Abstract
MsbA is a member of the ABC transporter superfamily and is homologous to ABC transporters linked to multidrug resistance. The nucleotide binding domains (NBDs) of these proteins include conserved motifs that are involved in ATP binding, including conserved SALD residues (D-loop) that are diagnostic in identifying ABC transporters but whose roles have not been identified. Within the D-loop, single point mutations L511P and D512G were discovered by random mutational analysis of MsbA to disrupt protein function in the cell [Polissi, A., and Georgopoulos, C. (1996) Mol. Microbiol. 20, 1221–1233] but have not been further studied in MsbA or in detail in any other ABC transporter. In these studies, we show that both L511P and D512G mutants of MsbA are able to bind ATP at near-wild-type levels but are unable to maintain cell viability in an in vivo growth assay, verifying the theory that they are dysfunctional at some point after ATP binding. An ATPase assay further suggests that the L511P mutation prevents effective ATP hydrolysis, and an ATP detection assay reveals that only small amounts of ATP are hydrolyzed; D512G is able to hydrolyze ATP at a rate 3-fold faster than that of the wild type. EPR spectroscopy studies using reporter sites within the NBDs also indicate that at least some hydrolysis occurs in L511P or D512G MsbA but show fewer spectral changes than observed for the same reporters in the wild-type background. These studies indicate that L511 is necessary for efficient ATP hydrolysis and D512 is essential for conformational rearrangements required for flipping lipid A.
ATP-binding cassette (ABC) transporters are a large and important superfamily of proteins that are involved in many human pathologies, including cystic fibrosis,2 multidrug resistance, 3 and Tangier’s disease.4 ABC transporters are responsible for the transport of a variety of substrates, including lipids, peptides, and drugs across the membrane. MsbA is homologous to bacterial ABC transporters, including LmrA,5 which can extrude drugs out of the cell.
MsbA is a 65 kDa protein found in the inner membrane of Gram-negative bacteria and is suggested to be a lipid flippase because of its ability to tranpsort lipid A across the inner membrane. MsbA is homologous to both bacterial and human proteins associated with multidrug resistance and is the only essential ABC transporter found in Escherichia coli;6 its knockout results in the toxic accumulation of lipid A within the inner leaflet of the inner membrane and cell death.7–9
The MsbA homodimer is comprised of two identical monomers, each with a cytosolic nucleotide binding domain (NBD) and a transmembrane domain (TMD) (Figure 1). The apo, MgADP/Vi-bound, and MgAMP-PNP-bound conformations of MsbA have been crystallized and show that the TMD includes a six-helix bundle and that the monomers are stably connected by an intertwining of the helical bundles at their periplasmic ends.10 The NBDs are responsible for ATP binding and hydrolysis, which is used as the energy to transport lipid A across the inner membrane of bacteria.
Figure 1.
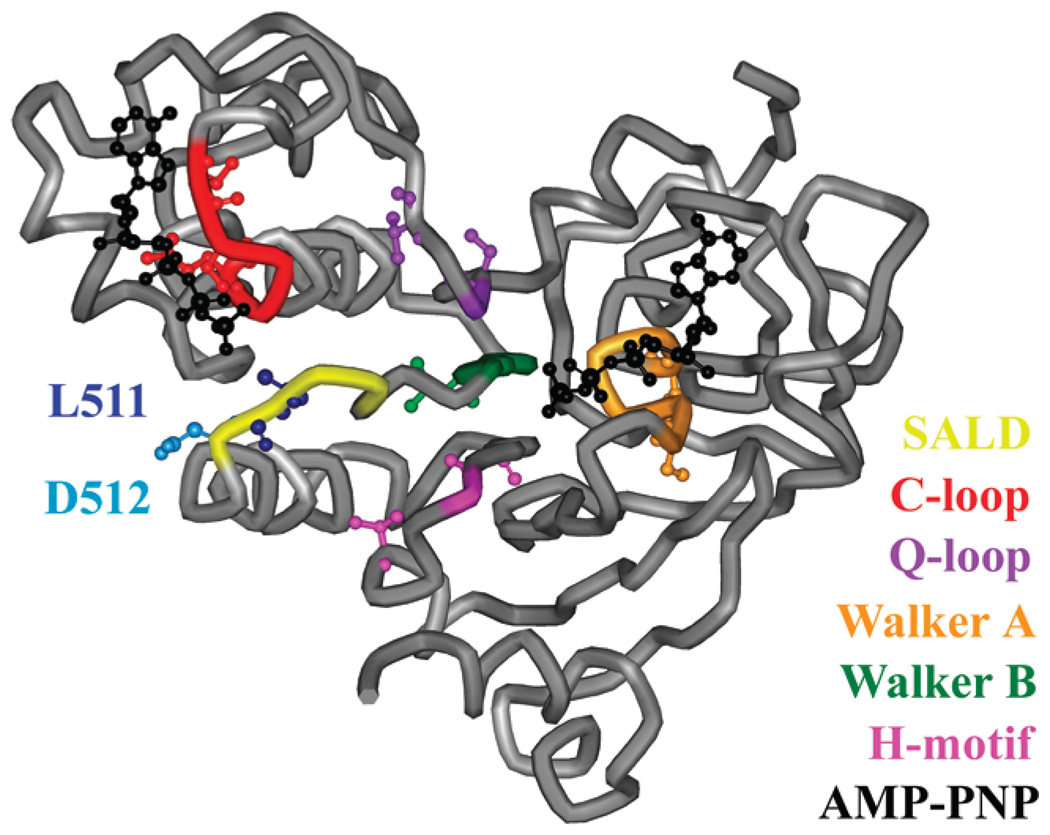
Nucleotide binding domain of the MsbA monomer. The SALD loop (yellow), including L511 (dark blue) and D512 (light blue), C-loop (red), Q-loop (purple), Walker A motif (orange), Walker B motif (green), H-motif (pink), and two ATP analogues (black) are highlighted on the MgAMP-PNP-bound structure (Protein Data Bank entry 3B60). The nine sites within and surrounding these motifs used in the spin labeling studies (S380, I385, S423, V426, S482, Q485, L504, V534, and T541) are represented as ball-and-stick models. The AMP-PNP shown nearest to the SALD loop (left) is the nucleotide associated with the opposing monomer (not shown).
Each NBD contains several motifs that are conserved throughout the ABC transporter superfamily. The C-loop, or LSGGQ signature sequence, Walker B motif, and Q- and H-motifs from one monomer come together with the Walker A motif from the opposite monomer to bind and/or hydrolyze ATP.11 This interaction, which involves opposite NBDs coming together to form two complete sites for the hydrolysis of ATP, has been described as a nucleotide sandwich.12 The Walker B motif (MsbA sites 501–505) has been thought to bind to the attacking water during hydrolysis and/or form a water-bridged contact with Mg2+. Following the Walker B motif is the D-loop (sites 509–512), with an SALD sequence, which is another conserved, diagnostic motif often used to determine membership in the ABC transporter superfamily.
The dysfunctional mutations studied here are the L511P and D512G point mutations in the D-loop identified by Polissi and Georgopoulis using random mutagenesis studies of the MsbA gene.1 Polissi and Georgopoulis first showed that these specific point mutations within the D-loop are not able to sustain cell growth in vivo but are likely able to allow binding to ATP. On the basis of these data, they suggested that L511P and D512G are unable to hydrolyze ATP in MsbA. Simulations of the NBDs of the maltose transporter (MalK)13 revealed probable interactions between the Walker A motif and H-motifs of one monomer and the D-loop of the opposite monomer, prompting speculation that the D-loop is involved in initiating dimer closure.14 Similarly, in HlyB-NBD, it has been suggested that the D-loop interacts with the H-motif from the opposite NBD.15,16 On the other hand, interactions observed in the MJ0796 crystal structure indicate that three of the D-loop residues (ALD) interact with the γ-Pi of the ATP and a nearby water molecule.12 Overall, the function of this motif is largely unknown and has yet to be investigated in detailed studies of ABC transporters.
In this study, we have used a variety of biochemical and biophysical approaches to characterize the L511P and D512G point mutations within E. coli MsbA. In vivo growth assays have been employed to monitor the ability of these mutant proteins, as well as more conservative amino acid substitutions such as L511I and D512E, to sustain cell growth and in vitro ATP binding. ATPase and ATP detection assays have been conducted to assess the ATP binding and hydrolysis capabilities of MsbA containing the previously identified L511P and D512G point mutations. In addition, site-directed spin labeling (SDSL) electron paramagnetic resonance (EPR) spectroscopy was employed to monitor the local mobility changes within this large membrane protein upon addition of nucleotide.17–19 Selected cysteine-substituted residues found in or around the C-loop, Walker A motif, Walker B motif, H-motif, and Q-loop were chosen to report on local changes within each motif resulting from the introduction of the L511P or D512G mutation. These studies show for the first time that L511 and D512 are highly essential sites within MsbA that do not generally tolerate substitutions without significant functional consequences.
EXPERIMENTAL PROCEDURES
Site-Directed Mutagenesis and Protein Purification
Amino acid point mutations were introduced into the Cys-less (C88S/C315S) MsbA gene within a pET28 (Novagen) plasmid using the QuikChange mutagenesis kit (Stratagene) and verified by sequencing (Retrogen) as described previously.20
MsbA containing an N-terminal 6x-His tag was overexpressed in E. coli NovaBlue cells (Novagen), purified by cobalt affinity chromatography (Talon resin, BD Biosciences Clontech), and spin-labeled with the cysteine-specific EPR probe MTSL (Toronto Research Chemicals), as described previously.20 Purified protein fractions were concentrated using Amicon Ultra YM-30 centrifugal filters, and final protein concentrations were determined using the detergent-compatible BCA protein assay kit (Pierce). For the EPR spectroscopy experiments, spin-labeled MsbA was reconstituted into 65:25:10 phosphatidylethanolamine (PE): phosphatidylglycerol (PG): cardiolipin (CL) inner membrane liposomes (Avanti Polar Lipids) at a 250:1 lipid: protein molar ratio, as described previously.21
In Vivo Growth Assay
pET28 plasmids encoding MsbA were individually electroporated into electrocompetent WD2 cells [a kind gift from C. Raetz (Duke University, Durham, NC) and W. Doerrler (Louisiana State University, Baton Rouge, LA)], which carry a chromosomal MsbA A270T mutation that causes the protein to become inactive at high temperatures.9 E. coli strains expressing WT and mutant MsbA protein were assayed for cell growth using a Thermo Varioskan Flash microplate reader as described previously.22 Cells expressing WT and Cys-less MsbA, or containing the empty pET28 plasmid, were run as positive and negative controls, respectively. Each mutant was run in triplicate at least twice using fresh WD2 cells and separate plasmid transformations. Cells expressing WT and normally functioning mutant MsbA typically grew to an OD600 of greater than ~0.5, while cells with no MsbA or completely inactive MsbA grew to an OD600 of no more than ~0.3.
ATP Binding Assay
Increasing concentrations of the fluorescently tagged ATP analogue TNP-ATP (0.02–1.0 µM) (Invitrogen) were added to 0.5 µM purified MsbA in 50 mM NaPO4 (pH 7), 0.01% DM, and 10% glycerol, and fluorescence spectra were recorded on a Photon Technology International spectrofluorometer using an excitation wavelength of 407 nm and an emission wavelength of 532 nm (Fobs). (Fobs − Fmax) / (Fmax − F0) was plotted versus TNP-ATP concentration23 in SigmaPlot (Systat Software) to determine Kd. Controls with and without 3 µM WT were used to determine Fmax and F0, respectively. Experiments were conducted in triplicate.
ATPase Assay
ATP hydrolysis was quantitated by measuring the release of γ-32Pi from ATP in the presence of MsbA, PE:PG: CL and CL lipids containing lipid A, and MgATP containing [γ-32P]ATP (Perkin-Elmer) at 37 °C at various time points over 2 min using Cherenkov counting in a Tri-Carb liquid scintillation counter (Perkin-Elmer) as previously described.22 Standard errors were generated from experiments conducted in triplicate and analyzed using SigmaPlot. Negative rates of hydrolysis arise from data points that appear to trend slightly downward over time, rather than upward, and are the result of inadequate, baseline hydrolysis.
ATP hydrolysis was also quantitated for WT, L511P, and D512G using the Enzchek Phosphate Assay kit (Molecular Probes) to determine Vmax and Km values for MsbA. The assays were conducted in a 96-well microplate following the manufacturer’s instructions for measuring the continuous release of Pi. MsbA (1 µM) and seven different concentrations (0–1 mM) of ATP were analyzed in triplicate in 200 µL reaction mixtures. A standard curve from measurements of 0–150 µM Pi was used to calibrate the data, which were recorded at 360 nm over 30 min using a ThermoScientific Varioskan Flash microplate reader and analyzed using SigmaPlot.
Nucleotide Detection Assay
The Kinase-Glo assay (Promega) utilizes luciferase and luciferin luminescence for the detection of ATP. For each experiment, excess MsbA and limiting ATP (10 µM MsbA and 5 µM ATP) or limiting MsbA and excess ATP (5 µM MsbA and 10 µM ATP) were analyzed along with standards containing 1–10 µM ATP (Sigma-Aldrich). Samples were incubated at room temperature for 30 min and then boiled for 5 min to denature the protein and release any bound nucleotide for detection in the assay. The assay was conducted according to the manufacturer’s protocol, and the luminescence was measured using a Thermo Scientific Varioskan Flash spectrophotometer with a LumiSens module.
Site-Directed Spin Labeling EPR Spectroscopy
Continuous wave (CW) EPR spectroscopy was conducted at room temperature on an X-band Bruker ELEXSYS 500 spectrometer equipped with a super high Q (SHQ) cavity (Bruker Biospin). Spectra were recorded under nonsaturating conditions over 100 G with a 1 G, 100 kHz field modulation. Samples were contained in a glass capillary and had volumes of 10–20 µL. The concentration of MsbA reconstituted into liposomes was approximately 200 µM in 50 mM NaPO4 (pH 7), and the mixture contained 20 mM MgCl2, 20 mM ATP or AMP-PNP, 2 mM EDTA, and 2 mM sodium orthovanadate (Vi), as indicated. The L504C/L511P and L504C/D512G spectra showed an increase in the small population of denatured protein (sharp peaks) during the freeze—thaw cycles used to ensure incorporation of ligand inside the lipid vesicles and also showed a small amount (<0.5 µM) of free spin-label released from the protein upon addition of ATP and MgATP/Vi, respectively, which was subtracted out for presentation in Figures 5 and 6. Spectra were analyzed using Multicomponent and dHpp software written in LabView by C. Altenbach (University of California, Los Angeles, CA).
Figure 5.
CW X-band EPR spectra of spin-labeled MsbA reconstituted into inner membrane lipids. Color coding is shown in the legend for each reporter residue in the Cys-less background and coupled with the L511P mutation without nucleotide (apo) or with ATP, MgATP/Vi, and MgATP. The motif in which each reporter cysteine is located is indicated above each set of spectra.
Figure 6.
CW X-band EPR spectra of spin-labeled MsbA reconstituted into inner membrane lipids. Color coding is shown in the legend for each reporter residue in the Cys-less background and coupled with the D512G mutation without nucleotide (apo) and with ATP, MgATP/Vi, and MgATP.
RESULTS
The in Vivo Growth Assay Confirms That Both Mutants Are Unable To Sustain Cell Growth
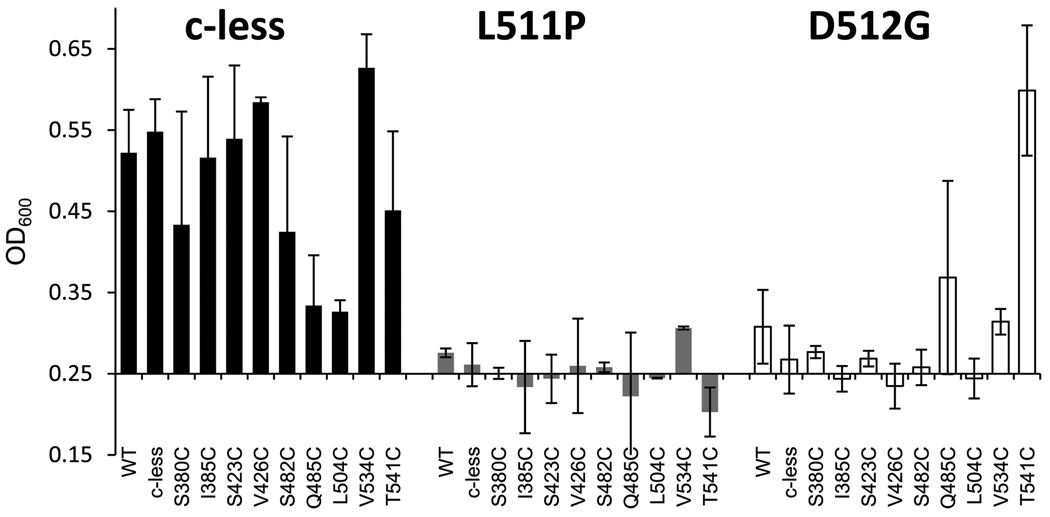
The data analysis of the in vivo growth assay of the L511P and D512G mutants confirmed the reduced cell viability originally reported by Polissi and Georgopoulos.1 E. coli strains expressing WT and Cys-less MsbA, and most of the reporter residues in the Cys-less background, grew to an OD600 of ≥0.50 (Figure 2), indicating normal cell viability.22 In contrast, L511P and D512G grew poorly in both WT and Cys-less backgrounds (Figure 2). When the L511P mutation is added to the cysteine reporters, the OD600 falls to 0.20–0.30, with L511P alone in a Cys-less background having an OD600 of 0.26 (Figure 2). This indicates that the L511P mutation in MsbA prevents cell growth and eliminates cell viability. When D512G is introduced in combination with each of the reporters, the OD600 varies between 0.23 and 0.6, with D512G in the Cys-less background having an OD600 of 0.27. These data indicate that the D512G mutation alone and coupled with most of the single-cysteine reporters prevents cell growth and eliminates cell viability. Interestingly, D512G in combination with either Q485C or T541C shows that cell growth and viability are intact. A lack of expression or insertion of the protein within the membrane could result in reduced cell viability, yet each of the MsbA mutants studied could be expressed and purified from the membrane fraction. Thus, loss of a specific protein function is being assayed here, and this could be due to a lack of or inefficient ATP binding, ATP hydrolysis, release of hydrolysis products, or lipid flipping.
Figure 2.
In vivo growth assay results for the NBD reporters in the Cys-less background (black22) and with the L511P (gray) and D512G (white) mutations. The in vivo growth assay signifies the ability of each plasmid-encoded MsbA mutant to maintain cell viability at 44 °C, after which the A270T-mutated chromosomal copy of MsbA is no longer functional. Error bars represent the standard deviation for data collected in triplicate twice. The WT data point listed with the Cys-less background series does in fact contain both native cysteine residues.
ATP Binding Intact
To further investigate the function of MsbA L511P and D512G, a fluorescent ATP analogue, TNP-ATP, was used to determine Kd values of ATP binding (Table 1). We find that L511P and D512G MsbA have Kd values for TNP-ATP binding of 0.33 and 0.81 µM, respectively, which is in good agreement with the WT MsbA value of 0.32 µM. Although the Kd value for D512G is slightly increased relative to the WT value, these data reveal that ATP binding is intact and at near normal levels for both point mutations. Because TNP-ATP is known to exhibit higher binding affinities than ATP, we also obtained Km values for ATP. L511P and D512G have Km values of 23 ± 12 and 122 ± 29 µM ATP, respectively. Upon comparison to the WT MsbA Km value of 117 ± 13 µM ATP, these data also suggest that ATP binding is indeed intact.
Table 1.
MsbA Dissociation Constants for TNP-ATP
| Kd (µM) | standard deviation | |
|---|---|---|
| WT | 0.32 | 0.05 |
| L511P | 0.33 | 0.16 |
| D512G | 0.81 | 0.15 |
ATP Is Hydrolyzed by MsbA D512G but Not L511P
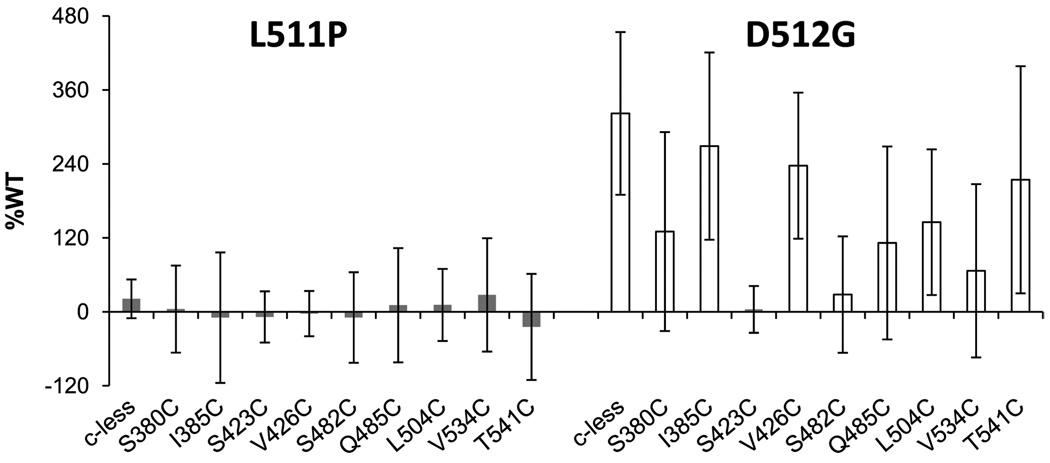
Given that L511P and D512G MsbA both bind ATP, we next examined their ability to hydrolyze ATP. Using a radioactive in vitro ATPase assay, we previously determined that the reporter residues in the Cys-less background maintain ATPase activity near or above that of WT, except for S482C and Q485C in the C-loop.22 In these studies, we find that L511P, in the Cys-less background or paired with each of the nine reporters, has a substantially decreased level of ATPase function (Figure 3). L511P in the Cys-less background has an ATPase rate that is 21% of that of WT, while L511P paired with each of the reporters has rates that vary between −21 and 28% of that of WT. In addition, full kinetic analysis showed that the maximal rate of hydrolysis for L511P is 5% of that of WT. These data clearly indicate that the L511P mutation drastically weakens MsbA’s ability to hydrolyze ATP.
Figure 3.
ATP hydrolysis rates as a percentage of WT activity over 2 min at 37 °C for L511P (gray) and D512G (white) in the Cys-less and single-cysteine reporter backgrounds, as indicated. Rates for the single-cysteine reporters in the Cys-less background have been published previously.22 Standard deviations are from experiments conducted in triplicate.
In contrast, D512G in the Cys-less background has an ATPase rate of hydrolysis that is 322% of that of WT (Figure 3). Also, full kinetic analysis gave a Vmax that was 165% of that of WT for D512G. When combined with the various reporter mutations, D512G has highly variable ATPase activity ranging from 4 to 269% of that of WT, with the majority of pairs exhibiting rates faster than that of WT. These data suggest that D512G has hyperactive ATPase activity itself and that hydrolysis rates can be reduced with the addition of the single cysteine mutations. Three double mutations have average rates of hydrolysis below that of WT. The S482C/D512G (28% WT) rate of hydrolysis is not considerably different from that observed for S482C in the Cys-less background (35% of that of WT), but the rates of hydrolysis for S423C/D512G (4% of that of WT) and V534C/D512G (67% of that of WT) fall significantly below the rates of the corresponding reporter mutations, which are 244 and 153% of that of WT, respectively.22 The ATPase data for L511P and D512G indicate for the first time that L511P has severely diminished ATPase activity and D512G is able to hydrolyze ATP at a much faster rate than WT or Cys-less MsbA.
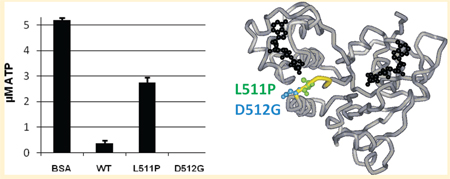
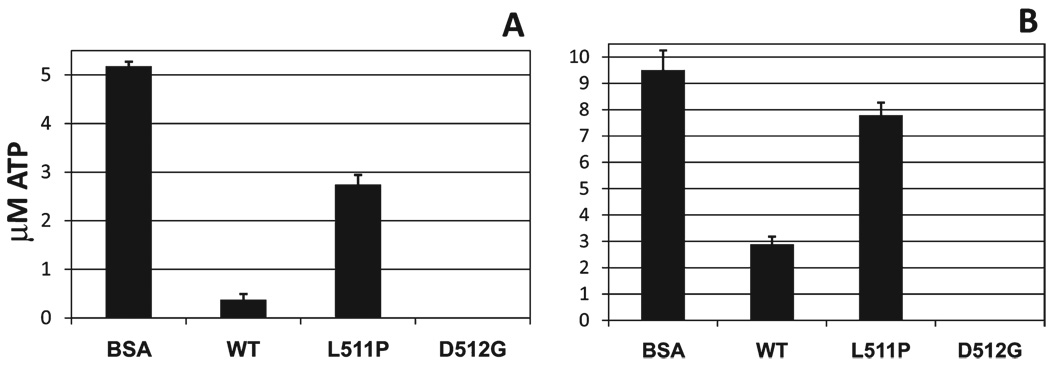
To further explore the ability of L511P and D512G MsbA to hydrolyze ATP, we conducted an ATP-detecting luminescence assay. WT MsbA and BSA were used as positive and negative controls, respectively. In Figure 4A, the experiment was performed with excess protein (10 µM) and a limiting amount of ATP (5 µM). Hydrolysis was allowed to proceed, and after 30 min, the remaining ATP was quantitated. The results show that, as expected, BSA does not hydrolyze ATP, with the 5 µM ATP added still present, and that WT MsbA does hydrolyze all but a small amount of ATP. L511P MsbA hydrolyzed approximately 2 µM ATP, while D512G hydrolyzed all of the available ATP. The fact that D512G hydrolyzed more ATP than WT is consistent with its faster rate of hydrolysis (Figure 3). Figure 4B shows the results of the experiment conducted with excess ATP (10 µM) and a limiting amount of protein (5 µM MsbA). Again, as expected, the BSA does not hydrolyze ATP and the WT hydrolyzes most of the available ATP. L511P again hydrolyzed approximately 2 µM ATP, and D512G once again hydrolyzed all of the ATP. These data confirm that on the time scale used here, D512G can hydrolyze ATP more efficiently than WT, and that L511P is inefficient at hydrolyzing ATP.
Figure 4.
ATP detection assay. L511P MsbA and D512G MsbA were analyzed using a luminescence assay to quantitate the ATP concentrations remaining after a 30 min room temperature incubation with MgATP. BSA and WT MsbA were analyzed as negative and positive controls, respectively: (A) 5 µM MgATP and 10 µM protein and (B) 10 µM MgATP and 5 µM protein. D512G data points for both panels are at 0 ± 0 µM.
CW EPR Spectroscopy Indicates Local Conformational Changes
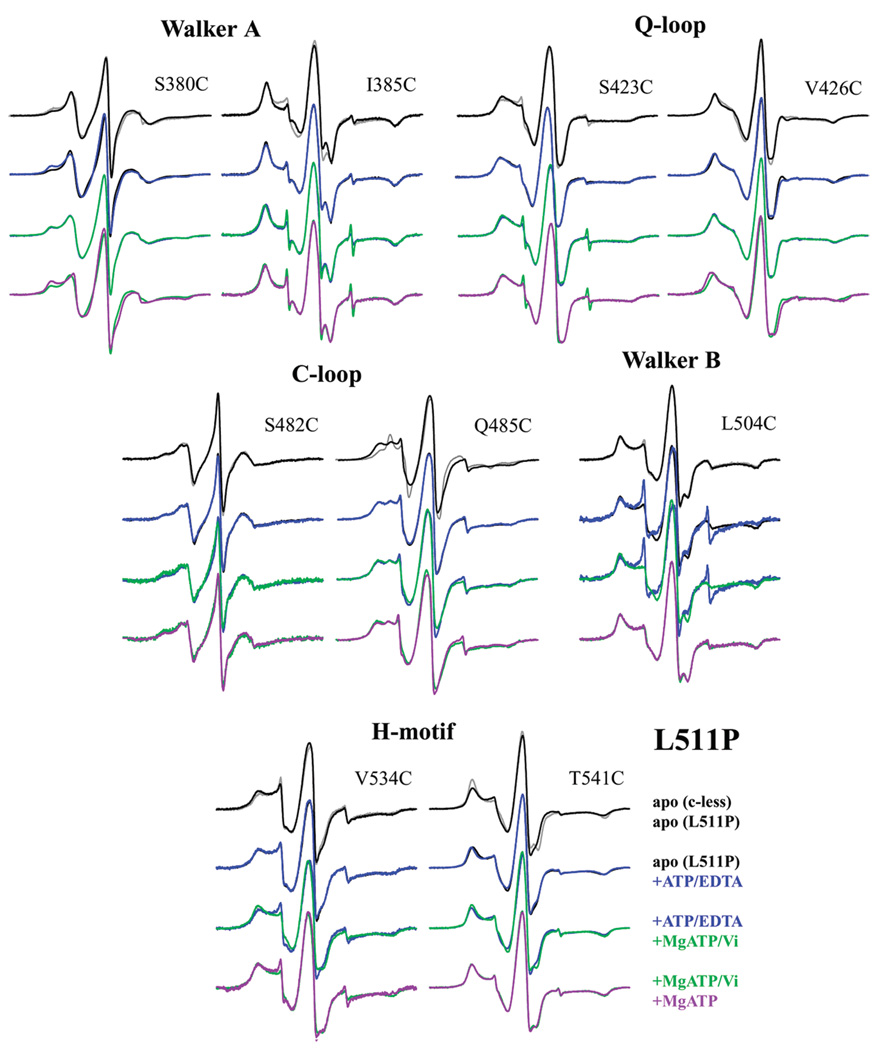
To assess the local changes within the MsbA NBD caused by the introduction of the L511P and D512G mutations, CW EPR spectroscopy was conducted using nine spin-labeled reporter sites, representing five of the conserved motifs, in the presence of each point mutation. Each reporter site (S380C, I385C, S423C, V426C, S482C, Q485C, L504C, V534C, and T541C) was selected from previous extensive characterization of spin-labeled cysteine sites within and surrounding the major conserved motifs21,22,24 based on its mobility changes upon the addition of specific ligands. Spectra of each reporter mutant pair were recorded for each of the four possible steps within the hydrolysis cycle: without ligand (apo) and with ATP, MgATP/Vi, and MgATP. The sample with ATP added also contained EDTA for the removal of any Mg, which prevents hydrolysis from proceeding and represents the ATP-bound state of the protein. The addition of MgATP allows hydrolysis to proceed if possible and potentially represents the MgADP-bound posthydrolysis state. The addition of MgATP and Vi together represents the immediate posthydrolysis state, if hydrolysis proceeds, trapping the MgADP/Vi ligands within the protein. All EPR data were collected on protein reconstituted into lipid bilayers.
L511P
The EPR spectrum of each of the spin-labeled reporter sites in the presence of the L511P point mutation showed mobility changes in the apo state when compared to the apo state of the reporter in the Cys-less background (Figure 5). Most of these changes were small but observable, while the spin-label at Q485C changed significantly because of the presence of the L511P mutation, indicating the local environment surrounding this site is altered solely because of the presence of this point mutation. However, no motional changes were observed for Q485C/L511P, or L504C/L511P, upon the addition of ligands.
The addition of ATP induced an increase in the more immobilized component of the spin-labeled S380C/L511P spectrum, as well as small decreases in mobility in the V426C/L511P and T541C/L511P spectra. The remaining sites were largely unchanged. This is similar to the changes observed upon ATP binding for S380C in the Cys-less background,21 which was likely attributed to direct interaction with ATP; however, the lack of change at I385C is inconsistent with its observed ATP-induced changes in the Cys-less background, indicating the ability of I385C to experience or report on changes upon ATP binding has been eliminated by the L511P mutation. On the other hand, the slight changes in mobility (outer splittings, 2Azz′, increase by 1 G) for V426C/L511P and T541C/L511P upon ATP binding were not observed in the Cys-less background. V426 is not near the bound ADP/Vi or L511 in the closed MsbA crystal structure (Protein Data Bank entry 3B5Z), though T541 is located on a helix neighboring L511; thus, it is possible that these subtle changes are the result of newly detected conformational rearrangements upon ATP binding caused by altered packing within the NBD because of the introduction of a proline residue.
Changes in mobility were more significant upon the addition of MgATP/Vi for the reporters in the Cys-less background (with the exception of the Walker A reporters).21 In the L511P background, some mobility changes were observed for S482C/L511P and V534C/L511P, as well as subtle changes for S385C, S423C, and T541C. Four of the reporters showed no changes upon addition of MgATP/Vi. This likely indicates that some hydrolysis may be occurring but not in a large population of the sample. Of the three samples that exhibited subtle changes upon addition of MgATP/Vi, the MgATP spectrum was identical in each case, suggesting that these sites do not report on changes caused by hydrolysis or the release of the γ-Pi but more likely on the small effect of Mg slightly increasing the amount of ATP bound.
Interestingly, S380C/L511P and V426C/L511P, both of which showed changes upon ATP binding, reveal a 1 G decrease in outer splittings upon addition of MgATP when compared to ATP or MgATP/Vi binding. S482C/L511P and V534C/L511P, both of which showed the most significant changes upon MgATP/Vi binding, show slight differences between the MgATP and MgATP/Vi spectra, indicating a motion more similar to that of the ATP-bound state and again suggesting a small amount of hydrolysis may be occurring.
Because of the observation of mobility changes in the L511P pairs, albeit much smaller in scale than those observed in the Cys-less background, in at least one stage of ligand binding in all motifs except the Walker B motif, it is suggested that limited hydrolysis does occur. This is consistent with all of our biochemical assays that show hydrolysis occurs but is severely limited in the L511P protein.
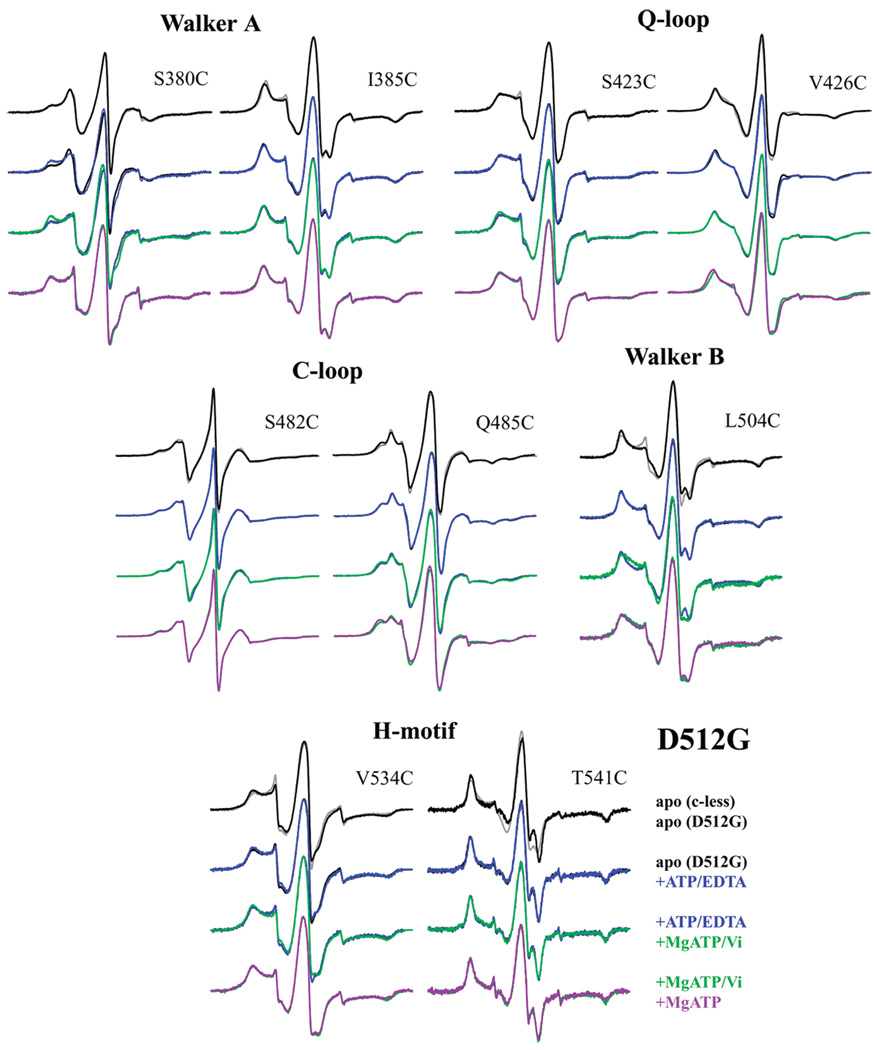
D512G
Most of the spin-labeled reporter sites, with the exception of S380C and S423C, showed slight changes in motion in their apo spectra in the presence of D512G when compared to the spectra observed in the Cys-less background (Figure 6), suggesting altered packing arrangements within the NBD due to elimination of the aspartate side chain. In contrast to the large motional change observed between Q485C/L511P and Q485C/Cys-less spectra (Figure 5), the Q485C/D512G spectrum was only slightly different from that in the Cys-less background, suggesting that the local environment surrounding Q485 is not significantly altered by the aspartate to glycine substitution at position 512. Overall, the changes induced in the apo form of the NBD due to the D512G mutation appear to be much more subtle and smaller in scale than those for the L511P mutation. However, changes observed upon MgATP and MgATP/Vi binding are less subtle.
Again, in D512G, ATP binding induced an approximately 25% shift from the mobile to the more immobilized component of the spin-labeled S380C spectrum, similar to the increase in the immobile component observed in both the Cys-less and L511P backgrounds. The remaining sites were not affected by ATP binding, which with the exception of I385C is consistent with the lack of changes observed upon ATP binding in the Cys-less background21,22,24 and with the hypothesis that ATP binds first to the Walker A motif.
In contrast, all motifs in the D512G background were affected by the addition of MgATP/Vi, albeit only slightly for S385C, S423C, Q485C, and V534. Again, these results are consistent with changes observed at this step in the Cys-less background, and the only difference is that these changes are not as large as those seen in the Cys-less background. Upon addition of MgATP/Vi, the spin-label at S380C-D512G exhibited an additional shift (approximately 30%) toward the immobilized population, which was not observed in the Cys-less background, and L504C/D512G exhibited a general broadening of the spectrum. These changes could be due to the higher affinity of these sites for MgATP versus ATP or due to tighter packing at the monomer interface as a result of the smaller side chain at D512G, resulting in greater immobilization of the Walker A site S380C mutation, which directly binds ATP, and the Walker B site L504C mutation, which is near D512.
For the majority of sites studied, the addition of MgATP induces the same mobility change observed for MgATP/Vi. Exceptions include V426C/D512G, which shows a more immobile spectrum in the presence of MgATP, and Q485C/D512G, which shows a slightly more immobile spectrum in the presence of MgATP. Evidently, these two sites, which are physically near the ATP molecule in the crystal structure, experience differences in mobility between the MgADP-bound and MgADP/Vi states, indicating that vanadate may slightly alter the conformation of nearby side chains in the D512G background.
The overall changes observed, especially upon addition of MgATP or MgATP/Vi, indicate that hydrolysis does occur in the D512G MsbA mutant and are consistent with the biochemical assay results presented above.
Mutational Analysis
Because leucine to proline and aspartate to glycine substitutions are rather extreme, more moderate substitutions at these two conserved D-loop sites were also created and evaluated for in vivo growth. Recently, we reported that L511C and D512C MsbA were able to sustain cell growth as well as or slightly better than WT MsbA but were unable to hydrolyze ATP.22 In this study, two similarly sized, but charge-altered, substitutions were made at each site (L511D and L511N or D512L and D512N), and the most conservative substitution according to size and charge (L511I and D512E), as well as a direct amino acid swap (L511D/D512L). Interestingly, neither L511 nor D512 is amenable to these non-cysteine substitutions, as observed by a complete lack of cell viability in the in vivo growth assay for each mutation listed above [OD600 < 0.3 (data not shown)], highlighting the fact that these are indeed highly critical conserved sites within the MsbA homodimer.
DISCUSSION
In these studies, the dysfunctional point mutations L511P and D512G in the nucleotide binding domain of the MsbA ABC transporter have been characterized utilizing a variety of biochemical and biophysical techniques. Mutations at these sites have been investigated for overall in vivo function in the regulatory sulfonylurea receptor SUR1 ATPase25 and when discovered in MsbA1 but otherwise have not been investigated further. In SUR1, L860 (equivalent to L511 in MsbA) retains full activity in its ability to regulate the Kir6.2 K+ channel when substituted with a cysteine in one NBD, similar to our findings, whereas in the opposing NBD, when I1512 (equivalent to L511 in the opposing MsbA NBD) is substituted with a cysteine, the activity is reduced to approximately half that of WT SUR1.25 Also, the equivalent D512C mutations in both domains of SUR1 are extremely detrimental to cell growth, unlike our findings that the D512C substitution is the only substitution identified so far in MsbA that is fully able to support cell growth in vivo. Though these two proteins do not perform similar functions beyond binding and hydrolyzing ATP, the fact that these conserved mutations tolerate substitutions differently emphasizes the sophistication and selectivity of ATP-binding proteins.
The inability of the L511P MsbA mutant to maintain cell viability is likely due to its inability to effectively hydrolyze ATP. Our data show that this mutant binds ATP at near WT levels but hydrolyzes only a small percentage of the ATP compared to WT. The ATP detection assay, which can determine if ATP is hydrolyzed to ADP, indicates that the L511P mutant can only hydrolyze fewer than one ATP per two homodimers on a longer time scale. This confirms that while the L511P mutant can bind ATP at WT levels, hydrolysis is not occurring at a sufficient pace to maintain cell viability in vivo.
On the other hand, the D512G point mutation in MsbA results in a protein that is able to hydrolyze more ATP and at a rate faster than that of WT yet is unable to sustain cell growth in vivo. These data suggest that this site is involved in communicating conformational changes to other parts of MsbA involved in rearrangements required for normal transport function. This is in agreement with the suggestions that the D-loop may be involved in communication with the opposing ATP binding site based on the structure of HlyB-NBD26 or in the initiation of NBD dimer closure based on simulations of MalK.14 Alternatively, because ATP hydrolysis is intact for D512G, which requires residues from both monomers to come together in a closed dimer, it is more likely that this motif is in communication with the TMDs for opening of the TMD dimer for lipid transport. The limited EPR spectral mobility changes observed for the D512G protein also indicate that large-scale conformational changes are not occurring. Additionally, the 3.2-fold increase in hydrolysis rate over the WT rate could be attributed to the fact that without these large conformational changes occurring during each cycle, ATP is more readily hydrolyzed.
It has been suggested that the D-loop sites interact with the Walker A motif and H-motif on the basis of MalK simulations and with the H-motif in HlyB-NBD. In our growth assay, we find that Q485C (C-loop) and T541C (near the H-motif) are the only two sites studied that can restore cell growth when coupled with D512G, which does not allow the cells to grow on their own. This indicates that interactions occur between the D-loop and both the C-loop and H-motif. In addition, spectral changes upon the addition of ATP are eliminated at position I385C (Walker A) when coupled with either the L511P or D512G mutation, and new spectral changes are observed at S380C (Walker A) when coupled with the D512G and S426C mutations (near the Q-motif) and at T541C (near the H-motif) when coupled with the L511P mutation. Plus, L504C (Walker B) is the only site studied that released free spin-label upon addition of ligand in the L511P and D512G backgrounds. These changes suggest that point mutations within the D-loop affect all of the other conserved motifs in some manner, including the Walker A motif, Walker B motif, H-motif, Q-motif, and C-loop. This likely accounts for the fact that most substitutions at the L511 and D512 positions in MsbA result in a loss of function in vivo. Thus, the highly conserved LD amino acids of the SALD loop appear to be more important than previously thought and deserve more thorough study in other transporters.
ACKNOWLEDGMENT
We thank Drs. Jimmy Feix and Neil Hogg for helpful discussions.
Funding Sources
This work was supported by the National Institutes of Health (GM070642).
ABBREVIATIONS
- ABC
ATP-binding cassette
- CL
cardiolipin
- CW
continuous wave
- DM
dodecyl maltopyranoside
- EPR
electron paramagnetic resonance
- MTSL
2,2,5,5-tetramethylpyrrolin-3-yl methanethiosulfonate spin-label
- NBD
nucleotide binding domain
- PE
phosphatidylethanolamine
- PG
phosphatidylglycerol
- SDSL
site-directed spin labeling
- TNP-ATP
2′ (3′)-O-(trinitrophenyl)-adenosine 5′-triphosphate, trisodium salt
- WT
wild-type
REFERENCES
- 1.Polissi A, Georgopoulos C. Mutational analysis and properties of the msbA gene of Escherichia coli, coding for an essential ABC family transporter. Mol. Microbiol. 1996;20:1221–1233. doi: 10.1111/j.1365-2958.1996.tb02642.x. [DOI] [PubMed] [Google Scholar]
- 2.Cotten JF, Welsh MJ. Covalent modification of the regulatory domain irreversibly stimulates cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 1997;272:25617–25622. doi: 10.1074/jbc.272.41.25617. [DOI] [PubMed] [Google Scholar]
- 3.Debenham PG, Kartner N, Siminovitch L, Riordan JR, Ling V. DNA-mediated transfer of multiple drug resistance and plasma membrane glycoprotein expression. Mol. Cell. Biol. 1982;2:881–889. doi: 10.1128/mcb.2.8.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodzioch M, Orso E, Klucken J, Langmann T, Bottcher A, Diederich W, Drobnik W, Barlage S, Buchler C, Porsch-Ozcurumez M, Kaminski WE, Hahmann HW, Oette K, Rothe G, Aslanidis C, Lackner KJ, Schmitz G. The gene encoding ATP-binding cassette transporter 1 is mutated in Tangier disease. Nat. Genet. 1999;22:347–351. doi: 10.1038/11914. [DOI] [PubMed] [Google Scholar]
- 5.van Veen HW, Venema K, Bolhuis H, Oussenko I, Kok J, Poolman B, Driessen AJ, Konings WN. Multidrug resistance mediated by a bacterial homolog of the human multidrug transporter MDR1. Proc. Natl. Acad. Sci. U.S.A. 1996;93:10668–10672. doi: 10.1073/pnas.93.20.10668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karow M, Georgopoulos C. The essential Escherichia coli msbA gene, a multicopy suppressor of null mutations in the htrB gene, is related to the universally conserved family of ATP-dependent translocators. Mol. Microbiol. 1993;7:69–79. doi: 10.1111/j.1365-2958.1993.tb01098.x. [DOI] [PubMed] [Google Scholar]
- 7.Doerrler WT, Gibbons HS, Raetz CRH. MsbA-dependent translocation of lipids across the inner membrane of Escherichia coli. J. Biol. Chem. 2004;279:45102–45109. doi: 10.1074/jbc.M408106200. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Z, White KA, Polissi A, Georgopoulos C, Raetz CR. Function of Escherichia coli MsbA, an essential ABC family transporter, in lipid A and phospholipid biosynthesis. J. Biol. Chem. 1998;273:12466–12475. doi: 10.1074/jbc.273.20.12466. [DOI] [PubMed] [Google Scholar]
- 9.Doerrler WT, Reedy MC, Raetz CRH. An Escherichia coli mutant defective in lipid export. J. Biol. Chem. 2001;276:11461–11464. doi: 10.1074/jbc.C100091200. [DOI] [PubMed] [Google Scholar]
- 10.Ward A, Reyes CL, Yu J, Roth CB, Chang G. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc. Natl. Acad. Sci. U.S.A. 2007;104:19005–19010. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davidson AL, Chen J. ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 2004;73:241–268. doi: 10.1146/annurev.biochem.73.011303.073626. [DOI] [PubMed] [Google Scholar]
- 12.Smith PC, Karpowich N, Millen L, Moody JE, Rosen J, Thomas PJ, Hunt JF. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol. Cell. 2002;10:139–149. doi: 10.1016/s1097-2765(02)00576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, Lu G, Lin J, Davidson AL, Quiocho FA. A tweezers-like motion of the ATP-binding cassette dimer in an ABC transport cycle. Mol. Cell. 2003;12:651–661. doi: 10.1016/j.molcel.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Oloo EO, Fung EY, Tieleman DP. The dynamics of the MgATP-driven closure of MalK, the energy-transducing subunit of the maltose ABC transporter. J. Biol. Chem. 2006;281:28397–28407. doi: 10.1074/jbc.M513614200. [DOI] [PubMed] [Google Scholar]
- 15.Zaitseva J, Jenewein S, Jumpertz T, Holland IB, Schmitt L. H662 is the linchpin of ATP hydrolysis in the nucleotide-binding domain of the ABC transporter HlyB. EMBO J. 2005;24:1901–1910. doi: 10.1038/sj.emboj.7600657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oswald C, Holland IB, Schmitt L. The motor domains of ABC-transporters. What can structures tell us? Naunyn-Schmiedeberg's Arch Pharmacol. 2006;372:385–399. doi: 10.1007/s00210-005-0031-4. [DOI] [PubMed] [Google Scholar]
- 17.Jeschke G, Bender A, Paulsen H, Zimmermann H, Godt A. Sensitivity enhancement in pulse EPR distance measurements. J. Magn. Reson. 2004;169:1–12. doi: 10.1016/j.jmr.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 18.Pannier M, Veit S, Godt A, Jeschke G, Spiess HW. Dead-time free measurement of dipole-dipole interactions between electron spins. J. Magn. Reson. 2000;142:331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- 19.Zhou Z, DeSensi SC, Stein RA, Brandon S, Dixit M, McArdle EJ, Warren EM, Kroh HK, Song L, Cobb CE, Hustedt EJ, Beth AH. Solution structure of the cytoplasmic domain of erythrocyte membrane band 3 determined by site-directed spin labeling. Biochemistry. 2005;44:15115–15128. doi: 10.1021/bi050931t. [DOI] [PubMed] [Google Scholar]
- 20.Buchaklian AH, Funk AL, Klug CS. Resting state conformation of the MsbA homodimer as studied by site-directed spin labeling. Biochemistry. 2004;43:8600–8606. doi: 10.1021/bi0497751. [DOI] [PubMed] [Google Scholar]
- 21.Buchaklian AH, Klug CS. Characterization of the Walker A motif of MsbA using site-directed spin-labeling electron paramagnetic resonance spectroscopy. Biochemistry. 2005;44:5503–5509. doi: 10.1021/bi047568v. [DOI] [PubMed] [Google Scholar]
- 22.Westfahl KM, Merten JA, Buchaklian AH, Klug CS. Functionally Important ATP Binding and Hydrolysis Sites in Escherichia coli MsbA. Biochemistry. 2008;47:13878–13886. doi: 10.1021/bi801745u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stewart RC, VanBruggen R, Ellefson DD, Wolfe AJ. TNP-ATP and TNP-ADP as probes of the nucleotide binding site of CheA, the histidine protein kinase in the chemotaxis signal transduction pathway of Escherichia coli. Biochemistry. 1998;37:12269–12279. doi: 10.1021/bi980970n. [DOI] [PubMed] [Google Scholar]
- 24.Buchaklian AH, Klug CS. Characterization of the LSGGQ and H motifs from the Escherichia coli lipid A transporter MsbA. Biochemistry. 2006;45:12539–12546. doi: 10.1021/bi060830a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masia R, Nichols CG. Functional clustering of mutations in the dimer interface of the nucleotide binding folds of the sulfonylurea receptor. J. Biol. Chem. 2008;283:30322–30329. doi: 10.1074/jbc.M804318200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanekop N, Zaitseva J, Jenewein S, Holland IB, Schmitt L. Molecular insights into the mechanism of ATP-hydrolysis by the NBD of the ABC-transporter HlyB. FEBS Lett. 2006;580:1036–1041. doi: 10.1016/j.febslet.2005.11.012. [DOI] [PubMed] [Google Scholar]