Abstract
Zinc finger nucleases (ZFN) have been used to direct precise modifications to the genetic information in living cells at high efficiency. An important consideration in the design of ZFNs is the number of zinc fingers that are required for efficient and specific cleavage. We examined dimeric ZFNs composed of 1+1, 2+2, 3+3, 4+4, 5+5 and 6+6 zinc fingers, targeting 6, 12, 18, 24, 30 and 36 bp, respectively. We found that 1+1 and 2+2 fingers supported neither in vitro cleavage nor single-strand annealing in a cell-based recombination assay. An optimum of ZFN activity was observed for 3+3 and 4+4 fingers. Surprisingly, 5+5 and 6+6 showed significantly reduced activity. While the extra fingers were not found to dramatically increase toxicity, directly inhibit recombination, or perturb the ZFN target site, we demonstrate the ability of subsets of three fingers in six-finger arrays to bind independently to regions of the target site, possibly explaining the decrease in activity. These results have important implications for the design of new ZFNs, as they show that in some cases an excess of fingers may actually decrease the performance of engineered multi-finger proteins. Maximum ZFN activity will require an optimization of both DNA binding affinity and specificity.
ZFNs consist of tandem arrays of engineered zinc finger proteins fused to a monomer of the dimeric nuclease FokI. When two such proteins bind to adjacent target sites, an active FokI dimer is constituted and creates a double-strand break (DSB) in the DNA. In cells, the DSB is rapidly targeted for repair by cellular factors using either non-homologous end joining (NHEJ) or homologous recombination (HR) pathways. This approach has been used to create targeted gene deletions both in cultured cells, including human embryonic and induced pluripotent stem cells, and in whole organisms such as fruit flies, nematodes, zebrafish, and rats, and to significantly improve the frequency of homologous recombination in gene therapy and genome engineering applications (1, 2).
An important consideration in the design of ZFNs is the number of zinc fingers (ZFs) that are required for efficient and specific cleavage. The ability to recognize 15–18 bp of DNA should be sufficient to specify a unique locus in a complex genome such as human. In principle, six fingers should provide such specificity since each zinc finger recognizes approximately 3–4 bp of DNA (3). Although no in vivo binding studies (i.e. chromatin immunoprecipitation followed by sequencing, ChIP-Seq (4)) have yet examined this proposition directly, some artificial transcription factors containing six engineered ZFs have been shown to regulate a single gene in human cells, suggesting high specificity (5). ZFNs consisting of two monomers of three ZFs each (3+3 fingers) can recognize two 9-bp zinc finger binding sites separated by several nucleotides that allow the FokI cleavage domains to dimerize and cleave (Figure 1A). The exact spacing between the two binding sites is dependent on the length and composition of the linker between the terminal zinc finger and the FokI domain, and the most commonly used linkers restrict the tolerated variation in spacer length to about one bp (6, 7). Therefore, ZFNs consisting of 3+3 fingers might be expected to be active only on unique target sites in the human genome. A recent study of a ZFN composed of 3+3 fingers at 141 potential off-target sites reported very weak but dose-dependent ZFN activity at 1% or less of examined sites (8). Other similar studies using ZFNs composed of four to six fingers per monomer appeared have reported similarly limited off-target events (9–11). However, the host organisms and composition of fingers were different, precluding a direct comparison of the effect of finger number on off-target events. A cytological analysis found ZFNs composed of 4+4, 6+6, and 3+3 fingers produced decreasing amounts of off-target cleavage events, respectively (12). However, the fingers differed not only in number but also in composition and recognition sequences, thus confounding a direct comparison between them.
Figure 1.
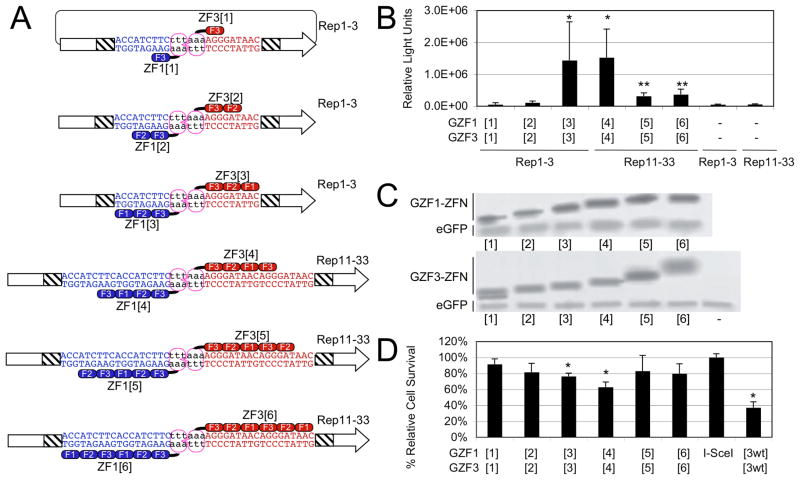
Effects of finger number on ZFN activity in cells. A) Schematic of experimental setup. A systematic series of multi-finger heterodimeric ZFNs and the various target DNA sites were studied. DNA-binding domains that mediate binding to the left (blue) and right (red) target subsite juxtapose the obligate heterodimeric FokI cleavage domains (purple ovals). The target sites were located between two fragments of a luciferase reporter (boxed arrows), which shared a region of homology (hatched region). The elements are not drawn to scale. A double-strand break at the target site in cells restores an active luciferase gene by SSA recombination. B) ZFN-stimulated SSA recombination assay. Luciferase activity induced by ZFN-mediated cleavage is shown as the combined results of six independent experiments. The significance of the measurements over no ZFN controls (indicated as “−“) was determined using a paired, one-tailed T-test (*, p<0.02; **, p<0.002). C) ZFN expression levels. HEK293T cells were co-transfected with ZFN expression vectors and pEGFP, and cell lysates probed with antibodies against the HA-tag and EGFP. “−“ indicates transfection with pEGFP only. D) Quantitative cytotoxicity assay. HEK293T cells were co-transfected with pEGFP and ZFN expression vectors as indicated below the graph. The columns represent the average fraction of EGFP-positive cells at day 5 as compared to the fraction at 30 h after transfection, and are shown relative to transfection with an expression vector encoding the endonuclease I-SceI. A statistically significant decrease in cell survival compared to the non-toxic I-SceI was determined using one-tailed T-test with unequal variance (*, p<0.002). “[3wt]”, expression of GZF1[3] and GZF3[3], which contain wild-type FokI domains and have been previously shown to be cytotoxic (14).
The intention of this study was to systematically investigate the relationship between the number of ZFs and ZFN activity. As one conceptual hypothesis, if the 3+3 was active, the 4+4 version might be expected to possess both greater affinity and greater specificity and thus be even more active. By this reasoning, the 6+6 version would be expected to have a combined recognition capability of 36 bp (or at least more specificity than the shorter zinc finger arrays) and be the most active. Here, a series of ZFNs consisting of one to six fingers per monomer were tested for activity in a plasmid-based single-strand annealing (SSA) recombination assay and an in vitro cleavage assay. We found that ZFNs containing any monomer with one or two ZFs were rarely active in any combination, while ZFNs containing monomers with three or four ZFs were active in all combinations. Surprisingly, ZFNs containing 5+5 or 6+6 ZFs showed significantly reduced activity in cells and failed to cleave target sites in vitro. This study therefore identifies a potential limitation to the use of arrays with more than four fingers for ZFN applications.
METHODS AND MATERIALS
Expression and reporter plasmids
The ZFN expression vectors pPGK.GZF1-N and pPGK.GZF3-N carrying the obligate heterodimer modifications RR and DD, respectively, have been described previously (13, 14). Additional fingers were digested with XhoI/AgeI and cloned between the XhoI/XmaI sites of these vectors so as to preserve canonical TGEKP linkers between all ZFs. Construction of the SSA luciferase reporter plasmid pSSA Rep 3-1 has been described previously (14). Construction of additional SSA reporters was performed similarly, using long oligonucleotides to introduce the target sequences described.
SSA assay
In 24-well plates, HEK 293T cells at 80% confluency in DMEM supplemented with 10% fetal calf serum were co-transfected with 200 ng of each ZFN monomer expression plasmid and 25 ng of SSA reporter plasmid using Lipofectamine 2000 (Invitrogen). Luciferase activity was determined 48 hours post-transfection using BrightGlo reagents (Promega) in a MD Analyst microplate luminometer (Molecular Devices).
Cell survival assay
The assay was basically performed as described previously (15). Briefly, HEK293T cells in 12-well plates were transfected with 1000 ng of the respective ZFN expression plasmid and 100 ng of pEGFP-C1 (Stratagene), and assessed by flow cytometry (FACSCalibur, BD Biosciences) after 2 and 5 days. For calculating cell survival, the ratio in the number of eGFP-positive cells at day 2 versus day 5 was determined and normalized to cells transfected with an I-SceI control expression vector.
In vitro cleavage assay
ZFNs were expressed in vitro using the TNT SP6 Quick Coupled Transcription/Translation System according to the manufacturer’s instructions (Promega) (16). The target DNA fragment was generated by PCR with Pfu polymerase (Stratagene) using plasmids pSSA Rep 3-1 or pSSA Rep 33-11 as templates and primers 5′-GCTGTTTCTGAGGAGCCTTC and 5′-CCCTTCTTGGCCTTTATGAG (Eurofins MWG Operon, Ebersberg, Germany) to amplify an 800-bp fragment. A target site for the restriction enzyme EcoRI was located next to the ZFN target site and used as a positive control. For in vitro cleavage, 1 Tl of each TNT lysate containing one ZFN subunit was mixed with 200 ng of the DNA template, 1 Tg of BSA, and NEBuffer 4 (50 mM potassium acetate, 20 mM Tris-acetate pH 7.9, 10 mM magnesium acetate, 1 mM dithiothreitol; New England Biolabs) in a total volume of 10 Tl. After incubation at 37°C for 90 min the reaction was analyzed on a 1% agarose gel.
Western blot
HEK293T cells in 6-well plates were transfected by polyethyleneimine (PEI)-mediated transfection. The transfection mix contained 800 ng of ZFN expression plasmid, 200 ng of pEGFP-N1 (Clontech), pUC118 to 5 μg, and PEI (0.1 g/l, pH 5.5) in 150 mM NaCl. Cells were harvested after 30 h, lysed in RIPA buffer (50 mM Tris, 150 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% deoxycholate), and 50 μg of lysate separated by SDS-PAGE. After transfer to polyvinylidene difluoride membranes, ZFN and EGFP expression was detected simultaneously with antibodies directed against the HA tag (NB600-363; Novus Biologicals) and EGFP (MAB3580; Millipore), and visualized by Infrared Imaging after incubation with secondary antibodies conjugated with either IR-Dyes 680 or 800CW (LI-COR Biosciences).
Electromobility shift assay (EMSA)
The LightShift Chemiluminescent EMSA Kit (Pierce, Rockford, IL) was used according to the manufacturer’s protocol. Complementary pairs of 5′-biotin labeled forward and 5′-poly T (to improve crosslinking to the membrane) reverse oligonucleotides were annealed to obtain double-stranded target DNAs. The sequences of all oligonucleotide targets are provided in the online supplemental information. Binding reactions were performed for 1 h at room temperature (22°C) in Zinc Buffer A (ZBA: 100 mM Tris base, 90 mM KCl, 1 mM MgCl2, and 90 Tm ZnCl2 at pH 7.5) containing 150 mM KCl, 5 mM DTT, 10% glycerol, 0.1 mg/ml BSA, 0.05% NP-40, 25–55 pM target DNA, and purified ZFPs with a concentration of 0.025 – 1000 nM. Gel electrophoresis was performed on a 10 % native polyacrylamide gel in 0.5× TBE buffer at 4°C. After blotting on a Biodyne B nylon membrane (Pierce) for 1 h at 100 V at 4°C, the DNA was cross-linked by a UV cross-linker (Stratagene) for 4 min. Equilibrium binding constants (KD) were calculated from protein titration experiments imaged on X-ray film. The reported values represent the results of at least two experiments, with a standard deviation of ±50%.
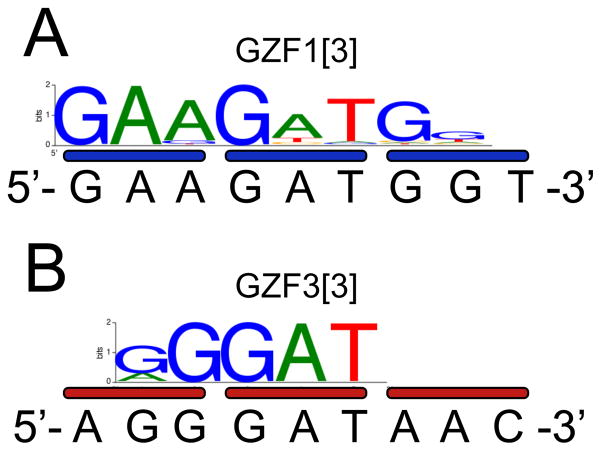
Binding site specificity assay (Bind-n-Seq)
Bind-n-Seq was performed essentially as described (17). Briefly, the coding regions for the DBDs were subcloned into pMAL-c2X (New England Biolabs), expressed in BL21 (DE3) E. coli (Invitrogen), and purified over amylose resin in ZBA and 10 mM maltose. The maltose was removed by overnight dialysis. Bar-coded 93-mer double-stranded oligonucleotide targets containing Illumina primer binding sites and a 21-nt random region were incubated with proteins at various concentrations and salt conditions. Bound complexes were enriched by six washing steps over amylose resin. Eluted DNA was sequenced on an Illumina Genome Analyzer. The motifs shown for GZF1[3] and GZF3[3] are based on approximately 100,000 reads that were enriched 12-fold and 7-fold over a non-selected background, respectively.
RESULTS
Number of zinc finger domains and activity in cells
In order to examine the relationship between the number of ZFs and ZFN activity in a systematic manner, we constructed a series of proteins based on the three-finger proteins GZF1 and GZF3, which have been extensively characterized as a ZFN pair (13, 14). ZFN pairs were constructed containing only one finger 3 of each protein (GZF1[1] and GZF3[1], with the number of ZFs indicated in brackets), fingers 2 and 3 of each protein (GZF1[2] and GZF3[2]), and all three fingers (GZF1[3] and GZF3[3]) (Figure 1A). To create proteins of four, five and six fingers, ZFN pairs were generated containing the three fingers of GZF1 or GZF3 and an additional finger 3 of each protein (GZF1[4] and GZF3[4]), additional fingers 2 and 3 of each protein (GZF1[5] and GZF3[5]), and an addition of all three fingers (GZF1[6] and GZF3[6]) (Figure 1A). The resulting six-finger proteins were therefore tandem dimers of the two three-finger arrays. All ZFNs contained FokI cleavage domains that carried the obligate-heterodimer modifications RR or DD, respectively (14). These variants restrict activity to only heterodimers, reducing cytotoxicity due to off-target DSBs.
A plasmid-based single-strand annealing (SSA) reporter assay in HEK293T cells was performed to assess the activity of the ZFN pairs (Figure 1A) (14). Briefly, a gene encoding luciferase was divided into two segments containing an 870-bp region of homology, separated by a stop codon and a ZFN target site. A ZFN-induced double-strand break (DSB) between the segments stimulates SSA homologous recombination, resulting in an active luciferase gene. Luciferase activity should therefore be proportional to ZFN activity. GZF1[3] and GZF3[3] were previously shown to have optimal activity in this assay on a heterodimeric target site consisting of 9-bp binding sites for each monomer in inverted orientation separated by a 6-bp spacer (Rep1-3, Figure 1A) (18). To accommodate the ZFN pairs containing four to six fingers, two tandem 9-bp binding sites for each monomer were constructed in inverted orientation separated by a 6-bp spacer (Rep11-33, Figure 1A). GZF1/GZF3 ZFN heterodimers consisting of [1]+[1] and [2]+[2] fingers did not stimulate SSA recombination, while [3]+[3] and [4]+[4] fingers stimulated robust recombination (Figure 1B). Although the protein expression levels were comparable (Figure 1C), [5]+[5] and [6]+[6] produced about 4-fold lower recombination than [3]+[3] and [4]+[4].
Some ZFNs have been shown to be cytotoxic, presumably due to numerous cleavages at off-target sites (13, 14, 19–21). Cytotoxicity may manifest as an apparent reduction of recombination in the SSA assay. To investigate if the reduced SSA activity was due to cytotoxicity, cell survival was examined using a ZFN-associated toxicity assay (14, 22). HEK293T cells were co-transfected with plasmids expressing the series of ZFNs and a green fluorescent marker protein. Cell survival frequency was determined by calculating the fraction of green fluorescent cells at day five as compared to 30 h post-transfection. We observed that cells expressing the highly active ZFN pairs [3]+[3] and [4]+[4] had similar or even slightly reduced viability compared to those expressing ZFNs [5]+[5] or [6]+[6] (Figure 1D). Overexpression of ZFN pairs [1]+[1] and [2]+[2] did not considerably affect viability of cells.
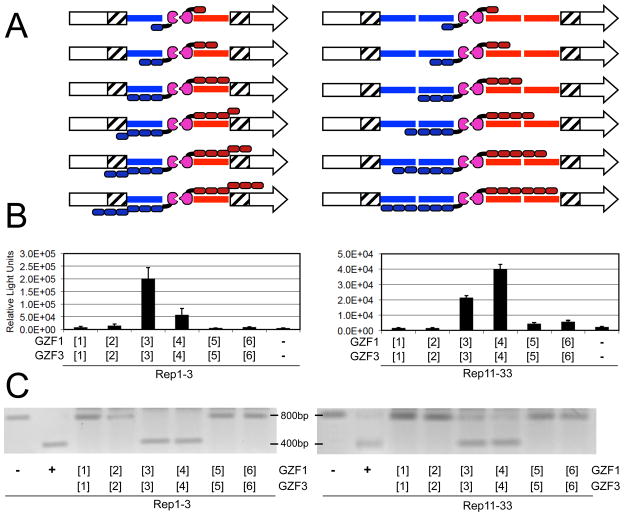
To further investigate the relationship between number of fingers and ZFN activity, we examined the effect of target site composition using the full series of ZFNs on either target Rep1-3 or Rep11-33 (Figure 2A). ZFN pairs with one to three fingers should be accommodated on the 9-bp monomer binding sites of Rep1-3. However, ZFNs with four to six fingers would not have binding sites for more than the three C-terminal fingers. In principle, these experiments could reveal an affect of unbound fingers (e.g. whether recombination is reduced if not all fingers have a cognate binding site). In contrast, all ZFN pairs should be accommodated on the 18-bp monomer binding sites of Rep11-33. As in our previous SSA experiment, we observed for both target Rep1-3 and Rep11-33 that GZF1/GZF3 heterodimers consisting of [1]+[1] and [2]+[2] fingers did not stimulate SSA recombination (Figure 2B). On Rep1-3, [3]+[3] appeared more active than [4]+[4], while on Rep11-33 the reverse was observed. In general, the activity of [3]+[3] and [4]+[4] seemed somewhat greater on Rep1-3 compared to Rep11-33. As observed previously, ZFNs [5]+[5] and [6]+[6] produced dramatically less recombination than [3]+[3] and [4]+[4].
Figure 2.
The effect of target site composition on ZFN activity. A) Cartoon of the experiments in which the full series of ZFNs were assayed on either target Rep1-3 (left) or Rep11-33 (right). Components are labeled as in Figure 1. B) ZFN activity in cells. The graph shows luciferase activity upon ZFN-mediated SSA using targets Rep1-3 (left) or Rep11-33 (right). C) In vitro cleavage activity. The activity of the ZFN pairs in an in vitro cleavage assay using a ~800-bp DNA containing the ZFN target site of Rep1-3 (left) or Rep11-33 (right). Cleavage produces two bands of ~400 bp. “−“, DNA with no ZFN; “+”, DNA digested by EcoRI, which cleaves just 3′ to the ZFN target site.
Number of zinc finger domains and activity in vitro
Since others have reported highly active ZFNs with six finger arrays (10, 12, 23), the reduced activity of GZF1/GZF3 [5]+[5] and [6]+[6] heterodimers was unexpected. To investigate if the reduced activity could be due to the inhibition of a cellular process (e.g. inhibition of the recombination mechanism), ZFN activities were examined using an in vitro cleavage assay (16). A segment containing the ZFN target site of Rep1-3 or Rep11-33 was amplified and exposed to in vitro translated ZFNs. Conversion of the original DNA to a lower molecular weight fragment should provide a direct measure of cleavage activity without potentially confounding cellular processes, such as recombination or cell viability. The observed in vitro cleavage pattern was nearly identical to the results obtained by the SSA assay (Figure 2C). In agreement with the SSA data, ZFN pairs [3]+[3] and [4]+[4] were able to cleave targets Rep1-3 and Rep11-33 in vitro, while ZFNs [1]+[1], [2]+[2], [5]+[5] and [6]+[6] did not reveal detectable activity.
The six-finger ZFN maintains expected target site spacing
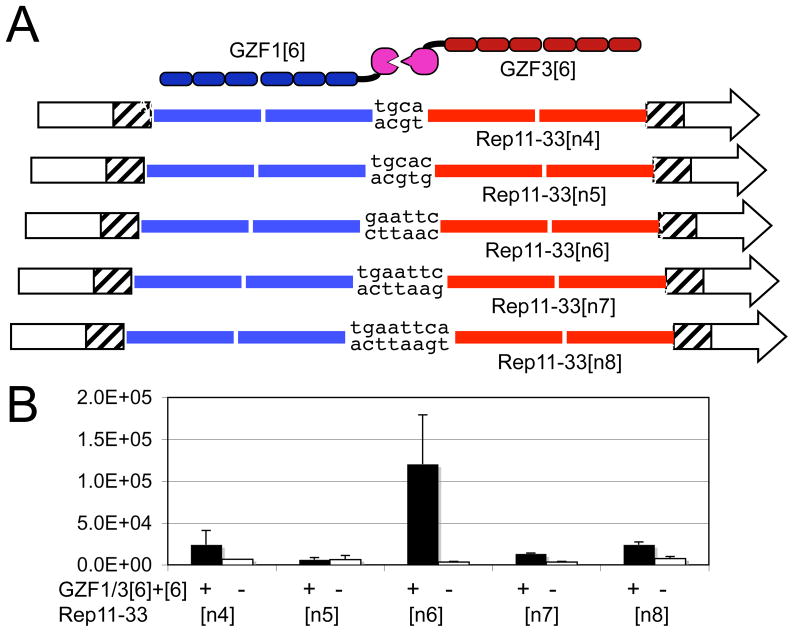
ZFs distort the B-form structure of the DNA double-helix upon binding (24), and it has been suggested that longer arrays of ZFs may create greater strains in the protein-DNA complex (25, 26). We have previously shown, using the same ZFN architecture and SSA assay as used here, that GZF1[3] and GZF3[3] heterodimers display a narrow peak of activity when their binding sites were separated by six bp (18). Spacers of five or seven bp resulted in a loss of SSA activity. To investigate if the reduced activity of GZF1/GZF3 [5]+[5] and [6]+[6] might be due to increased DNA distortions that change the spacing requirements of the target site, ZFN [6]+[6] was tested on a series of Rep11-33 variants in which the spacing between the 18-bp binding sites varied from four to eight bp (Figure 3A). We observed a narrow peak of recombination activity when the binding sites were separated by six bp (Figure 3B), consistent with the results previously obtained with ZFN [3]+[3].
Figure 3.
The effect of target site spacer length on ZFN [6]+[6] activity. A) Cartoon of the experiments in which the ZFN [6]+[6] was assayed on a series of Rep11-33 variants having four to eight base pairs between zinc finger binding sites. B) Luciferase activity upon ZFN-induced SSA recombination of the Rep11-33 series in the presence (“+”) or absence (“−“) of ZFN.
Reduced ZFN activity can be rescued by using fewer fingers on one side
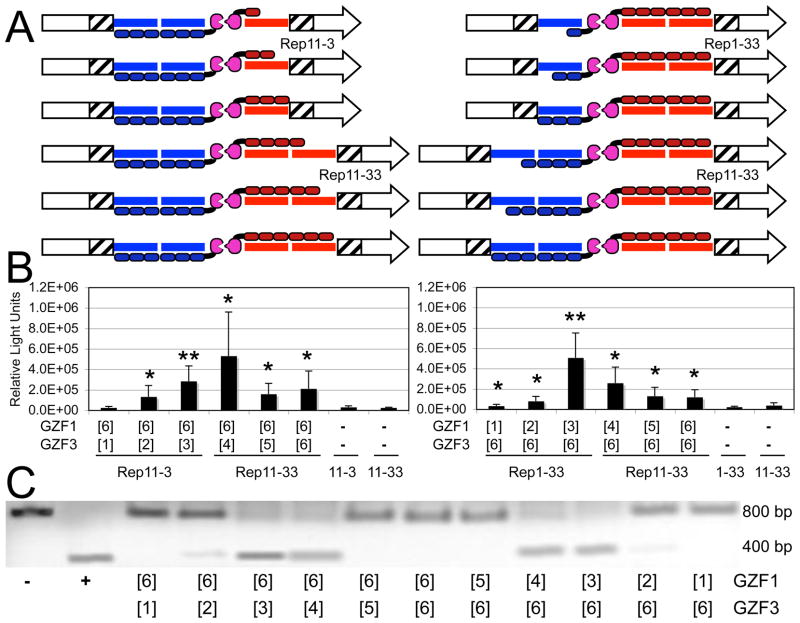
Since ZFN monomers of three or four fingers were active, we explored if the combination of an active monomer with an inactive monomer would rescue the activity of the resulting ZFN heterodimer. Combinations of GZF1[6] with the full series of GZF3 monomers (Figure 4A, left) and GZF3[6] with the full series of GZF1 monomers (Figure 4A, right) were examined using the SSA assay (Figure 4B) and the in vitro cleavage assay (Figure 4C). The results of the two assays were highly consistent. We observed that combined expression of GZF1/GZF3 [6]+[3] or [6]+[4] improved the recombination frequency as well as the cleavage activity as compared to [6]+[5] and [6]+[6]. ZFN [6]+[2] showed weak but observable activity, in contrast to the inactive [2]+[2] (Figures 1 and 2). Similarly, expression of GZF1/GZF3 [3]+[6] and [4]+[6] showed improved activity in the cellular recombination and the in vitro cleavage assay as compared to [5]+[6] and [6]+[6]. Although the rescue was only partial, a 2 to 5-fold increase in activity over [6]+[6] compared to a 7 to 20-fold increase in activity of [3]+[3] and [4]+[4] over [6]+[6] (Figures 1 and 2), these results demonstrated that the six-finger monomers used in this study had the capacity to be functional.
Figure 4.
Rescue of ZFN [6] activity by shorter arrays. A) Cartoon of the experiments in which full series of ZFN monomers were assayed with either GZF1[6] (left) or GZF3[6] (right). B) Luciferase activity upon ZFN-induced SSA recombination. Activity of the ZFN pairs in the SSA assay is shown as the combined results of five independent experiments. The significance of the measurements over no ZFN controls (indicated as “−“) was determined using a paired, one-tailed T-test (*, p<0.05; **, p<0.01). C) In vitro cleavage activity of ZFN. The activity of the ZFN pairs in an in vitro cleavage assay using a ~800-bp DNA containing the ZFN target site of Rep11-33. Cleavage produces two bands of ~400 bp. “−“, DNA with no ZFN; “+”, DNA digested by EcoRI, which cleaves just 3′ to the ZFN target site.
Number of zinc finger domains does not always correlate with binding affinity and specificity
Several possible models could explain the observations in this study, including the six-finger proteins binding to additional sites compared to the three-finger versions (i.e. reduced specificity), the six-finger proteins binding their targets less well (i.e. reduced affinity), the six-finger proteins binding their specific sites using alternative binding modes (i.e. destructive binding modes), or the six-finger proteins binding with such high affinity as to distort the DNA and render the target site resistant to nuclease activity (i.e. destructive high affinity). EMSA was used to investigate the binding affinity and specificity of the three and six-finger proteins. The affinity of GZF1[3] was similar to that of the well-characterized Zif268 protein for their respective 9-bp targets (Table 1, Kdspecific). The affinity of GZF3[3] was 30-fold lower. The addition of three fingers to GZF3[3] increased its affinity more than 80-fold. Unexpectedly, the addition of three fingers to GZF1[3] slightly decreased its affinity by about 2-fold. As one measure of specificity, binding of the six-finger proteins to targets containing only 9 bp of the 18-bp sites reduced their affinity only about 2-fold (Table 1, Kdhalf-site, and Figure S-1 in the online supplemental information). As another measure of specificity, each protein was assayed for non-specific binding the reciprocal 18-bp target (e.g. GZF1[6] on the target for GZF3[6], Table 1, Kdnon-specific). In most cases, binding to the non-specific target was below the measureable range of our assay, which had a 9,000-fold dynamic range. This corresponded to a ratio of non-specific to specific binding of >1,700. An important exception was GZF3[3], which had a specificity ratio of only 2.8. The low specificity of this protein was confirmed by target site selection analysis using the Bind-n-Seq method (17), which revealed that GZF3[3] recognizes only 4–5 bp of the intended 9-bp binding site (Figure 5). The low specificity of GZF3[3] in these assays is consistent with its known toxic activity as a homodimeric ZFN in vivo (13, 14). The results of these analyses therefore indicate that the additional fingers dramatically improved both the affinity and specificity of GZF3, while unexpectedly having a negligible or modestly impairing effect on the affinity and specificity of GZF1.
Table 1.
Affinity and specificity of three and six-finger proteins.
| Kdspecific (nM) | Kdhalf-site (nM) | Kdhalf-site/Kdspecific | Kdnon-specific (nM) | Kdnon-specific/Kdspecific | |
|---|---|---|---|---|---|
| GZF1[3] | 0.12 | >450 | >3846 | ||
| GZF1[6] | 0.26 | 0.62 | 2.4 | >450 | >1744 |
| GZF3[3] | 4.0 | 11 | 2.8 | ||
| GZF3[6] | <0.05 | 0.085 | 1.7 | >120 | >2400 |
| Zif268[3] | 0.34 |
Figure 5.
Binding specificity determined by the Bind-n-Seq target site selection assay. Binding motifs are shown for (A) GZF1[3] and (B) GZF3[3], with intended binding sites shown below.
DISCUSSION
The goal of this study was to systematically investigate the relationship between the number of ZFs and ZFN activity. We examined ZFN heterodimers containing one to six ZFs using a plasmid-based single-strand annealing (SSA) recombination assay and an in vitro cleavage assay. The in vitro data were in good correlation with the results obtained in cells, indicating that the phenomena we observed reflect the ability of the ZFN to cleave its target DNA rather than the effect of the ZFN on cellular processes, such as recombination or cell viability. In agreement with this hypothesis, a direct test of ZFN-induced cytotoxicity showed a weak inverse correlation between cell viability and ZFN activity.
We found that ZFNs composed of 1+1 and 2+2 fingers did not stimulate SSA recombination or cleave their targets in vitro. It is likely that these monomers lacked sufficient affinity to bind individually to their targets. Although the FokI dimerization interface could have enabled GZF1[2] and GZF3[2] to bind cooperatively, approximating a four-finger protein, this effect was not observed. The affinity of the wild type FokI dimerization interface is estimated to be about 100 nM (27), and the use of a modified obligate heterodimerization interface in this study may have further reduced this affinity (14). The 6+2 and 2+6 configurations displayed modest activity, suggesting a more stable partner could partially compensate the weaknesses of the shorter arrays.
ZFNs composed of 3+3 and 4+4 fingers were active in all assays, including assays in which the target site contained binding sites for only three of the four fingers. On Rep1-3, ZFN 4+4 appeared to have reduced activity compared to ZFN 3+3 in the SSA assay, particularly when compared to the increased activity of 4+4 compared to 3+3 on Rep11-33 in which binding sites were present for all four fingers. This suggests that binding of the terminal fourth finger to the target site was beneficial but not essential under the experimental conditions used.
An unexpected observation was that ZFNs composed of 5+5 or 6+6 ZFs showed significantly reduced activity compared to 3+3 and 4+4 fingers, while others have reported the use of five and six-finger monomers in highly active ZFNs (10, 12, 23, 28). One obvious possible explanation for the lower ZFN activity might be reduced DNA binding, which could be explained by either (i) context-dependent negative effects when adding additional fingers (29), or (ii) the use of the canonical “TGEKP” linker in six-finger arrays that may be too short for optimal binding, hence resulting in deformation of the DNA and making binding of longer arrays energetically less favorable (26, 30). However, in contrast to a decreased binding model, we observed that fusion of three additional fingers to GZF3[3] improved the affinity and specificity of the protein over 100-fold. This result is in agreement with observations made by Guo et al. who reported an increase in affinity when comparing related three (Kd = 35 nM), four (11 nM), five (4.2 nM), and six-finger (0.85 nM) arrays (28). Together, these data demonstrate that low-affinity binders can be rescued by the addition of further low-affinity ZFs and that linker modifications are not strictly required to generate highly affine ZF arrays.
A second potential explanation for decreased activity of six-finger ZFNs could be increased toxicity. However, in the current study the extra fingers were not found to dramatically affect ZFN-associated toxicity, directly inhibit recombination, perturb the optimal configuration on the ZFN target site, nor substantially reduce binding affinity or specificity. Interestingly, Guo et al. also reported a two-fold decrease in activity when comparing a high-affinity six-finger ZFN with lower-affinity four and five finger ZFNs (28), suggesting that high affinity does not necessarily translate in high activity. Cornu et al. showed that minor modifications to the DNA-binding domain of ZFN EB0, which reduced its affinity ~3-fold but increased its specificity ~14-fold, resulted in a ZFN of improved activity and significantly reduced toxicity (15). This suggests that not the number of fingers in a ZF array is important but rather the balance between affinity and specificity.
One difference between the current study and several others is that both six-finger monomers were actually tandem dimers of three-finger monomers. GZF1[6] and GZF3[6] were found to bind their half-sites with only 2-fold weaker affinity than for the 18-bp sites. In principle, the independent binding of subsets of fingers could affect ZFN activity by several mechanisms, including additional non-productive binding modes at the target site (suggested in Figure S-2 in the online supplemental information), or a reduced concentration of protein at the target site due to increased binding at off-target sites. The ability of subsets of fingers in multi-zinc finger proteins to bind independently has been well established for other natural and engineered DNA binding proteins. For instance, the capability of the human 11-finger protein CTCF to employ different combinations of zinc fingers to bind diverged promoter sequences was shown over a decade ago (31). The first engineered six-finger protein, C7-C7, was reported to have an affinity for a constituent 9-bp three-finger site that was only 10-fold weaker than for the 18-bp six-finger site (3). The first six-finger protein engineered to an endogenous site, E2C, was reported to have an affinity for a constituent 9-bp site that was only 2 to 3-fold weaker than for the 18-bp site (32). Yant et al. invoked alternative binding modes of the E2C protein at its target site as a mechanism to explain the increased number of transposition events into a mutated E2C target site (9 of 18 bp) compared to a perfect target site (18 bp) in the presence of an E2C-Sleeping Beauty chimeric transposase (33). Therefore these studies support a model in which subsets of fingers in a multi-domain zinc finger protein can bind with substantial affinity, resulting in unexpected binding behaviors that can adversely affect activity.
Such effects are likely to be observed only in cases for which multi-finger proteins contain subsets of fingers that can bind independently with high affinity. Three-finger proteins composed of fingers recognizing only 5′-GNN-3′ sites, such as GZF1 (5′-GAA GAT GGT-3′), have been reported to generally bind their target sites better than those containing fingers recognizing 5′-ANN-3′, 5′-CNN-3′, and 5′-TNN-3′ sites (34), such as GZF3 (5′-AGG GAT AAC-3′). The small number of high-affinity 5′-GNN-‘3 fingers in the 6+6 ZFN Chk2-ZFN1-MA (target site 5′-ACC CGG GTT CCC CTC GGG-3′) (12) might be anticipated to diminished the potential for independently binding subsets of fingers, possibly providing an explanation for the high activity of that 6+6 ZFN compared to those in our study. Similarly, we found that when proteins with a low probability for independently binding subsets (i.e. three and four fingers) were combined with proteins with a high probability (i.e. the five and six finger proteins used here), a partial rescue of ZFN activity was observed. It is possible that this rescue was dependent on the particular composition of fingers and conditions used here.
The current and above-mentioned studies identify an important potential limitation to the use of multi-finger proteins. They demonstrate that long arrays of zinc fingers have the potential to display complex binding behaviors, and that subsets of fingers within longer arrays can mediate efficient binding to DNA. Hence, more zinc fingers are not always better, and in some cases an excess of fingers may actually decrease the performance of engineered multi-finger proteins. The success of any particular array may depend less on the number of fingers and more on the composition of fingers. Maximal ZFN activity requires both an optimization of affinity as well as specificity. These results therefore have important implications for the design of new ZFNs, artificial transcription factors, and other zinc finger applications.
Supplementary Material
Acknowledgments
We thank Vincent Brondani, Eva Guhl, and Geoffrey Lovely for their technical contributions to this work.
ABBREVIATIONS
- DSB
double-strand break
- HR
homologous recombination
- NHEJ
non-homologous end-joining
- SSA
single-strand annealing
- ZFN
zinc finger nucleases
Footnotes
Funding for this project was provided by NIH grant GM077403 (DJS), and grants ZNIP-037783 and PERSIST–222878 of the 6th and 7th Framework Programme of the European Commission, respectively (TC).
SUPPORTING INFORMATION AVAILABLE
Figure S-1 shows electromobility shift assay (EMSA) results of the engineered zinc finger proteins. The sequences of all oligonucleotides used in EMSA studies are also provided. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 2.Handel EM, Cathomen T. Zinc-Finger Nuclease Based Genome Surgery: It’s all About Specificity. Curr Gene Ther. 2010;11:28–37. doi: 10.2174/156652311794520120. [DOI] [PubMed] [Google Scholar]
- 3.Liu Q, Segal DJ, Ghiara JB, Barbas CF., III Design of polydactyl zinc-finger proteins for unique addressing within complex genomes. Proc Natl Acad Sci U S A. 1997;94:5525–5530. doi: 10.1073/pnas.94.11.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 5.Tan S, Guschin D, Davalos A, Lee YL, Snowden AW, Jouvenot Y, Zhang HS, Howes K, McNamara AR, Lai A, Ullman C, Reynolds L, Moore M, Isalan M, Berg LP, Campos B, Qi H, Spratt SK, Case CC, Pabo CO, Campisi J, Gregory PD. Zinc-finger protein-targeted gene regulation: genomewide single-gene specificity. Proc Natl Acad Sci U S A. 2003;100:11997–12002. doi: 10.1073/pnas.2035056100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG, Chandrasegaran S. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol. 2001;21:289–297. doi: 10.1128/MCB.21.1.289-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Handel EM, Alwin S, Cathomen T. Expanding or restricting the target site repertoire of zinc-finger nucleases: the inter-domain linker as a major determinant of target site selectivity. Mol Ther. 2009;17:104–111. doi: 10.1038/mt.2008.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta A, Meng X, Zhu LJ, Lawson ND, Wolfe SA. Zinc finger protein-dependent and -independent contributions to the in vivo off-target activity of zinc finger nucleases. Nucleic Acids Res. 2010;39:381–392. doi: 10.1093/nar/gkq787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, Guschin DY, Rupniewski I, Waite AJ, Carpenito C, Carroll RG, Orange JS, Urnov FD, Rebar EJ, Ando D, Gregory PD, Riley JL, Holmes MC, June CH. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, Mitchell JC, Arnold NL, Gopalan S, Meng X, Choi VM, Rock JM, Wu YY, Katibah GE, Zhifang G, McCaskill D, Simpson MA, Blakeslee B, Greenwalt SA, Butler HJ, Hinkley SJ, Zhang L, Rebar EJ, Gregory PD, Urnov FD. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature. 2009;459:437–441. doi: 10.1038/nature07992. [DOI] [PubMed] [Google Scholar]
- 11.Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Amacher SL. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pruett-Miller SM, Connelly JP, Maeder ML, Joung JK, Porteus MH. Comparison of zinc finger nucleases for use in gene targeting in mammalian cells. Mol Ther. 2008;16:707–717. doi: 10.1038/mt.2008.20. [DOI] [PubMed] [Google Scholar]
- 13.Alwin S, Gere MB, Guhl E, Effertz K, Barbas CF, 3rd, Segal DJ, Weitzman MD, Cathomen T. Custom Zinc-Finger Nucleases for Use in Human Cells. Mol Ther. 2005;12:610–617. doi: 10.1016/j.ymthe.2005.06.094. [DOI] [PubMed] [Google Scholar]
- 14.Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 15.Cornu TI, Thibodeau-Beganny S, Guhl E, Alwin S, Eichtinger M, Joung JK, Cathomen T. DNA-binding specificity is a major determinant of the activity and toxicity of zinc-finger nucleases. Mol Ther. 2008;16:352–358. doi: 10.1038/sj.mt.6300357. [DOI] [PubMed] [Google Scholar]
- 16.Cathomen T, Sollu C. In vitro assessment of zinc finger nuclease activity. Methods Mol Biol. 2010;649:227–235. doi: 10.1007/978-1-60761-753-2_13. [DOI] [PubMed] [Google Scholar]
- 17.Zykovich A, Korf I, Segal DJ. Bind-n-Seq: high-throughput analysis of in vitro protein-DNA interactions using massively parallel sequencing. Nucleic Acids Res. 2009;37:e151. doi: 10.1093/nar/gkp802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimizu Y, Bhakta MS, Segal DJ. Restricted spacer tolerance of a zinc finger nuclease with a six amino acid linker. Bioorg Med Chem Lett. 2009;19:3970–3972. doi: 10.1016/j.bmcl.2009.02.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, Gregory PD, Pabo CO, Rebar EJ. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 20.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 21.Beumer K, Bhattacharyya G, Bibikova M, Trautman JK, Carroll D. Efficient gene targeting in Drosophila with zinc-finger nucleases. Genetics. 2006;172:2391–2403. doi: 10.1534/genetics.105.052829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cornu TI, Cathomen T. Quantification of zinc finger nuclease-associated toxicity. Methods Mol Biol. 2010;649:237–245. doi: 10.1007/978-1-60761-753-2_14. [DOI] [PubMed] [Google Scholar]
- 23.Santiago Y, Chan E, Liu PQ, Orlando S, Zhang L, Urnov FD, Holmes MC, Guschin D, Waite A, Miller JC, Rebar EJ, Gregory PD, Klug A, Collingwood TN. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci U S A. 2008;105:5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nekludova L, Pabo CO. Distinctive DNA conformation with enlarged major groove is found in Zn-finger-DNA and other protein-DNA complexes. Proc Natl Acad Sci U S A. 1994;91:6948–6952. doi: 10.1073/pnas.91.15.6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim JS, Pabo CO. Getting a handhold on DNA: design of poly-zinc finger proteins with femtomolar dissociation constants. Proc Natl Acad Sci U S A. 1998;95:2812–2817. doi: 10.1073/pnas.95.6.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore M, Klug A, Choo Y. Improved DNA binding specificity from polyzinc finger peptides by using strings of two-finger units. Proc Natl Acad Sci U S A. 2001;98:1437–1441. doi: 10.1073/pnas.98.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Catto LE, Ganguly S, Milsom SE, Welsh AJ, Halford SE. Protein assembly and DNA looping by the FokI restriction endonuclease. Nucleic Acids Res. 2006;34:1711–1720. doi: 10.1093/nar/gkl076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo J, Gaj T, Barbas CF., 3rd Directed evolution of an enhanced and highly efficient FokI cleavage domain for zinc finger nucleases. J Mol Biol. 2010;400:96–107. doi: 10.1016/j.jmb.2010.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imanishi M, Nakamura A, Morisaki T, Futaki S. Positive and negative cooperativity of modularly assembled zinc fingers. Biochem Biophys Res Commun. 2009;387:440–443. doi: 10.1016/j.bbrc.2009.07.059. [DOI] [PubMed] [Google Scholar]
- 30.Elrod-Erickson M, Rould MA, Nekludova L, Pabo CO. Zif268 protein-DNA complex refined at 1.6 A: a model system for understanding zinc finger-DNA interactions. Structure. 1996;4:1171–1180. doi: 10.1016/s0969-2126(96)00125-6. [DOI] [PubMed] [Google Scholar]
- 31.Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G, Neiman PE, Collins SJ, Lobanenkov VV. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol. 1996;16:2802–2813. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beerli RR, Segal DJ, Dreier B, Barbas CF., III Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc Natl Acad Sci U S A. 1998;95:14628–14633. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yant SR, Huang Y, Akache B, Kay MA. Site-directed transposon integration in human cells. Nucleic Acids Res. 2007;35:e50. doi: 10.1093/nar/gkm089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramirez CL, Foley JE, Wright DA, Muller-Lerch F, Rahman SH, Cornu TI, Winfrey RJ, Sander JD, Fu F, Townsend JA, Cathomen T, Voytas DF, Joung JK. Unexpected failure rates for modular assembly of engineered zinc fingers. Nature methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.