Abstract
Biofilms are complex communities of bacteria encased in a matrix composed primarily of polysaccharides, extracellular DNA, and protein. Staphylococcus aureus (S. aureus) can form biofilm infections, which are often debilitating due to their chronicity and recalcitrance to antibiotic therapy. Currently, the immune mechanisms elicited during biofilm growth and their impact on bacterial clearance remains to be defined. We utilized a mouse model of catheter-associated biofilm infection to assess the functional importance of Toll-like receptors 2 and 9 in the host immune response during biofilm formation, since ligands for both receptors are present within the biofilm. Interestingly, neither receptor impacted bacterial density or inflammatory mediator secretion during biofilm growth in vivo, suggesting that S. aureus biofilms circumvent these traditional bacterial recognition pathways. Several potential mechanisms were identified to account for biofilm evasion of innate immunity, including significant reductions in IL-1β, TNF-α, CXCL2, and CCL2 expression during biofilm infection compared to the wound healing response elicited by sterile catheters, limited macrophage invasion into biofilms in vivo, and a skewing of the immune response away from a microbicidal phenotype as evidenced by decreases in iNOS expression concomitant with robust arginase-1 induction. Co-culture studies of macrophages with S. aureus biofilms in vitro revealed that macrophages successful at biofilm invasion displayed limited phagocytosis and gene expression patterns reminiscent of alternatively activated M2 macrophages. Collectively, these findings demonstrate that S. aureus biofilms are capable of attenuating traditional host proinflammatory responses, which may explain why biofilm infections persist in an immunocompetent host.
Keywords: S. aureus, biofilm, macrophage, Toll-like receptor 2, Toll-like receptor 9, alternative activation, iNOS, arginase-1
INTRODUCTION
Biofilms are adherent communities of bacteria encased within a complex matrix that represents a considerable therapeutic challenge since organisms within these matrices are typically recalcitrant to conventional antibiotics. S. aureus is capable of causing biofilm infections on both natural body surfaces including the lung and heart as well as medical devices such as indwelling catheters and prostheses (1–3). Further complicating this issue is the recent emergence of community-associated methicillin-resistant S. aureus (CA-MRSA) strains that are able to cause serious infections in otherwise healthy individuals (4, 5). Although the majority of CA-MRSA isolates have been recovered from skin and soft tissue infections, the ability of S. aureus to form biofilms presents a significant concern for the diagnosis and therapeutic treatment of these infections.
Although host immune responses to planktonic S. aureus have been extensively investigated (6–10), little information is currently available regarding host immunity to S. aureus biofilms or how the bacteria modulate anti-microbial effector mechanisms when organized in this protective milieu. Due to the communal nature of biofilms, bacteria within them may exhibit distinct properties compared to planktonic growth phases of the same species, which may lead to differences in how biofilms are recognized by the host immune response. To date, the majority of studies investigating the innate immune response to biofilms have been performed with P. aeruginosa and S. epidermidis, where neutrophils have been shown to phagocytize biofilm-associated bacteria and produce oxidative bursts, albeit at reduced levels compared to planktonic bacteria (11–16). To our knowledge, only a few reports exist where leukocyte responses to S. aureus biofilm have been directly examined, most of which have been conducted in vitro (17–19). Therefore, additional mechanistic studies are needed to advance our understanding of the cross-talk between S. aureus biofilm and host innate immunity.
Currently, the majority of studies investigating innate immunity to S. aureus have focused on neutrophils (20–22), whereas macrophage responses to staphylococcal species have received relatively less attention. Although neutrophils are important anti-microbial effectors, their transcriptional capacity for inflammatory cytokine/chemokine production is limited and their short lifespan requires constant neutrophil recruitment due to cell turnover. In comparison, macrophages are more long-lived, produce high levels of proinflammatory mediators upon bacterial exposure that are critical for amplifying immune cell recruitment/activation cascades, and also exhibit potent phagocytic and anti-microbial effects (23, 24). In addition, resident macrophages are present in virtually all tissues and serve as a critical first line of defense against microbial invasion, in part, via bacterial recognition via Toll-like receptors (TLRs), which are discussed below. Therefore, macrophages represent another critical anti-microbial effector population; however, their responses to S. aureus biofilms remain to be defined.
Previous studies have demonstrated that bacterial biofilms, including those formed by S. aureus, are encased in a complex matrix composed of polysaccharides, extracellular DNA (eDNA), and proteins (25–28). It is well known that these microbial structural motifs are recognized by the innate immune system, in part, via the TLR family of pattern recognition receptors (PRRs). Upon activation, TLR signaling leads to the secretion of numerous proinflammatory mediators that serve to recruit and activate additional immune cell populations to sites of infection (29–35). TLR2 mediates recognition of several pathogen-associated molecular patterns (PAMPs) expressed by S. aureus that are released during normal bacterial growth and lysis (36–39) including lipoproteins, peptidoglycan (PGN), and lipoteichoic acids (LTA) (40–42). Numerous cell types express TLR2 including macrophages, dendritic cells, and neutrophils and recent reports have revealed that phagocytosis of S. aureus is required to trigger TLR2-dependent signaling within the phagosome and inflammasome activation (43, 44). TLR9 is an intracellular receptor that recognizes unmethylated CpG motifs characteristic of bacterial DNA (31, 45). Upon phagocytosis and digestion of S. aureus in the phagosome, bacterial DNA is liberated and engages TLR9. However, extracellular DNA (eDNA) can also trigger TLR9-dependent activation, which is relevant to biofilms due to the extensive amount of eDNA contained in the matrix (2, 25, 28). Although several studies have demonstrated that both TLRs are pivotal for host immune responses to planktonic S. aureus and associated PAMPs (7, 46, 47), to our knowledge the role of TLR2 and TLR9 in regulating host immunity to biofilm growth has not yet been examined in response to biofilm formed by any bacterial species.
Using a mouse model of catheter-associated biofilm infection, we demonstrate for the first time that S. aureus biofilms actively attenuate traditional anti-bacterial immune responses as demonstrated by significant reductions in cytokine/chemokine production associated with biofilm infected tissues compared to the wound healing response elicited by sterile catheters. In agreement with this apparent immune deviation, a role for either TLR2 or TLR9 in regulating biofilm infection in vivo could not be demonstrated. Several mechanisms were identified that could account for the persistence of biofilm infection including minimal iNOS induction concomitant with robust arginase-1 expression at the host-biofilm interface. In addition, although macrophage infiltration into biofilm infections was prominent, immunofluorescence staining revealed that few cells were able to migrate into the biofilm. Finally, biofilms induced significant and rapid cell death in those macrophages that invaded deep into the biofilm and programmed macrophages towards an alternatively activated M2 phenotype, suggesting additional mechanisms of immune deviation. Collectively, these studies demonstrate the unique properties of S. aureus biofilms to circumvent traditional anti-microbial effector pathways and persist in an immunocompetent host.
MATERIALS AND METHODS
Mouse strains
TLR2 and TLR9 KO mice (generously provided by Dr. Shizuo Akira, Osaka University, Japan) have been backcrossed with C57BL/6 mice for a total of eight generations and C57BL/6 mice (Charles River, Frederick, MD) were used as WT controls. Bone marrow-derived macrophages were generated from GFP transgenic mice (C57BL/6-Tg(CAG-EGFP)(Jackson Laboratories, Bar Harbor, ME). The animal use protocol, approved by the University of Nebraska Medical Center Animal Care and Use Committee, is in accord with the National Institutes of Health guidelines for the use of rodents.
S. aureus strains
S. aureus strain USA300 LAC was obtained from Dr. Frank DeLeo (NIAID Rocky Mountain Laboratories, Hamilton, MT) (48, 49). For in vivo imaging studies, USA300 LAC chromosomally transduced with the bacterial luciferase gene lux was used (Xen 29; Caliper Life Sciences, Hopkinton, MA; USA300 LAC::lux).
In vivo model of S. aureus biofilm infection
The impact of TLR2 or TLR9 on biofilm formation and growth in vivo was assessed using a mouse model of catheter-associated biofilm infection as previously described with minor modifications (50, 51). Briefly, WT, TLR2 KO, or TLR9 KO mice received subcutaneous implants of sterile 1 cm catheter segments in both flanks under general anesthesia. One catheter was inoculated with 20 μl (5 × 105 CFU) log-phase USA300 LAC::lux, whereas the other catheter received an equal volume of PBS to evaluate the foreign body response. The extent of biofilm formation was monitored longitudinally in the same cohort of mice using an In Vivo Imaging System (IVIS Spectrum; Caliper Life Sciences) under isoflurane anesthesia and separate groups of animals were sacrificed at days 3, 7, and 10 post-infection to determine absolute bacterial burdens associated with catheters and surrounding tissues. In experiments designed to assess the ability of identical S. aureus inoculums to establish biofilm versus subcutaneous infection in the absence of an indwelling device, mice received subcutaneous injections of USA300 LAC::lux (5 × 105 cfu in 20 μl) and processed as described above for biofilm infections.
Scanning electron microscopy (SEM)
Catheters and associated tissues were harvested from mice at day 10 post-infection and fixed with 0.1 M Sorensen’s phosphate buffer containing 2% glutaraldehyde and 2% paraformaldehyde. Fixed specimens were cut longitudinally and washed three times in 0.1 M Sorensen’s phosphate buffer. The specimens were dehydrated using a graded series of ethanol washes and critical pointed dried in a Pelco CPD2 critical point dryer (Ted Pella, Inc., Redding, CA). Dried specimens were mounted on aluminum stubs with carbon tabs and colloidal silver paste and sputter coated with gold-palladium using a Hummer VI sputter coater (Anatech, LTD, Battle Creek, MI). Samples were viewed using a Quanta 200 scanning electron microscope (FEI, Hillsboro, OR) operated at 25 Kv.
Flow cytometry
To compare the degree of macrophage influx into S. aureus biofilms versus subcutaneous abscesses, FACS analysis was performed. Briefly, abscesses and catheter-associated tissues were collected at days 3, 7, and 14 post-infection and minced in HBSS supplemented with 10% FBS (HyClone, Logan, UT) and filtered through a 70 μm nylon mesh cell strainer. The resulting slurry was digested for 30 min at 37° C in HBSS supplemented with 2 mg/ml collagenase type I and 28 U/ml DNAse I (both from Sigma-Aldrich, St. Louis, MO) to obtain a single-cell suspension. Following enzyme neutralization, cells were layered onto a discontinuous Percoll gradient (1.03 to 1.088 g/ml) and centrifuged at 2,400 rpm for 20 min at room-temperature in a swinging bucket rotor, whereupon the cell interface was collected. Following extensive washes and incubation in Fc Block™ (BD Biosciences, San Diego, CA) to minimize non-specific antibody binding to Fc receptors, cells were stained with F4/80-FITC (AbD Serotec, Raleigh, NC) to identify macrophages. Cells were analyzed using a BD FACSAria with compensation set based on the staining of each individual fluorochrome alone and correction for autofluorescence with unstained cells. Controls included cells incubated with isotype control antibodies to assess the degree of non-specific staining.
Immunofluorescence staining and confocal microscopy
Biofilm-associated and abscess tissues were collected at days 7 and 14 following infection, fixed in 10% formalin, and embedded in paraffin, whereupon paraffin sections (10 μm) were prepared by the UNMC Tissue Science Facility for H & E, Gram, and immunofluorescence staining. Sections were deparaffinized in xylene and a graded series of alcohols and antigen retrieval was performed by microwaving sections in 10 mM NaCi buffer, pH 6.0 for 14 min. Next, tissues were rinsed once in 1X PBS, pH 7.4 and stained with antibodies specific for arginase-1 (Santa Cruz; San Diego, CA), iNOS (Abcam, Cambridge, MA) and Iba-1 (macrophage-specific marker; Biocare Medical, Concord, CA). Staining was detected using either FITC-conjugated or biotinylated secondary antibodies (obtained from Jackson Immunoresearch, West Grove, PA), the latter of which was followed by a streptavidin-594 conjugate (Invitrogen). Confocal images were acquired using a Zeiss 510 META laser scanning confocal microscope (Carl Zeiss, Thornwood, NY) and staining specificity was confirmed by the absence of fluorescence signal following incubation of tissues with secondary antibodies alone.
Gentamicin protection assays (GPA)
To determine whether macrophages recovered from biofilm-infected tissues harbored viable S. aureus intracellularly, GPAs were performed at day 7 post-infection as previously described (52). Day 7 was the only interval examined as this represented the time point at which maximal catheter colonization had occurred. Following macrophage recovery by FACS, an aliquot of cells was taken to determine total bacterial counts (extra- and intracellular organisms). A second aliquot of sorted macrophages was exposed to 100 μg/ml gentamicin for 2 h at 37° C to kill extracellular organisms, whereupon macrophages were washed twice to remove residual gentamicin, lysed using sterile distilled water, and intracellular bacterial titers enumerated. To ensure that intracellular bacteria were sensitive to an antibiotic which can penetrate mammalian cell membranes, macrophages were incubated with 1 mg/ml rifampicin. Results are expressed as the number of viable intracellular S. aureus per 103 biofilm-associated macrophages.
ELISA
To compare the production of inflammatory mediators associated with biofilm infected versus sterile catheters of WT, TLR2 KO, and TLR9 KO mice, sandwich ELISA kits were utilized to quantitate TNF-α, IL-1β, and CCL2 (OptiEIA, BD Biosciences) or CXCL2 (DuoSet, R & D Systems) levels. Results were normalized to the amount of total protein recovered to correct for differences in tissue sampling size.
Generation of S. aureus static biofilms in vitro
The USA300 LAC strain was cured of its plasmid encoding erythromycin resistance (erm) to facilitate genetic manipulation. The erm-sensitive variant was transformed with plasmid pCM11 to express GFP driven by the sarA P1 promoter (USA300 LAC-GFP) and plasmid expression was maintained with erm selection (10 μg/ml) (53). Our initial investigation of macrophage-S. aureus biofilm interactions utilized trypticase soy broth (TSB). Although this broth formulation was compatible with biofilm growth, it induced changes characteristic of apoptosis in macrophages (data not shown). Therefore, we evaluated the growth kinetics of USA300 LAC-GFP in defined macrophage medium (i.e. RPMI-1640 supplemented with 10% heat-inactivated FBS) compared to TSB. Biofilm maturation was slightly delayed in macrophage medium compared to equivalent growth periods in TSB, and biofilms propagated for either 4 or 6 days were selected for further analysis based on their differing degrees of structural maturity. Specifically, 4 day-old biofilms were considered more immature in terms of average thickness (34 μm) and irregular density, whereas 6 day-old biofilms were classified as mature based on a relatively uniform thickness (52 μm average), presence of tower structures, and more consistent density (Supplemental Fig. 1). Biofilms were stained with TOTO3 or WGA-Alexa Fluor 633 (both from Molecular Probes, San Diego, CA) to visualize eDNA/dead cells or polysaccharide intercellular adhesion (PIA), respectively. TOTO3 staining revealed some eDNA/dead bacteria at the biofilm base with additional staining located throughout the structure in a punctate staining pattern (Supplemental Fig. 1 and data not shown). Both 4 and 6 day-old biofilms also expressed PIA as assessed by WGA staining (data not shown).
To generate static biofilms, sterile 2-well glass chamber slides (Nunc, Rochester, NY) were treated with 20% human plasma (kindly provided by Dr. Steve Carson, University of Nebraska Medical Center) in sterile carbonate-bicarbonate buffer (Sigma) overnight at 4° C to facilitate bacterial attachment (50). Starter cultures of USA300 LAC-GFP were prepared from a single bacterial colony and incubated overnight in macrophage medium supplemented with 10 μg/ml erm at 37° C while shaking at 250 rpm under aerobic conditions. The following day, plasma coating buffer was removed and each chamber inoculated with USA300 LAC-GFP (diluted to an OD600 of 0.050; 2 ml/chamber), whereupon bacteria were incubated at 37° C under static aerobic conditions for a period of up to 6 days. Medium was carefully replenished every 24 h to prevent disruption of the biofilm structure.
Preparation of bone marrow-derived and thioglycollate-elicited peritoneal macrophages
C57BL/6 mice were used to procure thioglycollate-elicited peritoneal macrophages as previously described (54). Elicited macrophages were washed extensively and labeled with 5 μM CellTracker Orange (CTO; Molecular Probes) for visualization according to the manufacturer’s instructions. Macrophage cell counts and viability following CTO staining were determined using Trypan blue dye exclusion. Initial optimization experiments verified that CTO labeling did not affect macrophage proinflammatory responses to planktonic S. aureus or cell viability (data not shown). For experiments to examine the impact of S. aureus biofilms on macrophage gene expression, bone marrow-derived macrophages (BMDM) were prepared from GFP Tg mice by flushing long bones with sterile 1X PBS and culturing cells in macrophage medium supplemented with 10 μg/ml macrophage colony-stimulating factor (M-CSF; Invitrogen). FACS analysis verified that > 95% of cells were macrophages based on F4/80 staining (data not shown).
Macrophage-biofilm co-culture models
CTO-labeled macrophages were added to biofilms at 107 cells/chamber prior to observation by confocal microscopy. Based on enumeration of bacterial densities within the biofilm, this equated to a multiplicity of infection (MOI) of approximately 10:1 (bacteria:macrophage). Slides were incubated with macrophages at 37° C under static aerobic conditions for either 1 or 24 h prior to observation. To determine how macrophage interactions may vary during biofilm versus planktonic growth, macrophages were also incubated with planktonic bacteria in identical glass chamber slides that had not been pre-coated with human plasma. In some experiments, TOTO3 staining was performed to discriminate between viable versus dead macrophages, which could be differentiated from dead bacteria based on size. Macrophage-biofilm or -planktonic interactions were visualized at the appropriate time point using a Zeiss laser scanning confocal microscope (LSM 510 META). The confocal pinhole was set to obtain an optical section thickness of 1 μm. Z-stacks (100–150 μm in thickness) were collected from beneath the glass slide extending to above the point where CTO-labeled macrophages could be detected. Three-dimensional images of biofilms and measurements to demonstrate the relative proximity of macrophages from the biofilm surface were made using Xen 2007 software (Zeiss). All confocal image files were subjected to a coding strategy to ensure that the observer performing quantitations was blinded to treatment group identity to avoid potential bias during analysis.
The impact of S. aureus biofilms on macrophage activation was evaluated by qRT-PCR. Briefly, GFP+ BMDM were incubated with USA300 LAC static biofilms for 2 h as described above or planktonic bacteria for comparison, followed by mechanical dissociation by trituration. Subsequently, both preparations were incubated with the vital dye 7-Aminoactinomycin D (7-AAD; eBiosciences, San Diego, CA) and viable macrophages (GFP+, 7-AAD−) were collected by FACS. Total RNA was isolated from sorted macrophages using a Taqman® Pre-Amp Cells-to-Ct™ kit (Applied Biosystems, San Diego, CA) for limiting cell numbers and qRT-PCR performed for iNOS, arginase-1, TNF-α, and IL-1β. Results were normalized against cycle thresholds for the housekeeping gene GAPDH and are presented as the fold-change (2ΔΔCt) of macrophages exposed to biofilms relative to planktonic bacteria (mean ± SEM from four independent experiments).
Phagocytosis Assay
To determine whether macrophages were capable of phagocytizing disrupted biofilm material, S. aureus biofilms were mechanically triturated and fragments incubated with macrophages for phagocytosis assays as previously described (55–57). Macrophages were exposed to planktonic bacteria at a 1:10 ratio as a control. Samples were incubated at 37°C for 30 min to allow bacterial uptake by macrophages, whereupon trypsin was added at a final concentration of 0.25% for 10 min to remove any residual bacteria at the macrophage surface. Macrophages were washed 3X with PBS to remove remaining bacteria, fixed to glass slides by cyto-centrifugation (CytoSpin, Thermo Scientific) and viewed under 40X oil immersion using a Zeiss Observer A.1 fluorescent microscope to evaluate the presence of intracellular GFP+ bacteria.
Statistics
Significant differences in bacterial titers and cytokine expression between experimental groups were determined using the Student’s t-test with Welch’s correction for unequal variances and FACS data was evaluated by One-way ANOVA followed by the Holm-Sidak method for multiple pair-wise comparisons. For all analyses, a p-value of less than 0.05 was considered statistically significant.
RESULTS
Establishment of S. aureus biofilms in vivo
Because bacterial biofilms are typically associated with chronic indolent infections they are presumed to evade the host immune response, yet few studies to date have provided scientific evidence to substantiate this phenomenon in vivo, which was the primary objective of this work. We utilized a mouse model where biofilms were established on surgically implanted catheters, which mimics biofilm formation associated with infected medical devices in humans (50, 51). Evaluation of tissues surrounding infected catheters by Gram-staining revealed evidence of biofilm growth within the catheter lumen as well as the catheter-tissue interface (Fig. 1A and B, respectively). At the ultrastructural level, scanning electron microscopy (SEM) also verified biofilm formation at the interface of the catheter and surrounding host tissue (Fig. 1C). Interestingly, based on size differentials, it was apparent that host cells were not capable of forming intimate associations with the biofilm (Fig. 1C, arrows). Detailed examination of the catheter lumen by SEM revealed a contiguous bacterial layer with the appearance of tower structures (Fig. 2A and B). The interior of these towers were largely hollow and higher magnification revealed the presence of an extensive fiber network, presumably matrix material that aggregated during the SEM dehydration process (Fig. 2C). These structural features are reminiscent of the tower structures observed during in vitro biofilm growth conditions (28, 58). Collectively, these findings demonstrate the formation of a bona fide S. aureus biofilm in vivo, allowing an assessment of the importance of innate immunity in regulating the host response to biofilm infection.
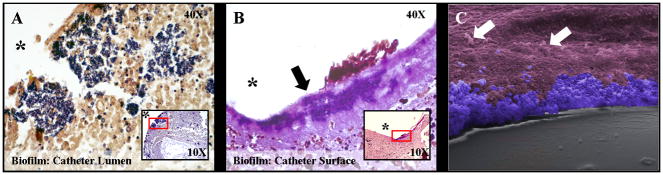
Figure 1. Characterization of S. aureus localization during biofilm development.
Catheters and surrounding tissues were isolated from wild type mice at day 10 following S. aureus infection and subjected to Gram-staining (A and B) or SEM (C). There was evidence of bacterial growth (purple) within the catheter lumen (A) as well as host tissue immediately surrounding the catheter (B, arrow). Insets in panels A & B depict 10X magnification with red rectangles indicating the area enlarged at 40X magnification and asterisks denoting the original location of infected catheters that were non-adherent to glass slides. A 400X magnification SEM image (C) has been pseudocolored to highlight S. aureus biofilm (blue) at the interface between the catheter (grey) and surrounding host tissue (purple). Host cells located at a distance from the biofilm are indicated by arrows.
Figure 2. Evidence of catheter-associated biofilm growth in vivo.
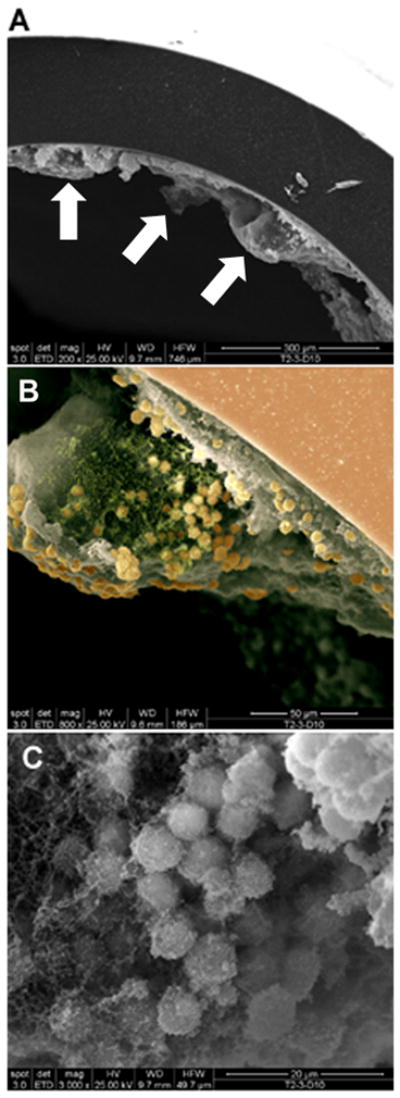
Catheters were isolated from wild type mice at day 10 following S. aureus infection and processed for SEM analysis. The smooth surface at the periphery of the images in A and B represents the catheter with biofilm visible on the internal face. (A), 200X magnification demonstrating the irregular undulating pattern of the biofilm surface (arrows indicate tower structures); (B), higher magnification of a tower from the image shown in (A) revealing a predominantly hollow interior with numerous cocci at the margins (800X magnification). The image has been pseudocolored to highlight S. aureus (gold) and presumably matrix material (green), which likely aggregated during the SEM dehydration process, from the remaining biofilm structure (grey; 800X magnification). The smooth surface at the upper right represents the catheter (salmon) with biofilm visible on the internal face. (C), 3000X magnification of a S. aureus cluster within the tower depicted in (B).
S. aureus biofilms evade TLR2 and TLR9 recognition in vivo
Since both TLR2 and TLR9 ligands are present within S. aureus biofilms (i.e. lipoproteins/PGN and eDNA, respectively), we next examined what functional role these receptors play in enabling biofilm recognition utilizing TLR2 and TLR9 KO mice. For these studies, we compared the establishment of catheter-associated biofilm formation versus abscesses induced by the subcutaneous inoculation of S. aureus in the absence of an indwelling device. The rationale for this approach was two-fold: first, it allowed us to evaluate whether TLR2 or TLR9 exert differential roles in bacterial clearance between biofilm versus subcutaneous infections; and second, it enabled an assessment of the importance of an indwelling device for bacterial persistence. Surprisingly, in vivo imaging studies and bacterial enumeration revealed that the degree of biofilm infection was qualitatively (Fig. 3) and quantitatively (Fig. 4) similar between TLR2 and TLR9 KO mice compared to WT animals. In addition, by comparing the ability of identical S. aureus inoculums to develop biofilm versus subcutaneous infection, we demonstrated an essential role for an indwelling catheter in establishing sustained infection. Specifically, prominent bioluminescent signals and bacterial burdens were detected in biofilm infected WT, TLR2 KO, and TLR9 KO mice, whereas minimal bacterial growth was detected in animals inoculated with S. aureus in the absence of an indwelling catheter (Fig. 3 and 4). Although previous studies have demonstrated that TLR2 and TLR9 are important for bacterial containment in other models of bloodstream or subcutaneous infection, these studies required extremely high bacterial inoculums to show any phenotypes (7, 35). In contrast, our experiments utilized a relatively low infectious dose (i.e. 5 × 105 cfu), which was rapidly cleared and implicates the involvement of alternative PRRs. Collectively, these findings highlight the ability of S. aureus to persist in the context of an indwelling device and circumvent recognition by traditional PRRs that would normally be triggered by planktonic bacteria. This speaks to the immune evasiveness of biofilms that is evident from a clinical perspective.
Figure 3. Visualization of S. aureus biofilm infection using IVIS.

Wild type (WT), TLR2 knockout (KO), and TLR9 KO mice were infected with 5 × 105 CFU of USA300 LAC::lux either in the lumen of surgically implanted catheters to establish biofilm infection or subcutaneously in the absence of any indwelling device. At the indicated time points post-infection, mice were subjected to IVIS imaging to visualize the extent of biofilm or subcutaneous infection. Images are presented from one mouse per group that was imaged at all time points to map the progression of biofilm infection.
Figure 4. Infection with an equivalent S. aureus dose leads to the establishment of catheter-related biofilm infection but rapid clearance from subcutaneous sites.
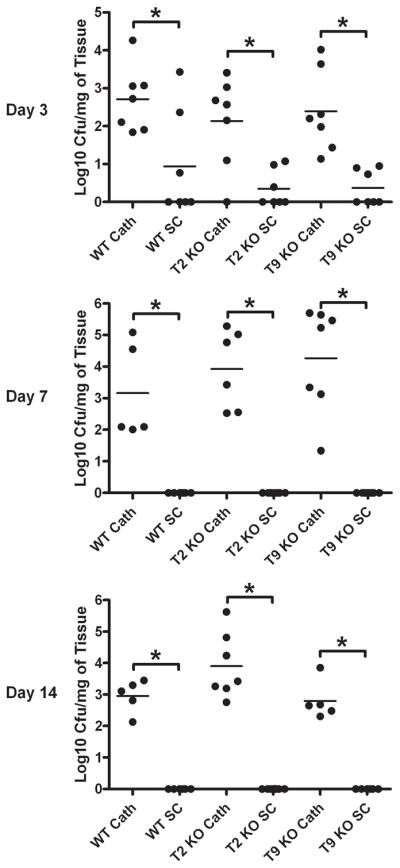
Wild type (WT), TLR2 knockout (KO), and TLR9 KO mice were infected with 5 × 105 CFU of USA300 LAC::lux either in the lumen of surgically implanted catheters (cath) to establish biofilm infection or subcutaneously (sc) in the absence of any indwelling device. Animals were sacrificed at the indicated days following S. aureus infection, whereupon host tissues surrounding infected catheters or s.c. injection sites were homogenized to quantitate bacterial burdens. Results are expressed as the number of CFU per mg host tissue to correct for differences in tissue sampling size. Significant differences in bacterial burdens between biofilm-infected versus s.c. injected mice are denoted by asterisks (*, p < 0.001).
S. aureus biofilms actively attenuate host proinflammatory responses
We next evaluated whether S. aureus biofilms would alter proinflammatory mediator secretion profiles in an attempt to explain why these infections persist in an immunocompetent host. Examination of several inflammatory signals responsible for macrophage and neutrophil recruitment (CCL2 and CXCL2, respectively) and activation (TNF-α and IL-1β) were significantly attenuated in biofilm-infected tissues compared to the wound healing response elicited following the insertion of sterile catheters (Fig. 5). Again, no differences in inflammatory mediator expression were observed between biofilm infected tissues from WT, TLR2 KO, or TLR9 KO mice (data not shown), which correlated with the failure to detect significant differences in bacterial burdens in these animals (Supplemental Figs. 2 and 3).
Figure 5. S. aureus biofilm attenuates inflammatory mediator expression compared to the wound healing response elicited following sterile catheter implantation.
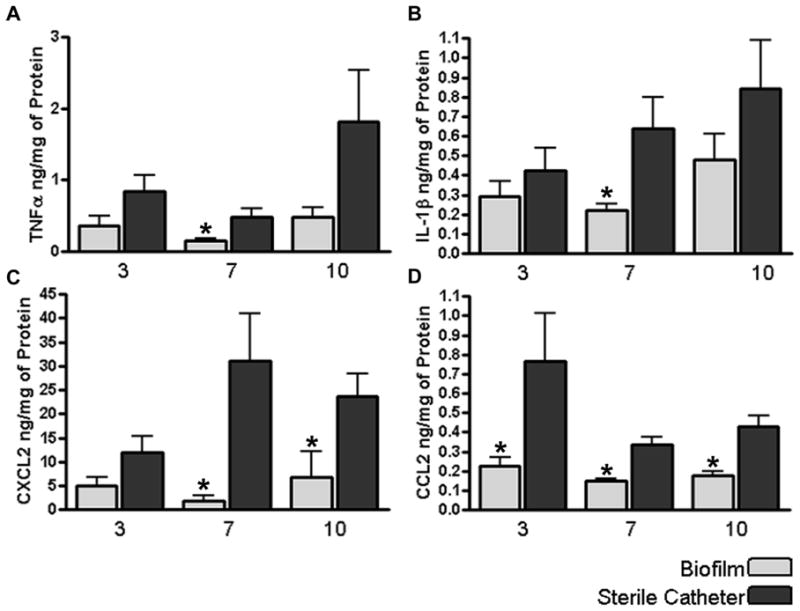
Tissues surrounding S. aureus infected or sterile catheters from wild type mice were collected and homogenized at the indicated time points to obtain supernatants to quantitate differences in TNF-α (A), IL-1β (B), CXCL2 (C), or CCL2 (D) expression by ELISA. Results were normalized to the amount of total protein recovered to correct for differences in tissue sampling size. Significant differences in inflammatory mediator expression between biofilm infected versus sterile catheters are denoted by asterisks (*, p < 0.05). Results are presented from individual animals combined from three independent experiments.
To further investigate mechanisms of immune deviation associated with biofilm infections, we compared the degree of macrophage influx into S. aureus-induced abscesses versus catheter-associated biofilms by FACS. Interestingly, macrophage accumulation into biofilm infected tissues was significantly increased at day 3 post-infection compared to abscesses (Fig. 6). Despite this early exaggerated macrophage infiltrate, immunofluorescence staining demonstrated that the majority of cells remained distant from the infection site, with only a few macrophages recruited to the biofilm surface (Fig. 7). Collectively, these findings demonstrate that S. aureus biofilms evade traditional TLR recognition and alter the immune response to infection.
Figure 6. S. aureus biofilms elicit exaggerated macrophage infiltrates compared to abscesses.
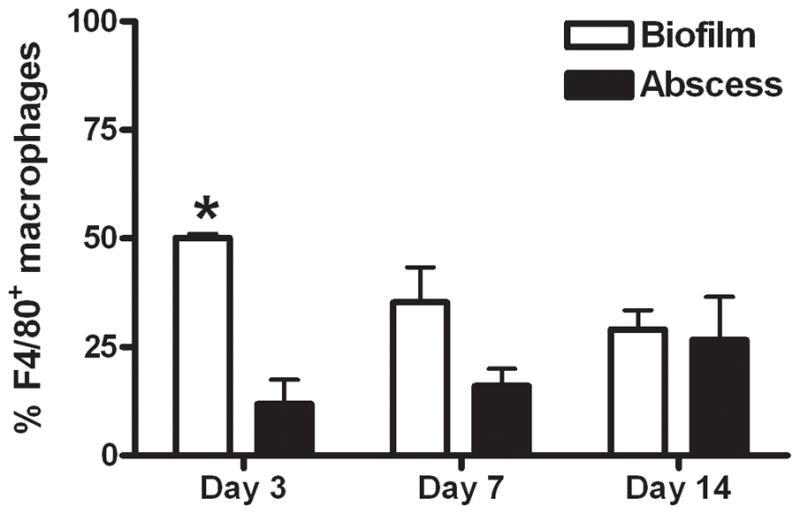
Wild type mice were infected with 5 × 105 CFU of USA300 LAC::lux either in the lumen of surgically implanted catheters or subcutaneously in the absence of any indwelling device to establish biofilm and abscess infections, respectively. Animals were sacrificed at days 3, 7, or 14 following S. aureus exposure, whereupon tissues surrounding infected catheters or s.c. injection sites were collected to quantitate macrophage infiltrates by FACS. Results are expressed as the percent of F4/80+ macrophages after correction for isotype control staining and represent the mean ± SEM of three independent experiments. Significant differences in macrophage infiltration into biofilm versus abscess infections are denoted by asterisks (*, p < 0.01).
Figure 7. Macrophage migration towards S. aureus biofilms in vivo is limited.

Tissues surrounding biofilm-infected catheters were collected from wild type mice at day 7 following S. aureus infection and subjected to immunofluorescence staining with Iba-1 to identify macrophages (red) and DAPI to demarcate nuclei (blue) by confocal microscopy (20X magnification). Asterisks indicate the location of infected catheters and the border between biofilm and surrounding host tissue is denoted by dashed lines. Results are representative of 6 individual animals.
Biofilm infections attenuate iNOS and augment arginase-1 expression in vivo
Reactive nitrogen intermediates represent a major microbicidal mechanism and are synthesized in macrophages by inducible nitric oxide synthase (iNOS)(59). On the other hand, arginase-1 leads to L-proline production, which is a precursor in the collagen biosynthetic pathway (60, 61). Both iNOS and arginase-1 compete for the same substrate (arginine) and, as such, are differentially expressed (62). Due to the fact that biofilm infections are typically associated with a fibrotic response and because of their chronic nature, we investigated whether differences in iNOS and arginase-1 expression would be evident. Indeed, iNOS expression associated with biofilm infections was reduced compared to abscesses and coincided with robust arginase-1 expression (Fig. 8). In addition, arginase-1 expression was detected in macrophages in closest proximity to the biofilm, although other cell type(s) were also arginase positive (Fig. 9). Arginase-1 expression in biofilm-associated macrophages in the absence of iNOS would be expected to skew the cellular response away from bacterial killing and could be another mechanism to account for the ability of biofilms to evade clearance.
Figure 8. S. aureus biofilms repress iNOS and augment arginase-1 expression.

Wild type mice were infected with 5 × 105 CFU of USA300 LAC::lux either in the lumen of surgically implanted catheters or subcutaneously in the absence of any indwelling device to establish biofilm and abscess infections, respectively. Animals were sacrificed at day 7 following S. aureus exposure, whereupon tissues surrounding infected catheters or s.c. injection sites were collected and subjected to immunofluorescence staining with arginase-1 (red) and DAPI to identify nuclei (blue) by confocal microscopy (20X magnification). Stars represent the location of material remaining from the catheter lumen and the diamonds denote the necrotic abscess core. The border between biofilm and surrounding host tissue is depicted by arrows. Results are representative of 6 individual animals.
Figure 9. A subset of biofilm-associated macrophages express arginase-1.
Tissues surrounding biofilm-infected catheters were collected from wild type mice at day 7 following S. aureus infection and subjected to immunofluorescence staining for the macrophage-specific marker Iba-1 (red) and arginase-1 (green) to determine co-localization patterns by confocal microscopy (arrows, 20X magnification). The position of the biofilm-infected catheter is denoted by asterisks. Results are representative of 6 individual animals.
Biofilms program macrophages towards an alternatively activated (M2) phenotype
Our in vivo studies had clearly demonstrated that S. aureus biofilms were capable of circumventing traditional anti-bacterial effector mechanisms. To further examine potential mechanisms responsible for this phenomenon, and to simplify the study of biofilm-macrophage interactions without confounds from other cell types, we next explored the effects of S. aureus biofilms on macrophage responses in vitro.
To date, few studies have examined the interactions between S. aureus biofilm and macrophages, which are an important anti-bacterial effector population. To determine whether biofilms influence macrophage activation, gene expression profiles were compared between macrophages incubated with either S. aureus biofilms or planktonic bacteria. Similar to our in vivo findings, macrophages co-cultured with biofilms demonstrated a gene expression profile reminiscent of alternatively activated (M2) cells, namely a reduction in iNOS coincident with a significant increase in arginase-1 expression, whereas no differences in TNF-α or IL-1β were observed (Fig. 10)(63). These results further support the conclusion that biofilm growth reprograms the macrophage response from a bactericidal M1 to a M2 phenotype, the latter of which is not optimal for maximal bacterial clearance (63, 64).
Figure 10. S. aureus biofilms skew macrophage gene expression towards an alternatively activated (M2) phenotype.
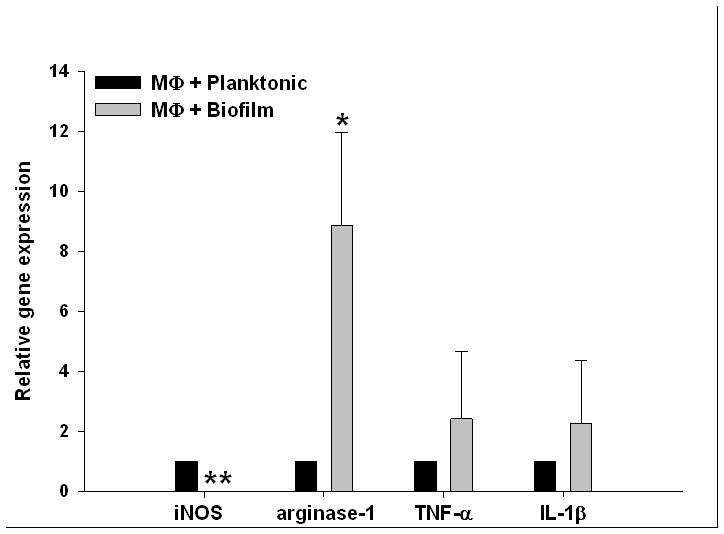
Bone marrow-derived macrophages from GFP Tg mice were incubated with 6 day-old biofilms or planktonic bacteria for 2 h, whereupon viable macrophages were purified by FACS and RNA immediately isolated for qRT-PCR analysis. Gene expression levels in macrophages exposed to S. aureus biofilms were calculated after normalizing signals against GAPDH and are presented as the fold-change relative to macrophages incubated with planktonic bacteria. Results represent the mean ± SEM of four independent experiments (*, p < 0.05; **, p < 0.001).
Macrophages actively phagocytize planktonic S. aureus but not biofilm-associated bacteria in vitro
Previous studies have suggested that neutrophils are capable of phagocytizing S. aureus biofilms (18, 19); however, less is known about the ability of macrophages to internalize bacteria in a biofilm state. To this end, we incubated primary mouse peritoneal macrophages with either 4 or 6 day-old S. aureus static biofilms and evaluated their phagocytic ability. Interestingly, macrophages incubated with S. aureus biofilms for either a 1 or 24 h period contained few internalized bacteria (Fig. 11A and data not shown). Similar findings were also obtained with bone marrow-derived macrophages (data not shown). In contrast to macrophages, intracellular bacteria were readily discernable in neutrophils co-cultured with either 4 or 6-day old biofilms (Supplemental Fig. 4), demonstrating the sensitivity of confocal analysis for visualizing intracellular organisms. The impact of S. aureus biofilms on neutrophil function will be the topic of a separate manuscript (Hanke and Kielian, in preparation). The inability of macrophages to phagocytize biofilm-associated bacteria was independent of the age of the biofilm in that intracellular bacteria could not be observed in macrophages incubated with either 4 or 6 day-old biofilms (Fig. 11A and data not shown). In addition, macrophages did not significantly impact the number of biofilm-associated bacteria during the 24 h co-incubation period (data not shown). These findings indicate that although macrophages are capable of infiltrating biofilms, phagocytic mechanisms are not discernable during this growth state. However, macrophages were capable of internalizing disrupted biofilm material (Fig. 11B) suggesting that the size and/or physical complexity of the intact biofilm is responsible for its inability to be phagocytized. As expected, macrophages were highly phagocytic towards planktonic S. aureus (Fig. 11A), indicating that they were capable of recognizing bacteria in this growth state.
Figure 11. Macrophages actively phagocytize planktonic S. aureus but not biofilm-associated bacteria.

(A) Primary macrophages labeled with CellTracker orange (orange) were exposed to 4 day-old USA300 LAC-GFP biofilms or planktonic bacteria (green) for a 24 h period, whereupon visualization of intracellular bacteria was evaluated by confocal microscopy (63X magnification, 1 μm slice). Biofilm-associated macrophages exhibited little evidence of phagocytosis, whereas intracellular bacteria were readily detectable in macrophages incubated with planktonic organisms. Results are representative of six independent experiments. In (B), four and six day-old USA300 LAC-GFP static biofilms were mechanically disrupted by triturating and incubated with macrophages for 1 h. Planktonic bacteria were included as a positive control. Fluorescent microscopy of cytospin preparations shows the ability of macrophages to phagocytize planktonic (left) as well as disassociated bacteria from four day-(center) and six day-old (right) biofilms. Results are representative of three independent experiments.
In contrast to these in vitro studies, macrophages recovered from biofilm infections in vivo harbored intracellular S. aureus as demonstrated by gentamicin protection assays (Fig. 12). The intracellular location of bacteria was confirmed by the ability of rifampicin, which is capable of crossing the eukaryotic cell membrane, to completely eliminate viable organisms (Fig. 12). However, this approach cannot discriminate between uptake of biofilm-associated versus planktonic bacteria, the latter of which occurs during biofilm dispersal in an attempt to colonize new sites of infection (28, 65, 66). Collectively, the fact that a large percentage of macrophages remain significantly distant from the biofilm surface in conjunction with reduced iNOS expression, suggest that even though macrophages exhibit a modest degree of phagocytosis, intracellular killing mechanisms are likely compromised.
Figure 12. Macrophages recovered from S. aureus biofilm infections harbor viable intracellular bacteria.
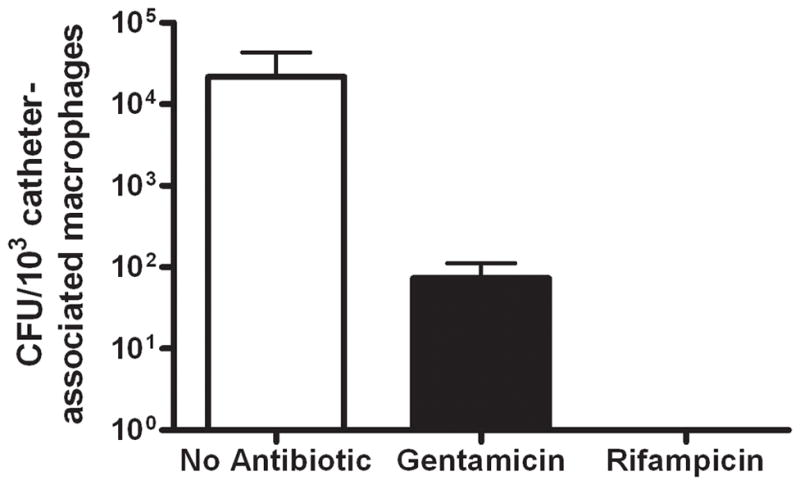
Viable macrophages (F4/80+, 7-AAD−) were recovered from tissues surrounding biofilm infections at day 7 post-infection by FACS, whereupon gentamicin protection assays were performed to evaluate the presence of viable intracellular bacteria. Controls included sorted macrophages without any antibiotic treatment (to assess extra- and intracellular bacteria) and macrophages treated with rifampicin, which is capable of penetrating the eukaryotic cell membrane. Results are expressed as the number of viable S. aureus (CFU per 103 biofilm-associated macrophages) and represent the mean ± SEM of five independent experiments.
Macrophage invasion into biofilms in vitro is associated with cell death
Based on the ineffectiveness of the host immune response to clear S. aureus biofilms in vivo, it is likely that the biofilm compromises numerous anti-bacterial effector mechanisms to persist in the host. To further investigate such possibilities, we next examined whether biofilms adversely affected macrophage survival. Interestingly, a striking difference in cell morphology was observed between macrophages incubated with 4 versus 6 day-old biofilms for a 24 h period. Namely, macrophages exposed to immature biofilms (i.e. 4 day-old) exhibited a typical rounded morphology, whereas macrophages incubated with mature biofilms (i.e. 6 day-old) appeared ghost-like with a dystrophic morphology as evidenced by their irregular shape and diminished CellTracker orange staining (Fig. 13). Since a mature (i.e. 6 day-old) biofilm represents a dense matrix network composed of polysaccharides, eDNA, and protein polymers, it is likely that this presents the macrophage with a substrate that is difficult to phagocytize, resulting in a phenomenon termed “frustrated phagocytosis” (67). Although macrophages exposed to immature biofilms retained a typical cell appearance, TOTO3 staining revealed that those macrophages most intimately associated with the biofilm were dead, whereas the majority of macrophages farther removed from the biofilm surface remained viable (Fig. 14). This live/dead cell analysis could not be conducted on macrophages incubated with 6 day-old biofilms due to the ghost-like dystrophic nature of the cells. Similar cell death patterns were observed during FACS studies with the vital dye 7-AAD, where approximately 55% of macrophages were dead within 2 h following biofilm co-culture (data not shown). The mechanisms responsible for this rapid induction of cell death are not known but may be influenced by toxin production, acidic pH, and/or anaerobic conditions within the biofilm proper. Collectively, these studies reveal that S. aureus biofilms induce macrophage dysfunction and cell death, which could contribute to biofilm evasion of host immunity and persistence in vivo.
Figure 13. Macrophages exposed to mature S. aureus biofilm exhibit a dystrophic morphology.

Primary macrophages labeled with CellTracker orange (CTO; yellow-orange) were exposed to 4 or 6 day-old USA300 LAC-GFP biofilms (green) for a 24 h period, whereupon visualization of macrophage morphology was evaluated by confocal microscopy (63X magnification, 1 μm slice). Macrophages infiltrating immature (4 day-old) biofilms retained a typical rounded morphology, whereas macrophages infiltrating mature biofilms (6 day-old) often exhibited a dystrophic ghost-like morphology with minimal retention of CTO (arrows). Results are representative of six independent experiments.
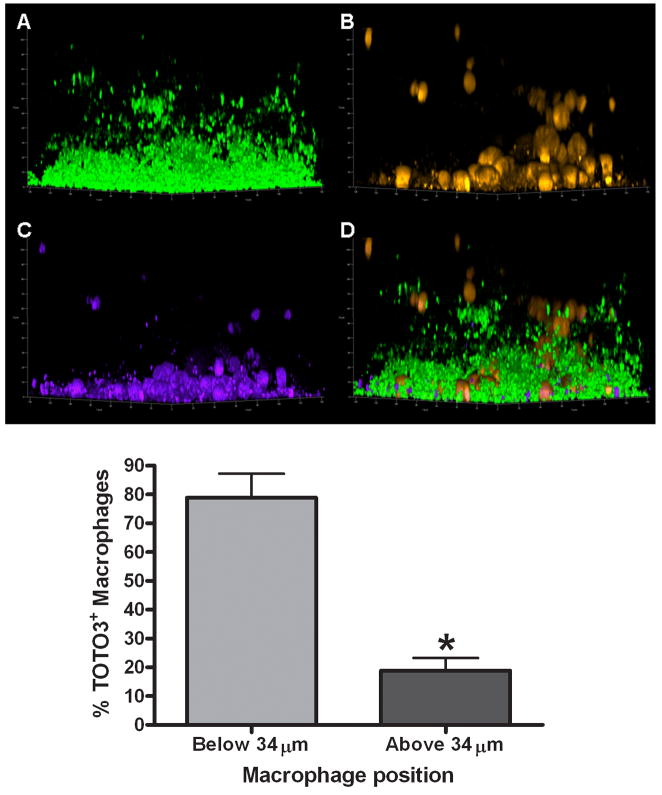
Figure 14. Macrophage viability is dictated by invasion into the biofilm proper.
(A) 4 day-old USA300 LAC-GFP biofilms (average height ~ 33 μm) were incubated with (B) primary macrophages labeled with CellTracker orange (yellow-orange) for a 24 h period, whereupon macrophage viability was determined by uptake of the live/dead stain TOTO3 (purple; C). The majority of dead (TOTO3+) macrophages are within 34 μm of the biofilm proper (p < 0.05) and a composite image of all staining is shown (D). Bottom; significant differences between the percentages of TOTO3+ macrophages based on location within the biofilm proper are denoted with an asterisk (*, p < 0.05; n = 218 cells).
DISCUSSION
To date, relatively little information is available regarding how innate immunity interfaces with S. aureus biofilms. Since S. aureus biofilms represent a serious clinical situation based on their recalcitrance to antibiotics, persistence, and the propensity of organisms to detach and colonize new infection sites (68, 69), understanding the dynamics between host immunity and S. aureus biofilms is an important issue that warrants further investigation.
To our knowledge, this study is the first to report that S. aureus biofilms actively modify the host innate immune response in vivo by attenuating proinflammatory cytokine production and circumventing traditional bacterial recognition pathways mediated by TLR2 and TLR9. In addition, iNOS levels were significantly reduced surrounding biofilm infections and instead were dominated by arginase-1 expression. In the context of biofilm infection, the skewing from iNOS to an arginase-1 prominent response is likely another explanation for why biofilm infections persist in an immunocompetent host. The innate immune alterations associated with biofilm infection identified in this study are summarized in Fig. 15.
Figure 15. S. aureus biofilms skew macrophage responses to favor bacterial persistence.

Despite the presence of TLR2 and TLR9 ligands within the biofilm (lipoproteins/PGN and eDNA, respectively), these traditional pattern recognition receptors to not impart macrophage responsiveness or bacterial clearance during biofilm infection (depicted by red “x” marks). Likewise, the expression of iNOS is reduced following biofilm formation concomitant with an increase in arginase-1. Collectively, these changes shift the immune response away from a bactericidal pathway and likely contribute to biofilm persistence.
The rationale for our investigation of TLR2 and TLR9 stemmed from the fact that their ligands are components of the biofilm (25–28). Although TLR2 has been shown to play a pivotal role in S. aureus recognition and clearance in several in vivo models of planktonic infection, it is clear that when S. aureus assumes a biofilm lifestyle the organism is capable of evading recognition by this classical PRR. This finding highlights the importance of bacterial growth state in dictating whether innate immune sensor mechanisms will be effective at clearing the infection. It was also unexpected that we did not observe a dramatic role for TLR9 in regulating biofilm growth since a recent report demonstrated that eDNA was a major proinflammatory stimulus of P. aeruginosa biofilms (70). However, this discrepancy may be attributed to several factors including making comparisons between in vitro and in vivo studies as well as divergent bacterial species. It is also possible that S. aureus biofilms may be recognized by alternative PRRs besides TLR2 or TLR9. For example, eDNA could also be sensed by other intracellular PRRs such as AIM2 or DNA-dependent activator of IFN-regulatory factors (DAI) (71, 72). The mechanism(s) responsible for TLR2/TLR9 evasion by S. aureus biofilms are not known but could also be explained by ligand inaccessibility. Indeed, our SEM data demonstrated that biofilms in vivo are encased within a complex three-dimensional structure with few free bacteria exposed at the outer surface (Fig. 2B), suggesting the inability to trigger PRRs expressed at the phagocyte surface. Alternatively, complex polysaccharide polymers that are known components of the biofilm matrix may interfere with optimal engagement of potential ligands with TLRs. However, these possibilities have yet to be investigated at the present time.
It is evident that S. aureus biofilms induce macrophage dysfunction at several levels, which provide potential explanations as to why these infections persist in an immunocompetent host. First, although biofilms induce greater macrophage influx during early infection compared to abscesses and harbor intracellular bacteria, the paucity of iNOS and robust arginase-1 expression suggest that bacteria may not be sufficiently cleared. A similar M2-like phenotype was observed in macrophages in vitro following biofilm co-culture. Second, although biofilms were associated with robust macrophage accumulation, only a small subset of cells migrated towards the biofilm surface and many that did expressed arginase-1, which skews macrophages for ineffective killing. Third, our findings clearly demonstrated that biofilms induced macrophage death by an as of yet unknown mechanism. Specifically, FACS staining with 7-AAD revealed significant macrophage cell death within 2 h following biofilm exposure. A longer co-incubation period (i.e. 24 h) led to dystrophic macrophages with a ghost-like appearance, whereas cells cultured with 4 day-old biofilms demonstrated a typical shape even though a high percentage of these macrophages were dead as revealed by TOTO3 staining. Importantly, macrophages were capable of phagocytizing bacteria from mechanically disrupted biofilms regardless of their maturation state, suggesting that the large size and/or inability to opsonize intact biofilms could explain the dystrophic macrophage phenotype. The latter possibility is supported by a recent study demonstrating that IgG and C3b deposition is reduced on the surface of biofilm-associated S. epidermidis compared to planktonic bacteria (16). Alternatively, PIA has anti-phagocytic properties, which could also interfere with phagocytic uptake of S. aureus biofilms. It is important to note that the USA300 LAC strain used in the current study expresses PIA as demonstrated by WGA staining, but lacks a capsule (73); therefore, capsule cannot account for the anti-phagocytic activity of the biofilm. The mechanism(s) that trigger macrophage cell death upon biofilm contact are not known but could be the result of acidic pH, bacterial toxins, and/or the hypoxic environment within the biofilm mass. Another question that remains is whether macrophage death following biofilm exposure is apoptotic or necrotic. Judging by the striking differences in macrophage morphology during incubation with immature vs. mature biofilms, we speculate that both types of cell death are operative. Specifically, macrophages maintained their characteristic shape following extended (i.e. 24 h) co-culture with immature biofilms, whereas those macrophages that were exposed to mature biofilms for the same interval exhibited a wispy, dystrophic morphology reminiscent of necrotic cell death. Whether apoptosis and/or necrosis are responsible for macrophage death requires clarification in future studies.
Our results demonstrating that macrophages are capable of infiltrating biofilms are similar to previous reports using mixed leukocyte and purified neutrophil populations with S. aureus biofilms (17–19). However, several differences were also noted between these earlier studies and our own, including the fact that we did not observe appreciable phagocytosis of biofilm-associated S. aureus by macrophages in vitro, whereas studies with neutrophils reported bacterial uptake within the biofilm, which we also observed in the current report (Supplemental Fig. 4) (18, 19). In addition, other studies have reported that S. aureus biofilms elicit proinflammatory mediator release, which was not observed in our experiments. The reasons to account for these differences are not known but could be influenced by several factors including the method of biofilm propagation (i.e. static versus flow cell), the type of immune cell population examined, and/or compatibility of medium formulations to achieve optimal survival of both the biofilm and immune cells. Indeed, the dynamics of the flow system used in the previously cited studies could lead to less compact biofilms and enhanced bacterial detachment that would render S. aureus more susceptible to phagocytosis. Although in vitro experiments demonstrated minimal S. aureus phagocytosis, in vivo studies revealed that macrophages associated with S. aureus biofilms in vivo are capable of limited bacterial uptake. However, when taken in the context of minimal iNOS expression and adverse effects on cell viability, the ability of macrophages to efficiently kill intracellular S. aureus is likely compromised. In addition, since bacteria regularly detach from biofilms as part of their normal growth cycle to seed new sites of infection, it is possible that the intracellular bacteria detected in macrophages originated from the internalization of planktonic bacteria. Unfortunately, we are not able to discriminate between the uptake of biofilm-associated versus detached planktonic bacteria with any available technology. Nonetheless, our results collectively suggest that biofilms re-program macrophage responsiveness towards a M2 phenotype that inherently possesses less anti-microbial activity (63, 64).
Another novel observation of the current study was the ability of S. aureus to establish a biofilm infection at a relatively low dose that was rapidly cleared following subcutaneous injection into sites lacking an indwelling device. The fact that fewer S. aureus are required to colonize foreign devices provides insights as to why indwelling devices are more prone to infection compared to natural body surfaces. It is likely that these artificial surfaces provide a niche for bacterial attachment either directly or indirectly when the foreign body becomes encased by host proteins that would facilitate the induction of biofilm growth. Collectively, these findings highlight the importance of studying indwelling device/biofilm-associated infections because of the blunted immune response and lower number of bacteria required to establish infection.
In summary, the current study demonstrates that S. aureus biofilms attenuate inflammatory mediator production and macrophage invasion into the biofilm. In addition, iNOS expression was attenuated and a coincident increase in arginase-1 suggests a skewing of the immune response towards a pro-fibrotic M2 phenotype. Another contributing factor is the ineffectiveness of classical TLR recognition pathways, which is likely dictated by the inability of tissue macrophages to significantly invade the biofilm proper in vivo. This result is in stark contrast with reports from other laboratories demonstrating an important role of TLR2 and TLR9 in regulating planktonic S. aureus infection (74, 75). Therefore, it is evident that by assuming a biofilm growth state, S. aureus actively subverts the resultant host immune response, which likely contributes to biofilm persistence.
Supplementary Material
Acknowledgments
The authors thank Dr. Shizuo Akira for providing the TLR2 and TLR9 KO mice, Debbie Vidlak and Shuliang Liu for excellent technical assistance, Tom Bargar for performing SEM analysis, Fang Yu for statistical analysis, and Jennifer Endres for generation of the USA300 LAC::lux construct. The authors also thank Dr. Charles Kuszynski, Megan Michalak, and Victoria Smith in the UNMC Cell Analysis Facility for assistance with FACS analysis and the UNMC Tissue Science Facility for preparing paraffin sections.
Footnotes
This work was supported by the NIH National Institute of Allergy and Infectious Disease (NIAID) P01 AI083211 Project 4 to T.K. and Project 3 to A.R.H.
References
- 1.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Otto M. Staphylococcal biofilms. Curr Top Microbiol Immunol. 2008;322:207–228. doi: 10.1007/978-3-540-75418-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fitzpatrick F, Humphreys H, O’Gara JP. The genetics of staphylococcal biofilm formation--will a greater understanding of pathogenesis lead to better management of device-related infection? Clin Microbiol Infect. 2005;11:967–973. doi: 10.1111/j.1469-0691.2005.01274.x. [DOI] [PubMed] [Google Scholar]
- 4.Burlak C, Hammer CH, Robinson MA, Whitney AR, McGavin MJ, Kreiswirth BN, Deleo FR. Global analysis of community-associated methicillin-resistant Staphylococcus aureus exoproteins reveals molecules produced in vitro and during infection. Cell Microbiol. 2007;9:1172–1190. doi: 10.1111/j.1462-5822.2006.00858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambers HF, Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 2009;7:629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voyich JM, Braughton KR, Sturdevant DE, Whitney AR, Said-Salim B, Porcella SF, Long RD, Dorward DW, Gardner DJ, Kreiswirth BN, Musser JM, DeLeo FR. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J Immunol. 2005;175:3907–3919. doi: 10.4049/jimmunol.175.6.3907. [DOI] [PubMed] [Google Scholar]
- 7.Takeuchi O, Hoshino K, Akira S. Cutting edge: TLR2-deficient and MyD88-deficient mice are highly susceptible to Staphylococcus aureus infection. J Immunol. 2000;165:5392–5396. doi: 10.4049/jimmunol.165.10.5392. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi SD, Braughton KR, Whitney AR, Voyich JM, Schwan TG, Musser JM, DeLeo FR. Bacterial pathogens modulate an apoptosis differentiation program in human neutrophils. Proc Natl Acad Sci U S A. 2003;100:10948–10953. doi: 10.1073/pnas.1833375100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foster TJ. Immune evasion by staphylococci. Nat Rev Microbiol. 2005;3:948–958. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- 10.Fournier B, Philpott DJ. Recognition of Staphylococcus aureus by the innate immune system. Clin Microbiol Rev. 2005;18:521–540. doi: 10.1128/CMR.18.3.521-540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walker TS, Tomlin KL, Worthen GS, Poch KR, Lieber JG, Saavedra MT, Fessler MB, Malcolm KC, Vasil ML, Nick JA. Enhanced Pseudomonas aeruginosa biofilm development mediated by human neutrophils. Infect Immun. 2005;73:3693–3701. doi: 10.1128/IAI.73.6.3693-3701.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mittal R, Sharma S, Chhibber S, Harjai K. Effect of macrophage secretory products on elaboration of virulence factors by planktonic and biofilm cells of Pseudomonas aeruginosa. Comp Immunol Microbiol Infect Dis. 2006;29:12–26. doi: 10.1016/j.cimid.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Chandra J, McCormick TS, Imamura Y, Mukherjee PK, Ghannoum MA. Interaction of Candida albicans with adherent human peripheral blood mononuclear cells increases C. albicans biofilm formation and results in differential expression of pro- and anti-inflammatory cytokines. Infect Immun. 2007;75:2612–2620. doi: 10.1128/IAI.01841-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen ET, Kharazmi A, Lam K, Costerton JW, Hoiby N. Human polymorphonuclear leukocyte response to Pseudomonas aeruginosa grown in biofilms. Infect Immun. 1990;58:2383–2385. doi: 10.1128/iai.58.7.2383-2385.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jesaitis AJ, Franklin MJ, Berglund D, Sasaki M, Lord CI, Bleazard JB, Duffy JE, Beyenal H, Lewandowski Z. Compromised host defense on Pseudomonas aeruginosa biofilms: characterization of neutrophil and biofilm interactions. J Immunol. 2003;171:4329–4339. doi: 10.4049/jimmunol.171.8.4329. [DOI] [PubMed] [Google Scholar]
- 16.Kristian SA, Birkenstock TA, Sauder U, Mack D, Gotz F, Landmann R. Biofilm formation induces C3a release and protects Staphylococcus epidermidis from IgG and complement deposition and from neutrophil-dependent killing. J Infect Dis. 2008;197:1028–1035. doi: 10.1086/528992. [DOI] [PubMed] [Google Scholar]
- 17.Leid JG, Shirtliff ME, Costerton JW, Stoodley AP. Human leukocytes adhere to, penetrate, and respond to Staphylococcus aureus biofilms. Infect Immun. 2002;70:6339–6345. doi: 10.1128/IAI.70.11.6339-6345.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gunther F, Wabnitz GH, Stroh P, Prior B, Obst U, Samstag Y, Wagner C, Hansch GM. Host defence against Staphylococcus aureus biofilms infection: phagocytosis of biofilms by polymorphonuclear neutrophils (PMN) Mol Immunol. 2009;46:1805–1813. doi: 10.1016/j.molimm.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 19.Guenther F, Stroh P, Wagner C, Obst U, Hansch GM. Phagocytosis of staphylococci biofilms by polymorphonuclear neutrophils: S. aureus and S. epidermidis differ with regard to their susceptibility towards the host defense. Int J Artif Organs. 2009;32:565–573. doi: 10.1177/039139880903200905. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi SD, Braughton KR, Palazzolo-Ballance AM, Kennedy AD, Sampaio E, Kristosturyan E, Whitney AR, Sturdevant DE, Dorward DW, Holland SM, Kreiswirth BN, Musser JM, DeLeo FR. Rapid neutrophil destruction following phagocytosis of Staphylococcus aureus. J Innate Immun. 2010;2:560–575. doi: 10.1159/000317134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graves SF, Kobayashi SD, DeLeo FR. Community-associated methicillin-resistant Staphylococcus aureus immune evasion and virulence. J Mol Med. 2010;88:109–114. doi: 10.1007/s00109-009-0573-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.S Anwar, Prince LR, Foster SJ, Whyte MK, Sabroe I. The rise and rise of Staphylococcus aureus: laughing in the face of granulocytes. Clin Exp Immunol. 2009;157:216–224. doi: 10.1111/j.1365-2249.2009.03950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silva MT. Macrophage phagocytosis of neutrophils at inflammatory/infectious foci: a cooperative mechanism in the control of infection and infectious inflammation. J Leukoc Biol. 2010 doi: 10.1189/jlb.0910536. [DOI] [PubMed] [Google Scholar]
- 24.Silva MT. When two is better than one: macrophages and neutrophils work in concert in innate immunity as complementary and cooperative partners of a myeloid phagocyte system. J Leukoc Biol. 2010;87:93–106. doi: 10.1189/jlb.0809549. [DOI] [PubMed] [Google Scholar]
- 25.Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, Bayles KW. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci U S A. 2007;104:8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 27.Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, Molin S, Givskov M, Tolker-Nielsen T. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol. 2006;59:1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- 28.Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, Tsang LH, Smeltzer MS, Horswill AR, Bayles KW. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One. 2009;4:e5822. doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 30.Hayashi F, Means TK, Luster AD. Toll-like receptors stimulate human neutrophil function. Blood. 2003 doi: 10.1182/blood-2003-04-1078. [DOI] [PubMed] [Google Scholar]
- 31.Bauer S, Kirschning CJ, Hacker H, Redecke V, Hausmann S, Akira S, Wagner H, Lipford GB. Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc Natl Acad Sci U S A. 2001;98:9237–9242. doi: 10.1073/pnas.161293498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hertz CJ, Kiertscher SM, Godowski PJ, Bouis DA, Norgard MV, Roth MD, Modlin RL. Microbial lipopeptides stimulate dendritic cell maturation via Toll-like receptor 2. J Immunol. 2001;166:2444–2450. doi: 10.4049/jimmunol.166.4.2444. [DOI] [PubMed] [Google Scholar]
- 33.Jones BW, Means TK, Heldwein KA, Keen MA, Hill PJ, Belisle JT, Fenton MJ. Different Toll-like receptor agonists induce distinct macrophage responses. J Leukoc Biol. 2001;69:1036–1044. [PubMed] [Google Scholar]
- 34.Kirschning CJ, Schumann RR. TLR2: cellular sensor for microbial and endogenous molecular patterns. Curr Top Microbiol Immunol. 2002;270:121–144. doi: 10.1007/978-3-642-59430-4_8. [DOI] [PubMed] [Google Scholar]
- 35.Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 36.Mercier C, Durrieu C, Briandet R, Domakova E, Tremblay J, Buist G, Kulakauskas S. Positive role of peptidoglycan breaks in lactococcal biofilm formation. Mol Microbiol. 2002;46:235–243. doi: 10.1046/j.1365-2958.2002.03160.x. [DOI] [PubMed] [Google Scholar]
- 37.Cerca N, Jefferson KK, Oliveira R, Pier GB, Azeredo J. Comparative antibody-mediated phagocytosis of Staphylococcus epidermidis cells grown in a biofilm or in the planktonic state. Infect Immun. 2006;74:4849–4855. doi: 10.1128/IAI.00230-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moscoso M, Garcia E, Lopez R. Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. J Bacteriol. 2006;188:7785–7795. doi: 10.1128/JB.00673-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qin Z, Ou Y, Yang L, Zhu Y, Tolker-Nielsen T, Molin S, Qu D. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology. 2007;153:2083–2092. doi: 10.1099/mic.0.2007/006031-0. [DOI] [PubMed] [Google Scholar]
- 40.Morath S, Stadelmaier A, Geyer A, Schmidt RR, Hartung T. Synthetic lipoteichoic acid from Staphylococcus aureus is a potent stimulus of cytokine release. J Exp Med. 2002;195:1635–1640. doi: 10.1084/jem.20020322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weber JR, Moreillon P, Tuomanen EI. Innate sensors for Gram-positive bacteria. Curr Opin Immunol. 2003;15:408–415. doi: 10.1016/s0952-7915(03)00078-5. [DOI] [PubMed] [Google Scholar]
- 42.Dziarski R. Recognition of bacterial peptidoglycan by the innate immune system. Cell Mol Life Sci. 2003;60:1793–1804. doi: 10.1007/s00018-003-3019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimada T, Park BG, Wolf AJ, Brikos C, Goodridge HS, Becker CA, Reyes CN, Miao EA, Aderem A, Gotz F, Liu GY, Underhill DM. Staphylococcus aureus evades lysozyme-based peptidoglycan digestion that links phagocytosis, inflammasome activation, and IL-1beta secretion. Cell Host Microbe. 2010;7:38–49. doi: 10.1016/j.chom.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ip WK, Sokolovska A, Charriere GM, Boyer L, Dejardin S, Cappillino MP, Yantosca LM, Takahashi K, Moore KJ, Lacy-Hulbert A, Stuart LM. Phagocytosis and phagosome acidification are required for pathogen processing and MyD88-dependent responses to Staphylococcus aureus. J Immunol. 2010;184:7071–7081. doi: 10.4049/jimmunol.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 46.Yoshimura A, Lien E, Ingalls RR, Tuomanen E, Dziarski R, Golenbock D. Cutting edge: recognition of Gram-positive bacterial cell wall components by the innate immune system occurs via Toll-like receptor 2. J Immunol. 1999;163:1–5. [PubMed] [Google Scholar]
- 47.Watanabe I, Ichiki M, Shiratsuchi A, Nakanishi Y. TLR2-mediated survival of Staphylococcus aureus in macrophages: a novel bacterial strategy against host innate immunity. J Immunol. 2007;178:4917–4925. doi: 10.4049/jimmunol.178.8.4917. [DOI] [PubMed] [Google Scholar]
- 48.Diep BA, Palazzolo-Ballance AM, Tattevin P, Basuino L, Braughton KR, Whitney AR, Chen L, Kreiswirth BN, Otto M, DeLeo FR, Chambers HF. Contribution of Panton-Valentine leukocidin in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. PLoS ONE. 2008;3:e3198. doi: 10.1371/journal.pone.0003198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kennedy AD, Otto M, Braughton KR, Whitney AR, Chen L, Mathema B, Mediavilla JR, Byrne KA, Parkins LD, Tenover FC, Kreiswirth BN, Musser JM, DeLeo FR. Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. Proc Natl Acad Sci U S A. 2008;105:1327–1332. doi: 10.1073/pnas.0710217105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cassat JE, Lee CY, Smeltzer MS. Investigation of biofilm formation in clinical isolates of Staphylococcus aureus. Methods Mol Biol. 2007;391:127–144. doi: 10.1007/978-1-59745-468-1_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rupp ME, Ulphani JS, Fey PD, Bartscht K, Mack D. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect Immun. 1999;67:2627–2632. doi: 10.1128/iai.67.5.2627-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kielian T, Cheung A, Hickey WF. Diminished virulence of an alpha-toxin mutant of Staphylococcus aureus in experimental brain abscesses. Infect Immun. 2001;69:6902–6911. doi: 10.1128/IAI.69.11.6902-6911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lauderdale KJ, Malone CL, Boles BR, Morcuende J, Horswill AR. Biofilm dispersal of community-associated methicillin-resistant Staphylococcus aureus on orthopedic implant material. J Orthop Res. 2009 doi: 10.1002/jor.20943. [DOI] [PubMed] [Google Scholar]
- 54.Liu S, Kielian T. Microglial activation by Citrobacter koseri is mediated by TLR4- and MyD88-dependent pathways. J Immunol. 2009;183:5537–5547. doi: 10.4049/jimmunol.0900083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thurlow LR, Thomas VC, Fleming SD, Hancock LE. Enterococcus faecalis capsular polysaccharide serotypes C and D and their contributions to host innate immune evasion. Infect Immun. 2009;77:5551–5557. doi: 10.1128/IAI.00576-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Drevets DA, Campbell PA. Macrophage phagocytosis: use of fluorescence microscopy to distinguish between extracellular and intracellular bacteria. J Immunol Methods. 1991;142:31–38. doi: 10.1016/0022-1759(91)90289-r. [DOI] [PubMed] [Google Scholar]
- 57.Drevets DA, Canono BP, Leenen PJ, Campbell PA. Gentamicin kills intracellular Listeria monocytogenes. Infect Immun. 1994;62:2222–2228. doi: 10.1128/iai.62.6.2222-2228.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lauderdale KJ, Boles BR, Cheung AL, Horswill AR. Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect Immun. 2009;77:1623–1635. doi: 10.1128/IAI.01036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kleinert H, Pautz A, Linker K, Schwarz PM. Regulation of the expression of inducible nitric oxide synthase. Eur J Pharmacol. 2004;500:255–266. doi: 10.1016/j.ejphar.2004.07.030. [DOI] [PubMed] [Google Scholar]
- 60.Curran JN, Winter DC, Bouchier-Hayes D. Biological fate and clinical implications of arginine metabolism in tissue healing. Wound Repair Regen. 2006;14:376–386. doi: 10.1111/j.1743-6109.2006.00151.x. [DOI] [PubMed] [Google Scholar]
- 61.Wynn TA. Cellular and molecular mechanisms of fibrosis. J Pathol. 2008;214:199–210. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hesse M, Modolell M, Flamme AC La, Schito M, Fuentes JM, Cheever AW, Pearce EJ, Wynn TA. Differential regulation of nitric oxide synthase-2 and arginase-1 by type 1/type 2 cytokines in vivo: granulomatous pathology is shaped by the pattern of L-arginine metabolism. J Immunol. 2001;167:6533–6544. doi: 10.4049/jimmunol.167.11.6533. [DOI] [PubMed] [Google Scholar]
- 63.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 64.Benoit M, Desnues B, Mege JL. Macrophage polarization in bacterial infections. J Immunol. 2008;181:3733–3739. doi: 10.4049/jimmunol.181.6.3733. [DOI] [PubMed] [Google Scholar]
- 65.Boles BR, Horswill AR. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 2008;4:e1000052. doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lauderdale KJ, Malone CL, Boles BR, Morcuende J, Horswill AR. Biofilm dispersal of community-associated methicillin-resistant Staphylococcus aureus on orthopedic implant material. J Orthop Res. 2010;28:55–61. doi: 10.1002/jor.20943. [DOI] [PubMed] [Google Scholar]
- 67.Bainton DF, Takemura R, Stenberg PE, Werb Z. Rapid fragmentation and reorganization of Golgi membranes during frustrated phagocytosis of immobile immune complexes by macrophages. Am J Pathol. 1989;134:15–26. [PMC free article] [PubMed] [Google Scholar]
- 68.Fatkenheuer G, Cornely O, Seifert H. Clinical management of catheter-related infections. Clin Microbiol Infect. 2002;8:545–550. doi: 10.1046/j.1469-0691.2002.00427.x. [DOI] [PubMed] [Google Scholar]
- 69.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 70.Fuxman Bass JI, Russo DM, Gabelloni ML, Geffner JR, Giordano M, Catalano M, Zorreguieta A, Trevani AS. Extracellular DNA: a major proinflammatory component of Pseudomonas aeruginosa biofilms. J Immunol. 2010;184:6386–6395. doi: 10.4049/jimmunol.0901640. [DOI] [PubMed] [Google Scholar]
- 71.Hornung V, Latz E. Intracellular DNA recognition. Nat Rev Immunol. 2010;10:123–130. doi: 10.1038/nri2690. [DOI] [PubMed] [Google Scholar]
- 72.Vilaysane A, Muruve DA. The innate immune response to DNA. Semin Immunol. 2009;21:208–214. doi: 10.1016/j.smim.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 73.Montgomery CP, Boyle-Vavra S, Adem PV, Lee JC, Husain AN, Clasen J, Daum RS. Comparison of virulence in community-associated methicillin-resistant Staphylococcus aureus pulsotypes USA300 and USA400 in a rat model of pneumonia. J Infect Dis. 2008;198:561–570. doi: 10.1086/590157. [DOI] [PubMed] [Google Scholar]
- 74.Takeuchi O, Takeda K, Hoshino K, Adachi O, Ogawa T, Akira S. Cellular responses to bacterial cell wall components are mediated through MyD88-dependent signaling cascades. Int Immunol. 2000;12:113–117. doi: 10.1093/intimm/12.1.113. [DOI] [PubMed] [Google Scholar]
- 75.Gillrie MR, Zbytnuik L, McAvoy E, Kapadia R, Lee K, Waterhouse CC, Davis SP, Muruve DA, Kubes P, Ho M. Divergent roles of Toll-like receptor 2 in response to lipoteichoic acid and Staphylococcus aureus in vivo. Eur J Immunol. 2010;40:1639–1650. doi: 10.1002/eji.200939929. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.