Abstract
The study of genetic variability within natural populations of pathogens may provide insight into their evolution and pathogenesis. We used a Mycobacterium tuberculosis high-density oligonucleotide microarray to detect small-scale genomic deletions among 19 clinically and epidemiologically well-characterized isolates of M. tuberculosis. The pattern of deletions detected was identical within mycobacterial clones but differed between different clones, suggesting that this is a suitable genotyping system for epidemiologic studies. An analysis of genomic deletions among an extant population of pathogenic bacteria provided a novel perspective on genomic organization and evolution. Deletions are likely to contain ancestral genes whose functions are no longer essential for the organism's survival, whereas genes that are never deleted constitute the minimal mycobacterial genome. As the amount of genomic deletion increased, the likelihood that the bacteria will cause pulmonary cavitation decreased, suggesting that the accumulation of mutations tends to diminish their pathogenicity. Array-based comparative genomics is a promising approach to exploring molecular epidemiology, microbial evolution, and pathogenesis.
Molecular genotyping is increasingly being used to track infectious diseases as they spread in human populations. For tuberculosis, such molecular epidemiologic approaches have provided answers to public-health-driven questions that have helped to confront the recent resurgence of disease in industrialized countries (Kato-Maeda and Small 2000). In addition, these studies have generated collections of exquisitely well-characterized mycobacteria that may serve as a foundation for exploring the nature and consequences of genetic variability within Mycobacterium tuberculosis (Small et al. 1994).
Bacterial population genetics has provided considerable insight into host–pathogen interactions and may provide empirical data relevant to understanding bacterial evolution (Musser 1996). This approach is likely to be particularly informative for pathogens that are difficult to manipulate and that have limited horizontal gene exchange and to address questions for which good experimental systems do not exist. Knowledge of the complete genomic sequence of one strain of M. tuberculosis, combined with DNA microarray technology, permits high-throughput whole-genome analysis (Cole et al. 1998; Behr et al. 1999; Winzeler et al. 1999) Here we describe the use of array-based comparative genomics to provide a snapshot of mycobacterial evolution and its pathogenesis.
RESULTS
We found that the pattern of deleted sequences was different in every clone examined, except for one clone in which no deletions were detected, making it indistinguishable from H37Rv (Fig. 1). In comparison with H37Rv, each clone was missing an average of 2.9 deleted sequences containing some or all of 17.2 open reading frames (ORFs). On average, clones were missing 0.3% (13,248 bp, range 0–31,581) of the H37Rv genome (Table 1). In contrast, polymorphisms were not detected between clonal isolates H37Ra and H37Rv or among three isolates from individuals involved in a chain of disease transmission. Among the 16 clones, we detected 25 different deleted sequences totaling 76,839 bp deleted, comprising 1.7% of H37Rv genome (Table 2). The deleted sequences included the partial or complete deletion of 93 ORFs and some or all of 22 intergenic regions. Eight of the deleted sequences were absent in more than one clone, such as the prophage phiRv1, which was absent in 11 clones. Three of the deleted sequences (DS5, DS10, DS21) were identical to those we previously reported for BCG, whereas one (DS13) had the same length as the corresponding deletion in BCG but was located 290 bp downstream from the corresponding deletion in BCG (Behr et al. 1999). Two deletion loci (DS6 and DS70) had different deletion lengths among isolates. Two (DS6L and DS6) were within the newly identified preferential insertion loci for IS6110 (ipl; Sampson et al. 1999) In addition to deleted sequences, we also detected two other types of genetic events: replacements of the deleted sequence with IS6110 (DS 4 and DS47) and deletions associated with local genomic rearrangements (DS9). Complex genomic rearrangements, similar but not identical to that which we observed in the region around DS9, have been described in detail elsewhere (Ho et al. 2000). The association of deleted sequences with IS6110 in five instances supports the contention that IS6110 is mechanistically involved in genomic rearrangements (Brosch et al. 2000; Ho et al. 2000).
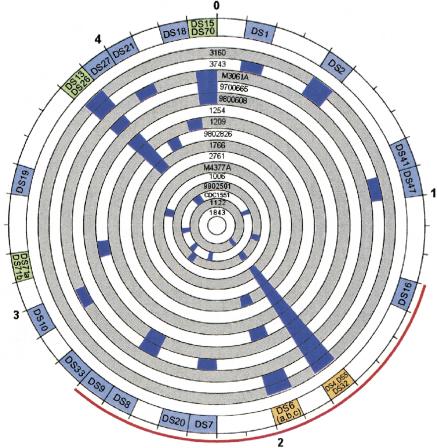
Figure 1.
Circular map of genomic deletions among Mycobacterium tuberculosis showing that the pattern of deletions differs between clones and is not spatially random. The outer numbers shows the scale in mega base pairs, with 0 representing the origin of replication. Each of the inner circles represents 1 of 16 clinical clones, labeled by isolate identification number. The blue regions denote the genomic locations of deleted sequences. The outer circle summarizes the sum of all detected deletions (each denoted by their identification number). Color of deletion regions varies by number of detected deletions within the 100-kb segment (light blue, 1 deletion; light green, 2 deletions; orange, 3 deletions). The thin red line spans the genomic region of the genome where the number of deletions detected is greater than that expected by chance alone.
Table 1.
Characteristics of the Clones Selected and Their Deletions
| Clone no. | No.pts | % pts with cavitary disease | % of contacts with positive tuberculin | % of contacts with active disease | No.deleted bp | No. of DS | No. of deleted ORFs |
|---|---|---|---|---|---|---|---|
| 3160* | 41 | 5.4 | 28.6 | 9.7 | 17862 | 4 | 25 |
| 3743 | 5 | 0.0 | 0.0 | 0.0 | 31581 | 6 | 37 |
| M3061A | 2 | 0.0 | 40.0 | 0.0 | 26481 | 6 | 38 |
| 9700665 | 23 | 13.0 | 27.3 | 4.2 | 22547 | 5 | 27 |
| 9800608 | 24 | 12.5 | 34.1 | 2.4 | 14973 | 3 | 20 |
| 1254 | 20 | 15.0 | 27.1 | 1.6 | 23344 | 4 | 27 |
| 1209 | 5 | 20.0 | 25.0 | 4.6 | 16556 | 3 | 23 |
| 9802826 | 6 | 33.3 | 82.4 | 5.9 | 3758 | 2 | 4 |
| 1766 | 10 | 40.0 | 55.7 | 2.4 | 18916 | 3 | 23 |
| 2761 | 2 | 50.0 | 71.4 | 0.0 | 9236 | 1 | 14 |
| M4377A | 2 | 0.0 | 10.0 | 0.0 | 2308 | 1 | 3 |
| 1006 | 5 | 60.0 | 9.7 | 0.0 | 2890 | 1 | 3 |
| 9802501 | 3 | 66.7 | 93.8 | 5.9 | 15817 | 3 | 20 |
| 1122† | 1 | — | — | — | 9236 | 1 | 14 |
| 1843‡ | 1 | — | — | — | 0 | 0 | 0 |
| CDC1551§ | — | 80.3 | 4.9 | 9718 | 6 | 14 | |
| H37Ra″ | — | — | — | — | 0 | 0 | 0 |
| Average | 13248.4 | 2.9 | 17.2 | ||||
| 2 STD | 19276.0 | 4.1 | 24.3 |
Isolates obtained from the previously reported outbreak: 3160 source case isolated in 1990, a second secondary case was diagnosed seven months later and another case in 1998. The following isolates were included because unusual characteristics: †, Rifampin-resistant with wild-type rpoB gene; ‡, Isoniazid-resistant with catalase-negative phenotype; §, Strain reported to have increased virulence12; ″, Attenuated variant of sequenced H37Rv strain.
Pts, patients; bp, base pairs; DS, deleted sequence; ORF, open reading frame; STD, standard deviation.
Table 2.
Characteristics of the Deleted Sequences
| Deleted sequence | Start | End | Length | No. of deleted ORFs | No. of deleted IGs |
|---|---|---|---|---|---|
| DS1 | 170013 | 173789 | 3776 | 5 | 0 |
| DS2 | 453365 | 455970 | 2605 | 3 | 1 |
| DS41 | 886541 | 887414 | 873 | 3 | 0 |
| DS47 | 931782 | 932199 | 417 | 0 | 1 |
| DS16 | 1310757 | 1311676 | 919 | 1 | 0 |
| DS4 | 1718911 | 1721219 | 2308 | 3 | 0 |
| DS32 | 1727659 | 1728461 | 802 | 1 | 1 |
| DS5* | 1779277 | 1788511 | 9234 | 14 | 0 |
| DS6L | 1986624 | 1987700 | 1076 | 2 | 1 |
| DS6† | 1989056 | 1998602 | 9546 | 8 | 4 |
| DS7‡ | 2225938 | 2228587 | 2649 | 2 | 2 |
| DS20 | 2381413 | 2383683 | 2270 | 2 | 0 |
| DS8 | 2585854 | 2588769 | 2915 | 3 | 0 |
| DS9 | 2627267 | 2632929 | 5662 | 4 | 2 |
| DS33 | 2704308 | 2704805 | 497 | 2 | 1 |
| DS10* | 2969982 | 2980968 | 10986 | 16 | 1 |
| DS71 | 3120465 | 3123063 | 2598 | 2 | 2 |
| DS19 | 3448494 | 3451384 | 2890 | 3 | 0 |
| DS13 | 3842306 | 3847231 | 4925 | 5 | 2 |
| DS26 | 3868777 | 3869666 | 889 | 1 | 0 |
| DS27 | 3955464 | 3956100 | 636 | 2 | 0 |
| DS21* | 4056943 | 4058393 | 1450 | 1 | 1 |
| DS18 | 4212648 | 4215043 | 2395 | 4 | 2 |
| DS15 | 4370421 | 4373243 | 2822 | 3 | 0 |
| DS70 | 4386638 | 4388337 | 1699 | 3 | 1 |
| SUM | 76839 | 93 | 22 | ||
| % OF | 1.7 | 2.4 | 3.0 | ||
| GENOME | |||||
| Average | 3074 | 4 | 1 | ||
| 2STD | 5818 | 8 | 2 |
Previously described in BCG:DS5 corresponds to RD3 in BCG, DS10 to RD13 and DS21 to RD9.
Different isolates had a deleted sequence in the same deletion loci but with different length: DS6b had a length of 20 bp and DS6c of 3677.
Another isolate had a deleted sequence in the same deletion loci but with different length: DS7a had a length of 517 bp.
ORF, open reading frame; IG, intergenic region; DS, deleted sequence; 2STD, 2 standard deviation.
The spatial distribution of deletion loci within the bacterial genome was not random (Fig. 1). The genomic region from 1.3 to 2.7 Mb indicated in red represents the region in which deletions were present more than expected if deletions were distributed randomly. A separate test of this region against remaining regions also showed a greater than expected number of deleted ORF (P = 0.018). One 100-kb segment was also statistically significant for more deletions than expected (1.7–1.8 Mb; P = 0.0001).
As a first step in understanding the factors that underlie genetic deletions, we explored the functional characteristics of the 93 ORFs that were deleted in one or more clones. Not surprisingly, insertion sequences and phages were deleted in excess of their presence in the genome (P < 0.001), whereas information pathway genes were deleted less frequently than would have been expected by chance alone (P = 0.02). At press time, the gene names or functions were annotated for 69 of the 93 ORFs, although in 17 the function assigned was only “possible”, “probable”, “similar,” or “putative”. From the 52 with gene names or specific functions annotated, 25 were phage-related genes, eight were insertion sequences and four were members of the PE or PPE gene families. Included among the remaining 15 were genes plausibly involved in pathogenicity or latency. Three were phospholipase-C genes, which influence the survival of Listeria monocytogenes in macrophages and thus has been proposed as a mycobacterial virulence gene (Smith et al. 1995). The deletion of a polyketide synthase gene (pks5) in the isolate M3061A is particularly intriguing in light of the recent finding that in Mycobacterium ulcerans, the product of this gene is the toxin responsible for cutaneous lesion (Table 3) (George et al. 1999).
Table 3.
List of Genes with Function Annotated
| Deleted sequence | Rv no. | Gene name | Function |
|---|---|---|---|
| DS1 | Rv0143c | Probable chloride channel | |
| DS1 | Rv0144 | Putative transcriptional regulator | |
| DS1 | Rv0147 | Aldehyde dehydrogenase | |
| DS2 | Rv0377 | Transcriptional regulator (LysR family) | |
| DS2 | Rv0378 | Possible PE member, similar to eg | |
| DS41 | Rv0792c | Transcriptional regulator (GntR family) | |
| DS41 | Rv0793 | Unknown but similar to Synechocystis sp. PCC6803 D90908 2 | |
| DS41 | Rv0794c | Dihydrolipoamide dehydrogenase | |
| DS4 | Rv1525 | wb12 | DTDP-rhamnosyl transferase |
| DS4 | Rv 1524 and Rv1526c | Possible rhamnosyl/glycosyl transferase | |
| DS32 | Rv1527c | pks5 | Polyketide synthase |
| DS5 | Rv1573–Rv1576c, Rv1578c–Rv1585c | phiRv1 phage related protein | |
| DS5 | Rv1577c | phiRv1 possible prohead protease | |
| DS5 | Rv1586c | phiRv1 integrase | |
| DS6L | Rv1755c | plcD | Partial CDS for phospholipase C |
| DS6abc | Rv1758 | Partial cutinase | |
| DS6a | Rv1765c | Almost 100% ID to MTV018.02c; not IS element ISB9 | |
| DS7 | Rv1984c | Probable secreted protein | |
| DS20 | Rv2124c | metH | 5-methyltetrahydrofolate-homocysteine methyltransferase |
| DS9 | Rv2349c | plcC | Phospholipase C precursor |
| DS9 | Rv2350c | plcB | Phospholipase C precursor |
| DS9 | Rv2351c | plcA | Phospholipase C precursor |
| DS33 | Rv2406c | Similar to YHCV BACSU P54606 | |
| DS33 | Rv2407 | = B1937 C1 163 | |
| DS10 | Rv2646 and Rv2659c | phiRv2 integrase | |
| DS10 | Rv2647, Rv2650c, Rv2652c–2656, Rv2658c | phiRv2 phage related protein | |
| DS10 | Rv2651c | phiRv2 prohead protease | |
| DS10 | Rv2657c | Similar to gp36 of mycobacteriophage L5 | |
| DS19 | Rv3083 | Probable monooxygenase | |
| DS19 | Rv3084 | lipR | Probable acetyl-hydrolase |
| DS19 | Rv3085 | Short chain alcohol dehydrogenase | |
| DS26 | Rv3448 | Probable membrane protein | |
| DS27 | Rv3518c | Probable Cytochrome P450 monooxygenase | |
| DS21 | Rv3617 | ephA | Probable epoxide hydrolase |
| DS18 | Rv3769 | Possible coiled-coil protein | |
| DS15 | Rv3887c | Probable membrane protein | |
| DS70 | Rv3901c | Membrane protein TM stretch |
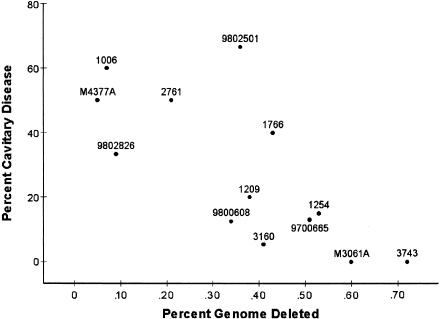
Because so little is known about the function of most mycobacterial genes, we used an agnostic approach, simply seeking correlation between the amount of genetic deletion and the phenotypic characteristics of the 13 clones from the San Francisco population. The results of correlation using four definitions (number of deleted base pairs, percentage of deleted genome, number of deleted sequences, and number of deleted ORFs) with the phenotypic characteristics were very similar; thus we will report only those using percentage of the genome deleted. No correlation was found between the percentage of the genome deleted and either the transmission or pathogenicity indexes. However, there was a statistically significant correlation between the percentage of the genome that was deleted from each clone and the percentage of the patients infected by that clone who had pulmonary cavitations revealed by chest radiography (R = −0.73; P = 0.0047; Fig. 2). The bootstrap estimates of bias and standard error were small (b = 0.02; SE = 0.03), indicating that the results for R were fairly accurate despite our analyzing only 13 clones and 148 patients. Human immunodeficiency virus (HIV) seropositivity was also correlated with cavitary disease (R = −0.58; P = 0.04). However, HIV seropositivity was not correlated with the percentage of the genome deleted (R = 0.50; P = 0.08) and thus was not confounding the correlation.
Figure 2.
Clinical phenotype correlated to genomic deletions showing that pathogenicity decreases as deletions accumulate (R = −0.73; P = 0.0047). The percentage of the mycobacterial sequence that is deleted from each clone (X-axis) is plotted against the percentage of persons infected with that clone who have cavitary pulmonary disease (Y-axis). Points are labeled by isolate identification number.
DISCUSSION
Investigating the genetic variability among natural populations of bacteria is a promising approach to understanding their evolution and pathogenesis. Whole-genome sequencing provides detailed information on genetic differences between bacteria. For example, two isolates of Helicobacter pylori varied due to single-nucleotide substitutions, repetitive elements, recombination, and insertions and deletions (Alm et al. 1999). Presently, however, this approach is prohibitively time consuming and expensive for comparing large numbers of isolates. We have used high-density oligonucleotide arrays, a relatively rapid and inexpensive approach, to detect small-scale genomic deletions among clinical isolates. On average, each clinical isolate was missing 0.3% of the genome, comprised of some or all of 17.2 ORFs that were present in the sequenced strain. Clearly, deletions represent only a subset of the total genetic variability; for example, they do not include sequence present in clinical isolates but absent from H37Rv. However, in clonal organisms such as M. tuberculosis, these detected deletions should also serve as markers for other mutations. Our results suggest that in M. tuberculosis, array-based analysis of small-scale genomic deletions is a suitable genotyping system for molecular epidemiologic studies and can provide a novel perspective on mycobacterial evolution and pathogenesis.
An ideal molecular epidemiologic genotyping system would be applicable to all isolates, polymorphic among unrelated isolates and yet remain recognizable over the period of investigation. In our study, DNA microarray analysis was easily performed on all isolates; polymorphisms were found among virtually all of the strains not known to be clonal and patterns remained unchanged over seven years of human passage. Perhaps the most important, but difficult to define, parameter of a genotyping system is the time period over which the degree of relatedness demonstrated is informative. The most widely used system for genotyping M. tuberculosis (IS6110) is clearly not informative over protracted periods. In contrast, the nature and structure of our data suggest that, similar to changes in human mitochondrial DNA, genomic deletions can be used to reconstruct meaningful phylogenetic trees. Such trees will permit the formal study of quantitative traits and geographic patterns of pathogen migration. Thus, while these preliminary observations must be further quantified in future studies, our data suggest that DNA microarray detected genomic deletions are promising markers for use in molecular epidemiologic studies.
There are limited data available regarding the magnitude of small-scale deletions among natural populations of other pathogens. However, available data suggests that, in comparison with other pathogens, there is relatively little variability within the species M. tuberculosis. Using less precise pulsed-field gel electrophoresis, the genomic size of clinical isolates of Escherichia coli has been estimated to vary by as much as 14% (Bergthorsson and Ochman 1995). Precise data obtained by sequencing two clones of H. pylori, which has a much smaller genome, showed 89 ORFs to be present in one but not the other strain (Alm et al. 1999). In contrast, we found that none of the 16 M. tuberculosis clones examined differed from the sequenced strain by more than 38 ORFs. Whereas the relative lack of genetic variability detected in M. tuberculosis may be a consequence of our inability to detect deletions smaller than 350 bp and other types of mutations, we suspect the paucity we observed is a consequence of mycobacterial population genetics. Sequence-based analysis of M. tuberculosis has shown that there are remarkably few single nucleotide polymorphisms within coding regions, including genes coding for targets of the host immune system, suggesting that it evolved from a progenitor species only 15,000 years ago (Sreevatsan et al. 1997; Musser et al. 2000) In addition, M. tuberculosis has not been shown to undergo horizontal gene exchange and thus has an extremely clonal population structure. Taken together, these observations could result in the relative lack of genetic deletions we detected.
By sampling the extant population, we are observing clonal differences that have been generated over evolutionary time frames. Thus, the analysis of their genomic deletions provides a novel perspective on genomic organization and a snapshot of mycobacterial evolution. The paucity of deletions close to the origin of replication suggests that genes in this region are relatively important. Deletions are likely to contain ancestral genes whose functions are no longer essential for the organism's survival. For example, strains 3743, CDC1551, and 1006 are all missing dehydrogenases, central enzymes in anaerobic metabolism (Murugasu-Oei et al. 1999). It is intriguing to speculate that these pathways were important for the survival of ancestral mycobacteria in the soil but had become superfluous now that the organism has evolved to thrive in the relatively well-oxygenated environment of its human host.
In the context of >7 yr of clinical and molecular epidemiologic data from San Francisco, the analysis of genomic deletions also provided a novel perspective on bacterial pathogenesis. This present work characterized 13 clones, which caused disease in 148 patients. Even this limited data set found that the likelihood that a clone of M. tuberculosis will cause pulmonary cavitation decreases as the amount of genomic deletion increases. We suggest that this observation be considered in the light of two facts. First, it is well accepted that most bacterial mutations are deleterious (Arber 2000). Second, the transmission from one host to the next is a key challenge facing all pathogens. For an airborne pathogen such as tuberculosis, the capacity to induce pulmonary cavities is an extremely effective way to meet this challenge, essentially converting a patient into a bacterial aerosolization device (Kline et al. 1995). Taken together, we propose that the accumulation of deletions among clinical isolates generally diminish their virulence. This observation is concordant with Muller's prediction that the accumulations of mutations in clonal organisms will results in genetic deterioration (Muller 1964). We attribute the lack of a correlation between deletions and transmission index to the limited statistical power of the current study. If so, larger studies will find correlations with additional clinical endpoints.
Closer scrutiny of this data suggests testable hypotheses about the specific genes involved in virulence. For example, the deletion of a polyketide synthase gene (pks5) with high homology to mycocerosic acid synthase is particularly intriguing because the product of this gene may be involved in the production of multimethylated branched lipids such as the phthioceranic acids that appear in sulfatides. Experiments from the 1960s have correlated variable expression levels of sulfatides with virulence of clinical strains in guinea pig models (Goren et al. 1974). Recently, the observation that sulfatides inhibit macrophage activation mediated by cytokines such as IFN-γ and TNF-α provides a possible mechanism by which sulfatides may impair bacterial killing (Brozna et al. 1991). Our data support the proposition that a more formal evaluation of polymorphisms in selected genes such as pks5 among large numbers of well-characterized natural populations of M. tuberculosis may identify mutations that attenuate or enhance the virulence of pathogens.
METHODS
Study Population
Between 1991 and 1998 clinical, epidemiologic, microbiologic, and genotypic data were collected on 1744 tuberculosis patients in San Francisco (Jasmer et al. 1999). In addition, we determined the rates of skin-test reactivity and active tuberculosis among the 14,293 contacts to these patients. Bacterial genotypes were determined by IS6110– and polymorphic GC rich sequence (PGRS)–based restriction-fragment-length polymorphism analysis (Yeh et al. 1998). A total of 1236 distinct mycobacterial genotypes were identified, among which 158 were isolated from more than one patient. From these data, for each genotype we calculated the number of tuberculosis cases in San Francisco, the percentage of contacts found to be tuberculin positive (transmission index), the percentage of patients with active disease (pathogenicity index), and the percentage of patients with pulmonary cavitation shown by chest radiography (Rhee et al. 1999).
For the present work, 15 isolates were selected from the San Francisco study population, each representing a different genotype and together spanning the observed variability in these characteristics (Table 1). Three of these isolates represented one genotype from a previously reported chain of transmission spanning 7.7 yr of human passage (the first case detected in 1990 and the last case in 1998; Small et al. 1994) We also selected two isolates from outside San Francisco because of their unique drug-susceptibility profiles, one isolate that was previously reported to be highly transmissible (CDC1551; Valway et al. 1998) and the attenuated variant of the strain that has been sequenced (H37Ra). A clone was defined as a group of isolates that share sufficient properties such that they are likely to represent progeny of the same progenitor (Orskov and Orskov 1983). Based on historical, clinical, epidemiological, and genotypic data, we considered H37Ra and H37Rv, as well as the isolates 3160, 1098, and 9802731, to be two clones and all others to represent unique clones. Thus, our 19 isolates represent 16 clones, 13 of which were sampled from the San Francisco population, in which they had caused 148 cases of tuberculosis and were epidemiologically implicated as the cause of infection in an additional 358 persons.
Deletion Detection
Genomic deletions were detected using a method fully described elsewhere. The analysis of CDC1551 showed that our approach was able to identify all deleted sequences longer than 350 bp (Salamon et al. 2000). In brief, an Affymetrix GeneChip, representing all 3924 ORF and 738 intergenic regions of H37Rv, was fabricated for the analysis of M. tuberculosis according to the published sequence (http://www.sanger.ac.uk/Projects/M_tuberculosis/). Twenty probe pairs (each 25 bp in length) were targeted to every ORF and intergenic region, and in total, the chip contained 118,180 probe pairs. Seventeen probe pairs were excluded from analysis because they failed to hybridize reliably with genomic DNA from H37Rv. In addition, hybridization was not expected to be informative from rRNA, and tRNA and from highly repetitive PE, PPE, leaving 111,488 probe pairs analyzed for each experiment. Mycobacteria were grown and DNA was extracted as previously described (Van Soolingen et al. 1991). Whole-genomic DNA was digested with DNaseI (GIBCO BRL Life Technologies), end-labeled with biotin-N6-dideoxyadenosine triphosphate (NEN Life Science Products), and hybridized to the array. Hybridized DNA was stained with phycoerythrin-streptavidin conjugate and fluorescent intensities were recorded using a confocal laser scanner. Data were analyzed using our Tandem Set Terminal Extreme Probability (TSTEP) algorithm in a semi-automated computational approach designed to identify putative genomic deletions (available at http://molepi.stanford.edu/TSTEP). Each putative deletion was confirmed by PCR amplification across the region in question, and the margins of the deletions were determined to the precise base pair using sequencing and blast analysis (http://www.sanger.ac.uk/Projects/M_tuberculosis/blast_server.html; Altschul et al. 1997). We used the term “deleted sequence” to refer to the genetic sequence that is present in H37Rv but missing from another isolate and the term “deletion locus” to refer to the region of the genome (according to the H37Rv map) from which this, or other, sequence is deleted.
To determine whether deleted sequences among the 16 mycobacterial clones were distributed randomly around the genome, we analyzed the distribution of deleted ORFs among 11 half-genome pairs, 22 quarter-genome sets, and 100-kb segments using χ2 tests and Bonferroni correction because of the multiple tests. To determine whether specific functional classes of genes (as assigned by Sanger Center) were disproportionately deleted or conserved, we analyzed each classification (excluding the PE and the PPE) against the remaining types using χ2 tests (Cole 1999). To investigate associations between genetic deletion data and epidemiologic and clinical characteristics, we analyzed the 13 clones from the San Francisco population. The Pearson's product-moment correlation coefficient, R, was calculated for the deletion sequence data and the phenotypic characteristics reported in Table 1 (S-Plus software, version 4, MathSoft). Because of the small sample size, we evaluated the accuracy of correlation with the bootstrap method using 100 replications to estimate the bias (b) and standard error (SE) of R.
Acknowledgments
We thank Julie Parsonnet for her many thoughtful comments; Kumiko Aman and Tamara van Gorkom for their assistance with PCR; Clifton Barry for sharing his expertise about polyketide synthase; and S. Cole, T. Shinnick, and E. Desmond for sharing H37Rv and clinical isolates CDC1551 and 1843, respectively. This study was supported by NIH grants TW00923, TW01135, and A134238.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL peter@molepi.stanford.edu; FAX 650-498-7011.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr166401.
REFERENCES
- Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, Smith DR, Noonan B, Guild BC, deJonge BL, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber W. Genetic variation: Molecular mechanisms and impact on microbial evolution. FEMS Microbiol Rev. 2000;24:1–7. doi: 10.1111/j.1574-6976.2000.tb00529.x. [DOI] [PubMed] [Google Scholar]
- Behr MA, Wilson MA, Gill WP, Salamon H, Schoolnik GK, Rane S, Small PM. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science. 1999;284:1520–1523. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
- Bergthorsson U, Ochman H. Heterogeneity of genome sizes among natural isolates of Escherichia coli. J Bacteriol. 1995;177:5784–5789. doi: 10.1128/jb.177.20.5784-5789.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch R, Gordon SV, Buchrieser C, Pym AS, Garnier T, Cole ST. Comparative genomics uncovers large tandem chromosomal duplications in Mycobacterium bovis BCG Pasteur. Yeast. 2000;17:111–123. doi: 10.1002/1097-0061(20000630)17:2<111::AID-YEA17>3.0.CO;2-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brozna JP, Horan M, Rademacher JM, Pabst KM, Pabst MJ. Monocyte responses to sulfatide from Mycobacterium tuberculosis: Inhibition of priming for enhanced release of superoxide, associated with increased secretion of interleukin-1 and tumor necrosis factor alpha, and altered protein phosphorylation. Infect Immun. 1991;59:2542–2548. doi: 10.1128/iai.59.8.2542-2548.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole ST. Learning from the genome sequence of Mycobacterium tuberculosis H37Rv. FEBS Lett. 1999;452:7–10. doi: 10.1016/s0014-5793(99)00536-0. [DOI] [PubMed] [Google Scholar]
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, III, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- George KM, Chatterjee D, Gunawardana G, Welty D, Hayman J, Lee R, Small PL. Mycolactone: A polyketide toxin from Mycobacterium ulcerans required for virulence. Science. 1999;283:854–857. doi: 10.1126/science.283.5403.854. [DOI] [PubMed] [Google Scholar]
- Goren MB, Brokl O, Schaefer WB. Lipids of putative relevance to virulence in Mycobacterium tuberculosis: Correlation of virulence with elaboration of sulfatides and strongly acidic lipids. Infect Immun. 1974;9:142–149. doi: 10.1128/iai.9.1.142-149.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TB, Robertson BD, Taylor GM, Shaw RJ, Young DB. Comparison of Mycobacterium tuberculosis genomes reveals frequent deletions in a 20 kb variable region in clinical isolates. Yeast. 2000;17:272–282. doi: 10.1002/1097-0061(200012)17:4<272::AID-YEA48>3.0.CO;2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasmer RM, Hahn JA, Small PM, Daley CL, Behr MA, Moss AR, Creasman JM, Schecter GF, Paz EA, Hopewell PC. A molecular epidemiologic analysis of tuberculosis trends in San Francisco, 1991-1997. Ann Intern Med. 1999;130:971–978. doi: 10.7326/0003-4819-130-12-199906150-00004. [DOI] [PubMed] [Google Scholar]
- Kato-Maeda M, Small PM. How molecular epidemiology has changed what we know about tuberculosis. West J Med. 2000;172:256–259. doi: 10.1136/ewjm.172.4.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline SE, Hedemark LL, Davies SF. Outbreak of tuberculosis among regular patrons of a neighborhood bar. N Engl J Med. 1995;333:222–227. doi: 10.1056/NEJM199507273330404. [DOI] [PubMed] [Google Scholar]
- Muller HJ. The relation of recombination to mutational advance. Mutat Res. 1964;1:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- Murugasu-Oei B, Tay A, Dick T. Upregulation of stress response genes and ABC transporters in anaerobic stationary-phase Mycobacterium smegmatis. Mol Gen Genet. 1999;262:677–682. doi: 10.1007/s004380051130. [DOI] [PubMed] [Google Scholar]
- Musser JM. Molecular population genetic analysis of emerged bacterial pathogens: Selected insights. Emerg Infect Dis. 1996;2:1–17. doi: 10.3201/eid0201.960101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser JM, Amin A, Ramaswamy S. Negligible genetic diversity of Mycobacterium tuberculosis host immune system protein targets. Evidence of limited selective pressure. Genetics. 2000;155:7–16. doi: 10.1093/genetics/155.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orskov F, Orskov I. From the National Institutes of Health. Summary of a workshop on the clone concept in the epidemiology, taxonomy, and evolution of the enterobacteriaceae and other bacteria. J Infect Dis. 1983;148:346–357. doi: 10.1093/infdis/148.2.346. [DOI] [PubMed] [Google Scholar]
- Rhee JT, Piatek AS, Small PM, Harris LM, Chaparro SV, Kramer FR, Alland D. Molecular epidemiologic evaluation of transmissibility and virulence of Mycobacterium tuberculosis. J Clin Microbiol. 1999;37:1764–1770. doi: 10.1128/jcm.37.6.1764-1770.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamon H, Kato-Maeda M, Drenkow J, Small PM, Gingeras TR. Detection of deleted genomic DNA using a semi-automated computational analysis of GeneChipTM data. Genome Res. 2000;10:2044–2054. doi: 10.1101/gr.gr-1529r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson SL, Warren RM, Richardson M, van der Spuy GD, van Helden PD. Disruption of coding regions by IS6110 insertion in Mycobacterium tuberculosis. Tuber Lung Dis. 1999;79:349–359. doi: 10.1054/tuld.1999.0218. [DOI] [PubMed] [Google Scholar]
- Small PM, Hopewell PC, Singh SP, Paz A, Parsonnet J, Ruston DC, Schecter GF, Daley CL, Schoolnik GK. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N Engl J Med. 1994;330:1703–1709. doi: 10.1056/NEJM199406163302402. [DOI] [PubMed] [Google Scholar]
- Smith GA, Marquis H, Jones S, Johnston NC, Portnoy DA, Goldfine H. The two distinct phospholipases C of Listeria monocytogenes have overlapping roles in escape from a vacuole and cell-to-cell spread. Infect Immun. 1995;63:4231–4237. doi: 10.1128/iai.63.11.4231-4237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreevatsan S, Pan X, Stockbauer KE, Connell ND, Kreiswirth BN, Whittam TS, Musser JM. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valway SE, Sanchez MP, Shinnick TF, Orme I, Agerton T, Hoy D, Jones JS, Westmoreland H, Onorato IM. An outbreak involving extensive transmission of a virulent strain of Mycobacterium tuberculosis. N Engl J Med. 1998;338:633–639. doi: 10.1056/NEJM199803053381001. [DOI] [PubMed] [Google Scholar]
- Van Soolingen D, Hermans PW, de Haas PE, Soll DR, van Embden JD. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: Evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA, Lee B, McCusker JH, Davis RW. Whole genome genetic-typing in yeast using high-density oligonucleotide arrays. Parasitology. 1999;118:S73–80. doi: 10.1017/s0031182099004047. [DOI] [PubMed] [Google Scholar]
- Yeh RW, Ponce de Leon A, Agasino CB, Hahn JA, Daley CL, Hopewell PC, Small PM. Stability of Mycobacterium tuberculosis DNA genotypes. J Infect Dis. 1998;177:1107–1111. doi: 10.1086/517406. [DOI] [PubMed] [Google Scholar]