Abstract
The dose-limiting side effect of the common colon cancer chemotherapeutic CPT-11 is severe diarrhea caused by symbiotic bacterial β-glucuronidases that reactivate the drug in the gut. We sought to target these enzymes without killing the commensal bacteria essential for human health. Potent bacterial β-glucuronidase inhibitors were identified by high-throughput screening and shown to have no effect on the orthologous mammalian enzyme. Crystal structures established that selectivity was based on a loop unique to bacterial β-glucuronidases. Inhibitors were highly effective against the enzyme target in living aerobic and anaerobic bacteria, but did not kill the bacteria or harm mammalian cells. Finally, oral administration of an inhibitor protected mice from CPT-11–induced toxicity. Thus, drugs may be designed to inhibit undesirable enzyme activities in essential microbial symbiotes to enhance chemotherapeutic efficacy.
Camptothecin, a potent antineoplastic compound, was added to the National Cancer Institute natural products screening set in 1966. It poisons the catalytic cycle of human topoisomerase I, which manages the super-helical tension associated with DNA metabolism and is preferentially active in rapidly dividing cells (1, 2). In preliminary clinical trials, camptothecin exhibited marked toxicity and poor bioavailability (3). Although its derivatives topotecan and CPT-11 (also called irinotecan) are now in clinical use (3), they still elicit pronounced side effects that limit efficacy. CPT-11 is one of the three commonly used chemotherapeutic agents for colon cancer, and it has also been used against lung and brain tumors as well as refractory forms of leukemia and lymphoma (4). It is a prodrug, with a carbamate-linked dipiperidino group that increases solubility and bioavailability (3); this moiety is removed in vivo to produce the active metabolite SN-38 (5) (Fig. 1A).
Fig. 1.
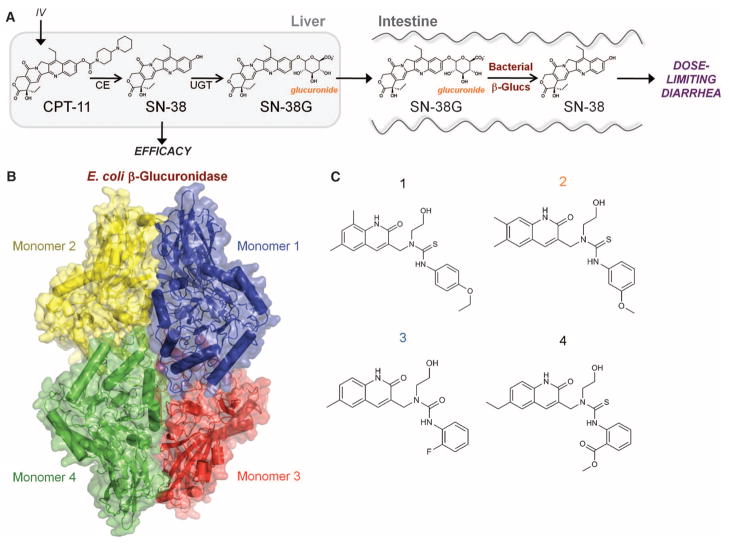
CPT-11 metabolism and E. coli β-glucuronidase. (A) Intravenously administered CPT-11 is activated by carboxylesterases (CE) to SN-38, an antineoplastic topoisomerase I poison. Liver SN-38 is inactivated via glucuronidation to SN-38G by UDP-glucuronosyltransferase (UGT) enzymes and sent to the intestines. β-Glucuronidases (β-glucs) in the symbiotic GI bacteria remove the glucuronide as a carbon source, and active SN-38 in the intestinal lumen generates dose-limiting diarrhea. (B) Crystal structure of the E. coli β-glucuronidase tetramer at 2.5 Å resolution. (C) Four selective bacterial β-glucuronidase inhibitors identified via high-throughput screening.
CPT-11 causes severe diarrhea generated by its complex activation and subsequent metabolism (Fig. 1A) (6, 7). SN-38 produced by carboxylesterases is glucuronidated in the liver by uridine diphosphate (UDP)–glucuronosyltransferase enzymes to form inactive SN-38G (8), which is excreted via the biliary ducts into the gastrointestinal (GI) tract (Fig. 1A). Once in the intestines, though, SN-38G serves as a substrate for bacterial β-glucuronidase enzymes in the commensal microbiota that remove the glucuronide group as a carbon source, producing reactivated SN-38 in situ (Fig. 1A) (9, 10). SN-38 levels in the intestinal lumen play an essential role in the delayed diarrhea that prevents dose intensification and efficacy in up to 40% of treated patients (11–13).
The feasibility of using antibiotics to reduce GI bacteria levels prior to CPT-11 treatment has been examined (14); however, this approach has several drawbacks. Intestinal biota play essential roles in carbohydrate metabolism, vitamin production, and the processing of bile acids, sterols, and xenobiotics (15, 16). Thus, the removal of GI bacteria is not recommended for patients already challenged by neoplastic growths and chemotherapy. In addition, elimination of symbiotic GI flora increases the chances of infections by pathogenic bacteria, including enterohemorrhagic Escherichia coli and Clostridium difficile (17–23).
β-Glucuronidase enzymes hydrolyze glucuronic acid sugar moieties from a variety of compounds (24), and their presence in a range of bacteria is exploited to detect bacterial contamination in commonly used water purity tests (25). The crystal structure of human β-glucuronidase was reported in 1996 (26), but no structure of a bacterial β-glucuronidase has been presented. In addition, only relatively weak inhibitors of β-glucuronidases have been described [inhibition constant (Ki) values of 25 μM to 2 mM] (27, 28). Thus, we used structural and chemical biology to identify potent and selective inhibitors of bacterial β-glucuronidases to eliminate the GI toxicity of CPT-11 without killing the bacterial symbiotes required for intestinal health.
Full-length E. coli β-glucuronidase was purified and shown to hydrolyze SN-38G to SN-38 in vitro (fig. S1). The enzyme was initially crystallized both alone and in complex with an established low-affinity inhibitor, glucaro-δ-lactam (GDL) (29), and data were collected to 2.5 and 2.4 Å resolution, respectively. Because molecular replacement using a previously reported human β-glucuronidase model [PDB ID 1BHG (30)] was unsuccessful, selenomethionine (SeMet)–substituted E. coli β-glucuronidase and single-wavelength anomalous dispersion x-ray data to 2.9 Å resolution were used for structure determination and refinement (PDB ID 3K4A). Molecular replacement using the SeMet model was then used to determine and refine the native (PDB ID 3K46), GDL-bound (PDB ID 3K4D), and Inhibitor 2 and Inhibitor 3 structures (PDB IDs 3LPF and 3LPG) (table S1).
The asymmetric unit of the E. coli β-glucuronidase structure contains two monomers of 597 ordered residues, and crystallographic symmetry generates the functionally relevant enzyme tetramer observed previously for the human enzyme (30) and confirmed by gel filtration chromatography for the E. coli form of the enzyme (Fig. 1B). The N-terminal 180 residues resemble the sugar-binding domain of family 2 glycosyl hydrolases (31), whereas the C-terminal domain (residues 274 to 603) forms an αβ barrel (31) and contains the active-site residues Glu413 and Glu504. The region between the N- and C-terminal domains exhibits an immunoglobulin-like β-sandwich domain consistent with other family 2 glycosyl hydrolases (31, 32) (fig. S2). The GDL inhibitor binds in a single orientation deep within the active site of the enzyme’s C-terminal domain and involves large (>14 Å) shifts in residue positions relative to the unliganded structure (figs. S3 and S4). Superimposing the E. coli β-glucuronidase structure on the human enzyme reveals a 1.4 Å root mean square deviation over 565 equivalent Cα positions with 45% sequence identity (figs. S5 and S6). The E. coli enzyme contains a 17-residue “bacterial loop” not found in the human ortholog, with the active site of each E. coli β-glucuronidase monomer containing “bacterial loops” from that monomer as well as a neighboring monomer within the enzyme tetramer (fig. S4).
Chemical library screening was conducted using a β-glucuronidase assay in which the conversion of 4-methylumbelliferyl-glucuronide (4-MUG) to 4-methylumbelliferone (4-MU) was monitored by measuring the increase in 4-MU fluorescence (excitation at 365 nm, emission at 450 nm) (fig. S7) (25). This assay exhibited robust characteristics when used with a 10,240-compound chemical library, with a screening Z-factor of 0.84 (33). The hit rate was 1%, with 100 compounds producing 90% inhibition or better and R2 values for inhibition curves of 0.99 or better. Four compounds were chosen for further investigation (Fig. 1C). Secondary β-glucuronidase assays were also used to examine inhibitor potency in vitro and in cell-based studies.
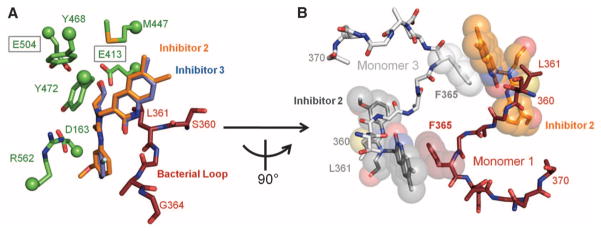
The GDL compound exhibited relatively weak in vitro enzyme inhibition [e.g., median inhibitory concentration (IC50) = 26.4 ± 4.83 μM] and did not disrupt β-glucuronidase activity in cultured E. coli cells (fig. S8). In contrast, the four compounds chosen from the high-throughput screen (Fig. 1C) were all potent in vitro inhibitors with submicromolar IC50 and Ki values and uncompetitive characteristics as assessed by their lack of impact on enzyme kcat/Km values (Table 1 and fig. S9). Crystal structures of E. coli β-glucuronidase in complexes with Inhibitors 2 and 3 were resolved to 2.3 and 2.4 Å resolution, respectively (table S1), and revealed that the inhibitors bound at the “bacterial loops” at the entrance to the active-site cavity, with the ethoxy groups extending to within 3.3 Å of the catalytic Glu413 residue (Fig. 2A). Primary contacts between the enzyme monomer and inhibitors involved the “bacterial loop” (Leu361) of the primary monomer, as well as the “bacterial loop” (Phe365) of an adjacent monomer within the tetramer (Fig. 2B and fig. S10). Thus, this loop may be essential to inhibitor efficacy.
Table 1.
In vitro and bacterial cell–based assays for β-glucuronidase activity and inhibitor efficacy. kcat, catalytic rate; Km, Michaelis constant; N/A, not applicable. N.E., not effective. Errors represent standard deviation, where N = 3.
|
E. coli β-glucuronidase in vitro
|
E. coli cell–based
|
|||
|---|---|---|---|---|
| IC50 (nM) | kcat/Km (s−1 μM−1) | Ki (nM) | EC50 (nM) | |
| No inhibitor | N/A | 0.134 ± 0.0123 | N/A | N/A |
| Glucaro-δ-lactam | 26400 ± 483 | N/A | 7750 ± 475 | N.E. |
| Inhibitor 1 | 283 ± 26.1 | 0.0987 ± 0.00621 | 164 ± 13.0 | 17.7 ± 7.42 |
| Inhibitor 2 | 369 ± 2.51 | 0.119 ± 0.00473 | 208 ± 25.4 | 28.3 ± 2.11 |
| Inhibitor 3 | 586 ± 31.1 | 0.136 ± 0.00633 | 684 ± 81.6 | 233.2 ± 2.99 |
| Inhibitor 4 | 1060 ± 3.54 | 0.122 ± 0.0205 | 1380 ± 166 | 1322.8 ± 1.15 |
Fig. 2.
Potent β-glucuronidase inhibitors. (A) Crystal structures of Inhibitors 2 and 3 bound to the active site of E. coli β-glucuronidase. (B) Inhibitors are observed to stack cooperatively between monomers in the E. coli β-glucuronidase tetramer. Amino acid abbreviations: D, Asp; E, Glu; F, Phe; G, Gly; L, Leu; M, Met; R, Arg; S, Ser; Y, Tyr.
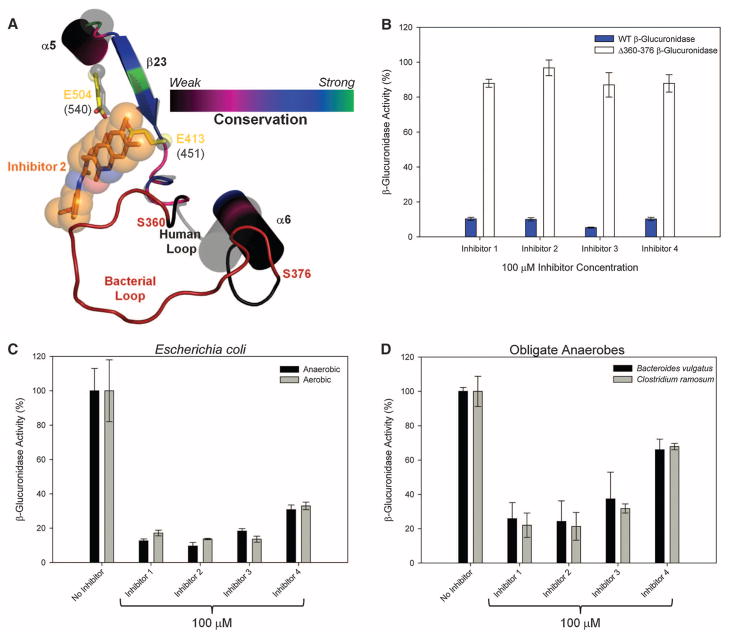
The specificity of the four characterized inhibitors for bacterial versus mammalian β-glucuronidases was assayed in vitro with the use of bovine liver β-glucuronidase. With a concentration range of 0 to 100 μM for each of the inhibitors, no effect was observed on the activity of this mammalian β-glucuronidase (figs. S11 and S12). The 17-residue loop that contacts the inhibitors in the crystal structures of E. coli β-glucuronidase (Fig. 2, A and B) is not present in mammalian forms of the enzyme (fig. S13), which act on larger substrates such as glycosaminoglycans (34). Of the 284 GI-associated bacterial species in the Human Microbiome Project database (35), 123 (43%) contained β-glucuronidases or candidate β-glucuronidases; of those, 121 (98%) maintained at least the N-terminal portion of the “bacterial loop,” and 110 (91%) contained the key residues at E. coli β-glucuronidase positions 361 and 365 capable of making the inhibitor contacts reported here (fig. S14; see also figs. S10 and S13 and table S2). To test the hypothesis that the “bacterial loop” is required for the efficacy of our inhibitors, we created a form of E. coli β-glucuronidase that lacks residues 360 to 376 (Fig. 3A). The Δ360-376 E. coli β-glucuronidase mutant was not inhibited even at 100 μM concentrations of the four compounds examined (Fig. 3B). Thus, we have identified inhibitors that are potent and selective for a bacterial β-glucuronidase.
Fig. 3.
Inhibitor selectivity for bacterial β-glucuronidase. (A) The 360–376 loop forms direct contact with the bound inhibitors in the E. coli β-glucuronidase structure. This loop is missing from the structure of human β-glucuronidase; thus, it is labeled the “bacterial loop.” (B) Elimination of the “bacterial loop” from E. coli β-glucuronidase produces an enzyme insensitive to inhibitor efficacy. (C) β-Glucuronidase inhibition in living E. coli cells grown under both aerobic and anaerobic conditions. (D) β-Glucuronidase inhibition in two obligate anaerobic bacteria. Error bars represent SD; N = 3.
We next examined the ability of the four lead compounds to inhibit β-glucuronidase activity in living bacterial cells. Whereas a ~26 μM in vitro inhibitor such as GDL has no effect against β-glucuronidase activity in cultured E. coli, each inhibitor tested was effective, with median effective concentration (EC50) values from 18 nM to 1.3 μM (Table 1 and fig. S15). Because >99% of the microbial species present in the GI tract are obligate anaerobes (36), we tested E. coli cells grown under anaerobic conditions and examined other relevant anaerobic bacterial species (35). The compounds were effective in E. coli grown under both aerobic and anaerobic conditions (Fig. 3C) and were effective against enzyme activity in two obligate anaerobes known to inhabit the mammalian GI, Bacteroides vulgatus and Clostridium ramosum (Fig. 3D). We also tested Lactobacillus reuteri and Bifidobacterium infantis, which do not contain the gene encoding β-glucuronidase (28, 37), and found no evidence of enzyme activity or inhibitor impact on cell viability for these or other strains (figs. S16 and S17). Consistent with their lack of effect on mammalian β-glucuronidase activity in vitro, inhibitors at relatively high concentrations (100 μM) had little to no effect on the survival of two human colon cancer cell lines (HCT116, Caco-2) and one murine colon cancer cell line (CMT93) (38) (fig. S18).
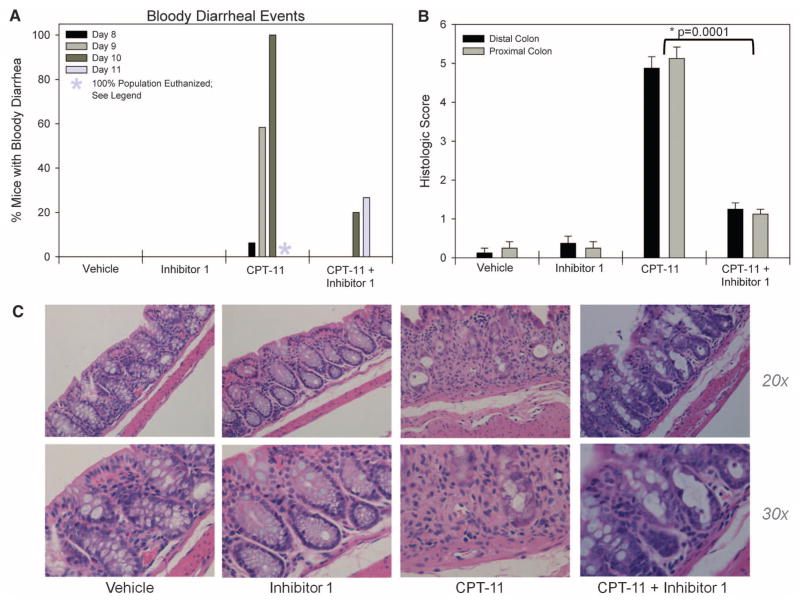
Finally, we examined the ability of Inhibitor 1 to eliminate the delayed diarrhea and intestinal damage caused by CPT-11 administration. Healthy 6- to 8-week-old Balb/cJ mice were divided into four groups of 16 animals each. Group 1 received 50 μl of double-distilled H2O intraperitoneally (i.p.) and 1% dimethyl sulfoxide by oral gavage twice daily. Group 2 received 10 μg of Inhibitor 1 via oral gavage twice per day, but no CPT-11. Group 3 received CPT-11 i.p. once daily, at a dose of 50 mg per kg of body weight, and no Inhibitor 1. Group 4 received CPT-11 and Inhibitor 1 with dose and schedule identical to animals in group 3 and group 2, respectively. GI symptoms appeared after 7 days. At days 8 to 10, all the animals on the CPT-11–only treatment (group 3) experienced both diarrhea and bloody diarrhea, whereas none of the animals receiving vehicle or inhibitor alone (groups 1 and 2, respectively) exhibited diarrhea at any point during the study (Fig. 4A and fig. S19). By day 11, all the group 3 animals had to be euthanized. In contrast, the animals receiving both CPT-11 and Inhibitor 1 (group 4), exhibited less diarrhea and bloody diarrhea than did the group 3 animals (Fig. 4A and fig. S19). Examination and scoring of colonic tissue samples from each group established that Inhibitor 1 protected the mouse GI epithelium from CPT-11–induced damage (Fig. 4, B and C) (39). Animals treated with vehicle and Inhibitor 1 exhibited healthy glandular structure and an intact epithelial layer; CPT-11 administration destroyed these tissues, eliminating the glands and causing a large influx of inflammatory cells (Fig. 4, B and C). In contrast, Inhibitor 1, when provided orally in combination with i.v. CPT-11, protected the glandular structure of these intestinal tissues (Fig. 4, B and C).
Fig. 4.
Alleviation of CPT-11 toxicity in mice. (A) CPT-11 produced bloody diarrhea starting after 8 days and peaking at 10 days, whereas oral administration of Inhibitor 1 with CPT-11 reduced the incidence of bloody diarrhea. Vehicle and Inhibitor 1 alone caused no bloody diarrhea. By day 8 to 11, mice in the CPT-11 group began to suffer from severe lethargy and lack of movement; by day 11, all mice in that group were euthanized according to AIC protocol 20070715. (B) Histologic score of the distal and proximal colon of animals in the four treatment groups. Error bars represent SD; N = 12. (C) Tissue histology of colons taken from mice from each treatment group show healthy glandular structure for both vehicle and Inhibitor 1 but highly disrupted tissues in the CPT-11 group. In contrast, Inhibitor 1 provided in combination with CPT-11 protects the colon from damage.
The lead compounds that we have characterized from high-throughput screening hits are all effective inhibitors of the β-glucuronidase enzyme in vitro (with Ki values of 160, 210, 680, and 1400 nM, respectively), and they maintain potent efficacy in living bacterial cells (EC50 values of 18, 28, 230, and 1300 nM, respectively) (Table 1) without affecting bacterial cell growth or survival under aerobic or anaerobic conditions (figs. S16 and S17) or killing mammalian epithelial cells (fig. S18). Lower EC50 values relative to the Ki values likely reflect relatively low β-glucuronidase concentrations in cells. Key regions of the “bacterial loop” identified in the E. coli β-glucuronidase crystal structure are present in 98% of the β-glucuronidases sequenced from human GI bacteria, and 91% of those sequences contain residues that appear capable of forming inhibitor contacts (figs. S13 and S14). Disruption of β-glucuronidase activity is also demonstrated in bacterial species beyond E. coli (Fig. 3D). Taken together, the data presented here strongly support the hypothesis that microbial β-glucuronidases can be inhibited to prevent the GI production of toxic CPT-11 metabolites. However, full pharmacokinetic studies demonstrating, for example, a reduction in GI SN-38G levels or improved CPT-11 efficacy will be required to unambiguously prove this hypothesis. In addition, the breadth of inhibitor efficacy on human GI bacteria requires further assessment. Still, these initial results involving the oral dosing of an unmodified lead compound are highly promising. If successfully translated to humans, leads like those described here could allow dose intensification of CPT-11, enabling studies of whether efficacy could be improved in relevant human cancers by reducing one of the current dose-limiting side effects. The strategy of selective targeting of an enzyme present in bacterial symbiotes to address a specific clinical problem could potentially be applied more broadly as we deepen our understanding of the essential and dynamic roles that commensal bacteria play in promoting human health.
Supplementary Material
Acknowledgments
We thank members of the Redinbo Laboratory at UNC Chapel Hill, the Mani lab and Einstein College of Medicine, and the BRITE Institute for experimental assistance and helpful discussions; B. Allard at UNC Chapel Hill for her expert help with the cytotoxicity assay; B. Sartor at UNC for providing bacterial cell lines; and E. Burgos in the Schramm lab at Einstein for use of the HPLC. Supported by NIH grant CA98468 (M.R.R.), the SPIRE Postdoctoral Fellowship Program at UNC Chapel Hill (K.T.L.), Damon Runyon Cancer Research Foundation grant CI15-02 and NIH grant CA127231 (S.M.), and a grant from the Golden Leaf Foundation and the State of North Carolina (L.-A.Y.). Atomic coordinates and structure factors have been deposited with the PDB (accession codes: 3K4A, 3K46, 3K4D, 3LPF, and 3LPG). The authors remember Lisa Benkowski and Stacey Micoli, for whom CPT-11’s efficacy was limited by its toxicity.
Footnotes
References and Notes
- 1.Hsiang YH, Hertzberg R, Hecht S, Liu LF. J Biol Chem. 1985;260:14873. [PubMed] [Google Scholar]
- 2.Redinbo MR, Champoux JJ, Hol WG. Curr Opin Struct Biol. 1999;9:29. doi: 10.1016/s0959-440x(99)80005-0. [DOI] [PubMed] [Google Scholar]
- 3.Pizzolato JF, Saltz LB. Lancet. 2003;361:2235. doi: 10.1016/S0140-6736(03)13780-4. [DOI] [PubMed] [Google Scholar]
- 4.Pommier Y. Nat Rev Cancer. 2006;6:789. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 5.Smith NF, Figg WD, Sparreboom A. Toxicol In Vitro. 2006;20:163. doi: 10.1016/j.tiv.2005.06.045. [DOI] [PubMed] [Google Scholar]
- 6.Mathijssen RHJ, et al. Clin Cancer Res. 2001;7:2182. [PubMed] [Google Scholar]
- 7.Ma MK, McLeod HL. Curr Med Chem. 2003;10:41. doi: 10.2174/0929867033368619. [DOI] [PubMed] [Google Scholar]
- 8.Nagar S, Blanchard RL. Drug Metab Rev. 2006;38:393. doi: 10.1080/03602530600739835. [DOI] [PubMed] [Google Scholar]
- 9.Tobin PJ, Dodds HM, Clarke S, Schnitzler M, Rivory LP. Oncol Rep. 2003;10:1977. [PubMed] [Google Scholar]
- 10.Stein A, Voigt W, Jordan K. Ther Adv Med Oncol. 2010;2:51. doi: 10.1177/1758834009355164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurita A, et al. Cancer Chemother Pharmacol. 2010 doi: 10.1007/s00280-010-1310-4. [DOI] [PubMed] [Google Scholar]
- 12.Hu ZP, et al. Toxicol Appl Pharmacol. 2006;216:225. doi: 10.1016/j.taap.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 13.See supporting material on Science Online.
- 14.Flieger D, et al. Oncology. 2007;72:10. doi: 10.1159/000111083. [DOI] [PubMed] [Google Scholar]
- 15.Cummings JH, Macfarlane GT. J Parenter Enteral Nutr. 1997;21:357. doi: 10.1177/0148607197021006357. [DOI] [PubMed] [Google Scholar]
- 16.Guarner F, Malagelada JR. Lancet. 2003;361:512. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- 17.Levy SB, Marshall B. Nat Med. 2004;10(suppl):S122. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 18.Nord CE, Kager L, Heimdahl A. Am J Med. 1984;76:99. doi: 10.1016/0002-9343(84)90250-x. [DOI] [PubMed] [Google Scholar]
- 19.Settle CD, Wilcox MH. Aliment Pharmacol Ther. 1996;10:835. doi: 10.1046/j.1365-2036.1996.79251000.x. [DOI] [PubMed] [Google Scholar]
- 20.Sears S, McNally P, Bachinski MS, Avery R. Gastrointest Endosc. 1999;50:841. doi: 10.1016/s0016-5107(99)70171-9. [DOI] [PubMed] [Google Scholar]
- 21.Stamp D. Med Hypotheses. 2004;63:555. doi: 10.1016/j.mehy.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 22.Yang L, Pei Z. World J Gastroenterol. 2006;12:6741. doi: 10.3748/wjg.v12.i42.6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen SH, et al. Infect Control Hosp Epidemiol. 2010;31:431. doi: 10.1086/651706. [DOI] [PubMed] [Google Scholar]
- 24.Basińska A, Floriańczyk B. Ann Univ Mariae Curie Sklodowska Med. 2003;58:386. [PubMed] [Google Scholar]
- 25.Farnleitner AH, Hocke L, Beiwl C, Kavka GG, Mach RL. Water Res. 2002;36:975. doi: 10.1016/s0043-1354(01)00288-3. [DOI] [PubMed] [Google Scholar]
- 26.Jain S, et al. Nat Struct Biol. 1996;3:375. doi: 10.1038/nsb0496-375. [DOI] [PubMed] [Google Scholar]
- 27.Fittkau M, Voigt W, Holzhausen HJ, Schmoll HJ. J Cancer Res Clin Oncol. 2004;130:388. doi: 10.1007/s00432-004-0557-8. [DOI] [PubMed] [Google Scholar]
- 28.Russell WM, Klaenhammer TR. Appl Environ Microbiol. 2001;67:1253. doi: 10.1128/AEM.67.3.1253-1261.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niwa T, Tsuruoka T, Inoue S, Naito Y, Koeda T. J Biochem. 1972;72:207. doi: 10.1093/oxfordjournals.jbchem.a129889. [DOI] [PubMed] [Google Scholar]
- 30.Jain S, et al. Nat Struct Mol Biol. 1996;3:375. [Google Scholar]
- 31.Jacobson RH, Zhang XJ, DuBose RF, Matthews BW. Nature. 1994;369:761. doi: 10.1038/369761a0. [DOI] [PubMed] [Google Scholar]
- 32.Marchler-Bauer A, et al. Nucleic Acids Res. 2009;37:D205. doi: 10.1093/nar/gkn845. (database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang JH, Chung TD, Oldenburg KR. J Biomol Screen. 1999;4:67. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 34.Ray J, Scarpino V, Laing C, Haskins ME. J Hered. 1999;90:119. doi: 10.1093/jhered/90.1.119. [DOI] [PubMed] [Google Scholar]
- 35.Turnbaugh PJ, et al. Nature. 2007;449:804. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sears CL. Anaerobe. 2005;11:247. doi: 10.1016/j.anaerobe.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 37.Grill JP, Manginot-Dürr C, Schneider F, Ballongue J. Curr Microbiol. 1995;31:23. doi: 10.1007/BF00294629. [DOI] [PubMed] [Google Scholar]
- 38.Brattain MG, Fine WD, Khaled FM, Thompson J, Brattain DE. Cancer Res. 1981;41:1751. [PubMed] [Google Scholar]
- 39.Siegmund B, et al. J Pharmacol Exp Ther. 2001;296:99. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.