Abstract
To efficiently manipulate large amounts of genotype data generated with fluorescently labeled dinucleotide markers, we developed a Microsoft Access database management system, named GenoDB. GenoDB offers several advantages. First, it accommodates the dynamic nature of the accumulations of genotype data during the genotyping process; some data need to be confirmed or replaced by repeat lab procedures. By using GenoDB, the raw genotype data can be imported easily and continuously and incorporated into the database during the genotyping process that may continue over an extended period of time in large projects. Second, almost all of the procedures are automatic, including autocomparison of the raw data read by different technicians from the same gel, autoadjustment among the allele fragment-size data from cross-runs or cross-platforms, autobinning of alleles, and autocompilation of genotype data for suitable programs to perform inheritance check in pedigrees. Third, GenoDB provides functions to track electrophoresis gel files to locate gel or sample sources for any resultant genotype data, which is extremely helpful for double-checking consistency of raw and final data and for directing repeat experiments. In addition, the user-friendly graphic interface of GenoDB renders processing of large amounts of data much less labor-intensive. Furthermore, GenoDB has built-in mechanisms to detect some genotyping errors and to assess the quality of genotype data that then are summarized in the statistic reports automatically generated by GenoDB. The GenoDB can easily handle >500,000 genotype data entries, a number more than sufficient for typical whole-genome linkage studies. The modules and programs we developed for the GenoDB can be extended to other database platforms, such as Microsoft SQL server, if the capability to handle still greater quantities of genotype data simultaneously is desired.
To search for genes underlying complex diseases, such as osteoporosis, schizophrenia, asthma, or diabetes, a genome-wide scan is necessary and often conducted (Lander and Schork 1994; Ghosh and Collins 1996; Hauser et al. 1996; Deng et al. 2001a). High-throughput genotyping capability is needed for genome-wide linkage studies, in which hundreds of markers often are genotyped for hundreds or even thousands of study subjects. Much progress in this area has been made. For example, automatic fluorescent detection systems and related software for fragment size analyses and data tracking make the high-throughput genotyping possible (Idury and Cardon 1997). Although some authors (Perlin et al. 1995; Hall et al. 1996) have realized that management of large amounts of genotype data is a bottleneck, little effort has been made to develop efficient genotype database management systems (DBMS) to support such large data flow during high-throughput genotyping.
There are two challenges for an efficient DBMS to support high-throughput genotyping. First, the genotype data are dynamic and can change over time. For example, the experiment (PCR and/or electrophoresis) for some particular markers for some particular DNA samples may fail, so that new data may be needed from repeat experiments. In addition, genotype data are not 100% accurate. Data may have to be replaced by results from repeated experiments, or eliminated if errors are detected. This procedure may be repeated several times and, consequently, the records of genotypes in the database then may need to be revised over time. Therefore, an ideal DBMS should be capable of managing the genotype data dynamically and also should provide functions for easy tracking of data or sample sources during data processing. When raw genotype data come from an experiment, the DBMS should be able to check if the same record exists in the database before a new record is entered. If a duplicate record exists, the DBMS also should have a function that decides whether or not the new record needs to replace the previous record.
Second, the accuracy of genotype data is important. Generally speaking, the raw genotype data obtained from automatic genotyping machines (e.g., ABI PRISM 310 genetic analyzer and ABI PRISM 377 DNA sequencer machine; Perkin-Elmer) are not completely accurate. There are many potential causes for genotype errors. For fluorescent genotyping (the method that we focus on in this paper), the causes may include incorrect allele sizing as a result of ambiguous sample peaks, misplacing PCR products during the process of loading samples for electrophoresis, DNA sample contamination, etc. Genotype errors should be detected and corrected before the data are used for formal genetic analyses.
An ideal genotype DBMS should have the functions to import raw data from automatic genotyping machines, process the raw data, detect potential errors, compile the data, and export the processed data to suitable programs for further genetic analyses (Hall et al. 1996). Although some software products (GENESCAN and GENOTYPER, Applied Biosystem Division/Perkin-Elmer) are available commercially, the process of managing raw data is considered to be only “semiautomatic” (Hall et al. 1996; Idury and Cardon 1997; Pálsson et al. 1999), and thus still time- and labor-consuming. Ideally, a genotype DBMS should have the capability of manipulating the genotype data nearly automatically to save time and labor in high-throughput genotyping.
In our study (Deng et al. 2001a), we are searching for major genes underlying osteoporosis by using the Linkage Mapping Set, version 2 (Perkin-Elmer Applied Biosystems). We are genotyping 435 microsatellite markers for 635 individuals. The total number of genotype data exceeds one-quarter million. To manipulate so many genotype data, which are typical in whole-genome scans for genes underlying complex traits, we developed a Microsoft (MS) Access application, which is named GenoDB (Genotype DBMS). This software can manipulate genotype data dynamically. Most functions provided by this software are performed automatically with only a few keystrokes. In addition, this software provides powerful built-in error-detecting functions (especially when used in conjunction with the Mendelian inheritance-check program PedCheck [O'Connell and Weeks 1998]), and a function that allows users to trace back easily to gel or sample sources for uncertain or erroneous genotype data. The latter function is useful for directing repeat experiments for uncertain or erroneous genotype data (Hall et al. 1996).
In this paper, we first will describe the database structure design for GenoDB. Then we will describe four essential modules for different functions of GenoDB:
- (1)
autocomparison of the data that are read by different persons for the same raw data, to minimize subjectivity in reading raw genotype data;
- (2)
autoadjustment of allele fragment sizes across platforms or across runs, to account for potential differences resulting from different gels run on the same machine or different machines;
- (3)
autobinning of alleles, to categorize alleles efficiently by their fragment sizes; and
- (4)
autocompilation of data in the format for PedCheck analyses (O'Connell and Weeks 1998).
The error detection functions built into GenoDB, the function for tracking genotype data sources, and several statistical reports will be discussed. We will then describe the user-friendly graphic interfaces of GenoDB. Finally, we will discuss the practical implications of GenoDB for high-throughput genotyping.
RESULTS
Database Design
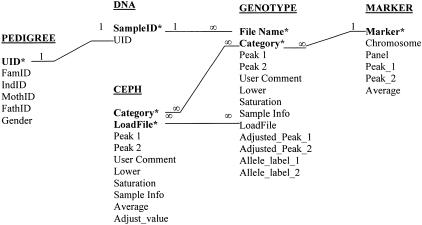
Throughout this paper, we use the small-cap font to indicate an entity of the database GenoDB. One may think of an entity as a table in MS Access, which stores information for a thing (e.g., a marker), a place, or a study subject in a database. Figure 1 is a simplified entity-relationship diagram (Chen 1976) that describes the database design of GenoDB in detail. The entity pedigree (Fig. 1) provides necessary family information of all the study subjects. The table pedigree may be imported from, or simply linked with, other phenotype DBMS. Because we concentrate on the topic of a genotype DBMS in this paper, we will not discuss database designs for phenotype DBMS that already have been dealt with by Cheung et al. (1996) and McMahon et al. (1998). Keeping pedigree information always updated is very important for GenoDB. Ideally, genotype and phenotype DBMSs should be combined so that pedigree information in both is consistent upon updating.
Figure 1.
Relational schema for GenoDB. (*), The field of a primary key of an entity; (1), the side of “one” relationship; (∞), the side of “many” relationship. The entity pedigree stores the pedigree information, including study subject unique identity ([UID]), family identity ([FamID]), study subject individual identity within a family ([IndID]), subject's mother's (father's) identity within a family ([MothID], [FathID]), and gender information ([Gender]). dna stores the DNA sample information, including DNA sample identity ([SampleID]) and corresponding study subject unique identity ([UID]). The fields [File Name], [Category], [Peak 1], [Peak 2], [User Comment], [Lower Signal], [Saturation], and [Sample Info] in both the entities ceph and genotype are defined the same as those used in the software GENOTYPER (version 2.1). The fields [LoadFile], [Adjusted_Peak_1], [Adjusted_Peak_1], [Allele_label_1], and [Allele_label_2] in the entity genotype are explained in the text. ceph stores the information of CEPH controls that run in a fixed place in each gel. The field [LoadFile] is explained in the text. [Average] stores the mean length of [Peak 1] and [Peak 2] of the entity ceph. [Adjust_value] stores the difference between the [Average] of CEPH control (in the ceph entity) obtained from experiments and the [Average] of CEPH standard (in the marker entity) obtained from published data from the web for the same dinucleotide marker. The fields [Marker], [Chromosome], and [Panel] in the entity marker store the information for marker name, the chromosome where a marker is located, and the panel where the marker is placed. The fields [Peak_1] and [Peak_2] store the published values of fragment length for CEPH 1347–02 from the Web site of Perkin-Elmer Applied Biosystems (http://www.pebio.com/ab/apply/dr/lmsv2). The field [Average] in the entity marker stores the mean of [Peak_1] and [Peak_2].
The entity genotype (Fig. 1) stores the information of genotype data, including raw data and processed data. The fields [Peak 1] and [Peak 2] contain the raw genotype data as real numbers representing the fragment lengths of two dinucleotide marker alleles that are obtained directly from experiments with automated machines. Throughout this paper, a bracket indicates an attribute of an entity. An attribute is a field of a table; therefore we will use “field” throughout this paper instead of “attribute”. The fields [Adjusted_Peak_1] and [Adjusted_Peak_2] contain the adjusted data for the raw data in [Peak 1] and [Peak 2], respectively. The method of adjustment is introduced in “auto-adjustment” in the “Functionality” section in the following. The fields [Allele_label_1] and [Allele_label_2] contain integer numbers (for two alleles of a dinucleotide marker of an individual) that are converted from [Adjusted_Peak_1] and [Adjusted_Peak_2] according to the binning criteria automatically generated by GenoDB.
The entity ceph (Fig. 1) stores genotype data of Centre d'Etude du Polymorphisme Humain (CEPH) that are used as controls run on each gel at a fixed location. The field [LoadFile] stores the names of electrophoresis gel files; while the field [Category] stores the names of dinucleotide markers. The combination of the fields [Category] and [LoadFile] are the primary key for ceph; therefore, the name of each gel file should be unique. A primary key is a field (attribute) or a combination of several fields that uniquely identifies each record stored in a table (entity). A primary key of a table guarantees that no redundant data (repeated records) exist in a table of a database. In our application of GenoDB, the format of gel files is “pt?-p?-date-?”. The question mark indicates an integer number. The first, second, and third question mark, respectively, indicate the identity of a numbered plate (each containing 96 DNA samples); the identity of a numbered marker panel (28 panels total for the whole genome in the ABI PRISM Linkage Mapping Set, version 2, Perkin-Elmer); and the identity of a numbered person who has created the file. For instance, “pt1-p2–062300–2” indicates that technician “2” created this file on Jun 23, 2000, for plate 1 and marker panel 2. The same file-naming format is used for the field [LoadFile] of the entity genotype. The unique name for every electrophoresis gel file renders it possible to keep track of the flow of data that are associated with gel files in GenoDB conveniently and easily.
The entity marker (Fig. 1) stores the information (fragment length, chromosome location, and panel information, etc.) for CEPH standard DNA that comes from the published data from Perkin-Elmer Applied Biosystems (www.pebio.com/ab/apply/dr/lmsv2). The entity dna (Fig. 1) stores the DNA sample information. Because we use CEPH DNA (that is duplicated on a specified location in each plate) as standard to control potential gel-to-gel variation and to detect potential DNA sample mislocation during loading samples for electrophoresis, we do not duplicate any DNA sample from study subjects in our experiments. Therefore, in our database, one record in the field [UID] (representing unique ID of each study subject for his/her pedigree information) of the entity pedigree only has one corresponding record in the field [SampleID] (identity of sample in DNA plates) of the entity dna.
Functionality
Overall Procedures
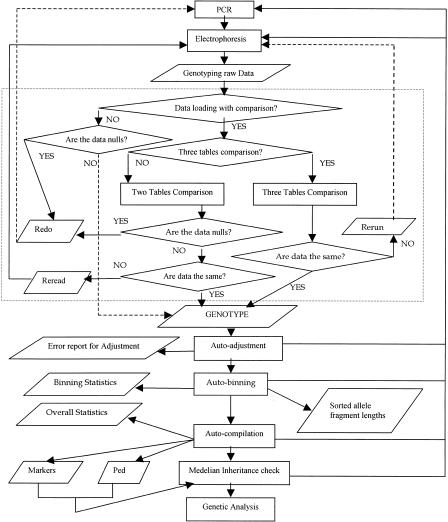
The procedure of GenoDB to manipulate genotype data is flexible. Figure 2 indicates the data flow of GenoDB. Essentially, there are four modules (functions) of GenoDB: loading raw genotype data from automated machines, adjustment of raw data by CEPH control DNA, allele binning, and compilation of genotype data for the Mendelian inheritance check of marker genotypes in pedigrees by PedCheck (O'Connell and Weeks 1998). These four functions can be performed in series or performed individually. During a genotyping project, one needs to load data after each gel file has been generated, and to check consistency of alleles labeled by different technicians with the data-loading module. Inconsistencies of allele labels by different technicians that are detected may not be settled immediately and may be set aside temporarily. Users can proceed to the next steps: adjustment of data obtained from different gel runs and/or different machines, allele binning, and data compilation for inheritance check. Errors may be detected during execution of each step. These errors may be stored in tables or recorded, and the erroneous genotypes may be set aside and not entered into the execution of the following steps. When accumulated to a certain extent, these error data will be subject to repeat experiments with PCR and/or electrophoresis and/or allele relabeling on the original gel files. Each functional step and associated algorithm will be described in detail below.
Figure 2.
Data flow of GenoDB. The rectangle indicates the process in the data flow chart; a diamond indicates a decision in the flowchart; a parallelogram indicates stored data. The section contained within the dotted lines indicates the data loading module.
Data Loading into GenoDB
After each electrophoresis, raw genotype data are sized by GENESCAN (version 3.1). One or two technicians then check genotype data using GENOTYPER (version 2.1). Manual checking by technician(s) for the resultant genotype data usually is necessary (Ghosh et al. 1997). GenoDB provides two ways to load raw genotyping data into the database, one with comparison (when two technicians check genotype data) and the other without comparison (when only one technician checks genotype data). The latter method (without comparison) may speed the process and does save time and labor; it may be adopted if the technician reading the gel data is very experienced. Otherwise, we recommend using the former method, as it may reduce subjective and/or uncertain judgement of allele peaks by different technicians upon data comparison.
Depending on whether one or two technicians read gel data, one or two tables—the primary check table (PCT) or PCT and the second check table (SCT)—are created in MS Excel format for each gel. These tables, together with the gel files, are recorded on a recordable CD, and may be stored in a safe place as permanent records. In the meantime, the MS Excel tables are transferred, through a local area network or the internet, to a personal computer that is dedicated to genotype data analyses. After loading and comparison of PCT and SCT, GenoDB identifies the inconsistent data between the PCT and SCT. These data then are subject to scrutiny by a third technician or discussion between the previous two technicians, which results in an additional MS Excel file, the third check table (TCT). Consistent data in the PCT, SCT, and TCT will be accepted, and inconsistent data will be subject to repeat experiments (PCR and/or electrophoresis, etc.).
Data Loading into GenoDB with Comparison
GenoDB compares the PCT and SCT automatically after the user locates and loads these two table files by using the browsing function built into GenoDB. After comparison, GenoDB accepts the same but not null records in the PCT and SCT. In the meantime, GenoDB generates two other reports, “redo” and “reread,” if there are any data needed for repeating experiments or for rereading the source gel files. All of the records without entry (no PCR products) in both PCT and SCT will appear in the report “redo.” The samples indicated in the report “redo” are subject to repeat experiments (PCR and electrophoresis). All the records that are different in both PCT and SCT appear in the report “reread.” These records are required to be read on the original gel file by a third technician or to be discussed by the previous two technicians, which will result in the TCT.
After the TCT is created, GenoDB compares TCT with PCT and SCT. If the record in TCT agrees with the record in either PCT or SCT, this record is accepted for entry into the database; otherwise, it is listed in the report, “rerun.” All of the DNA samples in “rerun” need to be repeated for electrophoresis.
Data Loading into GenoDB without Comparison
Without comparison, GenoDB loads all non-null records into the database, and leaves the null records in the report, “redo,” which requires repeat runs of PCR and electrophoresis.
Auto-Adjustment of Fragment Length Data across Runs and across Platforms
The same PCR products may yield different fragment lengths if run on different gels or different machines. Therefore, the data across runs (electrophoresis) and across platforms (different machines) need to be adjusted before being combined for further analyses (such as allele binning). We assume that a constant adjustment value can be applied for all PCR products on the same gel for the same marker. The adjustment value is the deviation of the value of the CEPH control sample on each gel for one marker from the standard published CEPH value of the same marker. We use CEPH 1347–02 DNA as a uniform control sample and run it on each gel in a fixed location (lane 25 for PE 377 DNA sequencer, or E7 for PE 310 DNA analyzer in our study). Although the results are similar, the adjustment algorithm developed here is unlike the one described in Ghosh et al. (1997). Ghosh et al. (1997) employed the average fragment lengths of all CEPH (1347–02) control values that are obtained from all running gels as the standard for adjustment. Thus, the adjustment value may change over time with addition of new gel data. In contrast, our algorithm takes advantage of the published value of CEPH (1347–02), which can be obtained from the Web site (http://www.pebio.com/ab/apply/dr/lmsv2). If the CEPH genotype is homozygous for a marker, we take the published fragment length of the marker directly as the standard for adjustment of gel data resulting from runs. If the CEPH genotype is heterozygous, the mean of the published fragment lengths of the two alleles is set as the standard for adjustment. In our algorithm, every record in the entity genotype has two values for each allele, an original value and an adjusted value. The adjusted values are used for further analysis, such as allele binning and conversion of allele sizes (as real numbers) to allele labels (as integers) according to the autoset binning criteria. The original values are useful for retrieval functions that may let user(s) trace back easily to the source gel files if necessary.
Compared with the Ghosh et al.'s (1997) algorithm for adjustment, ours is simple and can save computational time. Both algorithms can reach the same goal, namely, reducing the ranges of bins and increasing the interbin distances (Ghosh et al. 1997). The adjustment values of Ghosh et al. (1997) may change during genotyping projects with the addition of more data from more gels. Therefore, with their algorithm, the adjustment either may be done only at the end of a genotyping project, or the adjustment will change and need to be redone with every new additional set of data during a genotyping project. This is not a problem with our algorithm, in which the adjustment can be performed any time during a genotyping project and the adjustment for each record is final, as the adjustment standard does not change. The adjustment is performed automatically by GenoDB.
After autoadjustment of allele size, an error report of such adjustment is generated automatically if there is any error detected. Table 1 is an example of such a report. Using this report, some genotype errors may be detected. For example, misplacement of DNA samples when performing electrophoresis may be detected if the misplacement includes the CEPH control sample. GenoDB automatically compares the state of a locus of CEPH samples run on the gel with the published standard data. If the state is different, i.e., if the state is changed from heterozygous to homozygous, or vice versa, an error is indicated. This type of error is classified as error 1 in Table 1. Error 1 also may occur as a result of incorrect allele sizing for CEPH control data. Rereading the original gel files is required if an error 1 is detected. There are two other categories of adjustment errors. Error 2 (Table 1) reports the fact that the standard deviation (SD) of adjustment values for a marker across all gels is at least 0.2 for cross-runs with the same genotyping instrument platform. Usually, the SD should be <0.2; the SD of adjustment values for a marker across instrument platforms can be much larger (ABI PRISM 1997). Error 3 reports the fact that the adjustment value of a marker for a gel is beyond three SD from the mean adjustment value for that marker across all gels when error 2 is absent. The causes of errors 2 and 3 could be incorrect sizing for CEPH control samples, or misplacement of DNA samples, or other unknown reasons. Careful reanalysis of the data is required to detect the sources of specific genotype errors.
Table 1.
A Sample Report of Potential Genotype Errors Detected by the GenoDB Module for Adjustment of Allele Fragment Lengths across Runs and Across Platforms
| Marker | LoadFile | Error |
|---|---|---|
| D12S310 | pt1-p19-121099-1 | 1 |
| D12S324 | pt1-p19-121099-1 | 1 |
| D18S63 | All | 2 |
| D19S884 | All | 2 |
| D18S520 | Pt5-p24-102299-2 | 3 |
LoadFile is the name of electrophoresis gel files. There are three categories of potential genotype errors in this report; these are defined in the text.
Auto Allele Binning in GenoDB
The algorithm of allele binning in GenoDB is similar to that of Ghosh et al. (1997), and thus will not be elaborated. The values of allele fragment lengths used for binning are adjusted values. After allele binning, tables for sorted allele fragment sizes (adjusted) (e.g., Table 2) for each marker are created, and statistical report (e.g., Table 3) of auto allele binning is generated automatically.
Table 2.
Sorted Allele Fragment Lengths for Dinucleotide Marker D20S196
| Number | Sample ID | Allele size |
|---|---|---|
| 1 | f12-06 | 265.04 |
| 2 | b05-07 | 265.12 |
| 3 | a11-02 | 265.145 |
| 4 | d12-02 | 265.145 |
| 5 | g03-07 | 265.17 |
| — | — | — |
| 561 | f10-07 | 284.75 |
| 562 | a04-06 | 284.77 |
| 563 | h02-05 | 284.77 |
| 564 | h08-01 | 284.94 |
| 565 | c05-07 | 285.04 |
| 566 | b12-06 | 285.32 |
| 567 | a08-02 | 285.325 |
| 568 | f10-06 | 285.34 |
| 569 | a10-02 | 285.355 |
| 570 | h05-01 | 285.355 |
| — | — | — |
| 1250 | d12-05 | 296.195 |
| 1251 | b12-05 | 296.205 |
| 1252 | c12-05 | 296.205 |
| 1253 | b05-01 | 296.225 |
| 1254 | e12-05 | 296.34 |
This table represents a part of the genotype data for the marker D20S196. Sample ID is the identity of the DNA sample. Allele size is the fragment length in base pairs (after adjustment). The two boldface rows indicate the potential genotype errors that are detected by the auto allele-binning module of GenoDB. See text for details.
Table 3.
Example of a Statistical Report of Allele Binning (Marker D20S196)
| Bin | Start point | End point | BR | ABD | No. of alleles |
|---|---|---|---|---|---|
| 1 | 265.04 | 266.07 | 1.03 | — | 310 |
| 2 | 268.18 | 268.18 | — | 2.78 | 1 |
| 3 | 271.1 | 271.315 | 0.21 | 3.05 | 8 |
| 4 | 272.92 | 273.33 | 0.41 | 1.89 | 14 |
| 5 | 276.05 | 276.05 | — | 2.93 | 1 |
| 6 | 277.63 | 278.185 | 0.56 | 1.79 | 87 |
| 7 | 278.74 | 278.81 | 0.07 | 0.92 | 3 |
| 8 | 279.63 | 279.74 | 0.11 | 0.92 | 2 |
| 9 | 280.615 | 281.37 | 0.76 | 1.15 | 46 |
| 10 | 282.49 | 282.81 | 0.32 | 1.80 | 11 |
| 11 | 283.39 | 283.625 | 0.23 | 0.87 | 27 |
| 12 | 284.3 | 285.87 | 1.57 | 1.57 | 112 |
| 13 | 286.38 | 286.82 | 0.44 | 1.47 | 93 |
| 14 | 287.32 | 287.35 | 0.03 | 0.79 | 3 |
| 15 | 288.285 | 288.79 | 0.50 | 1.18 | 228 |
| 16 | 290.255 | 290.8 | 0.54 | 1.93 | 241 |
| 17 | 292.205 | 292.56 | 0.36 | 1.91 | 54 |
| 18 | 294.14 | 294.43 | 0.29 | 1.88 | 7 |
| 19 | 296.105 | 296.34 | 0.24 | 1.97 | 6 |
BR, bin range; ABD, adjacent bin distance. See text for definitions. Boldface numbers are cited in the text as examples of genotype error detection during allele binning. The start point is the smallest value of fragment lengths of a bin; end point is the largest value.
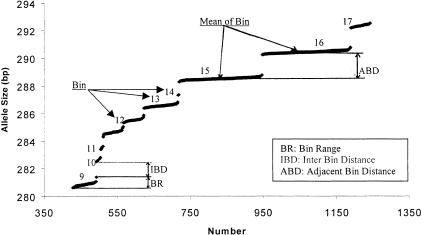
The histogram technique for visualizing allele binning is commonly used (e.g., Ghosh et al. 1997, ABI PRISM 1996). We developed an alternative technique — a plotting technique for visualizing allele binning, which is intuitive and informative in revealing bin range, interbin distance, and adjacent-bin distance (all defined below) that may reflect bin quality and suggest potential error in autoallele binning. GenoDB automatically generates tables for sorted allele sizes (e.g., Table 2) for each marker. A sorted table can be exported to MS Excel from MS Access. A scatter plot (Fig. 3) then can be drawn easily with MS Excel.
Figure 3.
A sorted allele fragment length scatter plot for the dinucleotide marker D20S196. Part of the genotype data of D20S196 is plotted. See text for the definition of BR, IBD, and ABD.
Interbin distance (IBD) is the difference between the longest fragment size of one bin and shortest fragment size of the adjacent bin that is larger in size (Fig. 3). The minimum value of IBD is called the tolerance level for creating a bin. Within a bin, the size difference between two sequentially sized alleles is always less than the tolerance level. In our study, the tolerance level is set at 0.5 bp, initially. One can reset this value before performing autobinning in GenoDB. Bin range (BR) is defined as the distance between the start point (the shortest fragment length) and the end point (the longest fragment length) of one bin. BR is usually <1.0 bp. From our experience in genotyping 635 individuals with 435 dinucleotide microsatellite markers, a potentially erroneous allele binning is signaled if BR is larger than 1.25 bp. Adjacent bin distance (ABD) is the difference between the mean fragment lengths of two adjacent bins. ABD should be close to an integer. Because we use dinucleotide markers in our experiment, ABD often is close to an even number. However, ABD may sometimes be close to an odd number, because some alleles of some markers may truly differ in size by odd numbers as a result of the plus-A phenomenon (Ghosh et al. 1997) that occasionally occurs. Significant deviation of ABD from an integer (>0.3 bp, by our experience) is a signal of bad allele binning, if the sample sizes of two adjacent bins are large enough (>20, by our experience). One or several alleles sometimes may be found with fragment lengths in the middle of two adjacent bins. These are largely a result of uncaught genotyping errors that are subject to further examination of the raw data or repeated experiments. With the aid of our plotting technique, these alleles can be identified easily, and elimination of these alleles is helpful in building good allele-binning criteria.
Table 3 and Figure 3 provide an example of detecting genotyping error problems in allele binning by using the above-defined parameters (BR and ABD) and our plotting technique. The fact that the BR of bin 12 (Table 3, automatically generated by GenoDB) is 1.57 bp (>1.25 bp) and the ABD is 1.57 bp (deviation from an integer 2 that is >0.3) indicate a potential error. Our plotting techniques are applied (Fig. 3) for exploring details associated with the potential errors signaled in Table 3. Two erroneous genotypes (h08–01 and c05–07 [DNA sample ID], which are highlighted in Table 2) then can be detected and identified for tracing back to the raw gel data for further analyses. After removing these two erroneous genotypes, bin 12 then can be divided into two bins. These two DNA samples with erroneous genotypes are subject to repeated experiments or relabeling in the original electrophoresis gel files and the resultant new records will be reloaded into GenoDB. Because the combination of the fields [Sample ID] (DNA sample Identity) and [Category] (marker name) is the primary key of the entity genotype and we have already known the DNA sample ID and marker name for these two errors; therefore, we can easily trace back to the records for them in the table genotype. Because every electrophoresis gel file has a unique name and the field [LoadFile] in the entity genotype contains the names of gel files, we can therefore easily track the gel file for a particular record of genotype data. The feature allowing easy tracking of source data in GenoDB is very useful for directing repeated experiments.
Data Compilation for a Suitable Program to Perform an Inheritance Check
Some genotyping error cannot be detected by previous built-in error-checking mechanisms in GenoDB until inheritance patterns of genotypes in pedigrees are examined by PedCheck (O'Connell and Weeks 1998). GenoDB can export two MS Excel tables, “Ped” and “Markers”, in the format (after converting to text files) for analyses by PedCheck. Errors detected then will be subjected to further analyses of raw data or repeated experiments.
GenoDB does not provide a function to generate directly the suitable files for genetic analysis, such as LINKAGE (Ott 1991), SOLAR (Almasy and Blangero 1998) and GENEHUNTER (Kruglyak et al. 1996). However, one can perform basic queries in MS Access to generate suitable files for particular types of genetic analyses from the table “Ped” (generated from GenoDB) and phenotype information from other phenotype DBMS.
GenoDB generates an overall statistical report (e.g., Table 4) automatically after data compilation, where the rates of data output, missing data, and mutations are listed. Data outputs are the records for those samples for which experimental data have been generated. The rate of data output bears information on genotyping speed and status. Missing data include those that have no data as a result of no PCR product output even after three repeated experiments. With correct pedigree information, some genotype data cannot pass the inheritance check, but the data are of high quality and are consistent even after three repeat experiments. These data are counted as mutations. Rates on missing data and mutations are important for assessment of genotyping quality and for genetic analysis concerning mutations.
Table 4.
Overall Statistic Report for Panel 25
| Marker | Data output (%) | Missing data (%) | Mutations (%) |
|---|---|---|---|
| D19S210 | 100 | 0.00 | 0.00 |
| D19S220 | 100 | 0.16 | 0.16 |
| D19S221 | 100 | 0.79 | 0.31 |
| D19S414 | 100 | 0.00 | 0.00 |
| D19S420 | 100 | 0.00 | 0.00 |
| D20S100 | 100 | 0.00 | 0.00 |
| D20S112 | 100 | 0.16 | 0.00 |
| D20S115 | 100 | 0.16 | 0.00 |
| D20S117 | 100 | 0.16 | 0.00 |
| D20S171 | 100 | 0.00 | 0.00 |
| D20S196 | 100 | 0.00 | 0.00 |
| D20S889 | 100 | 0.00 | 0.31 |
User-Friendly Graphic Interface
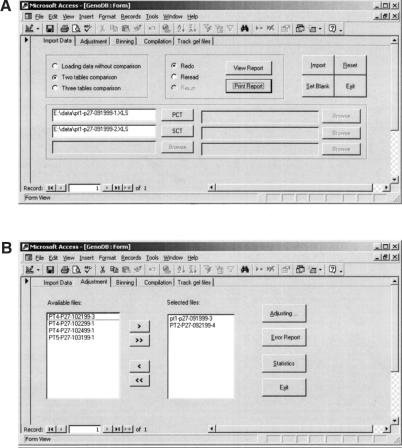
GenoDB offers a user-friendly graphic interface, which will be illustrated in this section by reproductions of actual screen images. GenoDB has five screen-view modes: auto-import view, auto-adjustment view, auto-binning view, auto-compilation view, and gel-file tracking view. Figure 4a shows the screen for importing genotype data into the database. In this example screen, the radio button, “Two table comparison,” is turned on and the paths of two files in MS Excel format are located by using the browse function. The user then can click the “Import” button to compare and import the genotype data automatically. After this procedure, the user can click the “View (Print) Report” button to view (print) a report “redo” or “reread.” For the three-table comparison mode, only one report, “rerun,” can be viewed or printed. If the radio button “Loading data without comparison,” is turned on, only one report, “redo,” will be generated.
Figure 4.
Screen views of GenoDB's user-friendly GUI. Plots (A), (B), (C), (D) and (E) are, respectively, the actual screen views for the following modules: data loading, adjustment of allele fragment lengths, allele binning, compilation of genotype data for Mendelian inheritance check for PedCheck, and the function of tracking the sources of genotype data and CEPH control that runs on each gel.
Figure 4b shows the screen for autoadjustment. The genotype data from one or more loading files can be adjusted simultaneously. After the user selects loading file names and clicks the “Adjusting” button, GenoDB automatically adjusts the fragment length values of alleles according to the CEPH standard. The error report for auto-adjustment can be viewed by clicking the “View error report” button after auto-adjustment.
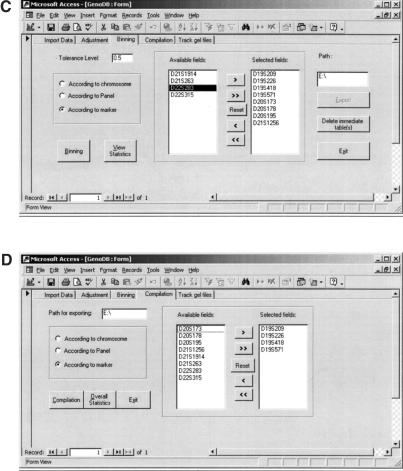
Figure 4c shows the screen for autobinning. The user can choose a tolerance level or simply use the default value of 0.5 bp. In this screen, there are four choices for allele binning all markers, according to either a chromosome, a marker panel, or an individual marker. After the user selects the proper chromosome(s), or panel(s), or marker(s), and clicks the “Binning” button, GenoDB will generate allele binning automatically. The statistics for auto-binning can be viewed by clicking the “View statistics” button. If one believes that there are some binning errors for one marker, he/she can highlight this marker in the selected list, and then click the “export” button. The table that contains all the records of allele sizes (sorted) will be exported in MS Excel format. The user can then draw a sorted allele size scatter plot with MS Excel.
Figure 4d shows the screen for autocompilation of the data for PedCheck analyses. There are four choices, the same as the screen for auto-binning. After the user selects the proper fields and clicks the “Compilation” button, GenoDB will automatically export two MS Excel formatting tables, Markers and Ped. After saving these two tables in text format, the user then can perform inheritance check using PedCheck. Clicking the “Overall statistic” button on this screen, the user can view the overall statistics.
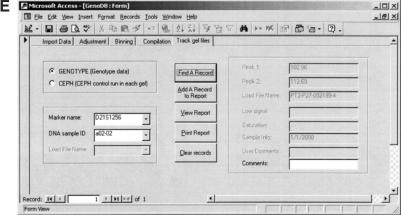
Figure 4e shows the screen for the function to track the source of genotype data and CEPH control that is run on each gel. There are two choices in this screen. One is for the function to track the source of raw genotype data according to a marker name and a DNA sample ID of a record of genotype data. Another is for the function to track the source of CEPH control DNA samples according to a marker name and the name of an electrophoresis gel file for a record of CEPH control. In this example screen, the radio button “GENOTYPE” is turned on. After the user selects a marker name and a DNA sample ID from the lists and clicks the button “Find A Record”, the source of the genotype data will appear in the proper fields that cannot be edited on this screen. However, the user can write some comments in the text box “Comments.” After the user clicks the button “Add a record to report”, the selected record together with the comments will be added to a report that can be viewed or printed by the functions that are provided on the screen. This procedure may be repeated until all records needed are added to the report. The information on this report is helpful for directing future repeat experiments. If the radio button “CEPH” is turned on, the source of the CEPH control runs in each gel can be tracked after the user selects a proper marker name and gel file name for a record of CEPH controls. This function is helpful for reanalyzing the genotype data of CEPH control DNA samples.
Application of GenoDB to a Whole-Genome Scan for Genes Underlying Osteoporosis
Osteoporosis is a major health problem largely characterized by low bone mass (Deng et al. 1998, 2000a,b). The risk of osteoporosis has been shown to have a very strong genetic component (Deng et al. 2000a,b). Extensive molecular genetic studies are being launched to search for genes underlying osteoporosis (Deng et al. 2001a,b). In a whole-genome scan for genes underlying osteoporosis that is being conducted in our center (Deng et al. 2001a), we applied GenoDB to process genotype data collected. In our practice, we set up one database for each marker panel so that the total number of genotype data in each database is generally <15,000. The capacity of GenoDB is more than sufficient to process such a large amount of data (see Discussion). For the 16 marker panels for which we have finished genotyping and data analyses, we performed PedCheck (O'Connell and Weeks 1998) for individual markers by each marker panel. We (Deng et al. 2001a) performed linkage analyses using the variance component approach (Almasy and Blangero 1998) by combining the genotype data with phenotype data via simple queries with MS Access.
For the 16 marker panels that we have completely finished thus far, our experience with GenoDB demonstrated that the autoallele-binning modules worked very well for almost all of the markers. GenoDB cannot properly set up bin criteria for some markers (e.g., D16S515 and D15S127) because the BR (bin range) is too large for one bin. This might be largely a result of the inherent difficulty in combining data from different runs and machines and GenoDB's inability to handle that difficulty for the few markers. We excluded all of the genotype data for these particular markers in our linkage analyses. There may be solutions for this binning problem, e.g., by employing the allelic ladder method (Puers et. al. 1993; Griffiths et al. 1998), which has been used by forensic community.
We used GenoDB to clean up and improve genotype data during our genotyping work with four cycles of procedures (Table 5). In each cycle, we applied the error detection methods in GenoDB to detect genotype errors. We also performed Mendelian inheritance check using PedCheck to detect genotype inconsistencies within pedigrees. In the first cycle, we did not perform Mendelian inheritance check because many genotype errors identifiable and potentially solvable by GenoDB may be detected before PedCheck. Example results (Table 6) of our genotyping procedures aided by GenoDB show that GenoDB can improve genotype data and clean up genotype error quite efficiently. Most of genotype errors can be detected in the 1st , 2nd, and 3rd cycles. After the 4th cycle, the unsolved genotype data rate (the number of unresolved genotype data /the total number of genotype data) is <0.2% in the final genotype database (Table 6). The genotype data in the final genotype database should be considered good enough to perform linkage analysis. For all those finished markers in our study, the unsolved genotype data rates range from 0.1% to 1% for various markers, with an average of 0.3%. The unsolved genotype data after the fourth cycle of genotyping procedures are set as missing data or mutation data as described earlier. The high-quality genotyping data of our finished marker panels are essential for the findings of significant and suggestive genomic regions containing genes for osteoporosis (Deng et al. 2001a).
Table 5.
Procedures in Four-Cycle Genotyping
| First cycle | 1. Genotyping for all DNA samples |
| 2. Importing genotype data into GenoDB | |
| 3. Detecting unresolved genotype data (including both missing data and genotype errors) by GenoDB | |
| Second cycle | 1. Re-genotyping for those unsolved genotype data in the first cycle |
| 2. Importing genotype data into GenoDB | |
| 3. Detecting unresolved genotype data by GenoDB | |
| 4. Detecting inconsistent genotype data by performing Medelian PedCheck | |
| Third cycle | 1. Re-genotyping for those unsolved and inconsistent genotype data in the second cycle |
| 2. Repeating steps 2, 3, and 4 of the second cycle | |
| Fourth cycle | 1. Repeating Steps 1, 2, 3, and 4 of the third cycle |
| 2. Unresolved genotype data are classified into two categories: missing data and mutations |
Table 6.
Cleaning Up and Improving Genotype Data by GenoDB for Three Marker Panels
| Marker | Total samples | 1st Cycle (unresolved/final) | 2nd Cycle (unresolved/final) | 3rd Cycle (unresolved/final) | 4th Cycle (unresolved/final) |
|---|---|---|---|---|---|
| Panel 18 | |||||
| D12S336 | 635 | 72/563 | 68/4 | 3/65 | 0/3 |
| D12S345 | 635 | 20/615 | 15/5 | 8/7 | 0/8 |
| D12S351 | 635 | 28/607 | 24/4 | 11/13 | 5/6 |
| D12S368 | 635 | 25/610 | 21/4 | 0/21 | 0/0 |
| D12S79 | 635 | 24/611 | 20/4 | 2/18 | 1/1 |
| D12S85 | 635 | 24/611 | 20/4 | 0/20 | 0/0 |
| D12S86 | 635 | 46/589 | 42/4 | 4/38 | 0/4 |
| D12S99 | 635 | 73/562 | 69/4 | 3/66 | 0/3 |
| D13S1265 | 635 | 28/607 | 24/4 | 3/21 | 0/3 |
| D13S156 | 635 | 98/537 | 96/2 | 10/86 | 0/10 |
| Subtotal | 6350 | 438/5912 | 399/39 | 44/355 | 6/38 |
| Panel 19 | |||||
| D12S310 | 635 | 144/491 | 138/6 | 18/120 | 0/18 |
| D12S324 | 635 | 109/526 | 103/6 | 7/96 | 0/7 |
| D12S326 | 635 | 24/611 | 18/6 | 3/15 | 0/3 |
| D12S352 | 635 | 77/558 | 74/3 | 6/68 | 0/6 |
| D12S364 | 635 | 145/490 | 142/3 | 10/132 | 0/10 |
| D13S153 | 635 | 149/486 | 144/5 | 10/134 | 1/9 |
| D13S158 | 635 | 94/541 | 92/2 | 10/82 | 1/9 |
| D13S159 | 635 | 50/585 | 47/3 | 18/29 | 7/11 |
| D13S171 | 635 | 68/567 | 62/6 | 4/58 | 0/4 |
| D13S173 | 635 | 96/539 | 90/6 | 7/83 | 1/6 |
| D13S265 | 635 | 82/553 | 82/0 | 3/79 | 0/3 |
| Subtotal | 6985 | 1038/5947 | 992/46 | 96/896 | 10/86 |
| Panel 25 | |||||
| D19S210 | 635 | 121/514 | 41/80 | 2/39 | 1/1 |
| D19S220 | 635 | 163/472 | 66/97 | 4/62 | 2/2 |
| D19S221 | 635 | 149/486 | 63/86 | 16/47 | 4/12 |
| D19S414 | 635 | 183/452 | 94/89 | 3/91 | 0/3 |
| D19S420 | 635 | 147/488 | 39/108 | 2/37 | 0/2 |
| D20S100 | 635 | 155/480 | 59/96 | 10/49 | 0/10 |
| D20S112 | 635 | 122/513 | 15/107 | 0/15 | 0/0 |
| D20S115 | 635 | 139/496 | 34/105 | 11/23 | 1/10 |
| D20S117 | 635 | 124/511 | 22/102 | 0/22 | 0/0 |
| D20S171 | 635 | 131/504 | 70/61 | 1/69 | 0/1 |
| D20S196 | 635 | 133/502 | 29/104 | 4/25 | 0/4 |
| D20S889 | 635 | 140/495 | 29/111 | 3/26 | 0/3 |
| Subtotal | 7620 | 1707/5913 | 561/1146 | 56/505 | 8/48 |
| Total | 20955 | 3183/17772 | 1952/1231 | 196/1756 | 24/172 |
“Unresolved” indicates that the genotype data errors have been detected but have not been corrected in this cycle. “Final” indicates the data contributing to the final database in this cycle. In each cycle, the error detection mechanisms described in the text are used in the GenoDB. The Mendelian inheritance check is also performed in second, third, and fourth cycle, but not in first cycle. After four cycles, unsolved data are set as either missing data or mutation data, as described in the text.
DISCUSSION
Whole-genome searches for genes underlying complex diseases in humans generally involve extensive genotyping. Management of large quantities of the resultant genotype data creates a bottleneck for high-throughput genotyping (Perlin et al. 1995; Hall et al. 1996). Developing a well-designed genotype DBMS helps to break down the bottleneck and thus speeds up genotyping. In a project underway in our center, we are genotyping 435 microsatellite markers for each of 635 individuals. The total number of genotype data exceeds 250,000, which is typical for whole-genome linkage studies for complex diseases. To manipulate efficiently the large quantities of genotype data that are generated by our experiment, we developed a genotype DBMS, GenoDB.
GenoDB has several strengths. First, almost all of the functions provided by GenoDB are automatic. These functions include autocomparison, autoadjustment of allele sizes across runs or across platforms, autobinning, and autocompilation of genotype data for suitable programs to perform inheritance checks in pedigrees. Automation greatly reduces time and labor and is a key to improving high-throughput genotyping. Ghosh et al. (1997) developed a software, ABAS (Automated Binning and Adjustment Software), which can perform part of the tasks in GenoDB (i.e., autobinning, autoadjustment of allele fragment lengths). However, according to P. Chines, a coauthor of Ghosh et al. (1997) who is also the system analyst of ABAS, the current version of ABAS “requires complex and expensive infrastructure (including Sybase)” and ABAS “is very difficult to install and set up properly.” A portable version of ABAS is being developed (P. Chines, pers. comm.).
Second, genotype databases are changed over time during a project and GenoDBcan manipulate genotype data dynamically. GenoDB automatically checks the whole database to see whether an incoming record of a dinucleotide marker exists for the same individual before the incoming raw genotype data enter into the database. If it exists, GenoDB automatically decides whether or not the incoming record should replace the previous one. We assume that the incoming record (if not null) obtained from a repeat experiment is always better than the previous one when a repeat experiment is required after a genotype error is detected with the previous record. Therefore, the incoming data replace the previous record in most situations. The previous record is kept in the table (entity) genotype only if the incoming record is a null record as a result of the failure of a repeat experiment. Hence, GenoDB updates the database without human intervention and the raw genotype data can be imported easily and continuously and incorporated into the database during the genotyping process that may continue over an extended period of time in large projects.
Third, GenoDB provides powerful genotype error detection functions. Generally speaking, the genotype data obtained from automated machines are not 100% accurate; therefore, genotype errors should be detected and corrected before the data can be used for formal genetic analysis. There are several useful mechanisms for detecting genotype errors in GenoDB. (1) Some genotype errors may be suggested by the reports generated by GenoDB (e.g., “redo,” “error report for auto-adjustment” [Table 1], and “statistical report of allele binning”[Table 3]). (2) Other genotype errors may be indicated by our plotting technique for visualizing allele binning (see section on auto-adjustment of fragment length data across runs and across platforms, above). (3) In conjunction with the PedCheck program (O'Connell and Weeks 1998), most genotype errors should be detected.
Fourth, GenoDB can process genotype data flexibly (see the discussion of overall procedures, above). This feature lets the user perform adjustment of fragment lengths, allele binning, and Mendelian inheritance checking for some of the markers of some of the collected pedigrees even if complete genotype data are not obtained for the whole experiment. The flexibility of the procedure is helpful for arranging repeat experiments because one can wait to perform them until a sufficient number of DNA samples and markers are accumulated for repeating.
Fifth, GenoDB provides a function to track electrophoresis gel files. If an uncertain or erroneous genotype datum has been detected, GenoDB lets the user easily trace back to the source of the raw genotype data in the original electrophoresis gel file. This function is helpful and very important for directing repeat experiments for those uncertain or erroneous genotype data.
Sixth, GenoDB offers a user-friendly graphical interface (GUI) that renders it fairly easy to learn to use and to work with.
We chose to use MS Access as the platform for GenoDB for the following reasons. First, MS Access is a commonly used software. Second, it is easy to design a user-friendly GUI with MS Access. Third, the cost for developing and using GenoDB is low with MS Access as the platform. The capacity of MS Access to manipulate large amounts of genotype data may be a concern. However, as tested, GenoDB can handle >500,000 genotype data at one time without any problem, although large amounts of genotype data will result in longer computer time for data manipulation. This capacity can easily handle almost all of the linkage analyses conducted so far, or those being conducted currently (e.g., Deng et al. 2001a), particularly when the whole genotype data are divided in the management by chromosome or by marker panel. In practice, because people usually perform Mendelian inheritance checking, or other genetic analysis for markers grouped by chromosome or panel, one may divide the whole genotype data set into dozens of groups according to chromosomes or panels. GenoDB handles only one such data group at a time. This procedure may greatly enhance the capability of GenoDB in handling large data sets by breaking them into smaller ones. In our project, we set up one GenoDB for every panel (28 panels total for the whole genome in the ABI PRISM Linkage Mapping Set, version 2), so that the number of genotype data that one single GenoDB deals with is usually <15,000. GenoDB can manage this amount of genotypes very efficiently. Importantly, the modules and algorithms we developed here for GenoDB can be extended to other database platforms, such as the MS SQL server to handle data sets that are each >500,000.
Although GenoDB can bridge the gap between genotyping and final genetic analyses and greatly speed up genotyping, it still depends upon other software, such as GENOTYPER, to obtain genotype data from automated genotyping machines. The limitation of GENOTYPER is that quite a large degree of human intervention is still required (Ghosh et al. 1997); therefore, use of GENOTYPER is still relatively time- and labor-consuming. A software program that could process electrophoresis gel files in an automated batch process without much manual editing is desirable. TrueAllele (TA) (Perlin 1994, 1995; Cybergenetics 2000) is an example of software for automated batch processing of genotype data, and Decode-GT represents another effort at improvement (Pálsson et al. 1999). The combination of software that automatically processes the electrophoresis gel files without much human intervention and a well-designed genotype DBMS that automatically and dynamically manipulates genotype data will be highly desirable for high-throughput genotyping.
Availability of Software
GenoDB 1.0 (beta version) is immediately available to academic groups upon request by contacting J.-L.L. at jllee@creighton.edu.
Acknowledgments
This study was supported in part by grants from the National Institutes of Health, Health Future Foundation, the State of Nebraska Cancer and Smoking Disease Research Program, U.S. Department of Energy, National Science Foundation of China, Creighton University, and HuNan Normal University. Technical assistance from W.-M. Chen and J. Li, and the user feedback on GenoDB from all the students, postdoctoral fellows, and technicians from the genetics lab of H.-W.D. is appreciated. We are grateful to two anonymous reviewers for their constructive comments that helped to improve the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL deng@creighton.edu; FAX (402) 280-5034.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.159701.
REFERENCES
- ABI PRISM. Genotyper version 2.1, user's manual. Foster City, CA: Perkin-Elmer Applied Biosystems; 1996. [Google Scholar]
- ABI PRISM. Linkage mapping set version 2, user's manual. Foster City, CA: Perkin-Elmer Applied Biosystems; 1997. [Google Scholar]
- Almasy L, Blangero J. Multipoint quantitative trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PP. The entity-relationship model — toward a unified view of data. ACM Trans Database Syst. 1976;1:9–36. [Google Scholar]
- Cheung KH, Nadkarni P, Silversten S, Kidd JR, Pakstis AJ, Miller P, Kidd KK. PhenDB: An integrated client/server database for linkage and population genetics. Comput Biomed Res. 1996;29:327–337. doi: 10.1006/cbmr.1996.0024. [DOI] [PubMed] [Google Scholar]
- Cybergenetics . Tutorial: Introducing TrueAllele technology. Pittsburgh, PA: Cybergenetics; 2000. [Google Scholar]
- Deng H-W, Li J-L, Li J, Davies MK, Recker RR. Heterogeneity of bone mass density across skeletal sites and its clinical implications. J Clinical Densitometry. 1998;1:339–353. doi: 10.1385/jcd:1:4:339. [DOI] [PubMed] [Google Scholar]
- Deng H-W, Chen W-M, Recker S, Stegman M-R, Li J-L, Davies KM, Zhou Y, Deng H-Y, Heaney R-R, Recker RR. Genetic determination of Colles' fractures and differential bone mass in women with and without Colles' fractures. J Bone Miner Res. 2000a;15:1243–1252. doi: 10.1359/jbmr.2000.15.7.1243. [DOI] [PubMed] [Google Scholar]
- Deng H-W, Chen W-M, Conway T, Zhou Y, Davies KM, Stegman MR, Deng HY, Recker RR. Determination of bone mineral density of the hip and spine in human pedigrees by genetic and life-style factors. Genet Epidemiol. 2000b;19:160–177. doi: 10.1002/1098-2272(200009)19:2<160::AID-GEPI4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Deng, H.-W., Xu, F.-H., Lai, D.-B., Johnson, M., Deng, H.-Y., Li, J.-L., Shen, H., Zhang, H.-T., Liu, Y.-J., Recker, R.R. 2001a A whole genome scan linkage study for QTLs underlying osteoporosis. In preparation for Nat. Genet..
- Deng, H.-W., Xu, F.-H., Conway, T., Deng, X.-T., Li, J.-L., Davies, K.-M., Deng, H.-Y., Johnson, M.,and Recker, R.R. 2001b. Is population BMD variation linked the marker D11S987 on chromosome 11q12–13. J. Clin. Endocrinol. Metab., in press. [DOI] [PubMed]
- Ghosh S, Karanjawala ZE, Hauser ER, Ally D, Knapp JI, Rayman JB, Musick A, Tannenbaum J, Te C, Shapiro S, et al. Method for precise sizing, automated binning of alleles, and reduction of error rates in large-scale genotyping using fluorescently labeled dinucleotide markers. Genome Res. 1997;7:165–178. doi: 10.1101/gr.7.2.165. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Collins FS. The geneticist's approach to complex disease. Annu Rev Med. 1996;47:333–353. doi: 10.1146/annurev.med.47.1.333. [DOI] [PubMed] [Google Scholar]
- Griffiths RAL, Barber MD, Johnson PE, Gillbard SM, Haywood MD, Smith CD, Arnold J, Burke T, Urquhart A, Gill P. New reference allelic ladders to improve allelic designating in a multiplex STR system. Int J Legal Med. 1998;111:267–272. doi: 10.1007/s004140050167. [DOI] [PubMed] [Google Scholar]
- Hall JM, LeDuc CA, Watson A R, Roter AH. An approach to high-throughput genotyping. Genome Res. 1996;6:781–790. doi: 10.1101/gr.6.9.781. [DOI] [PubMed] [Google Scholar]
- Hauser ER, Boehnke M, Guo S-W, Risch N. Affected-sib-pair interval mapping and exclusion for complex genetic traits: Sampling considerations. Genet Epidemiol. 1996;13:117–137. doi: 10.1002/(SICI)1098-2272(1996)13:2<117::AID-GEPI1>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Idury RM, Cardon LR. A simple method for automated allele binning in microsatellite markers. Genome Res. 1997;7:1104–1109. doi: 10.1101/gr.7.11.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES. Parametric and nonparametric linkage analysis: A unified multipoint approach. Am J Hum Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Schork N J. Genetic dissection of complex traits. Sciences. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- McMahon FJ, Thomas CJM, Koskela RJ, Breschel TS, Hightower TC, Rohrer N, Savino C, McInnis MG, Simpson SG, DePaulo JR. Integrating clinical and laboratory data in genetic studies of complex phenotypes: A network-based data management system. Am J Med Genet. 1998;81:248–256. [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE. PedCheck: A program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J. Analysis of human genetic linkage. Baltimore: The Johns Hopkins University Press; 1991. [Google Scholar]
- Pálsson B, Pálsson F, Perlin M, Gudbjartsson H, Stefánsson K, Gulcher J. Using quality measures to facilitate allele calling in high-throughput genotyping. Genome Res. 1999;9:1002–1012. doi: 10.1101/gr.9.10.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlin MW, Burks MB, Hoop RC, Hoffman EP. Toward fully automated genotyping: allele assignment, pedigree construction, phase determination, and recombination detection in Duchenne muscular dystrophy. Am J Hum Genet. 1994;55:777–787. [PMC free article] [PubMed] [Google Scholar]
- Perlin MW, Lancia G, Ng SK. Toward fully automated genotyping: genotyping microsatellite markers by deconvolution. Am J Hum Genet. 1995;57:1199–1210. [PMC free article] [PubMed] [Google Scholar]
- Puers C, Hammond H, Jin L, Caskey C, Schumm J. Identification of repeat sequence heterogeneity at the polymorphic short tandem repeat locus HUMTH01[AATG]n and reassignment of alleles in population analysis by using a locus-specific allelic ladder. Am J Hum Genet. 1993;53:953–958. [PMC free article] [PubMed] [Google Scholar]