Abstract
The single-copy hepatitis B virus transgene in the E36 transgenic mouse strain undergoes methylation changes in a parent-of-origin, tissue, and strain-specific fashion. In a C57BL/6 background, the paternally transmitted transgene is methylated in 30% of cells, whereas it is methylated in more than 80% of cells in (BALB/c x C57BL/6) F1 mice. We established previously that several genetic factors were likely to contribute to the transgene methylation profile, some with demethylating and some with de novo methylating activities. Using quantitative trait loci (QTL) mapping, we have now localized one major modifier locus on chromosome 13 (Mod13), which explains a 30% increase in the methylation level of this transgene with no effect on the flanking endogenous sequences. No other QTL could be identified, except for a demethylating activity of low significance located on chromosome 12. Recombinant inbred mice containing a BALB/c allele of Mod13 were then used to show that the presence of Mod13 is sufficient to induce de novo methylation. A segregation between de novo methylation and repression of transgene expression was uncovered, suggesting that this genetic system is also useful for the identification of factors that interpret methylation patterns in the genome.
CpG methylation of regulatory sequences is associated with the control of gene expression in vertebrates. Particular methylation patterns of genes are established during development by a combination of maintenance methylation by DNMT1 and de novo methylation by DNMT3a and DNMT3b (Bird and Wolffe 1999). Inactivation of these methyltransferase genes affects mouse development (Li et al. 1992; Okano et al. 1999), and mutations in Dnmt3a cause the ICF syndrome in human (Xu et al. 1999). In addition, the recently described demethylase may play a role in developmental and tissue-specific gene activation (Bhattacharya et al. 1999). Other factors that affect the methylation status of genes are largely unknown.
It has been proposed that cytosine methylation evolved in vertebrates as a host defense mechanism against mobile elements (Yoder et al. 1997). Transgenes carrying sequences of bacterial or viral origin and tandemly repeated sequences (Garrick et al. 1998) often appear to be the target for de novo methylation early in development. Strain-specific modifications of transgene methylation have been described (Allen et al. 1988; Engler et al. 1991; Weichman and Chaillet 1997) establishing a genetic system in which methylation modifiers can be characterized. Candidate genes for these modifiers include the members of the SWI/SNF family proteins, as reported for ATRX responsible for modifications of the methylation levels of repetitive elements (Gibbons et al. 2000).
We have previously described the effect of different genetic and environmental factors on the methylation status of the E36 transgene, consisting of one copy of a recombinant pBR322 plasmid bearing a portion of the hepatitis B virus genome (HBV; Babinet et al. 1985). On maternal transmission, the transgene becomes de novo methylated and inactivated (Hadchouel et al. 1987) in every genetic background tested. Following paternal transmission in a C57BL/6J background, the S gene remains undermethylated (Pourcel et al. 1990), whereas a single cross to a BALB/cJ female results in transgene hypermethylation and inactivation. We showed that several strain-specific genetic factors contributed to the transgene methylation profile, some with demethylating and some with de novo methylating activities (Schweizer et al. 1998). We have now performed a QTL analysis to genetically map the locus responsible for the transgene de novo methylation in the progeny of a cross between (BALB/cJ x C57BL/6J) F1 females and E36 (C57BL/6J) males (BCxE progeny).
RESULTS
The BALB/c Modifier Effect Is Restricted to the Transgene
The E36 transgenic line was in a C57BL/6J background at the beginning of this study, as the result of 12 successive backcrosses of trangenic males (originally C57BL/6J x SJL/J) with C57BL/6J females. E36 males were crossed to (BALB/cJ x C57BL/6J) F1 females, and the transgenic BCxE progeny was selected.
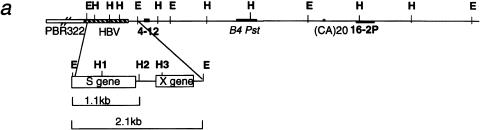
To analyze the transgene methylation levels, we used the methylation sensitive restriction enzyme HpaII, which cuts the HBV sequence at three sites. Site H1 is located in the S gene, whereas site H2 and site H3 are in the enhancer and X gene region, respectively (Fig. 1a). As there is only one copy of the transgene, the methylation level at these sites reflects cellular mosaicism. In a C57BL/6J background, site H1 is methylated in all the cells of non-liver tissues, whereas sites H2 and H3 are methylated in only about 30% of cells (Pourcel et al. 1990). In our previous study on a small set of BCxE mice, the transgene methylation level was measured in the liver, the organ in which the S gene is transcribed in response to tissue-specific and hormonal factors (Farza et al. 1987). To avoid any interference with liver-specific factors, and to specifically assess the effect of the BALB/c modifier on the transgene methylation, we performed the present study on DNA extracted from heart and lung of newborn mice. We analyzed the degree of methylation at site H2 following digestion of genomic DNA with EcoRI + HpaII, and hybridization with the HBV Eco-Ac probe (Fig. 1a). Figure 1b shows a representative blot displaying the transgene methylation profile of 10 BCxE mice from four litters. Only two bands were observed, the uncut 2.1-kb EcoRI fragment and a 1.1-kb fragment corresponding to digestion at site H2. As expected for non-liver tissue, there was no band corresponding to digestion at site H1 (producing fragments of 0.4 kb and 0.7 kb; see Fig. 4b). Probe Eco-Ac does not detect fragments produced by digestion at sites H2 and H3.
Figure 1.
Methylation levels of the transgene and endogenous sequences in 10 BCxE progeny. (a) Genetic organization and restriction map of HBV sequences (striped bar) and of 20-kb mouse genomic sequence flanking the transgene on one side. 4–12, B4 Pst and 16–2P (dark bars) are probes selected for methylation analysis. EcoRI (E) and HpaII (H) sites are shown. (b) Southern blot hybridization profile of genomic DNA digested with EcoRI + HpaII (samples 1–10) or EcoRI (sample 11), and hybridized successively with HBV Eco-Ac, 4–12 and 16–2P probes. The size of relevant restriction fragments are shown on the side. The transgene methylation levels calculated from the HBV hybridization are shown for samples 1–10.
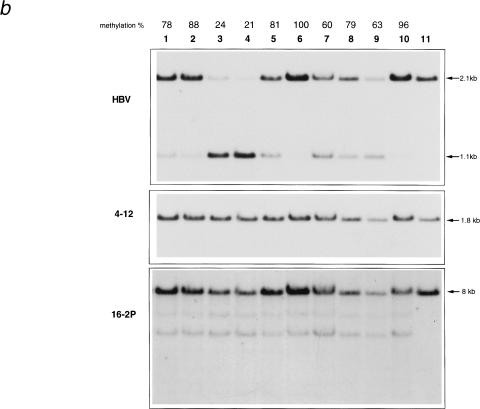
Figure 4.
Southern blot analysis of heart and lung DNA (a), or liver DNA (b), from the progeny of a E36 male crossed to BALB/cJ (B1, B2, BA1), C57BL/6J (C1, C2, CA1), and recombinant inbred mice CXB5 (5–1, 5–2, 5-A1, 5-A2), CXB7 (7–1, 7–2, 7A1, 7A2), CXB6 (6–1, 6–2) and CXB1 (1–1, 1–2). Samples were restricted with EcoRI and HpaII, except for sample Eco with EcoRI alone. (c) Northern blot analysis of the liver S mRNA in the progeny of a E36 male crossed to C57BL/6J (CA1, CA2), and recombinant inbred mice CXB5 (5-A1, 5-A2), CXB7 (7-A1, 7-A2), CXB6 (6–1, 6–2) and CXB1 (1–1, 1–2). Samples labeled A are from adults.
Hybridization with PBR322 sequences showed that they are methylated to the same degree as HBV sequences (data not shown). Thus, it appears that in a given cell all the transgene HpaII sites are either nonmethylated or methylated, with the exception of site H1, which requires transcription activation to be demethylated. The transgene methylation percentage was calculated as the ratio of the 1.1-kb signal to the 2.1-kb + 1.1-kb signals.
To investigate the methylation status of the whole insertion domain, we made use of several genomic probes, located in the vicinity of the transgene insertion. The E36 transgene insertion region (Hbvi) was previously assigned to chromosome 13, close to D13Mit64, by linkage analysis using a probe flanking the transgene (probe 4–12; Hadchouel et al. 1987). Analysis of a 30-kb genomic DNA region at the insertion site allowed the isolation of several unique probes. B4Pst1–2, localized 8 kb from the HBV insertion site, is a fragment of an expressed intronless gene and also cross-hybridizes to an intron-containing gene on chromosome 3 (Pourcel et al. 2000; AJ305034). The intronless gene can encode a protein, as shown by in vitro transcription-translation studies (Schweizer 1995). The analysis of the insertion locus also revealed the existence of a deletion of genomic DNA flanking the transgene. Probes 4–12 and B4Pst1–2 lie in hypermethylated domains, whereas probe 16–2P, localized 15 kb from HBV, is methylated in the testis only. The methylation level of these different probes was identical in BCxE mice with otherwise different HBV methylation levels (Fig. 1b; data not shown), indicating that the effect of the BALB/cJ modifiers was restricted to the transgene.
The Transgene Methylation Is Under Polygenic Control
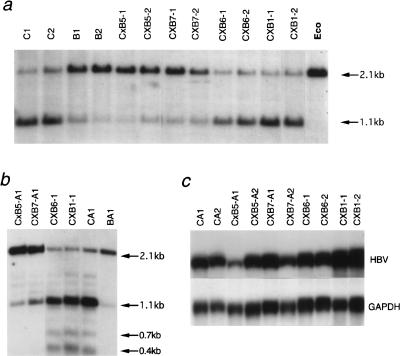
The transgene methylation level was measured on 256 BCxE samples, and the results are depicted in Figure 2. On Figure 2a, mice were ordered on the x-axis according to their transgene methylation percentage (y-axis). The methylation percentages show an even distribution between 3% and 100%, suggesting a polygenic control. About 15% of mice show a methylation level below that of the E36 (paternal) progenitors (30%), whereas 34% of mice show a methylation level above the (BALB/cJ x E36) F1 maternal progenitors (85%). On Figure 2b the data have been divided into categories (x-axis), and the number of mice in each category is shown on the y-axis. The two representations clearly show that the phenotypic distribution is not normal. These results from a large population confirm our previous hypothesis about the action of several trans-acting modifiers on the transgene, with methylating and demethylating activities segregating in BCxE mice (Schweizer et al. 1998).
Figure 2.
Distribution of the methylation percentages in 256 BCxE progeny. (a) BCxE mice (x-axis) were ordered according to their transgene methylation percentage (y-axis). (b) Mice have been grouped into categories (x-axis) from 0–10% methylation, 10–20%, etc. The number of mice in each category is shown on the y-axis.
A Major QTL Is Mapped on Chromosome 13
To analyze the linkage between the methylation level and the genotype, a whole genome scan was performed using 102 microsatellite markers from the selection of Schalkwyk et al. (1999). This corresponds to four to eight loci analyzed for each chromosome, with an average spacing on the mouse genetic map (http://www-genome.wi.mit.edu/) of 11cM.
Genetic distances between markers in the BCxE cross were determined using the Mapmaker 3.0 (Lander and Botstein 1989) analysis program with distances reported in Haldane centimorgan units. The maps we obtained were in good agreement with the consensus map of the mouse genome (Mouse Genome Database), taking into account the fact that the maps produced in the present work are female recombination maps.
A QTL analysis of the data for 88 mouse samples was first performed using the MultiQTL software package version 2.0 (http:\\www.multiqtl.com), identifying a region on chromosome 13 with a peak LOD score of four at position 27.5cM distal to D13Mit64. An extended analysis of chromosomes 2, 5, 6, 9, 12 and 18, for which weak LOD scores had been found, was performed with the total BCxE population (256 mice). The LOD score for the chr12 putative QTL, with a potential demethylating effect, reached 1.96. The significance of this LOD was evaluated by permutation tests to p = 0.015.
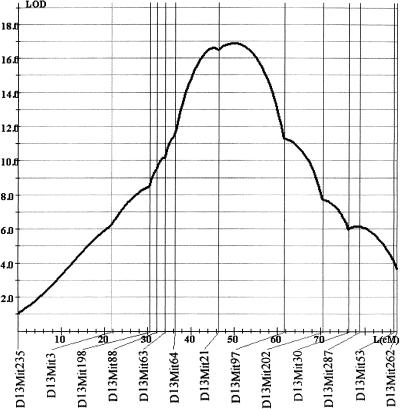
Additional informative markers (total of 15) were chosen for chr13, and the LOD score for the QTL reached 16.5 in the vicinity of markers D13Mit21-D13Mit97 (Fig. 3a). The presence of BALB/cJ alleles at this locus was responsible for an increase of the methylation level of 29% (95% confidence interval: 23% to 34%) when compared with mice homozygote for a C57BL/6J region. Thirty-three percent of the phenotypic variance (95% confidence interval: 22% to 44%) was explained by the presence of a QTL on chr 13. The 99.9% confidence interval for the location of the QTL is D13Mit64-D13Mit97, which includes the Hbvi locus.
Figure 3.
Localization of the chr 13 modifier. The LOD score peak is maximum between D13Mit21 and D13Mit97, but the 99.9% confidence interval includes Hbvi, the transgene insertion site.
Finally, because the effect of the chr 13 QTL is strong and may hide other, minor QTLs, we performed additional tests for linkage at a second locus (see Methods), and this showed no evidence of a second locus, whether or not interacting with the chr 13 locus.
The Effect of the chr13 QTL Is Confirmed Using Recombinant Inbred Mice
To test whether the presence of the BALB/c allele at the chr 13 modifier (named Mod13) is necessary and sufficient to induce a high level of transgene methylation, we sought to use recombinant inbred strains derived from the cross of parental BALB/cByJ (C) and C57BL/6ByJ (B) strains (Bailey 1971). We chose four strains of the CXB RI panel: two with a chr 13 region, encompassing the Mod13 QTL, of BALB/cByJ parental origin (CXB5 and CXB7), and two with a Mod13 region of C57BL/6ByJ origin (CXB1 and CXB6; Table 1). Because differences have been documented between the J and By substrains, and microsatellite genotyping reveal some degree of polymorphism (Panoutsakopoulou et al. 1997), we first checked the genotype of the four selected strains with markers D13Mit64, D13Mit21, D13Mit91, D13Mit97, and D13Mit202. No difference with the genotype of the BCXE parental strains could be observed.
Table 1.
Chr13 Genotype of the Four Recombinant Inbred Strains
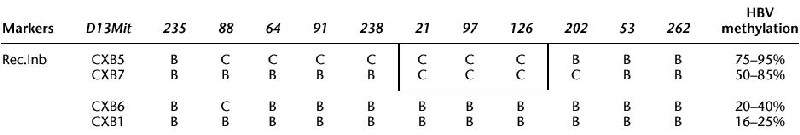 |
Markers are those from the literature and additional ones used in this study. The methylation percentage of hepatitis B virus sequences in (CXB x E36)F1 progeny is shown on the right side. (B) C57BL/6, (C) BALB/c.
The transgene methylation levels were thus analyzed in the heart and the lung of newborn transgenic progeny (Fig. 4a) and in the liver of adults (Fig. 4b). The transgene methylation in the CXB5 progeny was 80% (mean value from three newborn and two adult mice) and that of CXB7, 60% (mean value from two newborns and two adult mice). In the progeny of E36 x CXB1 and CXB6, the transgene methylation level was 30% (mean value from three newborn mice) and 20% (mean value from six newborn and four adult mice), respectively. Thus the effect of the BALB/cByJ Mod13 region provided by the recombinant inbred is similar to that of the BALB/cJ substrain. We then wished to analyze whether the level of S gene expression would be correlated with the degree of transgene methylation. Serum HBsAg was measured by ELISA (data not shown) in adult mice, and the liver S mRNA was analyzed by Northern blot (Fig. 4c). Interestingly, (CXB5 X E36) F1 and (CXB7 X E36) F1 mice had high levels of transgene expression, both at the RNA and protein level, similar to C57BL/6J E36 and (CXB6 X E36) F1 mice. (CXB1 X E36) F1 had the highest levels of expression.
DISCUSSION
By combining a QTL analysis and the use of recombinant inbred mice, we provide strong evidence of the existence on mouse chr 13 of a locus responsible for the E36 transgene de novo methylation. Examination of the alleles in the region encompassing D13Mit63 to D13Mit97 of the BCxE population shows that all but one of the mice with a transgene methylation level below 40% are C57BL/6 homozygous. Mice with a high transgene methylation levels have frequently BALB/c alleles at these loci, although there are exceptions showing that other loci can also induce de novo methylation. Crosses to recombinant inbred mice suggest that the existence of BALB/c alleles at the Mod13 locus on the maternally inherited chr 13 is sufficient to methylate the transgene sequences on the paternal chr 13 in more than 60% cells. Other factors may be necessary, in addition, to achieve the highest levels of methylation (80% and more).
The phenotypic distribution in the BCxE mice suggests the existence of several factors affecting the transgene methylation level. However, we have not found any strong evidence for the existence of other methylation QTL. A potential demethylating QTL may be present on chr 12 (on which Dnmt3a is localized), but the number of mice in the 0% to 30% methylation category, possibly affected by this QTL, is very small. Other studies will be necessary to confirm the existence of a demethylating activity.
As previously reported, the regulation of transgene methylation is site specific, and we show here that it does not affect the methylation of the endogenous insertion locus. This is different from the effect of the Ssm1 modifier, on the bacterial sequences present in the transgene pHRD (Engler et al. 1998). The transgene pHRD is highly methylated in 12 independent mouse lines when in the C57BL/6 strain background despite different integration sites (Engler et al. 1991). In E36, the transgene and the dominant modifier locus Mod13 are located in the same region on chr 13, although the genotyping and recombinant inbred data indicate that Mod13 may be distinct from Hbvi. In the present study, a microsatellite located 10 kb from Hbvi (CA 20; Fig. 1a) was mapped using a radiation hybrid panel near D13Mit91. The location of the modifier near the transgene insertion site may be coincidental and not functionally relevant. However, it is possible that the BALB/c region on the maternal chromosome interacts with the C57BL/6 paternal region bearing the transgene, resulting in a signal for the action of de novo methylation factors. This is reminiscent of the mechanism of ‘cross-talk’ proposed by M. Monk (1990) to explain both the maternal imprinting and strain-specific de novo methylation of transgenes. There is evidence for somatic pairing of homologous chromosomes in the mouse, and the existence of trans-allelic interactions has been described in several organisms (including maize, Drosophila, and mice). Diploid organisms may develop defense mechanisms that detect and block differences between two homologous chromosomes.
We previously postulated that B4Pst, a gene located near the transgene insertion site and weakly transcribed from the maternal allele during development, may influence the transgene methylation-imprinting (Schweizer 1995). This cannot explain, however, the promethylating effect of a modifier acting in trans.
The present study was conducted to map QTLs responsible for de novo methylation early in development and affecting the transgene to the same extent, in all the organs of the mouse. In previous analyses we showed that (BALB/cJ x E36) F1 mice with high transgene methylation levels (more than 80%) had little or no liver S gene expression (Schweizer et al. 1998). Interestingly, we found that the progeny of recombinant inbred strains CXB5 and CXB7 females crossed to a E36 male express large amounts of SmRNA and HBsAg, although the transgene methylation is above 80%. Thus, in this particular allelic combination, either 20% of liver cells can produce as much SmRNA as 70% cells in a C57BL/6 background, or methylation and repression are not directly coupled. It is possible that the By background of the RI parental strains is providing a particular control on the transcription level. Studies are in progress to test the basis for this regulation. In plants, the disruption of a member of the SWI/SNF family proteins, the MOM protein, releases transcriptional silencing of a methylated gene (Amedeo et al. 2000). Methyl-binding proteins, such as MeCP2 and the MBD protein family, involved in assembling transcriptional silencing complexes on methylated DNA (Bird and Wolffe 1999) may also be involved in this regulation.
This work suggests that several genes involved in methylation might possess alleles with different activities. This is important in view of the consequences of epigenetic modifications in cancer cells (Baylin and Herman 2000) and genetic diseases (Robertson and Wolffe 2000) or the possible role of methylation as a host defense mechanism (Bestor and Tycko 1996; Yoder et al. 1997).
METHODS
Transgenic Mice
The E36 transgenic mouse strain has been maintained by successive crosses of males to C57BL/6J females. In these mice the S gene encoding the surface antigen (HBsAg) is specifically transcribed in the liver in response to tissue-specific and hormonal factors (Farza et al. 1987), and HBsAg is secreted in the serum.
Recombinant inbred females were purchased from The Jackson Laboratory.
DNA and RNA Analyses
DNA was prepared from heart and lungs of one- or two-day-old newborn mice using the method described in Pourcel et al. (1990). For the methylation analysis, DNA was restricted with EcoRI and HpaII and run on 1% agarose gels. Southern blots were performed by capillarity on Genescreen membranes (NEN). The membranes were hybridized to a HBV EcoRI-AccI fragment (Eco-Ac probe) prepared from HBV-recombinant plasmid PCP10 (Dubois et al. 1980) or to mouse genomic fragments described in Pourcel et al. (2000). The intensity of each band was estimated by densitometric scanning using Storm Imager and Imagequant software (Molecular Dynamics). Total RNA was extracted using Tripure Isolation Reagent (Boehringer Mannheim) and analyzed on 1% formaldehyde agarose gels. The RNA transferred to nylon membranes (Hybond N Amersham France SA) was hybridized to the HBV Eco-Ac probe or a Gapdh probe as a control.
Genotyping and QTL Analysis
The PCR reactions were performed in part using forward primers labeled with fluorescein or FITC labels (TIB MOLBIOL, Berlin) and analyzed on a Pharmacia ALF Gel system using fragment manager software. For manual analysis, PCR products were separated by electrophoresis either on 4% agarose gels or on standard denaturing polyacrylamide gels followed by transfer to positive-charged nylon membrane (Pall). The membranes were hybridized with a primer labeled with alpha-32PdCTP using terminal transferase (Boehringer, Lewes). Genotypes were determined after autoradiography. QTL analysis was first performed with the MultiQTL program version 2.0 (http:\\www.multiQTL.com), using the interval analysis and marker restoration options. Five thousand permutations were run to estimate QTL significance.
Because of lack of normality of the phenotype distribution, a second analysis was performed by creating order categories for the methylation levels (x) into seven categories (x < 31%, 31% ≤ x < 45%, 45% ≤ x < 61%, 61% ≤ x < 78%, 78% ≤ x < 89%, 89% ≤7 x < 100%, x = 100%). We tested for a linear trend of genotype frequencies with respect to order categories. Additional tests of linear trend were performed separately in animals with heterozygous and homozygous genotypes at the marker on chromosome 13 most strongly linked to the trait
Acknowledgments
We thank Wendy Dean for advice and help with embryology, Xavier Montagutelli for helpful discussions and critical reading of the manuscript, and Jean-Paul Soulillou, Guillaume Dighiero, and Mark Lathrop for their support. Mark Lathrop performed the analysis to test for linkage at a second locus. This work was funded by grants from the EEC (BMH4-CT96–0050), the French Ministry ACCSV, the BBSRC, and MAFF. D.G. holds a Wellcome senior fellowship in basic biomedical science.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL cpourcel@pasteur.fr; FAX 33 (0) 1456-88951.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.163801.
REFERENCES
- Allen ND, Cran DG, Barton SC, Hettle S, Reik W, Surani MA. Transgenes as probes for active chromosomal domains in mouse development. Nature. 1988;333:852–855. doi: 10.1038/333852a0. [DOI] [PubMed] [Google Scholar]
- Amedeo P, Habu Y, Afsar K, Mittelsten Scheid O, Paszkowski J. Disruption of the plant gene MOM releases transcriptional silencing of methylated genes. Nature. 2000;405:203–206. doi: 10.1038/35012108. [DOI] [PubMed] [Google Scholar]
- Babinet C, Farza H, Morello D, Hadchouel M, Pourcel C. Specific expression of hepatitis B surface antigen (HBsAg) in transgenic mice. Science. 1985;230:1160–1163. doi: 10.1126/science.3865370. [DOI] [PubMed] [Google Scholar]
- Bailey DW. Recombinant inbred strains. Transplantation. 1971;11:325–327. doi: 10.1097/00007890-197103000-00013. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- Bestor TH, Tycko B. Creation of genomic methylation pattern. Nat Genet. 1996;12:363–367. doi: 10.1038/ng0496-363. [DOI] [PubMed] [Google Scholar]
- Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- Bird AP, Wolffe AP. Methylation-induced repression-belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- Dubois M-F, Pourcel C, Rousset S, Chany C, Tiollais P. Excretion of hepatitis B surface antigen particles from mouse cells transformed with cloned viral DNA. Proc Natl Acad Sci. 1980;77:4549–4553. doi: 10.1073/pnas.77.8.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler P, Doglio LT, Bozek G, Storb U. A cis-acting element that directs the activity of the murine methylation modifier locus Ssm1. Proc Natl Acad Sci. 1998;95:10763–10768. doi: 10.1073/pnas.95.18.10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler P, Haasch D, Pinkert CA, Doglio L, Glymour M, Brinster R, Storb U. A strain-specific modifier on mouse chromosome 4 controls the methylation of independent transgene loci. Cell. 1991;65:939–947. doi: 10.1016/0092-8674(91)90546-b. [DOI] [PubMed] [Google Scholar]
- Farza H, Salmon A-M, Hadchouel M, Moreau J-L, Babinet C, Tiollais P, Pourcel C. Hepatitis B surface antigen gene expression is regulated by sex-steroids and glucocorticoids in transgenic mice. Proc Natl Acad Sci. 1987;84:1187–1191. doi: 10.1073/pnas.84.5.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrick D, Fiering S, Martin DI, Whitelaw E. Repeat-induced gene silencing in mammals. Nat Genet. 1998;18:56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- Gibbons RJ, McDowell TL, Raman S, O R, DM, Garrick D, Ayyub H, Higgs DR. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat Genet. 2000;24:368–371. doi: 10.1038/74191. [DOI] [PubMed] [Google Scholar]
- Hadchouel M, Farza H, Simon D, Tiollais P, Pourcel C. Maternal inhibition of hepatitis B virus surface antigen gene expression correlates with de novo methylation. Nature. 1987;329:454–456. doi: 10.1038/329454a0. [DOI] [PubMed] [Google Scholar]
- Lander ES, Botstein D. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Monk M. Variation in epigenetic inheritance. Trends Genet. 1990;6:110–4. doi: 10.1016/0168-9525(90)90124-o. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Panoutsakopoulou V, Spring P, Cort L, Sylvester JE, Blank KJ, Blankenhorn EP. Microsatellite typing of CXB recombinant inbred and parental mouse strains. Mamm Genome. 1997;8:357–361. doi: 10.1007/s003359900441. [DOI] [PubMed] [Google Scholar]
- Pourcel C, Jaubert J, Hadchouel M, Xue W, Schweizer J. A new family of genes and pseudogenes potentially expressing testis-specific and brain-specific leucine zipper proteins in man and mouse. Gene. 2000;249:105–113. doi: 10.1016/s0378-1119(00)00158-x. [DOI] [PubMed] [Google Scholar]
- Pourcel C, Tiollais P, Farza H. Transcription of the S gene in transgenic mice is associated with hypomethylation at specific sites and with DNase I sensitivity. J Virol. 1990;64:931–935. doi: 10.1128/jvi.64.2.931-935.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KD, Wolffe AP. DNA methylation in health and disease. Nature Reviews. 2000;1:11–19. doi: 10.1038/35049533. [DOI] [PubMed] [Google Scholar]
- Schalkwyk LC, Jung M, Daser A, Weiher M, Walter J, Himmelbauer H, Lehrach H. Panel of microsatellite markers for whole-genome scans and radiation hybrid mapping and a mouse family tree. Genome Res. 1999;9:878–887. doi: 10.1101/gr.9.9.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer J, Valenza-Schaerly P, Goret F, Pourcel C. Control of expression and methylation of a Hepatitis B viral transgene by strain-specific modifiers. DNA Cell Biol. 1998;17:427–435. doi: 10.1089/dna.1998.17.427. [DOI] [PubMed] [Google Scholar]
- Schweizer JG. “Investigation of the mouse genomic integration locus and the genomic imprinting of a hepatitis B virus transgene.” Ph.D. thesis. Germany: Universitat Konstanz; 1995. [Google Scholar]
- Weichman K, Chaillet R. Phenotypic variation in a genetically identical population of mice. Mol Cel Biol. 1997;17:5269–5274. doi: 10.1128/mcb.17.9.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G-L, Bestor TH, Bourc'his D, Hsieh C-L, Tommerups N, Bugge M, Hulten M, Qu X, Russo JJ, Viegas-Péquignot E. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]