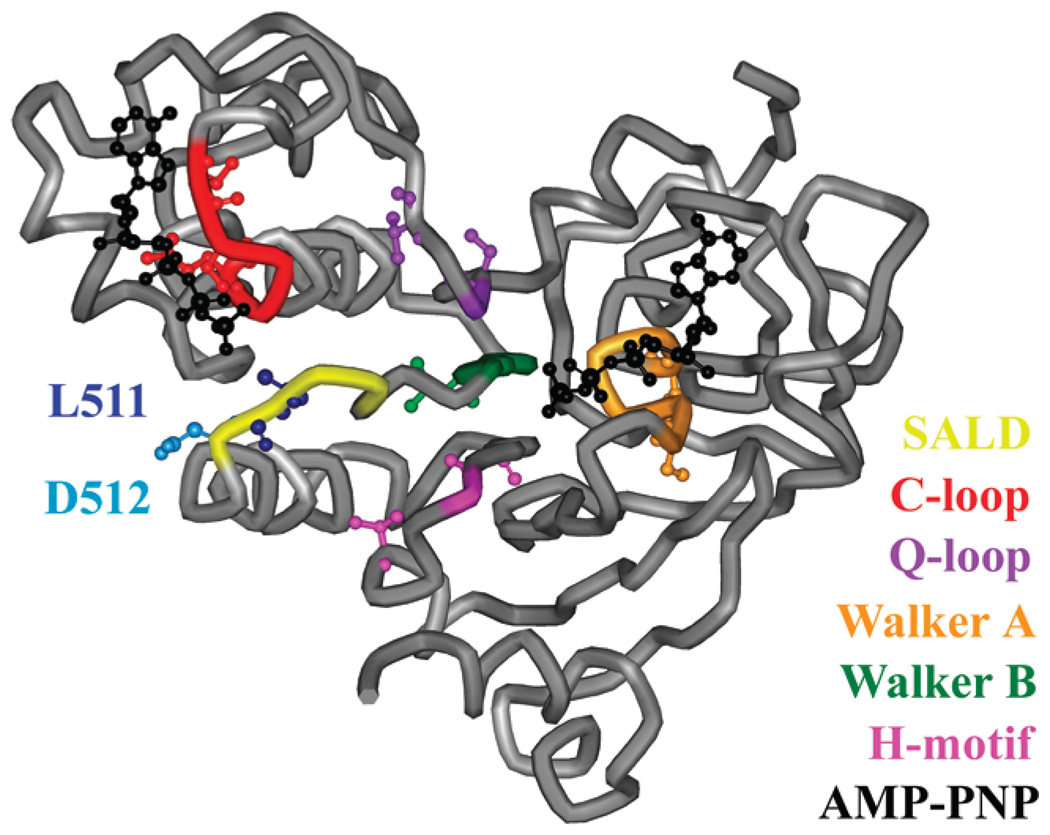
Figure 1.
Nucleotide binding domain of the MsbA monomer. The SALD loop (yellow), including L511 (dark blue) and D512 (light blue), C-loop (red), Q-loop (purple), Walker A motif (orange), Walker B motif (green), H-motif (pink), and two ATP analogues (black) are highlighted on the MgAMP-PNP-bound structure (Protein Data Bank entry 3B60). The nine sites within and surrounding these motifs used in the spin labeling studies (S380, I385, S423, V426, S482, Q485, L504, V534, and T541) are represented as ball-and-stick models. The AMP-PNP shown nearest to the SALD loop (left) is the nucleotide associated with the opposing monomer (not shown).