes: types I, II, and III PKSs. As in the nomenclature of fatty acid synthases (FASs), type I PKSs refers to covalently linked multifunctional enzymes, whereas the catalytic components in type II PKS are free-standing. Furthermore, type III PKSs are homodimeric multi-functional enzymes distinguished from types I and II by the use of malonyl-CoA rather than malonyl-S-pantetheinyl-thiolation domain species as substrates (Austin and Noel 2003). Apart from these three subtypes, PKSs are also categorized as iterative or noniterative, distinguished by whether the substrate is used for one or more than one round of elongation.
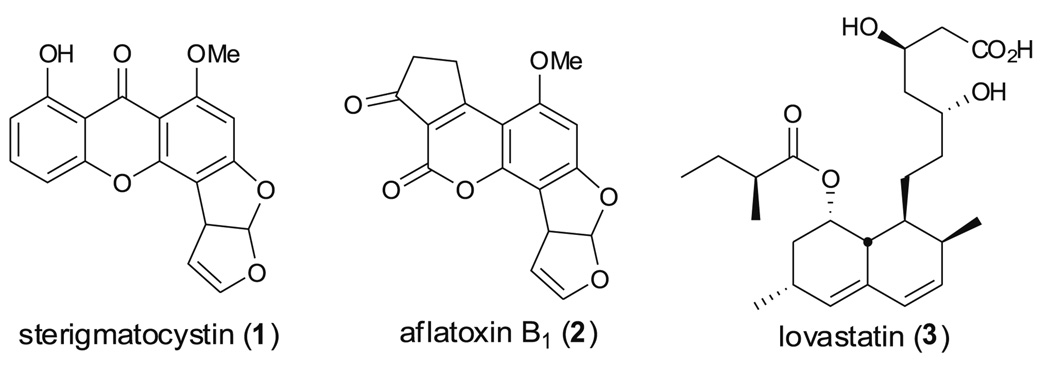
Polyketides produced from fungal species have drawn much attention due to their important role in human health. These compounds include toxins such as sterigmatocystin (1) and aflatoxin B1 (2); the virulence factor melanin; and the medicinally important anticholesterol drug lovastatin (3) (Fig. 1). Assembly of these fungal polyketide carbon skeletons are catalyzed by multi-domain type I polyketide synthases (PKSs) and all known fungal PKSs so far are iterative in nature except lovastatin diketide synthase (LovF) and its homolog compactin diketide synthase (MlcB) which are noniterative (vide infra and Abe et al. 2002). Recent fungal genome projects of Aspergillus species has revealed that these microorganisms have a surprisingly large number of secondary metabolite genes, the products of which are mostly currently unknown (Galagan et al. 2005). The sequencing of fungal genomes also revealed that biosynthesis genes of secondary metabolites in fungi are generally clustered (Galagan et al. 2005; Keller et al. 2005). For reasons still not well understood, it seems that most biosynthetic gene clusters are either silent or expressed at very low levels, and that their products are, thus, difficult to detect or identify. It appears that some chemical or environmental signals for triggering these “cryptic” pathways are missing in standard laboratory culture conditions (Bergmann et al. 2007; Chiang et al. 2008; Schroeckh et al. 2009). Finding the correct conditions to unlock cryptic natural product clusters will be important to decoding the complexity of the fungal secondary metabolome. Since reviews describing how to activate cryptic biosynthetic pathways have been published elsewhere (Chiang et al. 2009a; Scherlach and Hertweck 2009), this review will focus on the relationship of PKS domain architecture and the structure of the product of the PKS. Three excellent reviews concerning fungal PKSs have been published by Schumann and Hertweck; Cox; and Hoffmeister and Keller (Cox 2007; Hoffmeister and Keller 2007; Schumann and Hertweck 2006). We will try to extend previous understanding and highlight the most current knowledge of type I PKSs in Aspergillus species.
Fig. 1.
Three important secondary metabolites from Aspergillus
General domain architecture and reaction mechanisms of fungal PKSs
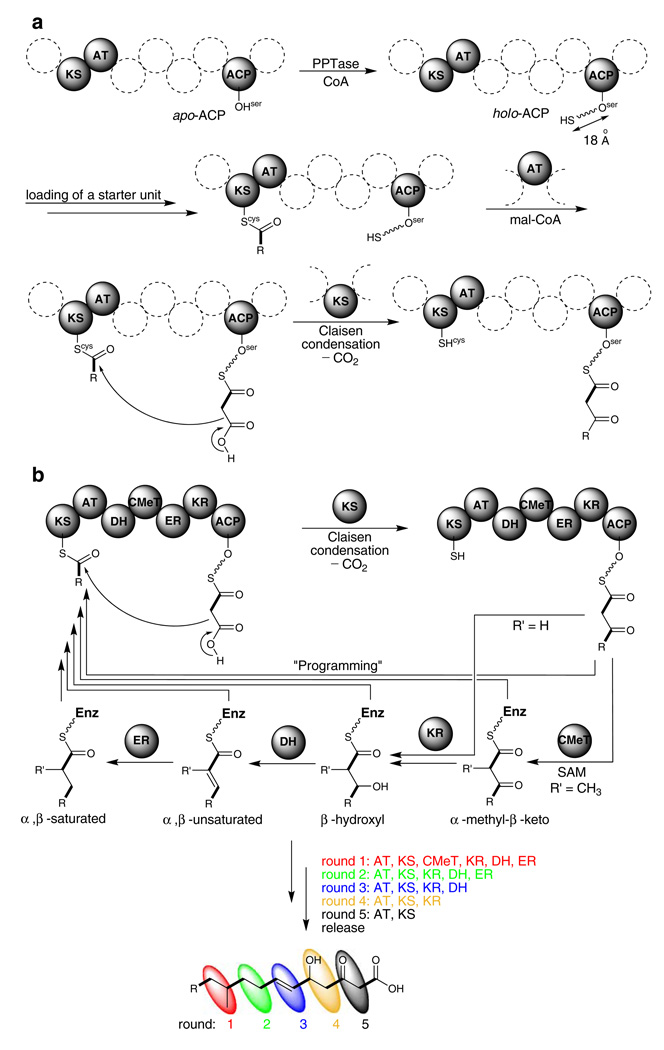
Type I multi-domain PKSs use substrates such as acetyl-CoA and malonyl-CoA (mal-CoA) in a similar fashion to multi-domain FASs but differ in the existence of incomplete reduction and/or dehydration steps. Table 1 lists the characterized polyketide genes and their cognate proteins, as well as domain organizations from Aspergillus species. These PKSs minimally contain ketosynthase (KS), acyl transferase (AT) and acyl carrier protein (ACP) domains (Fig. 2a). After translation, the ACP domain of PKSs requires post-translational modification through the addition of a phosphopantetheinyl group to the conserved Ser by phosphopantetheinyl transferase (PPTase). This generates a holo-ACP with an approximate 18 Å phosphopantetheine (PPT) arm from an apo-ACP. A single PPTase (CfwA/NpgA) has been identified and likely to be responsible for the activation of all PKSs in A. nidulans (Marquez-Fernandez et al. 2007). A malonyl extender unit is then transferred onto the terminal thiol of the phosphopantetheine prosthetic group of ACP, a reaction that is mostly catalyzed by the AT domain. In the presence of an acyl starter unit on a conserved cysteine residue of the KS domain, the KS catalyzes Claisen condensation of malonyl extender to generate a diketide. There are two major subclasses of fungal type I PKSs. They are non-reducing (NR) and highly reducing (HR) PKSs distinguished basically by the existence of domains that can catalyze the reduction and/or dehydration steps (Table 1). In HR-PKSs, the α-carbon of the diketide may be methylated to generate α-methyl-β-ketothioester by the C-methyltrasferase (CMeT) domain which uses S-adenosylmethionine (SAM) as a substrate. In addition, the β-keto group may further be reduced to β-hydroxyl, α,β-unsaturated carbonyl, or α,β-saturated carbonyl functionalities by the catalytic activities of keto reductase (KR), dehydratase (DH), and enoyl reductase (ER) domains (Fig. 2b). The processed diketide can then transfer back to the KS and be extended further. After several rounds of elongation, the polyketide chain is released from the PKS. Polyketide structural diversity is determined by the optionally “programmed” activity of CMeT, KR, DH, and ER domains during each round of extension. However, how those multi-functional enzymes control the product chain length and the site of “programming” is largely unknown.
Table 1.
Characterized multi-domain type I PKSs from Aspergillus
| Origin | Gene | Protein | No. of aad | Secondary metabolite | Domain architecturee | Ref. |
|---|---|---|---|---|---|---|
| NR-PKS | ||||||
| A. parasiticus | pksA | PksA (NSAS) | 2109 | Aflatoxin B1 (2) | SAT-KS-AT-PT-ACP-TE/CLC | (Chang et al. 1995; Trail et al. 1995) |
| A. parasiticus | pksL1 | PKSL1 (NSAS) | 2109 | Aflatoxin B1 (2) | SAT-KS-AT-PT-ACP-TE/CLC | (Feng and Leonard 1995) |
| A. nidulans | pksST, stcA | StcA (NSAS) | 2181 | Sterigmatocystin (1) | SAT-KS-AT-PT-ACP-ACP-TE/CLC | (Brown et al. 1996; Yu and Leonard 1995) |
| A. nidulans | wAb | WAb | 1986 | Isocoumarins (7–9) | SAT-KS-AT-PT-ACP-ACP | (Mayorga and Timberlake 1992; Watanabe et al. 1998) |
| A. nidulans | wA | WA | 2157 | YWA1 (10) | SAT-KS-AT-PT-ACP-ACP-TE/CLC | (Watanabe et al. 1999) |
| A. fumigatus | alb1 | Alb1p | 2146 | YWA1 (10) | SAT-KS-AT-PT-ACP-ACP-TE/CLC | (Tsai et al. 1998, 1999; Watanabe et al. 2000) |
| A. fumigatus | pksP | PksP | 2146 | YWA1 (10) | SAT-KS-AT-PT-ACP-ACP-TE/CLC | (Langfelder et al. 1998) |
| C. lagenariuma | PKS1 | PKS1 | 2187 | T4HN (11) | SAT-KS-AT-PT-ACP-ACP-TE/CLC | (Fujii et al. 1999, 2000; Takano et al. 1995) |
| A. nidulans | aptA | AptA | 1792 | Asperthecin (21) | SAT-KS-AT-PT-ACP | (Szewczyk et al. 2008) |
| A. nidulans | mdpG | MdpG | 1806 | Monodictyphenone (24) | SAT-KS-AT-PT-ACP | (Bok et al. 2009) |
| A. nidulans | orsA | OrsA | 2103 | Orsellinic acid (27) and F9775 (25 and 26) | SAT-KS-AT-PT-ACP-ACP-TE | (Bok et al. 2009; Sanchez et al. 2010; Schroeckh et al. 2009) |
| A. nidulans | afoE | AfoE | 2793 | Asperfuranone (30) | SAT-KS-AT-PT-ACP-CMeT-R | (Chiang et al. 2009b) |
| Acremonium strictuma | ASpks1 | MOS | 2729 | 3-Methylorcinaldehyde (32) | SAT-KS-AT-PT-ACP-CMeT-R | (Bailey et al. 2007) |
| HR-PKS | ||||||
| A. terreus | lovB | LovB | 3038 | Lovastatin (3) | KS-AT-DH-CMeT-(ER)-KR-ACP-C | (Hendrickson et al. 1999; Kennedy et al. 1999) |
| A. terreus | lovF | LovF | 2532 | Lovastatin (3) | KS-AT-DH-CMeT-ER-KR-ACP | (Kennedy et al. 1999) |
| A. nidulans | easB | EasB | 2534 | Emericellamides (35–39) | KS-AT-DH-CMeT-ER-KR-ACP | (Chiang et al. 2008) |
| A. nidulans | afoG | AfoG | 2527 | Asperfuranone (30) | KS-AT-DH-CMeT-ER-KR-ACP | (Chiang et al. 2009b) |
| Phoma sp. C2932a | PhPKS1 | SQTKS | 2714 | Squalestatin tetraketide (40) | KS-AT-DH-CMeT-ER-KR-ACP | (Cox et al. 2004) |
| Alternaria solania | alt5 | PKSN | 2551 | Alternapyrone (41) | KS-AT-DH-CMeT-ER-KR-ACP | (Fujii et al. 2005) |
| Alternaria solania | pksF | PKSF | 2260 | Aslanipyrone (42) and aslaniol (43) | KS-AT-DH-ER-KR-ACP | (Kasahara et al. 2006) |
| A. ochraceus | pksc | PKSc | 502 | Ochratoxin A (44) | (O’Callaghan et al. 2003) | |
| A. westerdijkiae | aoks1c | AoKS1c | 654 | Ochratoxin A (44) | (Bacha et al. 2009a) | |
| PR-PKS | ||||||
| A. terreus | atX | MSAS | 1803 | 6-MSA (46) | KS-AT-DH-Core-KR-ACP | (Fujii et al. 1996) |
| A. westerdijkiae | aomsas | MSAS | 1766 | 6-MSA (46), asperlactone (47) and isoasperlactone (48) | KS-AT-DH-Core-KR-ACP | (Bacha et al. 2009b) |
| PKS-NRPS | ||||||
| A. oryzae | cpaA | CpaA | 3907 | CPA (49) | KS-AT-(DH)-(CMeT)-(ER)-(KR)-ACP-C-A-PCP-DKC | (Tokuoka et al. 2008) |
| A. flavus | cpaA | CpaA | 3906 | CPA (49) | KS-AT-(DH)-(CMeT)-(ER)-(KR)-ACP-C-A-PCP-DKC | (Chang et al. 2009; Seshime et al. 2009) |
| Beauveria bassianaa | tenS | TENS | 4239 | Tenellin (51) | KS-AT-DH-CMeT-(ER)-KR-ACP-C-A-PCP-DKC | (Eley et al. 2007; Halo et al. 2008) |
| A. nidulans | apdA | ApdA | 3930 | Aspyridones (56 and 57) | KS-AT-DH-CMeT-(ER)-KR-ACP-C-A-PCP-DKC | (Bergmann et al. 2007) |
| A. fumigatus | psoA | PsoA | 3991 | Pseurotin A (59) | KS-AT-DH-CMeT-KR-ACP-C-A-PCP-R | (Maiya et al. 2007) |
Identified by heterologous expression in A. oryzae M-2-3
67 aa C-terminal truncated WA
Partial DNA sequences
Predicted no. of aa if studied from genome mining
Domains in parenthesis are non-functional
Fig. 2.
General reaction mechanisms catalyzed by iterative fungal PKSs. a Minimal fungal PKS containing KS, AT, and ACP domains. b Programming of CMeT, KR, DH, and ER domains in HR-PKS
Two major subclasses of type I PKSs were also observed by Kroken et al. who performed phylogenetic analyses on amino acid sequences of KS domains of characterized fungal PKS genes and putative PKS genes from genomic sequences (Kroken et al. 2003). They showed that fungal NR-and HR-PKSs are the two main clades of microbial type I PKSs. They also proposed that NR-PKSs evolved by losing DH-ER-KR domains while gaining the TE/CLC (thioesterase/Claisen cyclase) domain from their “reducing” PKSs ancestors. Moreover, the Townsend group has recently identified SAT (starter unit-ACP transacylase) and PT (product template) domains that control the structural outcome of NR-PKSs. We will describe their elegant work before going on to the next section.
Discovery of SAT and PT domains
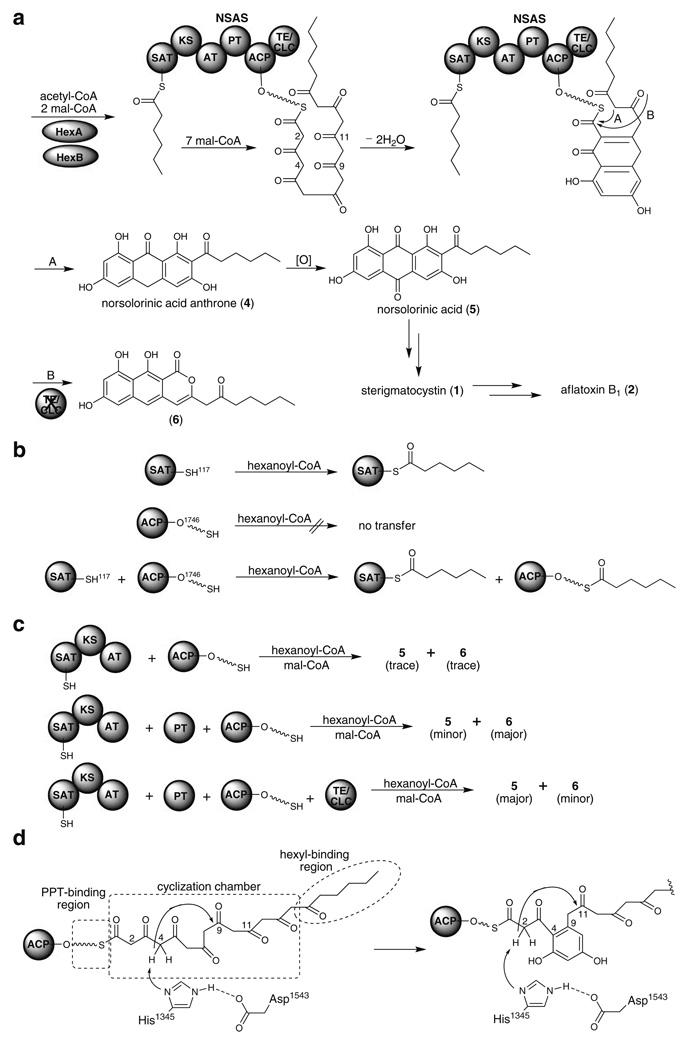
Fungal PKSs are large multi-domain proteins which make overexpression, functional purification and accurate examination of individual reactions difficult. In order to dissect the function of individual domains, Udwary, Merski, and Townsend developed an algorithm, UMA (Udwary-Merski algorithm) that combines primary sequence similarity, predicted secondary structure, and local hydrophobicity to predict interdomainal linker regions (Udwary et al. 2002). By using UMA, they identified two previously unidentified domains, SAT and PT domains, from the norsolorinic acid synthase (NSAS) family of NR-PKSs (Table 1). NSAS is a family of octaketide synthases that catalyze the assembling of a hexanoyl starter unit with seven mal-CoA followed by dehydration/aromatization and Claisen cyclization to generate norsolorinic acid anthrone (4) (Fig. 3a). It was found that 4 oxidized to norsolorinic acid (5) spontaneously during workup, although this oxidation might be catalyzed enzymatically in vivo, as is known to occur in emodin biosynthesis (Chen et al. 1995). After purifying different domains as recombinant proteins from E. coli, the Townsend group showed that hexanoyl-CoA could be transferred to the SAT domain directly but could only be transferred to the ACP domain in the presence of the SAT domain (Fig. 3b) (Crawford et al. 2008a). Ma et al. also noticed that the AT domain of NSAS could only transfer malonate from mal-CoA to ACP but not hexanoate or acetate from the cognate CoA (Ma et al. 2006). Thus, the SAT domain located in the N terminus of NSAS with high similarity to the mal-CoA/ACP transferase domain is responsible for selecting the hexanoate starter unit. The same group further aligned SAT domains of 76 NSAS family PKSs identified from Blastp searches of the NCBI non-redundant protein sequence database and realized that SAT domains are widespread among fungal NR-PKSs (Crawford et al. 2008b). Forty-nine protein sequences harbored the expected GXCXG active-site motif in the SAT domain, where the active site Cys tethers to the starter unit. Twelve sequences contained a GXSXG motif, which presumably use oxyester rather than thioester chemistry for transfering the starter unit. Fourteen sequences had a GXGXG motif, which might be inactive or have an unknown function. Interestingly, the residue directly upstream of the active site His in the catalytic dyad of SAT domain contains a conserved Ala for the known hexanoyl acceptors and has either a bulky Phe or Tyr for the known acetyl acceptors. Thus, the SAT domains might use a steric approach to “selecting” the acyl starter unit.
Fig. 3.
SAT and PT domains of NSAS in aflatoxin B1 biosynthetic pathway. a Biosynthetic pathway of norsolorinic acid (5), sterigmatosystin (1), and aflatoxin B1 (2) (pathway A); and naphthopyrone (6) formation in the absence of TE/CLC domain (pathway B). b In vitro hexanoyl transfer activity in the presence of SAT, ACP, or SAT and ACP monodomains. c Products detected after reconstituting different domain (s) of NSAS in vitro. d Proposed cyclization mechanism catalyzed by the NSAS PT domain
The PT domain which is located between the AT and ACP domains has a critical role in specific aldol cyclization and aromatization. Without the PT domain, the SAT-KS-AT tridomain and the holo-ACP monodomain in NSAS can still produce polyketides of the correct chain length (5 and 6) but in very small amounts (Fig. 3c). However, the products markedly increase in the presence of the PT domain that drives the irreversible dehydration/aromatization reaction (Crawford et al. 2008a). Without the TE/CLC monodomain, 5 is the minor product and naphthopyrone 6 is the major product (Fig. 3a, pathway B). In the presence of all domains including the TE/CLC monodomain, 5 becomes the major product, supporting the idea that TE/CLC functions as a Claisen cyclase (Fig. 3a, pathway A). The crystal structure of the NSAS PT monodomain has recently been solved and displays a distinct “double hot dog” (DHD) fold (Crawford et al. 2009). The rigid octaketide binding pocket in the PT domain extends 30 Å from the surface to the bottom and can be divided into three regions; the PPT-binding region, cyclization chamber, and hexyl-binding region (Fig. 3d). The PPT-binding region extends 14 Å from the protein surface into the pocket and is proposed to bind the ~ 18 Å PPT arm. The cyclization chamber (8×13.5 Å) can accommodate two aromatic rings and contains the proposed catalytic dyad His1345 and Asp1543. The hydrophobic hexyl-binding region (6×6 Å) lies at the bottom and is perfectly adapted to accept the substrate hexanoyl starter unit.
NR-PKSs containing the TE/CLC releasing domain
Aflatoxins are a group of potent environmental toxins and carcinogens produced by Aspergillus species and their biosynthesis has drawn much attention. Chang et al. and Feng et al. identified the PKS gene from A. parasiticus responsible for aflatoxin biosynthesis (the same gene was given different designations, pksA and pksL1, by the two groups) (Chang et al. 1995; Feng and Leonard 1995). Yu et al. identified pksST from A. nidulans required for sterigmatocystin (1) biosynthesis (Yu and Leonard 1995), originally designated pksST but renamed stcA by Brown et al. in order to simplify the nomenclature and reflect the fact that genes required for a specific secondary metabolite are usually clustered (Brown et al. 1996). The PKS responsible for sterigmatocystin (1) and aflatoxin biosynthesis, NSAS as discussed above, contains SAT, KS, AT, PT, ACP, and TE/CLC domains (Fig. 3). It is interesting to note that StcA contains two ACP domains (Table 1). NSAS forms a complex with HexA/HexB (α and β components of fungal FAS) and produces norsolorinic acid (5) in the presence of acetyl-CoA, mal-CoA, and NADPH. Watanabe et al. showed that hexanoyl-CoA is not a free intermediate produced by the NSAS/HexA/HexB complex suggesting that hexanoate must be passed directly to the SAT domain of NSAS after being synthesized by HexA/HexB (Watanabe and Townsend 2002).
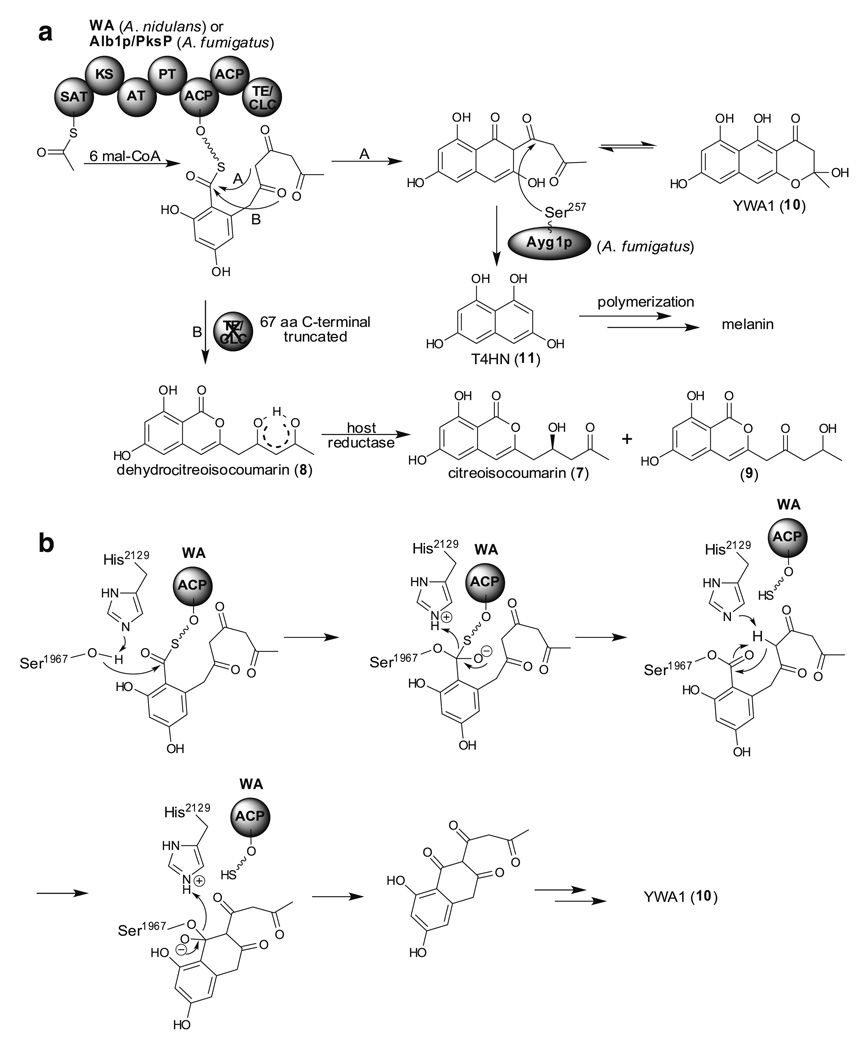
Many fungi produce melanins, which are dark brown or black pigments formed by oxidative polymerization of phenolic compounds. Melanin is known to contribute to survival, cell wall rigidity and impermeability, and most importantly, is a virulence factor for pathogenic fungi (Gomez and Nosanchuk 2003). In contrast to melanin biosynthesis in brown and black fungi, little is known about pigment biosynthesis in green and bluish-green fungi. In A. nidulans, wA gene was identified to encode a PKS required of green pigment biosynthesis (Mayorga and Timberlake 1992). Watanabe et al. expressed the wA gene in A. oryzae strain M-2-3 and this led to the isolation of citreoisocoumarin (7), and its derivatives dehydrocitreoisocoumarin (8) and compound 9, thus establishing that WA is a heptaketide synthase (Fig. 4a, pathway B) (Watanabe et al. 1998). Since there is no KR domain in WA, the secondary alcohol in the side chain of 7 and 9 is presumably reduced by a host reductase. However, it was later realized that the wA gene originally identified was missing one base pair thus caused a frame shift and produced a 67 aa C-terminal truncated protein in their heterologous expression experiment (Fig. 4a, pathway B). The full length WA was later heterologously expressed and naphthopyrone YWA1 (10) was re-identified to be the real metabolite (Fig. 4a, pathway A) (Watanabe et al. 1999). This suggested that SAT, KS, AT, PT, and two ACP domains of WA produce a heptaketide and catalyze the first aromatic ring formation, and the C-terminal region of WA is involved in the cyclization of the second aromatic ring of YWA1 (10). Thus, the Ebizuka group first demonstrated that the C terminus of WA turns out to function as a Claisen cyclase (CLC) by using a heterologous expression system. A detailed functional analysis by a series of C-terminal deletions and site-directed mutagenesis showed that Ser1967 and His2129 residues in CLC domain of WA are responsible for naphthopyrone (10) formation and this leads to the proposed mechanism shown in Fig. 4b. Another feature of WA is that it contains two ACP domains. Site-directed mutagenesis of the Ser residue, a known phosphopantetheine anchor site, to Cys in either ACP domain produced YWA1 (10). This suggests that only a single ACP is necessary to catalyze the heptaketide formation (Fujii et al. 2001).
Fig. 4.
TE/CLC containing NR-PKSs involved in melanin biosynthesis. a Biosynthetic pathway of YWA1 (10), T4HN (11), and melanin (pathway A); and isocoumarin formation in the absence of TE/CLC domain (pathway B). b Proposed mechanism of Claisen cyclization by TE/CLC domain of WA
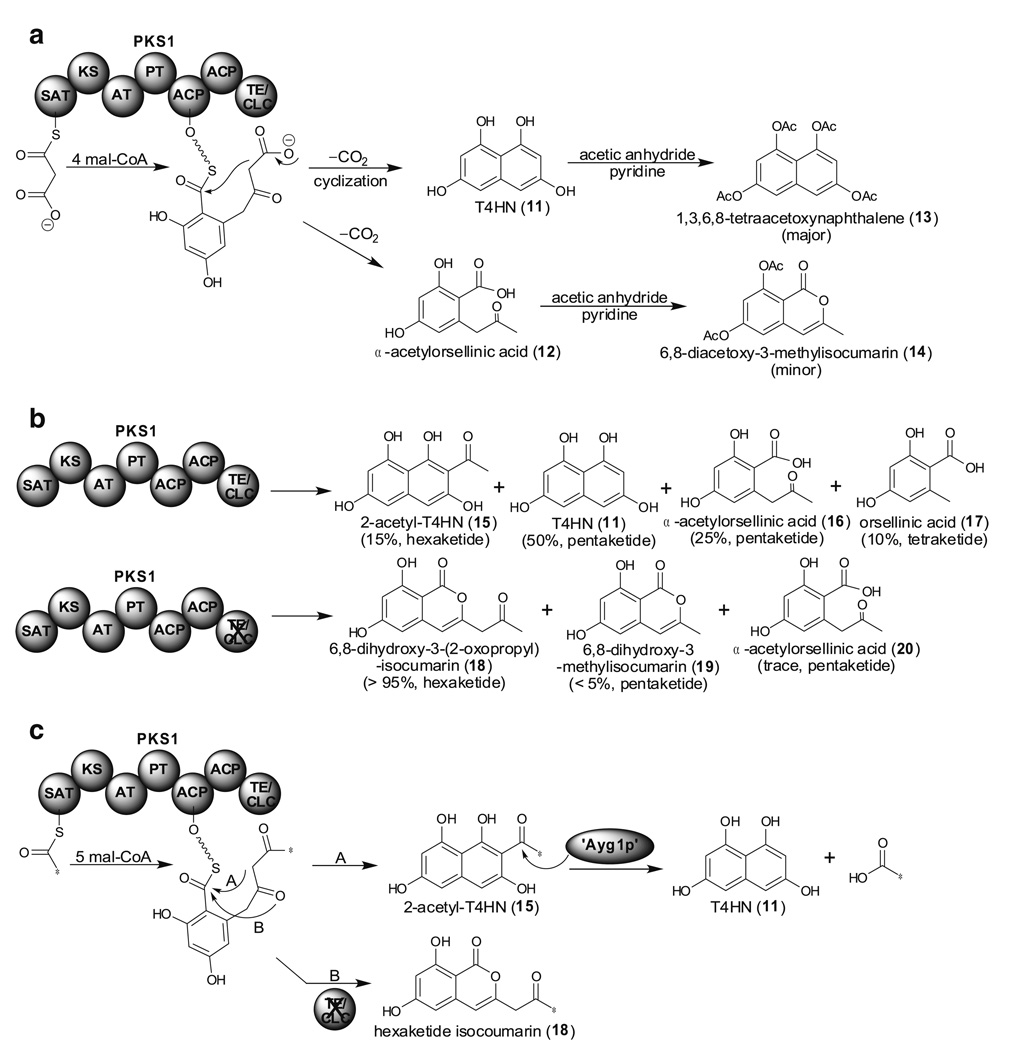
A pentaketide, 1,3,6,8-tetrahydroxynaphthalene (T4HN, 11), is an initial precursor of melanin. In 1998, Tsai et al. and Langfelder et al. identified the PKS gene from A. fumigatus necessary for melanin biosynthesis (the same gene was designated alb1 and pksP by the two groups) (Langfelder et al. 1998; Tsai et al. 1998). Heterologous expression of the alb1 gene in A. oryzae M-2-3 resulted in the isolation of a heptaketide YWA1 (10) but not the pentaketide T4HN (11) (Watanabe et al. 2000). It was later realized that an enzyme, Ayg1p, catalyzed chain-length shortening to produce T4HN (11) (Fig. 4a, pathway A) (Fujii et al. 2004; Tsai et al. 2001). Notably, another PKS gene, PKS1, responsible for melanin biosynthesis was identified from Colletotrichum lagenarium in 1995 (Takano et al. 1995). When PKS1 was heterologously expressed in A. oryzae M-2-3, a major pentaketide T4HN (11) together with other minor pentaketides including α-acetylorsellinic acid (12) were isolated as their acetylated derivatives, 13 and 14, after acetylation in order to avoid oxidative polymerization (Fig. 5a) (Fujii et al. 1999). When isotope-labeled acetyl-CoA or mal-CoA was fed into a cell-free extract containing PKS1, only mal-CoA was able to incorporate into T4HN (11) and this led to the hypothesis that mal-CoA but not acetyl-CoA serves as the starter unit (Fig. 5a) (Fujii et al. 2000). The detailed product distribution of this heterologous expression system was later revisited by using LC-ESI-MS (liquid chromatography-electrospray ionization-mass spectrometry) (Fig. 5b) (Watanabe and Ebizuka 2004). Interestingly, when the TE/CLC domain was inactivated, PKS1 produced mainly 6,8-dihydroxy-3-(2-oxopropyl)-isocumarin (18), a hexaketide. The authors thus concluded that the TE/CLC domain has a critical role in chain-length determination. However, Crawford et al. demonstrated that when recombinant SAT domain of PKS1 was purified from E. coli, it was able to accept acetyl-CoA as a starter unit (Crawford et al. 2008c). Crawford et al. thus proposed an alternative mechanism, that PKS1 incorporates one acetyl-CoA and five mal-CoA to generate 2-acetyl-T4HN (15) which is later shortened by an ‘Ayg1p’ like enzyme in the heterologous host (Fig. 5c). This alternative mechanism also fulfills the observed phenomenon that isotope labeled acetyl-CoA is not incorporated into T4HN since it has been cleaved by the ‘Ayg1p’ like enzyme to become acetic acid. As a consequence, both chain-length control and dehydration/aromatization control would be carried out prior to the TE/CLC catalyzed Claisen cyclization. This provides a general pattern for iterative systems that contain SAT domains and a biochemical rationale for the classical acetyl “starter unit effect”. The function of each domain will promise to become clearer as data are accumulated in future experiments.
Fig. 5.
Two different proposed T4HN biosynthetic pathways catalyzed by PKS1. a Proposed biosynthetic pathway using mal-CoA as a starter unit and metabolites isolated from hetrologous expression of PKS1 in A. oryzae M-2-3. b Revisit the product distribution produced by wild type and TE/CLC less PKS1 by LC-ESI-MS. c Alternative proposed mechanism of T4HN (11) formation using acetyl-CoA as a starter unit. *: 14C-labeled carbon form acetyl-CoA
NR-PKSs with no releasing domain
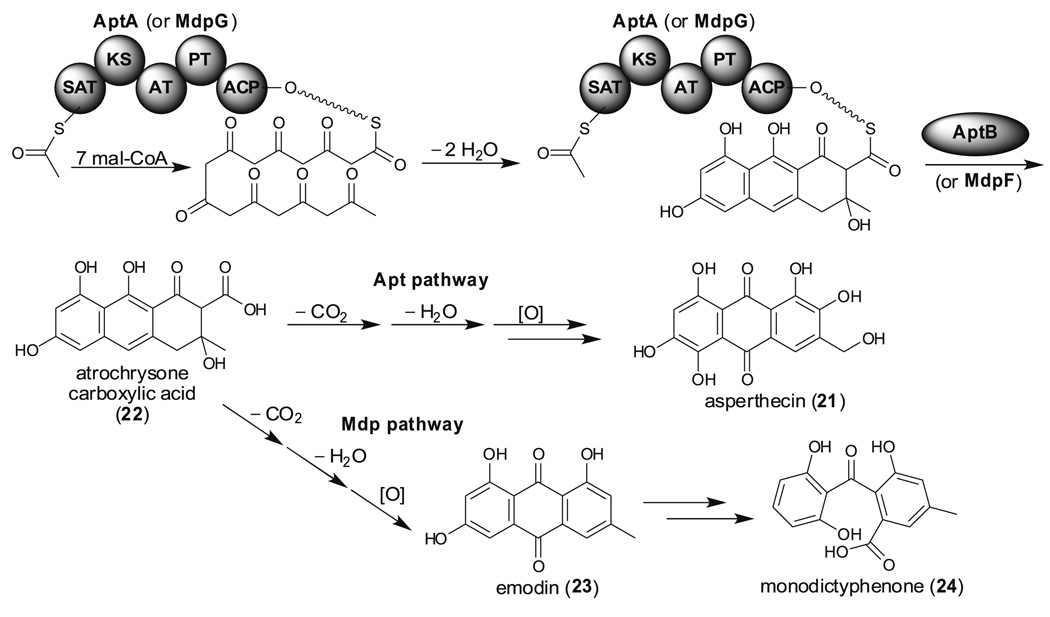
Since most biosynthetic gene clusters are cryptic, we were curious whether SUMOylation might play a part in secondary metabolite regulation. Deletion of the sumo gene in A. nidulans dramatically increases asperthecin (21) production and this led us to identify the apt gene cluster through a series of targeted deletions (Fig. 6, Apt pathway) (Szewczyk et al. 2008). AptA is a TE/CLC less NR-PKS that catalyzes octaketide formation from one acetyl-CoA and seven mal-CoA. The lack of a TE/CLC domain in AptA and the presence, near aptA in the genome, of a β-lactamase, aptB, which is also essential for asperthecin biosynthesis led us to propose that AptB is responsible for releasing the octaketide from the NR-PKS, AptA. This hypothesis has recently been validated biochemically by Awakawa et al. who showed that atrochrysone carboxylic acid (22) is an unstable metabolite released from ACAS (atrochrysone carboxylic acid synthase) in the presence of ACTE (atrochrysone carboxyl ACP thioesterase, a β-lactamase-like protein) (Awakawa et al. 2009). Subsequent decarboxylation, dehydration, and oxidation at various positions resulted in the formation of 21.
Fig. 6.
TE less NR-PKSs involved in asperthecin (21, Apt pathway) and monodictyphenone (24, Mdp pathway) biosynthesis
In response to the accruing evidence that a significant portion of the regulation of the expression of secondary metabolite gene clusters occurs at the chromatin level (Cichewicz 2010), we deleted the A. nidulans cclA gene, an ortholog of the bre2 gene of Saccharomyces cerevisiae (Bok et al. 2009). Bre2 encodes a critical member of the COMPASS (complex associated with Set1), a conserved eukaryotic transcriptional effector both facilitating and repressing chromatin-mediated processes through methylation of lysine 4 of histone H3 (Mueller et al. 2006; Sims and Reinberg 2006). Chemical profiling of a cclA deletant followed by genetic analysis led us to identify two silent gene clusters, one involved in monodictyphenone biosynthesis (Fig. 6, Mdp pathway) and the other required for F-9775 biosynthesis (vide infra). MdpG which has high similarity with AptA is also a TE/CLC less NR-PKS. MdpF, a β-lactamase-type TE nearby, thus presumably catalyzes product release. Subsequent decarboxylation, dehydration, and oxidation generate emodin (23) which is further processed by downstream enzymes to produce monodictyphenone (24).
NR-PKS containing the TE releasing domain
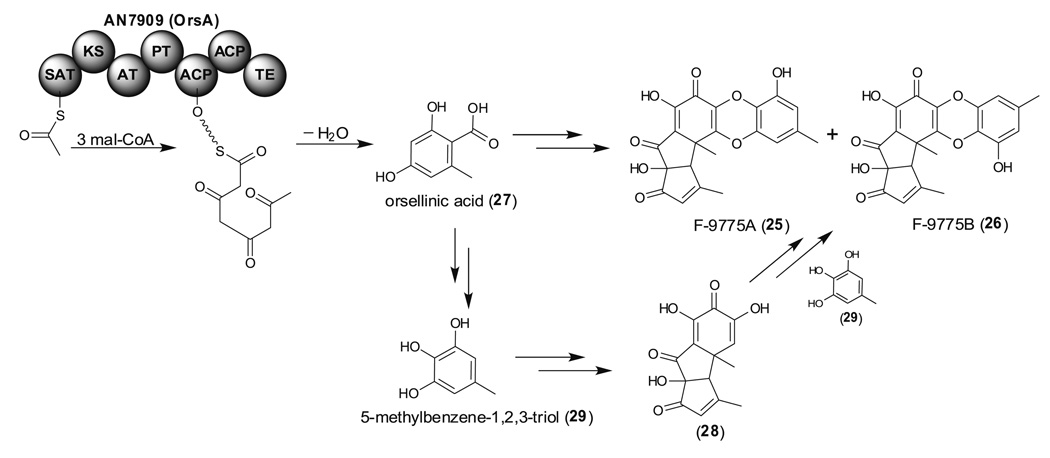
In addition to MdpG, another PKS, AN7909, responsible for F-9775 biosynthesis has also been identified from a cclA deletion strain (Bok et al. 2009). Notably, F-9775A and B (25 and 26) together with orsellinic acid (27) can be induced by growth in Czapek's media even in a wild-type cclA background and deletion of AN7909 not only eliminates 25 and 26 but also 27 production (Fig. 7) (Sanchez et al.). These results together with the structures of F-9775A and B, suggested that these compounds are derived from orsellinic acid (27). Interestingly, a theacitrin derivative (28) has been proposed to be generated from two molecules of a pyrogallol derivative (29) after oxidative dimerization, intramolecular cyclization and rearrangement (Davis et al. 1997). F-9775A and B, thus, could be biosynthesized from three molecules of 27. Remarkably, co-cultivated A. nidulans with the bacterium Streptomyces hygroscopicus also resulted in the activation of AN7909 and the production of 25–27 (Schroeckh et al. 2009). Both approaches demonstrated that AN7909 encodes a long-searched for orsellinic acid synthase, OrsA.
Fig. 7.
Proposed F9775 (25 and 26) and orsellinic acid (27) biosynthetic pathway catalyzed by TE containing NR-PKS, OrsA
NR-PKSs containing the reductase releasing domain
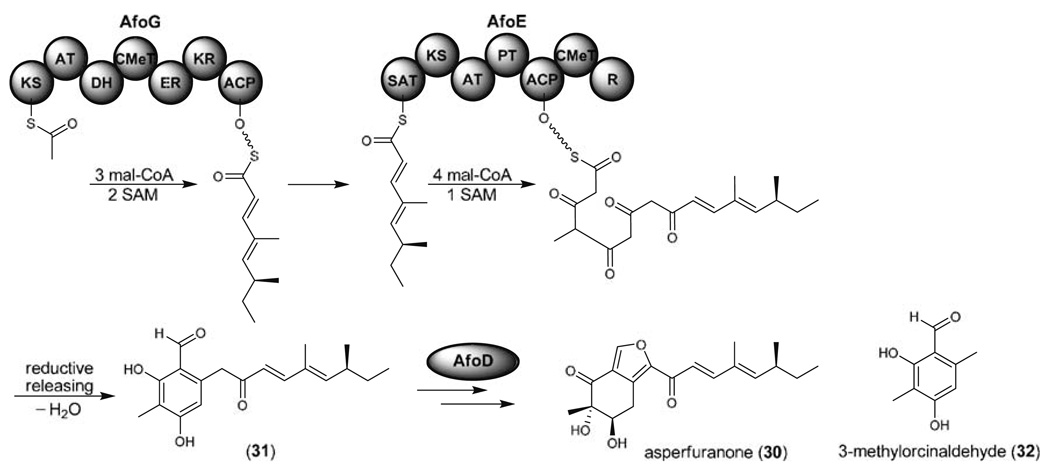
The presence of cluster-specific regulatory activators is a common feature of fungal secondary metabolite biosynthetic gene clusters (Chiang et al. 2009a). Bergmann et al. first demonstrated that ectopic overexpression of apdR, a regulatory gene within a cryptic hybrid PKS-non-ribosomal peptide synthetase (NRPS) gene cluster, resulted in the concerted activation of the gene cluster and production of two new cytotoxic metabolites (Fig. 13c, vide infra). Using a similar approach, the endogenous promoter of a regulatory gene within the afo gene cluster was replaced with an inducible promoter (Chiang et al. 2009b). This led us to discover a new compound asperfuranone (30) and two PKSs, one HR-PKS (AfoG) and one NR-PKS (AfoE), necessary for asperfuranone biosynthesis (Fig. 8). Deletion of a key hydroxylase (afoD) in the asperfuranone biosynthetic pathway resulted in the accumulation of benzaldehyde derivative 31 suggesting that AfoG is responsible for catalyzing the biosynthesis of a tetraketide and then transfering the reduced tetraketide to the SAT domain of AfoE to generate the reduced and non-reduced hybrid octaketide. Interestingly, the C-terminal region of AfoE contains a reductase (R) domain and this matches with the reductive release mechanism to generate aldehyde 31. The reductive release mechanism was first described by the Cox group who isolated 3-methylorcinaldehyde (32) by heterologous expression of 3-methylorcinaldehyde synthase (MOS) in A. oryzae M-2-3 (Bailey et al. 2007).
Fig. 13.
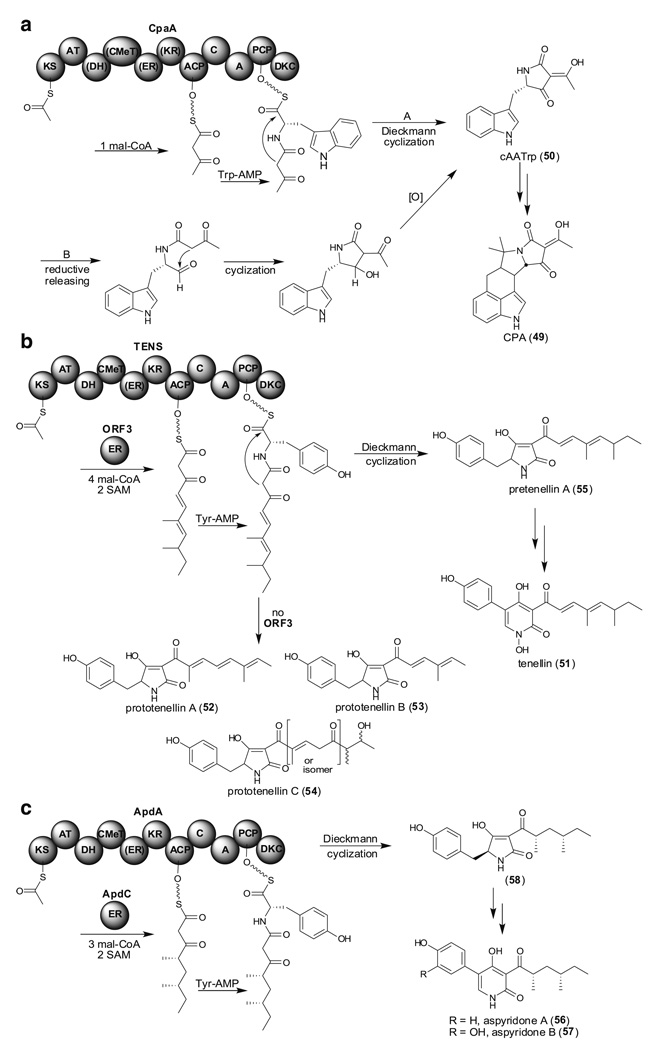
Fungal PKS-NRPSs containing the Dieckman cyclase (DKC) releasing domain. a Two possible product release mechanisms in cAATrp (50) formation in the CPA (49) biosynthetic pathway. Dieckman cyclization/releasing (pathway A) and reductive releasing/cyclization/reoxidation (pathway B). b Major compounds isolated from A. oryzae M-2-3 transformed to express tenS (52–54) or to co-express tenS and orf3 (55). c Proposed biosynthesis and production of aspyridones (56 and 57) after activation of the regulator in apd gene cluster
Fig. 8.
Biosynthetic pathway of asperfuraone (30) mediated by one HR-PKS (AfoG) and one NR-PKS (AfoE) that containing the reductase (R) releasing domain
HR-PKSs
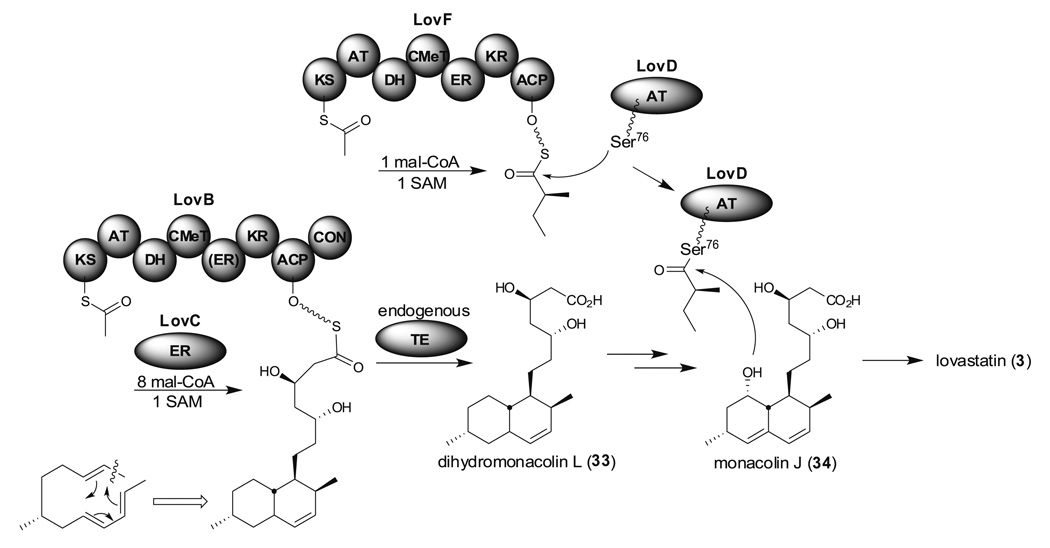
HR-PKSs found in filamentous fungi are the most interesting but the least understood of the PKS family members. Lovastatin (3, Mevacor, Merck) is a cholesterol lowering drug produced by A. terreus. It is also a precursor of a more potent anticholesterol drug simvastatin (Zocor, Merck) which was the second largest selling cholesterol lowering drug in the world before losing US patent protection (Maggon 2005). The biosynthesis of lovastatin has been extensively reviewed by Hill and Cox (Cox 2007; Hill 2006). Two HR-PKSs, LovB (lovastatin nonaketide sythases) and LovF, are involved in the formation of the lovastatin core structure. LovB and LovF both contain KS, AT, DH, CMeT, ER, KR, and ACP domains (Fig. 9). Unlike NR-PKSs, HR-PKSs analyzed to date do not contain SAT domains. It is likely that fungal HR-PKSs initiate polyketide synthesis through decarboxylation of malonyl-ACP or direct transfer of an acetyl unit, but with less efficiency, by the AT domain (Ma and Tang 2007). Interestingly, the ER domain of LovB is nonfunctional and needs to be compensated by the free-standing ER protein, LovC, to produce dihydromonacolin L (33). There is a condensation (CON) domain in the C terminus of LovB and this domain is hypothesized to be involved in product release. In vitro studies of LovB have been hampered by difficulties in purifying sufficient amounts of functional LovB from either A. terreus or a heterologous Aspergillus host. Recently, the Vederas and Tang groups were able to purify the 335-kD LovB by using an efficient expression system from an engineered Saccharomyces cerevisiae strain, BJ5464-NpgA, which contains a chromosomal copy of the A. nidulans PPTase gene (npgA) (Ma et al. 2009). Reconstitution experiments indicated that the CON domain is not involved in product release but does play a crucial role in the formation of 33. This also suggested that other endogenous TEs might be involved in product release (Fig. 9). LovF is the only known noniterative fungal PKS identified from Aspergillus so far. It has similar domain architecture to LovB but there is no CON domain. The Tang group investigated the mechanism of product release from LovF and found that LovD, an acyl transferase, interacts with LovF and transfers (S)-2-methylbutyrate to monacolin J (34) to produce 3 (Xie et al. 2009).
Fig. 9.
Lovastatin (3) biosynthesis mediated by one iterative HR-PKS (LovB) and one noniterative HR-PKS (LovF)
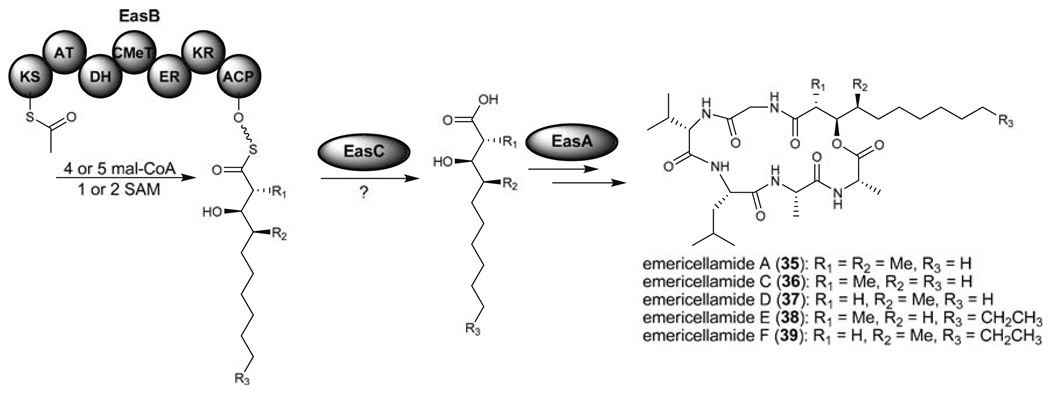
In the course of screening for NRPS gene deletion mutants, EasB was identified to be involved in the biosynthesis of the side chains of emericellamides (35–39) (Fig. 10) (Chiang et al. 2008). Interestingly, the lack of a product release domain in EasB as well as the necessity of a free-standing acyl transferase (EasC) for emericellamide biosynthesis nearby EasB in the genome might indicate that EasC is involved in the product release. The long chain fatty acids released from EasB then incorporate into EasA, an NRPS, to generate 35–39. Interestingly, when comparing EasA with AfoG, the HR-PKS responsible for asperfuranone biosynthesis (Fig. 8), AfoG does not have a product release domain. Notably, the absence of a free-standing acyl transferase in the afo gene cluster suggests that the tetraketide synthesized by AfoG might transfer directly to the SAT domain of AfoE.
Fig. 10.
Proposed functions of EasB (HR-PKS), EasC (AT), and EasA (NRPS) involved in the biosynthesis of emericellamides (35–39)
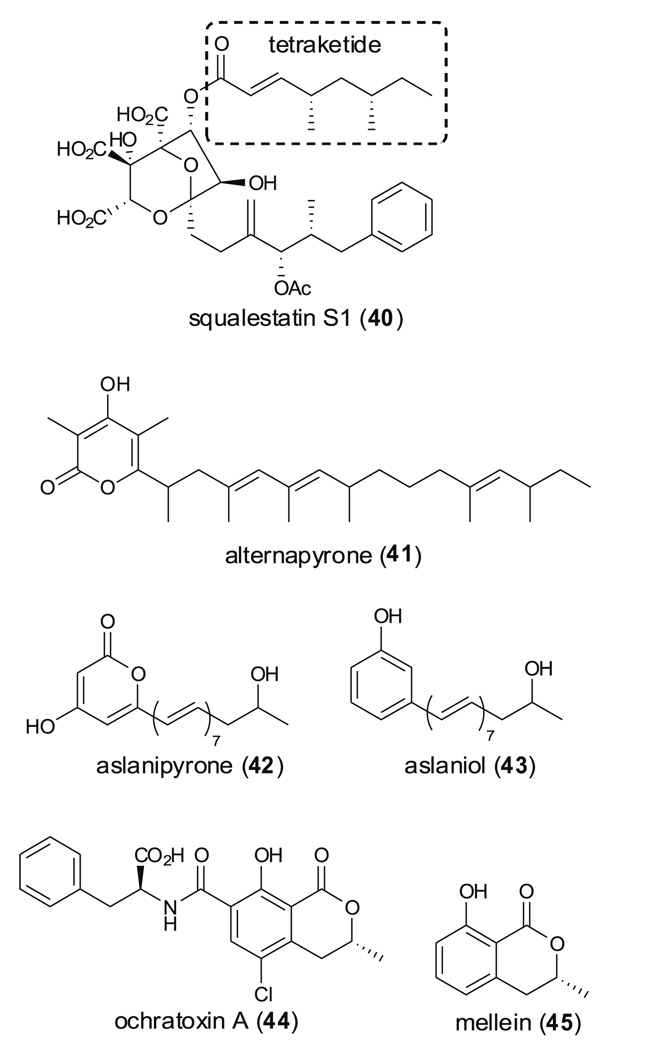
Critical insights by the Cox and Fujii groups have been obtained from heterologous expression of HR-PKSs using A. oryzae as a host. Examples include the tetraketide side chain of squalestatin S1 (40) biosynthesized by SQTKS (squalestatin tetraketide synthase) from Phoma sp. C2932; alternapyrone (41) catalyzed by PKSN; and aslanipyrone (42) and aslaniol (43) biosynthesized by PKSF from Alternaria solani (Table 1 and Fig. 11) (Cox et al. 2004; Fujii et al. 2005; Kasahara et al. 2006). Partial sequences of two different PKS genes, pks and aoks1 involved in ochratosin A (44) biosynthesis, have been obtained by O'Callaghan et al. and Bacha et al.; and their roles in ochratosin A biosynthesis have been demonstrated through disruption of these genes (Bacha et al. 2009a; O'Callaghan et al. 2003). In combination, these studies indicate that two different PKSs are involved in the biosynthesis of 44. Bacha et al. also showed that disruption of apks1 did not abolish mullein (45) production arguing against the previous hypothesis that mullein (45) is a precursor of 44.
Fig. 11.
Chemical structures of compounds 40–45 synthesized by fungal HR-PKSs
Partially reducing PKSs
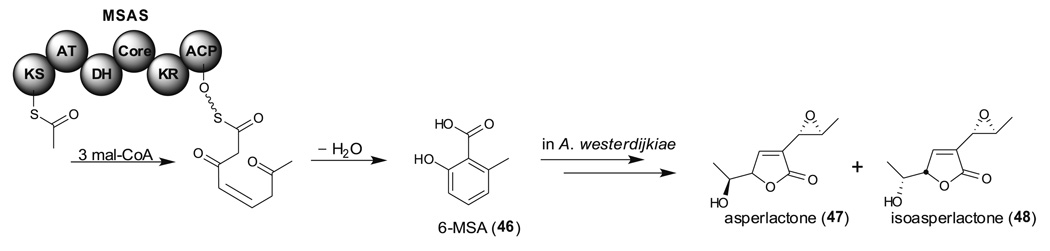
The 6-methylsalicylic acid synthase (MSAS) was the first PKS to be purified from Penicillium patulum (Dimroth et al. 1970) and also the first fungal PKS gene to be sequenced (Beck et al. 1990). By using the KS region of MSAS as a probe, Fujii et al. cloned a PKS gene, atX, from A. terreus (Fujii et al. 1996). Heterologous expression of atX in A. nidulans led to the isolation of 6-methylsalicylic acid (6-MSA, 46) confirming that atX encodes MSAS (Fig. 12). MSAS has been comprehensively reviewed by Cox and it should be note that MSAS does not contain SAT, PT, and TE/CLC domains when compared with NR-PKSs (Cox 2007). It is not clear how the acetyl starter unit loads to the KS domain and how the 6-MSA off-loads from the ACP domain. Recently, the biosynthesis of two potent anti-bacterial and anti-fungal compounds, asperlactone (47) and isoasperlactone (48), identified from A. westerdikiae has been linked to the 6-MSA biosynthesis pathway (Fig. 12) (Bacha et al. 2009b).
Fig. 12.
RP-PKS domain architecture of 6-MSA synthase (MSAS) and the connection of asperlactone (47) and isoasperlactone (48) biosynthesis in A. westerdijkiae
PKS-NRPS hybrids containing the Dieckmann cyclase domain
As discussed above, structural diversity in Aspergillus secondary metabolites is often created by mixing PKSs with FASs, other PKSs, and NRPSs in a single biosynthesis pathway. Many filamentous fungi sequenced to date also contain at least one hybrid PKS-NRPS where a single enzyme contains both PKS and NRPS domains. α-Cyclopiazonic acid (CPA, 49) is a pentacyclic indole tetramic acid mycotoxin produced by several Aspergillus and Penicillium strains. Because the dimethylallyltransferase involved in CPA biosynthesis is located between the aflatoxin biosynthesis gene cluster and the adjacent telomere of A. oryzae, Tokuoka et al. were able to identify cpaA, the PKS-NRPS gene responsible for CPA biosynthesis, by comparing the subtelomeric region of the CPA nonproducing strain A. oryzae RIB40 with that of the CPA-producing strains A. oryzae NBRC4177 and A. flavus NRRL3357 (Tokuoka et al. 2008). They found that NBRC 4177 has an additional 17–18 kb beyond the region corresponding to the telomere repeat in RIB40. The subtelomeric region of RIB40 near the aflatoxin biosynthesis gene cluster encodes only a portion of the 5′ region of the cpaA gene, but not the entire gene, and it could consequently be deduced that cpaA is necessary for CPA biosynthesis. The remaining genes involved in CPA biosynthesis are next to the aflatoxin biosynthesis gene cluster. (Chang et al. 2009; Tokuoka et al. 2008). Heterologous expression of CpaA in A. oryzae M-2-3 led to the isolation of cyclo-acetoacetyl-l-tryptophan (cAATrp, 50) (Fig. 13a) (Seshime et al. 2009). This suggests that the PKS portion of CpaA catalyzes diketide formation and DH, CMeT, ER, and KR domains are nonfunctional. The NRPS portion of CpaA containing condensation (C), adenylation (A), and peptidyl carrier protein (PCP) domains that catalyze the condensation of diketide and Trp. Two possible mechanisms of 50 release by the R domain located on the C terminus of CpaA have been proposed, including Dieckman cyclization (Fig. 13a, pathway A) and reductive releasing/cyclization/reoxidation (Fig. 13a, pathway B). By using recombinant PTP-R didomain, PTP, and R monodomain proteins, the Walsh group demonstrated that the R domain of CpaA does not function as a reductase because the Tyr in Ser-Tyr-Lys catalytic triad for the reduction is mutated to Leu (Liu and Walsh 2009). This provides direct evidence that the C-terminal R domain actually functions as a Dieckmann cyclase (DKC) but not a reductase. They also showed that the Tyr in Ser-Tyr-Lys catalytic triad in the R domain of TENS and ApdA is mutated to Phe. Therefore, Dieckman cyclization is the mechanism of releasing products from TENS and ApdA (vide infra).
TenS (tenellin synthetase) and the adjunct genes involved in tenellin (51) biosynthesis have been identified from the insect pathogenic fungus Beauveria bassiana by using degenerate primers for the CMeT domain (Fig. 13b) (Eley et al. 2007). Heterologous expression of tenS without orf3, encoding a trans ER, in A. oryzae M-2-3 led to the isolation of prototenellins (52–54) (Halo et al. 2008). This validated that the C-terminal domain of TENS does not function as a reductase but, rather, as a DKC. Coexpression of tenS with orf3 led to the production of pretenellin A (55) suggesting that the ER activity is required at the first round of polyketide synthesis to generate a correctly elaborated polyketide.
By using a genomic-driven approach, Bergmann et al. induced aspyridone A (56) and B (57) production via the activation of the apd gene cluster regulator, ApdR, in A. nidulans (Fig. 13c) (Bergmann et al. 2007). Because ApdA lacks a functional ER domain, the free-standing ER, ApdC, is likely involved in the formation of the tetraketide that is then linked to Tyr by the NRPS portion of ApdA. Similar to TENS, a Dieckmann cyclization is likely to occur to generate intermediate 58, which then converts to 56 and 57 after oxidative ring expansion.
PKS-NRPS hybrid containing the reductase domain
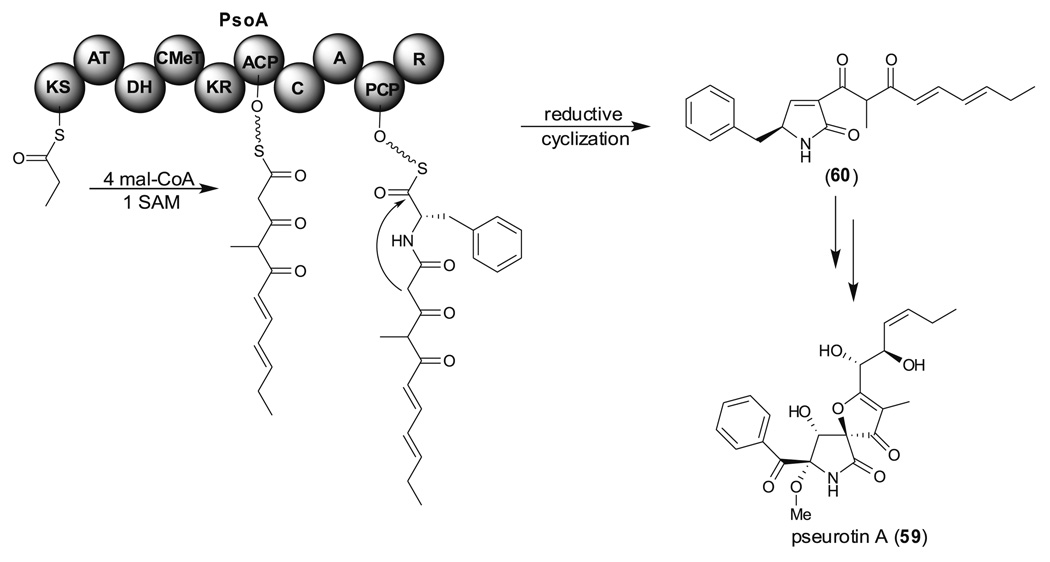
Pseurotin A (59) is a competitive inhibitor of chitin synthase and an inducer of nerve-cell proliferation. Maiya et al. identified a PKS-NRPS, PsoA which is involved in 59 biosynthesis, from A. fumigatus (Fig. 14) (Maiya et al. 2007). The PKS portion of PsoA contains KS, AT, DH, CMeT, KR, and ACP domains that catalyze pentaketide formation, presumably using a propionate starter. The NRPS portion then condenses l-Phe with the pentaketide. The R domain which has correct catalytic triad in the C terminus of PsoA might catalyze a reductive cyclization (intramolecular Knoevenagel condensation) to generate the first putative intermediate, 60, that then serves as a precursor of 59.
Fig. 14.
Proposed function of PsoA, a PKS-NRPS containing the reductase (R) releasing domain, involved in pseurotin A (49) biosynthesis Appl Microbiol Biotechnol
Perspective
Continued advances in genome sequencing and molecular genetic technology will ensure that reverse genetic approaches, i.e., analyzing the chemotype of a specific PKS null mutant, will continue to play a crucial role in elucidating secondary metabolite biosynthesis. Since the complexity in chemical structures seen in secondary metabolites is generated by enzymes and ultimately genes, scientists worldwide are now trying to predict the structure of the secondary metabolites from their cognate genes. A central question is how the “programming”, i.e., chain length determination, sites of reduction and cyclization, are achieved in an “iterative” system. By using heterologous expression systems and/or reconstitution of the catalytic domains, deduced with the UMA algorithm, as individual enzymes, it is now possible to dissect these megasynthases and understand the function of each domain(s). Taking advantage of fungal genome projects and modern molecular genetic techniques, fungal natural product researchers are now entering an exciting new era in understanding the logic of polyketide biosynthesis and in engineering the biosynthetic pathways to produce novel chemical entities. It should be noted that there are no SAT and PT domains in PR- and HR-PKSs, and thus, the basis for their complex programming are unclear. Since fungal genome projects have made the sequences of hundreds of PKS genes available, analyzing PKS architecture or each PKS domain provides an avenue to discover new biosynthetic pathways.
Acknowledgments
The project described was supported by Grants Number PO1GM084077 and R01GM031837 from the National Institute of General Medical Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Contributor Information
Yi-Ming Chiang, Graduate Institute of Pharmaceutical Science, Chia Nan University of Pharmacy and Science, Tainan 71710, Taiwan, Republic of China; Department of Pharmacology and Pharmaceutical Sciences, University of Southern California, School of Pharmacy, 1985 Zonal Avenue, Los Angeles, CA 90089, USA.
Berl R. Oakley, Department of Molecular Biosciences, University of Kansas, 1200 Sunnyside Ave, Lawrence, KS 66045, USA
Nancy P. Keller, Department of Medical Microbiology and Immunology, University of Wisconsin-Madison, Madison, WI 53706, USA Department of Bacteriology, University of Wisconsin-Madison, Madison, WI 53706, USA.
Clay C. C. Wang, Email: clayw@usc.edu, Department of Pharmacology and Pharmaceutical Sciences, University of Southern California, School of Pharmacy, 1985 Zonal Avenue, Los Angeles, CA 90089, USA; Department of Chemistry, University of Southern California, College of Letters, Arts, and Sciences, Los Angeles, CA 90089, USA.
Reference
- Abe Y, Suzuki T, Ono C, Iwamoto K, Hosobuchi M, Yoshikawa H. Molecular cloning and characterization of an ML-236B (compactin) biosynthetic gene cluster in Penicillium citrinum. Mol Genet Genomics. 2002;267:636–646. doi: 10.1007/s00438-002-0697-y. [DOI] [PubMed] [Google Scholar]
- Austin MB, Noel JP. The chalcone synthase superfamily of type III polyketide synthases. Nat Prod Rep. 2003;20:79–110. doi: 10.1039/b100917f. [DOI] [PubMed] [Google Scholar]
- Awakawa T, Yokota K, Funa N, Doi F, Mori N, Watanabe H, Horinouchi S. Physically discrete beta-lactamase-type thioesterase catalyzes product release in atrochrysone synthesis by iterative type I polyketide synthase. Chem Biol. 2009;16:613–623. doi: 10.1016/j.chembiol.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Bacha N, Atoui A, Mathieu F, Liboz T, Lebrihi A. Aspergillus westerdijkiae polyketide synthase gene “aoks1” is involved in the biosynthesis of ochratoxin A. Fungal Genet Biol. 2009a;46:77–84. doi: 10.1016/j.fgb.2008.09.015. [DOI] [PubMed] [Google Scholar]
- Bacha N, Dao HP, Atoui A, Mathieu F, O'Callaghan J, Puel O, Liboz T, Dobson AD, Lebrihi A. Cloning and characterization of novel methylsalicylic acid synthase gene involved in the biosynthesis of isoasperlactone and asperlactone in Aspergillus westerdijkiae. Fungal Genet Biol. 2009b;46:742–749. doi: 10.1016/j.fgb.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Bailey AM, Cox RJ, Harley K, Lazarus CM, Simpson TJ, Skellam E. Characterisation of 3-methylorcinaldehyde synthase (MOS) in Acremonium strictum: first observation of a reductive release mechanism during polyketide biosynthesis. Chem Commun (Camb) 2007:4053–4055. doi: 10.1039/b708614h. [DOI] [PubMed] [Google Scholar]
- Beck J, Ripka S, Siegner A, Schiltz E, Schweizer E. The multifunctional 6-methylsalicylic acid synthase gene of Penicillium patulum. Its gene structure relative to that of other polyketide synthases. Eur J Biochem. 1990;192:487–498. doi: 10.1111/j.1432-1033.1990.tb19252.x. [DOI] [PubMed] [Google Scholar]
- Bergmann S, Schumann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat Chem Biol. 2007;3:213–217. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- Bok JW, Chiang YM, Szewczyk E, Reyes-Domingez Y, Davidson AD, Sanchez JF, Lo HC, Watanabe K, Strauss J, Oakley BR, Wang CC, Keller NP. Chromatin-level regulation of biosynthetic gene clusters. Nat Chem Biol. 2009;5:462–464. doi: 10.1038/nchembio.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DW, Yu JH, Kelkar HS, Fernandes M, Nesbitt TC, Keller NP, Adams TH, Leonard TJ. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc Natl Acad Sci USA. 1996;93:1418–1422. doi: 10.1073/pnas.93.4.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PK, Cary JW, Yu J, Bhatnagar D, Cleveland TE. The Aspergillus parasiticus polyketide synthase gene pksA, a homolog of Aspergillus nidulans wA, is required for aflatoxin B1 biosynthesis. Mol Gen Genet. 1995;248:270–277. doi: 10.1007/BF02191593. [DOI] [PubMed] [Google Scholar]
- Chang PK, Horn BW, Dorner JW. Clustered genes involved in cyclopiazonic acid production are next to the aflatoxin biosynthesis gene cluster in Aspergillus flavus. Fungal Genet Biol. 2009;46:176–182. doi: 10.1016/j.fgb.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Chen Z-G, Fujii I, Ebizuka Y, Sankawa U. Purification and characterization of emodinanthrone oxygenase from Aspergillus terreus. Phytochemistry. 1995;38:299–305. [Google Scholar]
- Chiang YM, Szewczyk E, Nayak T, Davidson AD, Sanchez JF, Lo HC, Ho WY, Simityan H, Kuo E, Praseuth A, Watanabe K, Oakley BR, Wang CC. Molecular genetic mining of the Aspergillus secondary metabolome: discovery of the emericellamide biosynthetic pathway. Chem Biol. 2008;15:527–532. doi: 10.1016/j.chembiol.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YM, Lee KH, Sanchez JF, Keller NP, Wang CC. Unlocking fungal cryptic natural products. Nat Prod Commun. 2009a;4:1505–1510. [PMC free article] [PubMed] [Google Scholar]
- Chiang YM, Szewczyk E, Davidson AD, Keller N, Oakley BR, Wang CC. A gene cluster containing two fungal polyketide synthases encodes the biosynthetic pathway for a polyketide, asperfuranone, in Aspergillus nidulans. J Am Chem Soc. 2009b;131:2965–2970. doi: 10.1021/ja8088185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichewicz RH. Epigenome manipulation as a pathway to new natural product scaffolds and their congeners. Nat Prod Rep. 2010;27:11–22. doi: 10.1039/b920860g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RJ. Polyketides, proteins and genes in fungi: programmed nano-machines begin to reveal their secrets. Org Biomol Chem. 2007;5:2010–2026. doi: 10.1039/b704420h. [DOI] [PubMed] [Google Scholar]
- Cox RJ, Glod F, Hurley D, Lazarus CM, Nicholson TP, Rudd BA, Simpson TJ, Wilkinson B, Zhang Y. Rapid cloning and expression of a fungal polyketide synthase gene involved in squalestatin biosynthesis. Chem Commun (Camb) 2004:2260–2261. doi: 10.1039/b411973h. [DOI] [PubMed] [Google Scholar]
- Crawford JM, Thomas PM, Scheerer JR, Vagstad AL, Kelleher NL, Townsend CA. Deconstruction of iterative multidomain polyketide synthase function. Science. 2008a;320:243–246. doi: 10.1126/science.1154711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford JM, Vagstad AL, Ehrlich KC, Townsend CA. Starter unit specificity directs genome mining of polyketide synthase pathways in fungi. Bioorg Chem. 2008b;36:16–22. doi: 10.1016/j.bioorg.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford JM, Vagstad AL, Whitworth KP, Ehrlich KC, Townsend CA. Synthetic strategy of nonreducing iterative polyketide synthases and the origin of the classical “starter-unit effect”. Chembiochem. 2008c;9:1019–1023. doi: 10.1002/cbic.200700702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford JM, Korman TP, Labonte JW, Vagstad AL, Hill EA, Kamari-Bidkorpeh O, Tsai SC, Townsend CA. Structural basis for biosynthetic programming of fungal aromatic polyketide cyclization. Nature. 2009;461:1139–1143. doi: 10.1038/nature08475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AL, Lewis JR, Cai Y, Powell C, Davis AP, Wilkins JPG, Pudney P, Clifford MN. A polyphenolic pigment from black tea. Phytochemistry. 1997;46:1397–1402. [Google Scholar]
- Dimroth P, Walter H, Lynen F. Biosynthesis of 6-methylsalicylic acid. Eur J Biochem. 1970;13:98–110. doi: 10.1111/j.1432-1033.1970.tb00904.x. [DOI] [PubMed] [Google Scholar]
- Eley KL, Halo LM, Song Z, Powles H, Cox RJ, Bailey AM, Lazarus CM, Simpson TJ. Biosynthesis of the 2-pyridone tenellin in the insect pathogenic fungus Beauveria bassiana. Chembiochem. 2007;8:289–297. doi: 10.1002/cbic.200600398. [DOI] [PubMed] [Google Scholar]
- Feng GH, Leonard TJ. Characterization of the polyketide synthase gene (pksL1) required for aflatoxin biosynthesis in Aspergillus parasiticus. J Bacteriol. 1995;177:6246–6254. doi: 10.1128/jb.177.21.6246-6254.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- Fujii I, Ono Y, Tada H, Gomi K, Ebizuka Y, Sankawa U. Cloning of the polyketide synthase gene atX from Aspergillus terreus and its identification as the 6-methylsalicylic acid synthase gene by heterologous expression. Mol Gen Genet. 1996;253:1–10. doi: 10.1007/s004380050289. [DOI] [PubMed] [Google Scholar]
- Fujii I, Mori Y, Watanabe A, Kubo Y, Tsuji G, Ebizuka Y. Heterologous expression and product identification of Colletotrichum lagenarium polyketide synthase encoded by the PKS1 gene involved in melanin biosynthesis. Biosci Biotechnol Biochem. 1999;63:1445–1452. doi: 10.1271/bbb.63.1445. [DOI] [PubMed] [Google Scholar]
- Fujii I, Mori Y, Watanabe A, Kubo Y, Tsuji G, Ebizuka Y. Enzymatic synthesis of 1, 3, 6, 8-tetrahydroxynaphthalene solely from malonyl coenzyme A by a fungal iterative type I polyketide synthase PKS1. Biochemistry. 2000;39:8853–8858. doi: 10.1021/bi000644j. [DOI] [PubMed] [Google Scholar]
- Fujii I, Watanabe A, Sankawa U, Ebizuka Y. Identification of Claisen cyclase domain in fungal polyketide synthase WA, a naphthopyrone synthase of Aspergillus nidulans. Chem Biol. 2001;8:189–197. doi: 10.1016/s1074-5521(00)90068-1. [DOI] [PubMed] [Google Scholar]
- Fujii I, Yasuoka Y, Tsai HF, Chang YC, Kwon-Chung KJ, Ebizuka Y. Hydrolytic polyketide shortening by ayg1p, a novel enzyme involved in fungal melanin biosynthesis. J Biol Chem. 2004;279:44613–44620. doi: 10.1074/jbc.M406758200. [DOI] [PubMed] [Google Scholar]
- Fujii I, Yoshida N, Shimomaki S, Oikawa H, Ebizuka Y. An iterative type I polyketide synthase PKSN catalyzes synthesis of the decaketide alternapyrone with regio-specific octa-methylation. Chem Biol. 2005;12:1301–1309. doi: 10.1016/j.chembiol.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Basturkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, D'Enfert C, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, Yu J, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Peñalva MA, Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, Birren BW. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- Gomez BL, Nosanchuk JD. Melanin and fungi. Curr Opin Infect Dis. 2003;16:91–96. doi: 10.1097/00001432-200304000-00005. [DOI] [PubMed] [Google Scholar]
- Halo LM, Marshall JW, Yakasai AA, Song Z, Butts CP, Crump MP, Heneghan M, Bailey AM, Simpson TJ, Lazarus CM, Cox RJ. Authentic heterologous expression of the tenellin iterative polyketide synthase nonribosomal peptide synthetase requires coexpression with an enoyl reductase. Chembiochem. 2008;9:585–594. doi: 10.1002/cbic.200700390. [DOI] [PubMed] [Google Scholar]
- Hendrickson L, Davis CR, Roach C, Nguyen DK, Aldrich T, McAda PC, Reeves CD. Lovastatin biosynthesis in Aspergillus terreus: characterization of blocked mutants, enzyme activities and a multifunctional polyketide synthase gene. Chem Biol. 1999;6:429–439. doi: 10.1016/s1074-5521(99)80061-1. [DOI] [PubMed] [Google Scholar]
- Hertweck C. The biosynthetic logic of polyketide diversity. Angew Chem Int Ed Engl. 2009;48:4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- Hill AM. The biosynthesis, molecular genetics and enzymology of the polyketide-derived metabolites. Nat Prod Rep. 2006;23:256–320. doi: 10.1039/b301028g. [DOI] [PubMed] [Google Scholar]
- Hoffmeister D, Keller NP. Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat Prod Rep. 2007;24:393–416. doi: 10.1039/b603084j. [DOI] [PubMed] [Google Scholar]
- Kasahara K, Fujii I, Oikawa H, Ebizuka Y. Expression of Alternaria solani PKSF generates a set of complex reduced-type polyketides with different carbon-lengths and cyclization. Chembiochem. 2006;7:920–924. doi: 10.1002/cbic.200600034. [DOI] [PubMed] [Google Scholar]
- Keller NP, Turner G, Bennett JW. Fungal secondary metabolism—from biochemistry to genomics. Nat Rev Microbiol. 2005;3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- Kennedy J, Auclair K, Kendrew SG, Park C, Vederas JC, Hutchinson CR. Modulation of polyketide synthase activity by accessory proteins during lovastatin biosynthesis. Science. 1999;284:1368–1372. doi: 10.1126/science.284.5418.1368. [DOI] [PubMed] [Google Scholar]
- Kroken S, Glass NL, Taylor JW, Yoder OC, Turgeon BG. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc Natl Acad Sci USA. 2003;100:15670–15675. doi: 10.1073/pnas.2532165100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder K, Jahn B, Gehringer H, Schmidt A, Wanner G, Brakhage AA. Identification of a polyketide synthase gene (pksP) of Aspergillus fumigatus involved in conidial pigment biosynthesis and virulence. Med Microbiol Immunol. 1998;187:79–89. doi: 10.1007/s004300050077. [DOI] [PubMed] [Google Scholar]
- Liu X, Walsh CT. Cyclopiazonic acid biosynthesis in Aspergillus sp.: characterization of a reductase-like R* domain in cyclopiazonate synthetase that forms and releases cyclo-acetoacetyl-l-tryptophan. Biochemistry. 2009;48:11032–11044. doi: 10.1021/bi901123r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma SM, Li JW, Choi JW, Zhou H, Lee KK, Moorthie VA, Xie X, Kealey JT, Da Silva NA, Vederas JC, Tang Y. Complete reconstitution of a highly reducing iterative polyketide synthase. Science. 2009;326:589–592. doi: 10.1126/science.1175602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma SM, Tang Y. Biochemical characterization of the minimal polyketide synthase domains in the lovastatin nonaketide synthase LovB. FEBS J. 2007;274:2854–2864. doi: 10.1111/j.1742-4658.2007.05818.x. [DOI] [PubMed] [Google Scholar]
- Ma Y, Smith LH, Cox RJ, Beltran-Alvarez P, Arthur CJ, Simpson FRST. Catalytic relationships between type I and type II iterative polyketide synthases: the Aspergillus parasiticus norsolorinic acid synthase. Chembiochem. 2006;7:1951–1958. doi: 10.1002/cbic.200600341. [DOI] [PubMed] [Google Scholar]
- Maggon K. Best-selling human medicines 2002–2004. Drug Discov Today. 2005;10:739–742. doi: 10.1016/S1359-6446(05)03468-9. [DOI] [PubMed] [Google Scholar]
- Maiya S, Grundmann A, Li X, Li SM, Turner G. Identification of a hybrid PKS/NRPS required for pseurotin A biosynthesis in the human pathogen Aspergillus fumigatus. Chembiochem. 2007;8:1736–1743. doi: 10.1002/cbic.200700202. [DOI] [PubMed] [Google Scholar]
- Marquez-Fernandez O, Trigos A, Ramos-Balderas JL, Viniegra-Gonzalez G, Deising HB, Aguirre J. Phosphopantetheinyl transferase CfwA/NpgA is required for Aspergillus nidulans secondary metabolism and asexual development. Eukaryot Cell. 2007;6:710–720. doi: 10.1128/EC.00362-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayorga ME, Timberlake WE. The developmentally regulated Aspergillus nidulans wA gene encodes a polypeptide homologous to polyketide and fatty acid synthases. Mol Gen Genet. 1992;235:205–212. doi: 10.1007/BF00279362. [DOI] [PubMed] [Google Scholar]
- Mueller JE, Canze M, Bryk M. The requirements for COMPASS and Paf1 in transcriptional silencing and methylation of histone H3 in Saccharomyces cerevisiae. Genetics. 2006;173:557–567. doi: 10.1534/genetics.106.055400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Callaghan J, Caddick MX, Dobson AD. A polyketide synthase gene required for ochratoxin A biosynthesis in Aspergillus ochraceus. Microbiology. 2003;149:3485–3491. doi: 10.1099/mic.0.26619-0. [DOI] [PubMed] [Google Scholar]
- Sanchez JF, Chiang YM, Szewczyk E, Davidson AD, Ahuja M, Oakley CE, Bok JW, Keller NP, Oakley BR, Wang CC. Molecular genetic analysis of the orsellinic acid/F9775 gene cluster of Aspergillus nidulans. Mol Biosyst. 2010;6:587–593. doi: 10.1039/b904541d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherlach K, Hertweck C. Triggering cryptic natural product biosynthesis in microorganisms. Org Biomol Chem. 2009;7:1753–1760. doi: 10.1039/b821578b. [DOI] [PubMed] [Google Scholar]
- Schroeckh V, Scherlach K, Nutzmann HW, Shelest E, Schmidt-Heck W, Schuemann J, Martin K, Hertweck C, Brakhage AA. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc Natl Acad Sci USA. 2009;106:14558–14563. doi: 10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann J, Hertweck C. Advances in cloning, functional analysis and heterologous expression of fungal polyketide synthase genes. J Biotechnol. 2006;124:690–703. doi: 10.1016/j.jbiotec.2006.03.046. [DOI] [PubMed] [Google Scholar]
- Seshime Y, Juvvadi PR, Tokuoka M, Koyama Y, Kitamoto K, Ebizuka Y, Fujii I. Functional expression of the Aspergillus flavus PKS-NRPS hybrid CpaA involved in the biosynthesis of cyclopiazonic acid. Bioorg Med Chem Lett. 2009;19:3288–3292. doi: 10.1016/j.bmcl.2009.04.073. [DOI] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Reinberg D. Histone H3 Lys 4 methylation: caught in a bind? Genes Dev. 2006;20:2779–2786. doi: 10.1101/gad.1468206. [DOI] [PubMed] [Google Scholar]
- Szewczyk E, Chiang YM, Oakley CE, Davidson AD, Wang CC, Oakley BR. Identification and characterization of the asperthecin gene cluster of Aspergillus nidulans. Appl Environ Microbiol. 2008;74:7607–7612. doi: 10.1128/AEM.01743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano Y, Kubo Y, Shimizu K, Mise K, Okuno T, Furusawa I. Structural analysis of PKS1, a polyketide synthase gene involved in melanin biosynthesis in Colletotrichum lagenarium. Mol Gen Genet. 1995;249:162–167. doi: 10.1007/BF00290362. [DOI] [PubMed] [Google Scholar]
- Tokuoka M, Seshime Y, Fujii I, Kitamoto K, Takahashi T, Koyama Y. Identification of a novel polyketide synthase-nonribosomal peptide synthetase (PKS-NRPS) gene required for the biosynthesis of cyclopiazonic acid in Aspergillus oryzae. Fungal Genet Biol. 2008;45:1608–1615. doi: 10.1016/j.fgb.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Trail F, Mahanti N, Rarick M, Mehigh R, Liang SH, Zhou R, Linz JE. Physical and transcriptional map of an aflatoxin gene cluster in Aspergillus parasiticus and functional disruption of a gene involved early in the aflatoxin pathway. Appl Environ Microbiol. 1995;61:2665–2673. doi: 10.1128/aem.61.7.2665-2673.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HF, Chang YC, Washburn RG, Wheeler MH, Kwon-Chung KJ. The developmentally regulated alb1 gene of Aspergillus fumigatus: its role in modulation of conidial morphology and virulence. J Bacteriol. 1998;180:3031–3038. doi: 10.1128/jb.180.12.3031-3038.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HF, Wheeler MH, Chang YC, Kwon-Chung KJ. A developmentally regulated gene cluster involved in conidial pigment biosynthesis in Aspergillus fumigatus. J Bacteriol. 1999;181:6469–6477. doi: 10.1128/jb.181.20.6469-6477.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HF, Fujii I, Watanabe A, Wheeler MH, Chang YC, Yasuoka Y, Ebizuka Y, Kwon-Chung KJ. Pentaketide melanin biosynthesis in Aspergillus fumigatus requires chain-length shortening of a heptaketide precursor. J Biol Chem. 2001;276:29292–29298. doi: 10.1074/jbc.M101998200. [DOI] [PubMed] [Google Scholar]
- Udwary DW, Merski M, Townsend CA. A method for prediction of the locations of linker regions within large multifunctional proteins, and application to a type I polyketide synthase. J Mol Biol. 2002;323:585–598. doi: 10.1016/s0022-2836(02)00972-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A, Ebizuka Y. Unprecedented mechanism of chain length determination in fungal aromatic polyketide synthases. Chem Biol. 2004;11:1101–1106. doi: 10.1016/j.chembiol.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Watanabe A, Ono Y, Fujii I, Sankawa U, Mayorga ME, Timberlake WE, Ebizuka Y. Product identification of polyketide synthase coded by Aspergillus nidulans wA gene. Tetrahedron Lett. 1998;39:7733–7736. [Google Scholar]
- Watanabe A, Fujii I, Sankawa U, Mayorga ME, Timberlake WE, Ebizuka Y. Re-identification of Aspergillus nidulans wA gene to code for a polyketide synthase of naphthopyrone. Tetrahedron Lett. 1999;40:91–94. [Google Scholar]
- Watanabe A, Fujii I, Tsai H, Chang YC, Kwon-Chung KJ, Ebizuka Y. Aspergillus fumigatus alb1 encodes naphthopyrone synthase when expressed in Aspergillus oryzae. FEMS Microbiol Lett. 2000;192:39–44. doi: 10.1111/j.1574-6968.2000.tb09356.x. [DOI] [PubMed] [Google Scholar]
- Watanabe CM, Townsend CA. Initial characterization of a type I fatty acid synthase and polyketide synthase multienzyme complex NorS in the biosynthesis of aflatoxin B1. Chem Biol. 2002;9:981–988. doi: 10.1016/s1074-5521(02)00213-2. [DOI] [PubMed] [Google Scholar]
- Xie X, Meehan MJ, Xu W, Dorrestein PC, Tang Y. Acyltransferase mediated polyketide release from a fungal megasynthase. J Am Chem Soc. 2009;131:8388–8389. doi: 10.1021/ja903203g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JH, Leonard TJ. Sterigmatocystin biosynthesis in Aspergillus nidulans requires a novel type I polyketide synthase. J Bacteriol. 1995;177:4792–4800. doi: 10.1128/jb.177.16.4792-4800.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]