Abstract
We found that among four master epithelial-to-mesenchymal transition (EMT)-inducing genes (ZEB1, SIP1, Snail, and Slug) ZEB1expression was most significantly correlated with the mesenchymal phenotype (high Vimentin and low E-cadherin expression) in non-small cell lung cancer (NSCLC) cell lines and tumors. Furthermore, ZEB1 knockdown with RNA interference in three NSCLC cell lines with high ZEB1 expression suppressed to varying degrees mass culture growth and liquid colony formation but in all cases dramatically suppressed soft agar colony formation. In addition, ZEB1 knockdown induced apoptosis in one of the three lines, indicating that the growth inhibitory effects of ZEB1 knockdown occurs in part through the activation of the apoptosis pathway. These results suggest that inhibiting ZEB1 function may be an attractive target for NSCLC therapeutic development.
Keywords: Lung cancer, Epidermal growth factor receptor, Anchorage-independent growth, EMT, MicroRNA, RNA interference
1. Introduction
Epithelial-to-mesenchymal transition (EMT) is an embryonic developmental program involving changes in cell morphology and expression of EMT-associated genes [1,2]. EMT also occurs during the progression of several types of human cancer and confers motility and invasiveness on cancer cells, leading them to acquire ability to metastasize to distant sites. Genetics of early development discovered a number of EMT-inducing genes encoding transcription factors capable of inducing EMT when ectopically expressed in epithelial cells. Several EMT-inducing genes that have essential roles in EMT are called master EMT genes, including Twist, ZEB1, SIP1, Snail, Slug, and Goosecoid [1,2]. These genes function as transcriptional repressors of the cell–cell adhesion glycoprotein, E-cadherin whose functional loss is one of the hallmarks of EMT [3]. Among these master EMT genes, Snail was shown to repress E-cadherin and to induce EMT in cancer cells [4,5], while Twist was demonstrated to promote breast cancer metastasis [6]. Increased expression of Twist and Snail have been shown in hepatocellular, breast, colorectal, and gastric cancers, often correlating with poor prognosis [7–10].
Lung cancer is the leading cause of cancer deaths, killing over 1 million people every year worldwide [11]. It develops through a multi-step process involving accumulation of multiple genetic and epigenetic changes that confer growth advantages on normal lung epithelial cells, leading them to transform to clinically evident lung cancer cells [12,13]. Analyzing a large number of lung tumor specimens, Prudkin et al. showed that the majority of primary lung cancers and even premalignant lesions have the mesenchymal phenotype as characterized by down-regulation of E-cadherin and up-regulation of Vimentin [14]. Although several master EMT genes have been shown to directly contribute to tumor progression in breast, colon, and pancreatic cancers, very little is known about functional roles of master EMT genes in lung cancer progression.
In addition to Snail and Twist, recently ZEB1 has emerged as a key player in cancer progression. ZEB1 promotes tumor metastasis in colon and breast cancer [15], is associated with resistance to conventional chemotherapy in pancreatic cancer [16,17], and potentially has a predominant role in inducing EMT in NSCLC. First, among several transcription factors including ZEB1, Snail, and β-catenin, ZEB1 protein expression showed the most significant inverse correlation with E-cadherin in NSCLC and mesothelioma cell lines [18]. Second, prostaglandin E2 was shown to exert its ability to suppress E-cadherin through inducing ZEB1 and Snail in lung cancer cell lines [19]. Third, ZEB1 has been shown to suppress the Semaphorin 3F tumor suppressor gene in lung cancer cells [20]. These suggest relevant roles of ZEB1 as an EMT-inducer as well as an oncogene in lung cancer.
Thus, we performed this study aiming to evaluate the association between ZEB1 expression and the mesenchymal phenotype in lung cancer, and to test the effects of ZEB1 knockdown with RNA interference on the growth of lung cancer cells. We found that ZEB1 expression significantly correlates with increased Vimentin and decreased E-cadherin expression in lung cancer, while knockdown of ZEB1 resulted in dramatic growth inhibition in lung cancer cell lines. These results suggest that ZEB1 is a promising therapeutic target for lung cancer.
2. Materials and methods
2.1. Cell lines and primary tumor tissues
NSCLC cell lines used in this study were purchased from American Type Culture Collection or obtained from the Hamon Center collection (University of Texas Southwestern Medical Center). These cells include PC9, A549, NCI-H157, NCI-H460, NCI-H820, NCI-H838, NCI-H1155, NCI-H1299, NCI-H1666, NCI-H1650, NCI-H1975, NCI-H3255, HCC44, HCC827, HCC2279, HCC2935, HCC4006, and HCC4011 (cells with mutation in epidermal growth factor receptor (EGFR) gene are underlined) [21]. A mesothelioma cell line, ACC-MESO-1, which was used as positive control for western blot of cleaved caspase-3, was established by ourselves [22]. Cells were cultured with RPMI 1640 (Sigma–Aldrich Corp, MO, USA) supplemented with 10% fetal bovine serum. Surgically resected 32 primary tumor specimens (19 adenocarcinomas and 13 squamous cell carcinomas) were obtained from patients at the Nagoya University Hospital, Nagoya First Japan Red Cross Hospital, Nagoya Second Japan Red Cross Hospital, Kasugai Municipal Hospital and Chukyo Hospital in Nagoya, Japan. Before tissue samples were collected ethical approval of the each institute and fully informed written consents from all patients were obtained. We previously analyzed EGFR mutation status of these samples and used the data of the analysis for the present study [23].
2.2. RNA isolation and quantitative real-time PCR analysis
For mRNA analysis, 5 μg of total RNA isolated using Trizol (Invitrogen Corp., CA, USA) were reverse transcribed with Super script III First-Strand Synthesis System using Random primer system (Invitrogen Corp.). Quantitative real-time PCR (qRT-PCR) analysis of E-cadherin, Vimentin, ZEB1, SIP1 Snail, and Slug, was performed as described previously using the standard Taqman assay-on-demand PCR protocol in a reaction volume of 20 μL, including 50 ng cDNA [24]. We used the comparative Ct method to compute relative expression values. For microRNA analysis, 10 ng of total RNA isolated using mirVana miRNA Isolation Kit (Applied Biosystems, CA, USA) were reverse transcribed with TaqMan MicroRNA Reverse Transcription Kit using a primer set specific for each of microRNAs (miR-200a, miR-200b, miR-200c, and miR-205) studied (Applied Biosystems). qRT-PCR analysis of microRNA was done as described above. We used GAPDH (Applied Biosystems assay-on-demand) for mRNA analysis and U6 small nuclear (sn) RNA for microRNA analysis as internal controls.
2.3. Western blot analysis
Western blot analysis was done as described previously using whole cell lysates [24]. Primary antibodies used were mouse monoclonal anti-E-cadherin, anti-Vimentin (BD Bioscience, NJ, USA), goat polyclonal anti-ZEB1 (Santa Cruz biotech., CA, USA), and rabbit polyclonal anti-cleaved caspase-3 (Cell Signaling Tech., MA, USA). Actin protein levels were used as a control for adequacy of equal protein loading. Anti-rabbit, anti-mouse (GE healthcare, Buckinghamshire, England), or anti-goat antibody (R&D Systems, MN, USA) was used at 1:2000 dilution as a secondary antibody.
2.4. Transfection of short interfering RNA
The 4.5 × 105 of H1299 and H157 or 9.0 × 105 of H460 cells were plated in 6-well plates. Next day, cells were transiently transfected with either 10 nM predesigned short interfering RNA (siRNA) (Stealth Select RNAi) targeting ZEB1 or control siRNA purchased from Invitrogen using Lipofectamine RNAiMAX (Invitrogen Corp.) according to the manufacturer’s protocol. After 48 h, the transfected cells were harvested for further analyses or plated for cell growth assays.
2.5. Cell growth assays
Colorimetric proliferation assay was performed using WST-1 assay kit (Roche, Basel, Switzerland) according to manufacturer’s instruction. Liquid and soft agar colony formation assays were done as described previously [24].
2.6. Cell cycle analysis
Cells were harvested 48 h after the transfection of siRNA oligos. Cells were, fixed, treated with RNase A, stained with propidium iodide using BD Cycletest Plus Reagent Kit (BD Bioscience) according to the instructions of the manufacture, and analyzed by flow cytometry for DNA synthesis and cell cycle status [FACSCalibur instrument, (Becton Dickinson) with BD CellQuest™Pro Ver.5.2.1 (BD Bioscience)].
2.7. Senescence associated β-galactosidase staining
Cells were stained with β-galactosidase using Senescence β-Galactosidase Staining Kit (Cell Signaling Tech.), and cells stained blue were counted under a microscope (200× total magnification).
2.8. Statistics
SPSS ver.17 software was used for all statistic analyses in this study. Spearman’s correlation coefficients with associated P values were calculated between mRNA expression of Vimentin, E-cadherin, and four EMT-inducing genes, microRNA expression of the four microRNAs, and the ratio of Vimentin to E-cadherin expression (RVE). Mann–Whitney U test was used for analyzing difference between two groups.
3. Results
3.1. Lung cancer cell lines can be divided into epithelial and mesenchymal phenotypes based on the expression status of E-cadherin and Vimentin
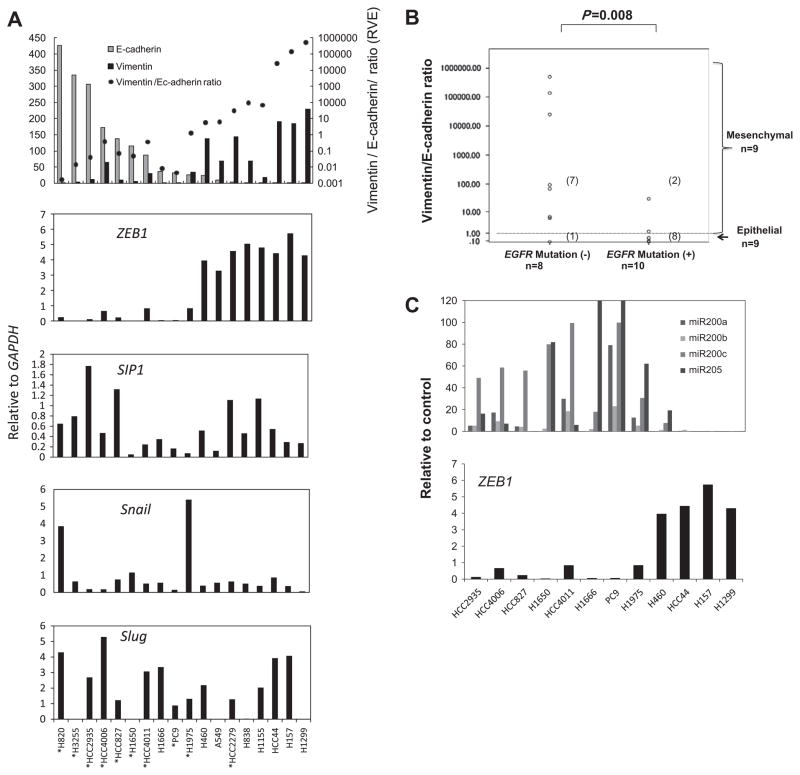
We first examined whether lung cancer cell lines can be classified into mesenchymal and epithelial phenotypes based on expression status of E-cadherin and Vimentin, which are markers for epithelial and mesenchymal phenotypes, respectively. qRT-PCR analysis of E-cadherin and Vimentin in 18 non-small cell lung cancer cell (NSCLC) lines including 10 with mutations in the epidermal growth factor receptor (EGFR) gene showed that most predominantly expressed either E-cadherin or Vimentin (Fig. 1A). We classified NSCLCs as either “epithelial” (high E-cadherin/low Vimentin) or “mesenchymal” (high Vimentin/low E-cadherin) according to the expression status of E-cadherin and Vimentin. We quantified the ratio of Vimentin to E-cadherin expression (RVE) as an index that represents the degree of mesenchymal phenotype, with NSCLCs showing RVE ≥ 1.0 classified as mesenchymal phenotype (n = 9 NSCLCs) and those with RVE < 1.0 as epithelial phenotype (n = 9 NSCLCs). Notably, RVEs of EGFR wild-type NSCLCs were significantly higher than those of EGFR mutant NSCLCs (Fig. 1B) (Median; 80.0 vs. 0.06, P = 0.008, Mann–Whitney U test). All but one EGFR wild-type NSCLC lines were “mesenchymal” while 8 of 10 EGFR mutant NSCLCs were “epithelial”, suggesting that EGFR mutation is associated with epithelial characteristics (Fig. 1B).
Fig. 1.
ZEB1 expression significantly correlates with E-cadherin, Vimentin and two microRNAs, miR-200c and miR-205, known ZEB1 repressor in lung cancer cell lines. (A) Quantitative real-time PCR (qRT-PCR) analysis of E-cadherin, Vimentin and four master EMT genes, ZEB1, SIP1, Snail, and Slug in 18 lung cancer cell lines. The cell lines are aligned by expression levels of E-cadherin from high (left) to low (right). The results are average of two independent PCR experiments done in duplicated reactions. Statistic values of correlations between these genes are shown in Table 1. *Cells with mutation in the epidermal growth factor receptor (EGFR) gene. (B) The association between the ratio of Vimentin to E-cadherin (RVE) and mutation status of EGFR in lung cancer cell lines. (C) qRT-PCR analysis of four microRNAs known as repressors of ZEB1. The expression levels of miR-200c and miR-205 were significantly negatively associated with ZEB1 expression (Spearman’s correlation coefficients: −0.71, P = 0.01 for miR-200c and −0.79, P = 0.002 for miR-205).
3.2. Among four master EMT genes only ZEB1 expression was significantly correlated with both Vimentin and E-cadherin expression in lung cancer cell lines
To identify master EMT genes whose expressions are significantly associated with the mesenchymal phonotype in lung cancer cell lines, we analyzed the expression levels of four master EMT genes (ZEB1, SIP1, Snail, and Slug) (Fig. 1A, Table 1). ZEB1 expression was inversely correlated with E-cadherin expression (Spearman’s correlation coefficient = −0.82, P < 0.001), and positively correlated with Vimentin expression (Spearman’s correlation coefficient = 0.80, P < 0.001) (Table 1), resulting in highly significant association between ZEB1 expression and RVE (Spearman’s correlation coefficient = 0.88, P < 0.001) (Table 1). By contrast, SIP1, Snail, and Slug expression were not correlated with E-cadherin expression, Vimentin expression, or RVE (Table 1). These results suggested that ZEB1 may induce EMT in lung cancer.
Table 1.
Correlations between mRNA expression of master EMT genes, E-cadherin, and Vimentin in 18 non-small lung cancer cell lines.
| E-cadherin | Vimentin | RVE | ZEB1 | SIP1 | Snail | |
|---|---|---|---|---|---|---|
| E-cadherin Vimentin | −0.74 <0.001 |
|||||
| RVE | −0.88 <0.001 |
0.92 <0.001 |
||||
| ZEB1 | −0.82 <0.001 |
0.80 <0.001 |
0.88 <0.001 |
|||
| SIP1 | 0.22 0.38 |
−0.05 0.85 |
−0.06 0.81 |
−0.06 0.80 |
||
| Snail | 0.30 0.23 |
−0.22 0.38 |
−0.26 0.30 |
−0.23 0.36 |
−0.03 0.92 |
|
| Slug | 0.09 0.72 |
0.07 0.79 |
−0.04 0.88 |
0.20 0.46 |
0.24 0.34 |
−0.06 0.82 |
Spearman’s correlation coefficients (upper row) and statistic values (lower row) are shown. Statistically significant correlations (P < 0.01) are in bold. RVE; the ratio of Vimentin to E-cadherin.
3.3. The expression levels of four miroRNAs known as repressors of ZEB1 were negatively correlated with ZEB1 expression in NSCLCs
Recently, several groups have shown that ZEB1 expression is down-regulated by three members of microRNA-200 family (miR-200a, miR-200b, miR-200c) and miR-205 in different types of cancer cells [25–28]. To confirm these findings in a panel of lung cancer cell lines, we performed qRT-PCR of the four microRNAs in 12 NSCLC cell lines and correlated their expression with ZEB1 expression (Fig. 1C). We found that the expression levels of miR-200c and miR-205 were negatively correlated with ZEB1 expression (Spearman’s correlation coefficients: −0.71, P = 0.01 for miR-200c, −0.79, P = 0.002 for miR-205), while miR-200a and miR-200b were not correlated with ZEB1 expression. These results suggest that miR-200c and miR-205 may have a major role in regulating ZEB1 expression in lung cancer (Fig. 1C).
3.4. ZEB1 and Snail expression were significantly correlated with RVE in primary lung tumor tissues
Next, we analyzed the expression of E-cadherin, Vimentin, and the four master EMT genes in 32 NSCLC tumor specimens and found that ZEB1 expression was highly correlated with Vimentin expression (Spearman’s correlation coefficient = 0.92, P < 0.001) (Table 2). ZEB1 (Spearman’s correlation coefficient = 0.51, P = 0.003), and Snail (Spearman’s correlation coefficient = 0.45, P = 0.01) expression were significantly correlated with RVE. However, we note that overall in the tumor specimens in contrast to the NSCLC lines, E-cadherin and Vimentin expression were positively correlated (Spearman’s correlation coefficient = 0.63, P < 0.001) (Table 2). This may be because the primary tumor specimens were macrodissected and thus included transcripts from both tumor and non-tumorous lung tissue while the NSCLC lines only reflected tumor expression. Probably because of this reason, unlike in the NSCLC lines, EGFR mutation status was not correlated with epithelial characteristics (high E-cadherin and low Vimentin) in the primary tumors (data not shown).
Table 2.
Correlations between mRNA expression of master EMT genes, E-cadherin, and Vimentin in 32 non-small lung cancer tumor tissues.
| E-cadherin | Vimentin | RVE | ZEB1 | SIP1 | Snail | |
|---|---|---|---|---|---|---|
| E-cadherin Vimentin |
0.63 <0.001 |
|||||
| RVE | −0.26 0.15 |
0.53 0.002 |
||||
| ZEB1 |
0.57 <0.001 |
0.92 <0.001 |
0.51 0.003 |
|||
| SIP1 |
0.81 <0.001 |
0.73 <0.001 |
0.09 0.61 |
0.64 <0.001 |
||
| Snail | 0.32 0.07 |
0.71 <0.001 |
0.45 0.01 |
0.69 <0.001 |
0.49 0.004 |
|
| Slug | 0.29 0.11 |
0.39 0.03 |
0.31 0.08 |
0.41 0.02 |
0.43 0.01 |
0.26 0.15 |
Spearman’s correlation coefficients (upper row) and statistic values (lower row) are shown. Statistically significant correlations (P < 0.01) are in bold. RVE; the ratio of Vimentin to E-cadherin.
3.5. Knockdown of ZEB1 induced significant suppression of anchorage-independent cell growth in lung cancer cell lines
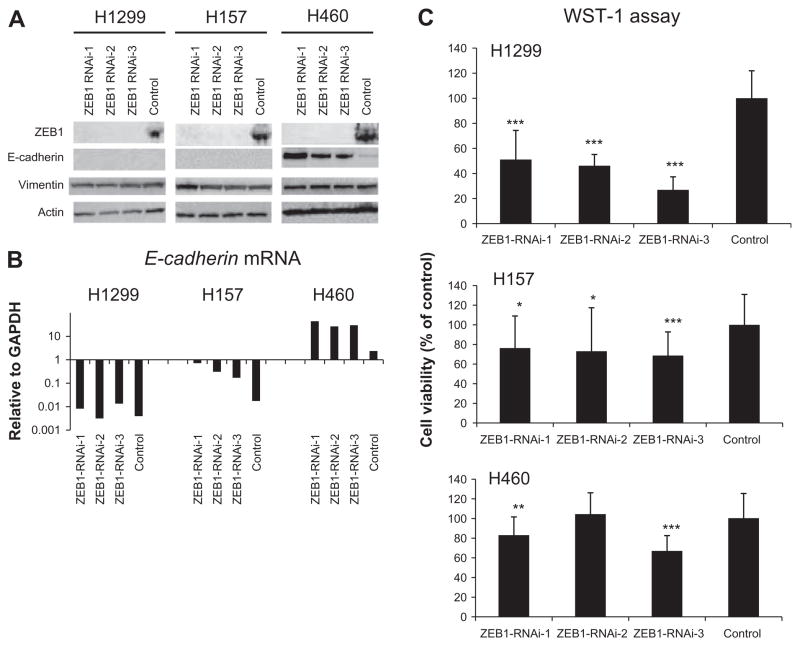
The results presented above suggest that ZEB1 plays a dominant role in maintaining the mesenchymal phenotype in NSCLCs. To test the therapeutic potential of ZEB1 we performed RNA interference (RNAi)-mediated gene silencing against ZEB1. To minimize the possibility of “off target effect”, we used low dose (10 nM) Stealth Select RNAi (Invitrogen), which includes three short interfering RNA (siRNA) oligos with non-overlapping sequences targeting ZEB1. We studied three NSCLC cell lines, H1299, H460, and H157, all of which express high levels of ZEB1 mRNA (Fig. 1A). These cell lines were transiently transfected with each of the three ZEB1 siRNAs or control oligos and harvested for analyses 48 h after transfection. Western blot of ZEB1 showed that with all three oligos clear suppression of ZEB1 protein was obtained in all cell lines (Fig. 2A). Western blot of E-cadherin, which is a direct negative transcription target of ZEB1, showed reexpression of E-cadherin protein in H460 but not in H1299 or H157 after the transfection (Fig. 2A). This was likely to reflect the difference in basal expression of E-cadherin protein between the cells; H460 but not H1299 or H157 expressed detectable levels of E-cadherin protein (Fig. 2A). However, we also considered the possibility that ZEB1 knockdown did not upregulate E-cadherin mRNA in H1299 or H157 and thus performed qRT-PCR analysis of E-cadherin. The analysis revealed that after the ZEB1 knockdown transfection E-cadherin mRNA significantly increased in H157 and H460 but remained unchanged in H1299, indicating that in H1299 E-cadherin mRNA was not upregulated by ZEB1 knockdown (Fig. 2B). Small or no changes in Vimentin protein expression were seen after the transfection of ZEB1 knockdown oligos (Fig. 2A). Finally, we did not see significant morphologic changes suggestive of the cells undergoing EMT (data not shown).
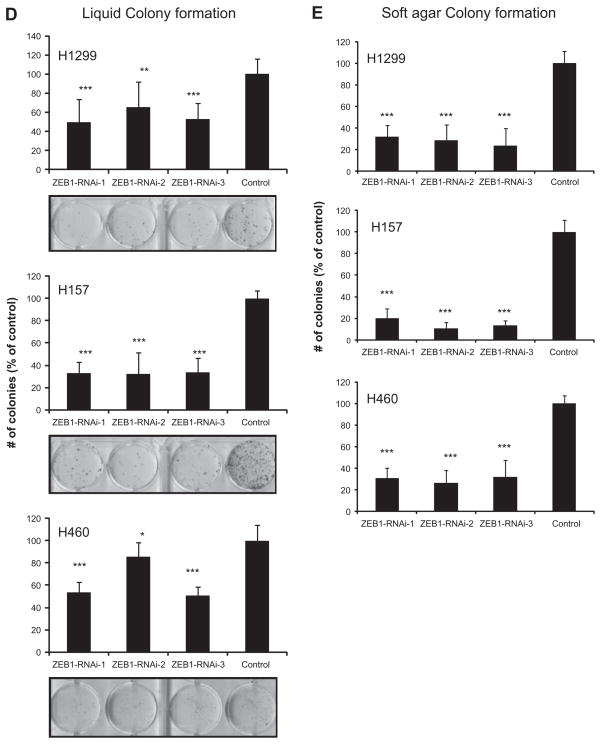
Fig. 2.
RNAi-mediated knockdown of ZEB1 suppresses cell proliferation, liquid colony formation (anchorage-dependent condition), and soft agar colony formation (anchorage-independent condition) in lung cancer cells. (A) Western blot of ZEB1, E-cadherin, and Vimentin for lung cancer cell lines transfected with ZEB1 knockdown or control oligos. Actin was used as a loading control. The result is a representative result of two independent experiments. (B) qRT-PCR analysis of E-cadherin for lung cancer cell lines transfected with ZEB1 knockdown or control oligos. The result is an average of three independent experiments done in duplicates, (C) Cell proliferation (WST-1) assay of lung cancer cell lines transfected with ZEB1 knockdown or control oligos. Cells were transfected with three different RNAi oligos targeting ZEB1 or control oligos. Forty-eight hours after the transfection cells were counted and 1000 cells were plated in 96-well plates. Absorbance values were determined 96 h after the transfection. Results are from three independent experiments in eight replicates and shown as mean ± SD. Values of cells transfected with control oligos are set as 100%. (D) Liquid colony formation assay for lung cancer cell lines transfected with ZEB1 knockdown or control oligos. Forty-eight hours after the transfection cells were counted and 500 or 1000 cells were plated in 6-well plates in triplicates, cultured for two weeks, and stained with methylene blue. Colonies >2 mm were counted. Results are from three independent experiments and shown as mean ± SD. Colony numbers of cells transfected with control oligos are set as 100%. (E) Soft agar colony formation assay for lung cancer cell lines transfected with ZEB1 knockdown or control oligos. Forty-eight hours after the transfection cells were counted and 1000 cells were suspended in 0.37% SeaKem GTG Agarose (Lonza, Rockland, ME, USA) in triplicate 12-well plates. About 2 weeks later, colonies (>50 cells) were counted. Results are from three independent experiments and shown as mean ± SD. Colony numbers of cells transfected with control oligos are set as 100%. In Fig. 2C–E, *, **, and *** indicate P < 0.05, P < 0.01, and P < 0.001 (Mann–Whitney U test), respectively.
We found ZEB1 knockdown to have modest effects suppressing mass culture and liquid colony formation growth but much greater effects suppressing anchorage-independent growth in soft agar (Fig. 2C–E). This result indicates that the ability of these cells to grow in soft agar is highly dependent on ZEB1 expression.
3.6. Growth inhibitory effect of ZEB1 knockdown in lung cancer was caused in part by apoptosis but not by induction of senescence
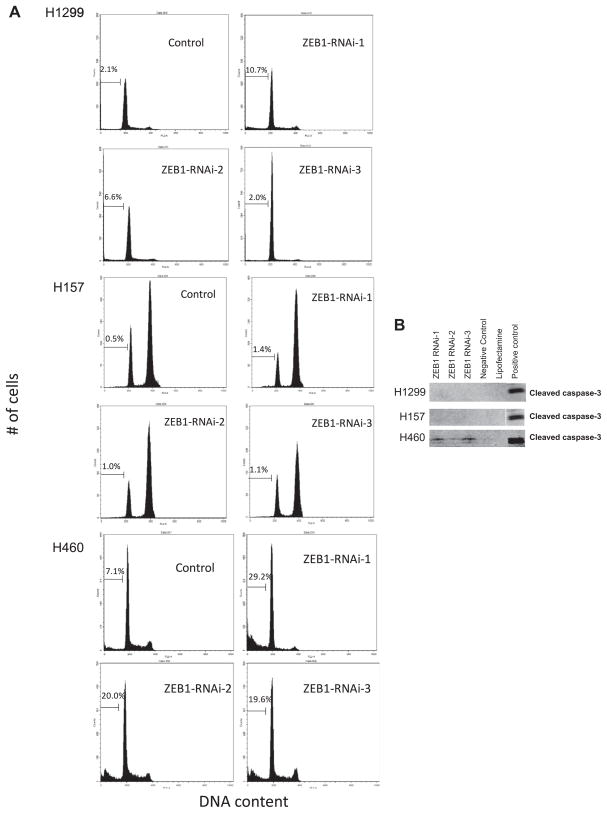
The NSCLC cell lines varied in the induction of apoptosis following ZEB1 knockdown with H460 cells showing the largest amount of ZEB1 knockdown-induced apoptosis (Fig. 3A and B). One study has showed that mouse embryonic fibroblasts derived from Zeb1 −/− mice underwent premature senescence, suggesting that ZEB1 may function as inhibitor of senescence [29]. Nevertheless, we did not see any increase in the number of cells exhibiting morphologic changes or β-galactosidase staining suggestive of senescence, in any of cells transfected with ZEB1 knockdown oligos (data not shown).
Fig. 3.
ZEB1 knockdown induces apoptosis in H460. (A) Flow cytometry analysis for lung cancer cell lines transfected with ZEB1 knockdown or control oligos. Forty-eight hours after the transfection cells were stained with propidium iodine and 20,000 cells were analyzed for cell cycle profiling. Representative results in three independent experiments are shown. (B) Western blot of cleaved caspase-3 for lung cancer cell lines transfected with ZEB1 knockdown or control oligos. ACC-MESO-1 mesothelioma cell line treated with cisplatin was used as a positive control.
4. Discussion
In the present study, we have shown that ZEB1 expression was correlated with the mesenchymal phenotype in NSCLC, and that its depletion with RNA interference suppressed anchorage-independent growth, thus providing a rationale for developing therapeutics targeting ZEB1 function in lung cancer. Consistent with reports in other cancers, we also found that miR-200c and miR-205 expression were inversely correlated with ZEB1 expression in NSCLCs, suggesting potential use as therapy targeting ZEB1.
Consistent with the literature [30], we found that EGFR mutations were associated with epithelial characteristics (E-cadherin expression) in NSCLCs. The literature study showed that EGFR mutant genotype is an independent predictor for the epithelial phenotype in logistic regression analysis, excluding the possibility that observed high frequency of the epithelial phenotype in EGFR mutants was due to other factors that may be associated with or may cause epithelial phenotype. Such factors include early stage disease and non- or low smoking history. The finding that EGFR mutant cells frequently have an epithelial phenotype seems to be unexpected because signaling pathways activated by EGFR including RAS, AKT, and SRC pathways can induce EMT [31]. It is possible that EGFR mutant cells are resistant to EMT-inducing signals. Alternatively, it is also possible that mutant cells specifically express genes that retain cells to the epithelial state. It will be important to further elucidate the molecular mechanisms leading to epithelial phenotype in EGFR mutant cells because molecules involved in such mechanisms could serve as therapeutic targets.
We found that in NSCLC cell lines ZEB1 expression (in contrast to othermaster EMT genes) was significantly correlated with both Vimentin and E-cadherin expression. In addition, we saw that ZEB1 expression was very strongly correlated with Vimentin expression in primary tumors. Two previous studies also reported that ZEB1 expression was most significantly correlated with E-cadherin expression in lung cancer among several EMT-inducing genes [18,32]. Although ZEB1 has been shown to directly repress transcription of E-cadherin in several different types of cells [33,34], it remains unknown whether ZEB1 up-regulates Vimentin expression. In our ZEB1 knockdown experiments, we did not see a significant down-regulation of Vimentin in any of the three lines studied, suggesting that ZEB1 does not directly up-regulate Vimentin expression. However, the observed strong correlation between ZEB1 and Vimentin expression in both NSCLC cell lines and tumors suggests that ZEB1 may indirectly up-regulate Vimentin possibly through some of its downstream targets. Collectively, our results, along with those of others, suggest a dominant role of ZEB1 in maintaining the mesenchymal phenotype of lung cancer.
ZEB1 knockdown induced E-cadherin mRNA expression in H157 and H460 but not in H1299. There is one possible explanation for this. In H1299, the promoter region of E-cadherin is known to be heavily methylated, causing its silenced expression [35]. Thus, it is possible that transient knockdown of ZEB1 was unable to overcome methylation- mediated gene silencing of E-cadherin in H1299, resulting in the unchanged E-cadherin mRNA. Long-term ZEB1 knockdown could reverse methylation of E-cadherin, and thus it wound be interesting to see the effects of stable ZEB1 knockdown on methylation status of E-cadherin in H1299.
ZEB1 knockdown inhibited NSCLC growth most dramatically noted in soft agar colony formation assay. Since the ability of transformed cells to grow under anchorage-independent condition is the most reliable predictor for tumorigenicity and metastatic potential, this strong growth inhibitory effect of ZEB1 knockdown in anchorage-independent condition suggests that ZEB1 expression contributes to maintaining aggressive phenotype of lung cancer cells.
Studies have shown that Twist, Snail and SIP1 show antiapoptotic effects when ectopically expressed [36–38]. Nevertheless, to our knowledge, only one study has demonstrated anti-apoptotic effect of a master EMT gene in a gene-silencing experiment; the study demonstrated that Twist knockdown induced apoptosis in human neuroblastoma cells [39]. We also observed that ZEB1 knockdown induced apoptosis in H460. These results including ours demonstrate that in some cancer cells survival signals are dependent on expression of master EMT genes such as Twist and ZEB1. Cancer cells are thought to acquire the ability to evade apoptosis in early stage of progression, and thus these finding suggest that master EMT genes may play an important role not only in late stage but also in early stage of carcinogenesis.
In conclusion, we have shown that ZEB1 expression is very well correlated with the mesenchymal phenotype of NSCLC, and that its removal induces significant growth inhibition in NSCLC partially through induction of apoptosis. These results suggest that ZEB1 is a promising therapeutic target for lung cancer.
Acknowledgments
This work was supported by Grant-in-Aid for Scientific Research (C) 20590919 (to M.S.), Grant-in-Aid for Scientific Research (C) 20590918 (to M.K), and Grant-in-Aid for Scientific Research (B) 21390257 (to Y.H.) from Japan Society for the Promotion of Science, Global COE program in Nagoya University Graduate School of Medicine funded by Japan’s Ministry of Education, Culture, Sports, Science and Technology, NCI Special Program of Research Excellence in lung Cancer (SPORE P50CA70907), and DOD PROSPECT (to JDM and AFG). We thank Dr. David S. Shames for his helpful comments and discussion on this study.
Footnotes
5. Conflict of interest
We have no conflict of interest to be disclosed.
References
- 1.Christiansen JJ, Rajasekaran AK. Reassessing epithelial to mesenchymal transition as a prerequisite for carcinoma invasion and metastasis. Cancer Res. 2006;66:8319–8326. doi: 10.1158/0008-5472.CAN-06-0410. [DOI] [PubMed] [Google Scholar]
- 2.Klymkowsky MW, Savagner P. Epithelial–mesenchymal transition: a cancer researcher’s conceptual friend and foe. Am J Pathol. 2009;174:1588–1593. doi: 10.2353/ajpath.2009.080545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vandewalle C, Van Roy F, Berx G. The role of the ZEB family of transcription factors in development and disease. Cell Mol Life Sci. 2009;66:773–787. doi: 10.1007/s00018-008-8465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A. The transcription factor snail is a repressor of Ecadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 5.Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial–mesenchymal transitions by repressing Ecadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 6.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Miyoshi A, Kitajima Y, Kido S, Shimonishi T, Matsuyama S, Kitahara K, Miyazaki K. Snail accelerates cancer invasion by upregulating MMP expression and is associated with poor prognosis of hepatocellular carcinoma. Br J Cancer. 2005;92:252–258. doi: 10.1038/sj.bjc.6602266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan-Qi Z, Xue-Yan G, Shuang H, Yu C, Fu-Lin G, Fei-Hu B, Shi-Ren S, Xu-Feng W, Jie D, Dai-Ming F. Expression and significance of TWIST basic helix-loop-helix protein over-expression in gastric cancer. Pathology. 2007;39:470–475. doi: 10.1080/00313020701570053. [DOI] [PubMed] [Google Scholar]
- 9.Vesuna F, van Diest P, Chen JH, Raman V. Twist is a transcriptional repressor of E-cadherin gene expression in breast cancer. Biochem Biophys Res Commun. 2008;367:235–241. doi: 10.1016/j.bbrc.2007.11.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valdes-Mora F, Gomez del Pulgar T, Bandres E, Cejas P, Ramirez de Molina A, Perez-Palacios R, Gallego-Ortega D, Garcia-Cabezas MA, Casado E, Larrauri J, Nistal M, Gonzalez-Baron M, Garcia-Foncillas J, Lacal JC. TWIST1 overexpression is associated with nodal invasion and male sex in primary colorectal cancer. Ann Surg Oncol. 2009;16:78–87. doi: 10.1245/s10434-008-0166-x. [DOI] [PubMed] [Google Scholar]
- 11.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 12.Sato M, Shames DS, Gazdar AF, Minna JD. A translational view of the molecular pathogenesis of lung cancer. J Thorac Oncol. 2007;2:327–343. doi: 10.1097/01.JTO.0000263718.69320.4c. [DOI] [PubMed] [Google Scholar]
- 13.Sekido Y, Fong KM, Minna JD. Molecular genetics of lung cancer. Annu Rev Med. 2003;54:73–87. doi: 10.1146/annurev.med.54.101601.152202. [DOI] [PubMed] [Google Scholar]
- 14.Prudkin L, Liu DD, Ozburn NC, Sun M, Behrens C, Tang X, Brown KC, Bekele BN, Moran C, Wistuba Epithelial-tomesenchymal transition in the development and progression of adenocarcinoma and squamous cell carcinoma of the lung. Mod Pathol. 2009;22:668–678. doi: 10.1038/modpathol.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spaderna S, Schmalhofer O, Wahlbuhl M, Dimmler A, Bauer K, Sultan A, Hlubek F, Jung A, Strand D, Eger A, Kirchner T, Behrens J, Brabletz T. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res. 2008;68:537–544. doi: 10.1158/0008-5472.CAN-07-5682. [DOI] [PubMed] [Google Scholar]
- 16.Arumugam T, Ramachandran V, Fournier KF, Wang H, Marquis L, Abbruzzese JL, Gallick GE, Logsdon CD, McConkey DJ, Choi W. Epithelial to mesenchymal transition contributes to drug resistance in pancreatic cancer. Cancer Res. 2009;69:5820–5828. doi: 10.1158/0008-5472.CAN-08-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, VandenBoom TG, II, Kong D, Wang Z, Ali S, Philip PA, Sarkar FH. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69:6704–6712. doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohira T, Gemmill RM, Ferguson K, Kusy S, Roche J, Brambilla E, Zeng C, Baron A, Bemis L, Erickson P, Wilder E, Rustgi A, Kitajewski J, Gabrielson E, Bremnes R, Franklin W, Drabkin HA. WNT7a induces E-cadherin in lung cancer cells. Proc Natl Acad Sci USA. 2003;100:10429–10434. doi: 10.1073/pnas.1734137100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dohadwala M, Yang SC, Luo J, Sharma S, Batra RK, Huang M, Lin Y, Goodglick L, Krysan K, Fishbein MC, Hong L, Lai C, Cameron RB, Gemmill RM, Drabkin HA, Dubinett SM. Cyclooxygenase-2-dependent regulation of E-cadherin: prostaglandin E(2) induces transcriptional repressors ZEB1 and snail in non-small cell lung cancer. Cancer Res. 2006;66:5338–5345. doi: 10.1158/0008-5472.CAN-05-3635. [DOI] [PubMed] [Google Scholar]
- 20.Clarhaut J, Gemmill RM, Potiron VA, Ait-Si-Ali S, Imbert J, Drabkin HA, Roche J. ZEB-1, a repressor of the semaphorin 3F tumor suppressor gene in lung cancer cells. Neoplasia. 2009;11:157–166. doi: 10.1593/neo.81074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gandhi J, Zhang J, Xie Y, Soh J, Shigematsu H, Zhang W, Yamamoto H, Peyton M, Girard L, Lockwood WW, Lam WL, Varella-Garcia M, Minna JD, Gazdar AF. Alterations in genes of the EGFR signaling pathway and their relationship to EGFR tyrosine kinase inhibitor sensitivity in lung cancer cell lines. PLoS One. 2009;4:e4576. doi: 10.1371/journal.pone.0004576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Usami N, Fukui T, Kondo M, Taniguchi T, Yokoyama T, Mori S, Yokoi K, Horio Y, Shimokata K, Sekido Y, Hida T. Establishment and characterization of four malignant pleural mesothelioma cell lines from Japanese patients. Cancer Sci. 2006;97:387–394. doi: 10.1111/j.1349-7006.2006.00184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yokoyama T, Kondo M, Goto Y, Fukui T, Yoshioka H, Yokoi K, Osada H, Imaizumi K, Hasegawa Y, Shimokata K, Sekido Y. EGFR point mutation in non-small cell lung cancer is occasionally accompanied by a second mutation or amplification. Cancer Sci. 2006;97:753–759. doi: 10.1111/j.1349-7006.2006.00233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sato M, Vaughan MB, Girard L, Peyton M, Lee W, Shames DS, Ramirez RD, Sunaga N, Gazdar AF, Shay JW, Minna JD. Multiple oncogenic changes (K-RAS(V12), p53 knockdown, mutant EGFRs, p16 bypass, telomerase) are not sufficient to confer a full malignant phenotype on human bronchial epithelial cells. Cancer Res. 2006;66:2116–2128. doi: 10.1158/0008-5472.CAN-05-2521. [DOI] [PubMed] [Google Scholar]
- 25.Hurteau GJ, Carlson JA, Spivack SD, Brock GJ. Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Res. 2007;67:7972–7976. doi: 10.1158/0008-5472.CAN-07-1058. [DOI] [PubMed] [Google Scholar]
- 26.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 28.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, El-Naggar S, Darling DS, Higashi Y, Dean DC. Zeb1 links epithelial–mesenchymal transition and cellular senescence. Development. 2008;135:579–588. doi: 10.1242/dev.007047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng QF, Zhou CC, Su CX. Clinicopathological features and epidermal growth factor receptor mutations associated with epithelial–mesenchymal transition in non-small cell lung cancer. Respirology. 2009;14:371–376. doi: 10.1111/j.1440-1843.2009.01496.x. [DOI] [PubMed] [Google Scholar]
- 31.Larue L, Bellacosa A. Epithelial–mesenchymal transition in development and cancer: role of phosphatidylinositol 3′ kinase/AKT pathways. Oncogene. 2005;24:7443–7454. doi: 10.1038/sj.onc.1209091. [DOI] [PubMed] [Google Scholar]
- 32.Witta SE, Gemmill RM, Hirsch FR, Coldren CD, Hedman K, Ravdel L, Helfrich B, Dziadziuszko R, Chan DC, Sugita M, Chan Z, Baron A, Franklin W, Drabkin HA, Girard L, Gazdar AF, Minna JD, Bunn PA., Jr Restoring E-cadherin expression increases sensitivity to epidermal growth factor receptor inhibitors in lung cancer cell lines. Cancer Res. 2006;66:944–950. doi: 10.1158/0008-5472.CAN-05-1988. [DOI] [PubMed] [Google Scholar]
- 33.Grooteclaes ML, Frisch SM. Evidence for a function of CtBP in epithelial gene regulation and anoikis. Oncogene. 2000;19:3823–3828. doi: 10.1038/sj.onc.1203721. [DOI] [PubMed] [Google Scholar]
- 34.Guaita S, Puig I, Franci C, Garrido M, Dominguez D, Batlle E, Sancho E, Dedhar S, De Herreros AG, Baulida J. Snail induction of epithelial to mesenchymal transition in tumor cells is accompanied by MUC1 repression and ZEB1 expression. J Biol Chem. 2002;277:39209–39216. doi: 10.1074/jbc.M206400200. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki M, Sunaga N, Shames DS, Toyooka S, Gazdar AF, Minna JD. RNA interference-mediated knockdown of DNA methyltransferase 1 leads to promoter demethylation and gene reexpression in human lung and breast cancer cells. Cancer Res. 2004;64:3137–3143. doi: 10.1158/0008-5472.can-03-3046. [DOI] [PubMed] [Google Scholar]
- 36.Kajita M, McClinic KN, Wade PA. Aberrant expression of the transcription factors snail and slug alters the response to genotoxic stress. Mol Cell Biol. 2004;24:7559–7566. doi: 10.1128/MCB.24.17.7559-7566.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vega S, Morales AV, Ocana OH, Valdes F, Fabregat I, Nieto MA. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 2004;18:1131–1143. doi: 10.1101/gad.294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sayan AE, Griffiths TR, Pal R, Browne GJ, Ruddick A, Yagci T, Edwards R, Mayer NJ, Qazi H, Goyal S, Fernandez S, Straatman K, Jones GD, Bowman KJ, Colquhoun A, Mellon JK, Kriajevska M, Tulchinsky E. SIP1 protein protects cells from DNA damage-induced apoptosis and has independent prognostic value in bladder cancer. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0902042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valsesia-Wittmann S, Magdeleine M, Dupasquier S, Garin E, Jallas AC, Combaret V, Krause A, Leissner P, Puisieux A. Oncogenic cooperation between H-Twist and N-Myc overrides failsafe programs in cancer cells. Cancer Cell. 2004;6:625–630. doi: 10.1016/j.ccr.2004.09.033. [DOI] [PubMed] [Google Scholar]