Abstract
The ongoing revolution in microarray technology allows biologists studying gene expression to routinely collect >105 data points in a given experiment. Widely accessible and versatile database software is required to process this large amount of raw data into a format that facilitates the development of new biological insights. Here, we present a novel microarray database software system, named Argus, designed to process, analyze, manage, and publish microarray data. Argus imports the intensities and images of externally quantified microarray spots, performs normalization, and calculates ratios of gene expression between conditions. The database can be queried locally or over the Web, providing a convenient format for Web-publishing entire microarray data sets. Searches for regulated genes can be conducted across multiple experiments, and the integrated results incorporate images of the actual hybridization spots for artifact screening. Query results are presented in a clone- or gene-oriented fashion to rapidly identify highly regulated genes, and scatterplots of expression ratios allow an individual ratio to be interpreted in the context of all data points in the experiment. Algorithms were developed to optimize response times for queries of regulated genes. Supporting databases are updated easily to maintain current gene identity information, and hyperlinks to the Web provide access to descriptions of gene function. Query results also can be exported for higher-order analyses of expression patterns. This combination of features currently is not available in similar software. Argus is available at http://vessels.bwh.harvard.edu/software/Argus.
Over the last five years, large-scale sequencing projects have produced a flood of data, and it has been challenging to characterize and assimilate this new information concerning tens of thousands of known and novel genes (Ermolaeva et al. 1998; Claverie 1999). Even more recently, there has been an explosion in the amount of gene expression data available regarding this immense sequence database. Rather than adding to the complexity of genome analysis, high-throughput expression data holds the promise of providing a simplifying, functional framework for seemingly chaotic genomes. Applying various forms of pattern recognition to these data sets is a tool for generating new hypotheses about the vast number of transcribed sequences.
Although new array substrates and protocols are constantly evolving, the microarray technology used for high-throughput expression studies is conceptually a scaled-up version of a traditional DNA dot blot. Thousands or tens of thousands of cDNAs, cDNA fragments, or oligonucleotides are immobilized on a substrate and hybridized with labeled nucleic acid derived from RNA samples of interest. The relative expression levels of all of these gene fragments then are measured in parallel and compared across multiple RNA samples. For the first time, the biologist studying gene expression can routinely collect far more data than can be analyzed comfortably using conventional spreadsheets. Database software capable of dealing with larger volumes of numeric and image data is required. All data must be transformed into a format centered around biological questions, while at the same time reducing distraction from the artifacts that inevitably appear whenever many measurements are made.
To meet these needs, the typical process for analysis of microarray data is as follows: A scanned image of a microarray reflecting radioactive or fluorescent intensity is imported into software programmed to recognize which spots correspond to which cDNA or oligo. The “raw intensity” (or “unnormalized intensity”) of each spot is quantified using various image processing methods to locate the center of the spot, quantify its intensity, and subtract the background. Hybridizations tend to vary somewhat in their overall signal strength due to factors that are difficult to control, such as labeling efficiency. To adjust for these differences in overall intensity, which are assumed to be nonbiological, the intensity measurements from each microarray are normalized using one of a variety of methods. Normalization methods have not yet been standardized, but common methods include dividing all raw intensity values by the average or median of all data points. The resulting normalized intensity can be compared with the results from other hybridizations to the same array, or from hybridizations to identically prepared batches of arrays. (For an assessment of inter- vs. intra-array comparisons, see Aach et al. 2000.)
The simplest measure of gene regulation is to divide the normalized intensity of a spot in one condition by the normalized intensity of the corresponding spot in a reference condition, producing a ratio representing the fold-change of the expression level of the gene. However, the responsiveness of this ratio as measured by microarray often is dampened compared to that observed on a Northern blot (Livesey et al. 2000) or using real-time RT-PCR (G. García-Cardeña and J. Comander, unpubl.). Therefore, genes whose ratios are far from unity often represent highly regulated genes, potentially having biological significance. Ratios far from unity are also less likely to be due to random noise in measurement, and the noise level itself often is influenced by the intensity of the measurements from which the ratio was derived. Thus, the researcher needs to select a particular minimum ratio and intensity combination as being worthy of further review, or “significant”. The “significant ratio” and intensity cutoffs should be based on the observed noise level in the experiment (Manduchi et al. 2000) and, separately, a judgment of what levels of regulation would be biologically interesting in the final analysis.
Expression ratios can be calculated between all pairs of experimental conditions, and all genes with significantly regulated ratios can be displayed as a large list. Stopping the analysis at this point, however, could fail to identify higher-order patterns in the data. A variety of multivariate data analysis techniques can be used to find such patterns. For example, clustering algorithms can be used to group genes based on similar expression patterns across the conditions measured (Eisen et al. 1998; Tamayo et al. 1999). These clusters sometimes are enriched for genes with certain biological functions, which can identify the biological pathways that were being modulated in the experiments under study. Once a cluster has been associated with a biological function, it is possible to make new hypotheses about the functions of unidentified genes that also are contained in that cluster. In organisms in which the genome sequence is known, the promoter regions of genes in these clusters can be searched for shared, novel regulatory elements (Roth et al. 1998; Tavazoie et al. 1999; Iyer et al. 1999, 2001; Hughes et al. 2000; Aach et al. 2001). In addition to techniques based on clustering, more complicated statistical algorithms such as singular value decomposition can be used to identify higher-order descriptors of variance in the data (Alter et al. 2000; Holter et al. 2000). Techniques for classifying biological samples also are being developed (Golub et al. 1999; Furey et al. 2000).
The number of such analyses available and the number of yet-to-be-standardized permutations possible for each analysis makes the integration of a microarray database and microarray data analysis difficult. In addition, expression data are being collected from a variety of microarray types, and even from experiments that do not use arrays, such as those using SAGE (Velculescu et al. 1995). Importing and integrating expression data from all of these disparate sources remains a challenge (Kuo et al., in prep.). Integration across data sets has been accomplished for several yeast data sets (Aach et al. 2000) and may become centralized through projects such as the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/). Despite the above conceptual difficulties, database development for microarrays has progressed rapidly for certain array technologies and computer platforms (for review, see Bowtell 1999 and http://www.gene-chips.com/Datamining, http://www.biologie.ens.fr/en/genetiqu/puces/bddeng.html, http://genome-www4.stanford.edu/MicroArray/SMD/restech.html).
Nevertheless, there is a significant need for widely accessible and versatile microarray data management and analysis tools. Here, we present microarray database and analysis software named Argus, after the mythical Greek watchman with 100 eyes who could keep a lookout in all directions simultaneously. Argus imports intensities, images, and quality control values of microarray spots that have been quantified in an external program. Argus then performs normalization of intensities between arrays, calculates ratios of gene expression between conditions, and generates scatterplots of all expression ratios. A preprocessing step allows implementation of a database structure and search algorithm that optimize complex queries over multiple experiments, resulting in fast response times. The preprocessed database can be used locally, or it can be transferred as a stand-alone unit to a remote Web server for queries over the Internet. Searches for highly regulated genes can be conducted across multiple experiments and multiple arrays, and all results are integrated into a single results page. The results page incorporates images of the actual hybridization spots to allow visual screening for artifacts, and expression ratios are displayed in the context of all other data from that clone or gene for confirmation of reproducibility and analysis across experimental conditions. Scatterplots of experiment pairs can be used for quality control purposes, statistical significance estimation, and direct retrieval of data for specific spots. Query results also can be exported to other software packages for higher-order analyses, such as those described above. Hyperlinks to other databases of biological information allow direct access to the latest gene information. Thus, Argus is a stand-alone application that combines and preprocesses data from multiple sources and produces as its output an interactive Web site used to analyze the microarray data. Argus has been successfully used in our laboratory to analyze and publish microarray data (García-Cardeña et al. 2001, http://vessels.bwh.harvard.edu/papers/PNAS2001).
RESULTS
Database Software Platform
A distinctive characteristic of Argus is that it takes advantage of the built-in database and Web server features of Microsoft Windows products (Microsoft Corporation). Users of Windows operating systems (including Windows ME, NT 4.0 Server and 2000 Server) can use Argus without purchasing any additional software. Although proprietary, these products are commonly available in biological laboratories. In contrast, some microarray analysis/database packages require the purchase of an expensive database management system that often requires specialized maintenance.
A major advantage of using this platform is that the resulting output files can be transferred to a remote (i.e., centrally managed) Microsoft Web server. There, they form a stand-alone database that can be searched locally or over the Web by using a standard Web browser. This format is well-suited for Web publishing microarray data sets. For this use, Argus functions in a client/server mode in which all data and specialized software are kept on the central server. Alternatively, users without access to a central Web server can freely download the Microsoft Personal Web Server and use all Argus features from their personal computers, avoiding the need to send data to remote facilities.
Data Flow
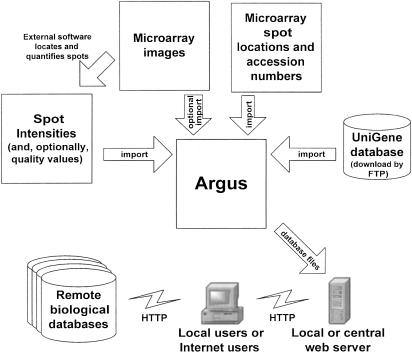
The data flow diagram (Fig. 1) shows the various data sources that Argus integrates into the creation of the final product, a Web site containing all of the data and analysis tools. An external array analysis program processes the original microarray image to locate the spot centers and quantify their intensity. Any array analysis program that can export the spot quantification and accession numbers as a text file can be used. In cases in which two-color arrays are used, Argus can split the data into two one-color files for further processing. Alternatively, Argus can import a ratio for each spot instead of an intensity. (In this case, the intensity cutoff feature would not apply.) With some array quantification programs, it is possible to access supporting information linked to the spot quantification—the raw images of the spots themselves and quality control values describing the confidence in the quantification. This additional information, when available, allows for efficient detection, notation, and filtering of artifacts, as discussed below. In addition, Argus incorporates updated gene identity information from the latest version of the UniGene database (Boguski and Schuler 1995). This allows the user to have the latest, most accurate information about a particular clone in situations in which an unidentified gene finally has been assigned a name, when a particular cDNA clone has been reassigned to a different gene, or when the name of a gene has changed. All data from the database also can be exported to a text file for cluster analysis and other higher-order analyses by other programs.
Figure 1.
Data flow diagram. First, microarray spots are quantified using an external program (arrow, top left). Argus then imports all data necessary to analyze a set of microarray experiments (counterclockwise from right): the latest version of the UniGene database, a description of the location of each clone on the arrays, the actual images of the scanned microarrays, and the intensity and quality value of each spot on the arrays. Argus processes these data and produces a database and a set of supporting files that are transferred (arrow, bottom right) to a local or centrally managed Web server. Users (bottom center), whether local or at a remote location, can access all analysis features of the interactive database using a Web browser and also can conveniently access remote biological databases (bottom left) for additional gene information.
Query Example
Figure 2A shows the database query form, which allows searching for genes using a variety of criteria. The user first chooses the reference (or control) condition. The normalized intensities for this condition will be used for the denominator of every ratio shown in the results. Then, the user selects which experimental condition(s) will be compared to the reference condition to search for highly regulated ratios. The user can specify whether the expression data from all experimental conditions in the database should be displayed in the results or just those conditions that were used to search for regulated ratios. The algorithm can be set to search for genes that were up-regulated, down-regulated, or either up-regulated or down-regulated, by at least a certain ratio. An intensity cutoff also is provided to avoid the relatively noisy quantifications of the faintest spots on the array. Finally, the user has the option of restricting the query to specific accession numbers or to a gene name (or partial gene name). A more complex version of the query page with additional features is also available (see Argus Web site: http://vessels.bwh.harvard.edu/software/Argus).
Figure 2.
A typical query for regulated genes. (A) The search form is configured to retrieve clones that were down-regulated at least twofold when comparing condition B to condition A and that have a minimum intensity of 3000, from any array or experiment in the database. (B) Three clones meet these criteria for at least one of their replicate measurements. If desired, ratios from all conditions can be shown on this page. Clicking on the accession number next to fibronectin 1 retrieves all data from that clone (C), including thumbnail images of the actual hybridization spots. Clicking on the image of a spot produces a scatterplot (D) highlighting that data point, in this case showing that the point is outside the scatter of points around the unity line.
To show a basic query using a Web site produced by Argus, we created a sample data set from four different RNA samples, labeled Condition A through Condition D. (In a real data set, the conditions can be given more informative names, such as “Control cells” and “Cells plus drug”.) Microarrays with labeled cDNA from each condition were quantified using Pathways 3.0 (Research Genetics) and imported into Argus for creation of the analysis Web site (see Methods). A set of three separate nylon arrays was probed for each condition. Figure 2A displays a query for genes that were down-regulated at least twofold when comparing Condition B to Condition A, and whose spot intensities met a relatively stringent cutoff of 3000. Searches across multiple conditions can be performed simply by checking more than one condition box on the query form, and the results page integrates results from every selected condition across all replicate experiments and array sets in the database. In our experience, this feature is not generally available in other analysis packages.
Figure 2B shows the results from executing the above query. Three clones were identified as meeting the criteria across any of the three arrays analyzed. The row(s) that met the search criteria for each clone can be located by looking for (1) a bright blue (down-regulated) block in the Condition B column and (2) a condition A intensity (in the Ref Intensity column) or condition B intensity of at least 3000. The clones are listed in descending order of the maximum regulation observed for the clone in all search conditions and replicates. A ratio of 0.5 corresponds to a twofold down-regulation. In this data set, all of the mRNA samples were derived from a single experiment, so every value in the Experiment column is 1. If replicate experiments are performed, additional values will appear in the Experiment column. The Array column indicates on which of the arrays the data was found, in this example either GF200, GF204, or GF211. Clicking on the array value brings up an extended description of that array in a pop-up window. Multiple rows with the same experiment number and array represent clones that were spotted multiple times on the same array. For the first two clones, the ratio appears to depend on which array the measurement was made. Several technical factors may account for this observation (see Discussion).
By clicking on the accession number for the clone labeled fibronectin 1, a detail window appears that contains further information about this clone and the results from all experiments in the database (Fig. 2C). In each box, the normalized intensity of the spot is displayed below the ratio. Using this display, the value of a particular ratio can be viewed in the context of ratios from all other experiments. In this case, relative to Condition A, fibronectin 1 was down-regulated in Condition B but was up-regulated in Conditions C and D. Using the drop-down menu, searches on the Web can be initiated quickly to obtain more information about the particular clone displayed. For example, the user can directly perform a BLASTN search to verify that UniGene correctly identified the gene from which the cloned cDNA fragment is derived. Information about the gene's function in the OMIM database (McKusick 2000) can be accessed through the results page of the Search UniGene link.
The clone-oriented view in Figure 2C shows a particular ratio in the context of the clone's ratios in all other conditions and experiments. A complementary array-oriented perspective would show the ratio of a particular clone compared to the ratios of all other clones on the same array. This view is obtained by inspecting an intensity scatterplot in which each point represents a spot on an array. The x value of a point is its normalized intensity in one condition, and the y value is its normalized intensity in the other condition. Clicking on the image of any spot in Figure 2C brings up such a scatterplot. For example, clicking on the image next to 0.473 brings up a scatterplot of spots found on the GF200 array in Conditions A and B (Fig. 2D). The point corresponding to the 0.473 ratio is highlighted by a blue cross. This point falls outside the scatter of points near the unity line, making it more likely that this observed ratio is not due to noise.
The user also can use the scatterplots as a starting point for an analysis, as they can be an effective form of quality control. Scatterplots with most points clustered near the unity line generally indicate an experiment of low biological and analytical noise. Generally, if the scatter of points is severely curved it may indicate a problem with the hybridization, whereas if many points are far from the unity line, then there is a large amount of analytical noise or the transcriptional profiles of the two biological conditions are very different. The user also can click on outlying spots in the scatterplot to directly display results for that clone.
The ratios on the results page are grouped by clone, but in theory other cDNA fragments from the same gene should show similar results. This gene-oriented view is obtained by clicking on the UniGene Cluster name from the details page of any clone. Another search is executed that displays all of the clones on the arrays that are pieces of the same gene, according to the UniGene database. (If the UniGene Cluster name on the details page is not clickable, there are no additional clones available from the same gene.) For example, Figure 3 shows a gene name query in which fibroblast growth factor receptor 3 was similarly regulated in two nonidentical clones found on various arrays, increasing confidence in the measurements.
Figure 3.
Display of multiple clones from the same gene. From the details page of any clone, a new search can be initiated that displays all clones with the same gene name. In both the top and bottom clones, fibroblast growth factor receptor 3 was not regulated in condition B but was down-regulated in conditions C and D. The top clone was nearly five times as intense as the bottom clone, as seen in the Ref Intensity column, yet the intensity ratios between conditions are very similar.
Benchmarks
To provide an estimate of the computing power and memory required to run Argus on data sets containing multiple arrays and experiments, we measured the response time and memory usage for queries of highly regulated genes. The database files and Web browser for analysis were both on the same computer. On a PC running Windows NT 4.0 Server and Internet Explorer 5.0 with a 550-MHz Pentium III, a 7400-rpm IDE drive, and 756 MB of RAM, a simple query (default search settings, searching one condition) for regulated genes across the sample data set of 59,344 spots required <2 sec of processing time, and a complex query (clicking “relax all restrictions”) required <3 sec of processing time. (As expected for a Web site, the first search of a session takes several additional sec while files are cached into memory.) Loading Internet Explorer and performing several complicated searches decreased the amount of free RAM by <30 MB. When the same queries were requested from a remote machine, the decrease in free RAM was 21.8 MB.
When the size of the database was increased 89% to 112,370 spots, memory usage increased only 2.7% to 22.4 MB. A simple query on the larger database required <2 sec, and a complex query required <4 sec. These modest increases in time and memory with a near doubling of database size are a result of the preprocessing algorithm and suggest that handling even larger data sets may be practical. We anticipate that Argus, as written, will be useful for data sets containing tens but not hundreds of arrays. Note that when cross-comparison between sets of arrays is not needed (i.e., sets from conceptually different biological experiments), an unlimited number of data sets can be processed separately, and all can be installed on the same Web server for concurrent access.
To achieve these fast response times, preprocessing is required once for each new data set. Argus processed the complete human UniGene database in 5.5 min. Three hours were required to preprocess the 112,370 data point data set described above. Maximum RAM usage of the program was 100 MB. This extended processing time is composed of several computationally demanding tasks, including creating thumbnail images of each spot from a large image and creating the query lookup tables that allow fast query execution times.
DISCUSSION
Collection and analysis of microarray data, from raw image files to biological interpretation, is a long and complex procedure, and various software packages emphasize different stages in this process. Some microarray databases are designed to be archives of very large amounts of gene expression data (Gene Expression Omnibus), whereas others focus on integrating large amounts of data from various sources (ArrayDB). Argus is designed to automate and standardize common steps in the routine processing and interpretation of microarray data. We envision the typical Argus user to be a researcher who needs to efficiently import a batch of microarray data, use scatterplots to perform quality control on the experiments, exclude artifacts, produce lists of regulated genes, and investigate these genes over the Web, and make these results available to other researchers.
Data Reproducibility and Statistics
The current version of the Argus query page asks users to specify fixed ratio and intensity cutoffs to find significantly regulated genes. An estimate of the statistical significance of a particular ratio can be obtained by looking at the location of the point on a scatterplot of all the intensities from the array pair from which the ratio was derived. An isolated point that is far beyond the scatter of points around the unity line is less likely to be due to noise, whether biological or analytical.
Ideally, a statistical model would be more useful than using fixed ratio and intensity cutoffs combined with significance estimation using scatterplots. A user would specify what false-positive rate is acceptable for the intended downstream application of the results, and the program would return a set of results expected to meet the specified false-positive rate. The ratios would be presented with confidence intervals. Much of the early work in the microarray field was performed without replicates, which makes it extremely difficult to predict false-positive rates or to estimate confidence intervals for any particular intensity or ratio. Methods for approximating confidence intervals appropriate for small numbers of replicates and for specifying false-positive rates have been developed only recently (Claverie 1999; Manduchi et al. 2000; Kadota et al. 2001) or are in development (Dudoit et al. 2000; Newton et al. 2000).
One complexity of using such statistical techniques is that the proper technique is likely to be highly dependent on the arrays themselves and on the particular source of the biological sample. For example, duplicate spots on arrays from one manufacturer could be so reproducible that it would be optimal to average their intensities and compute a confidence interval. An array from a different manufacturer, however, could contain occasional blank spots due to failed PCR reactions or inaccuracies in spotting a fixed amount of DNA, and calculating a mean and confidence interval of such replicate spots would not be ideal. Important sources of variance are likely to be different between technologies, or even between laboratories or biological experiments. One study has determined that expression data with higher variance at low intensities requires a “shift” technique to produce optimal results, although if the variance does not follow this pattern the shift is not needed (Grant et al. 2000). Different sensitivity and cross-hybridization characteristics have been found for certain implementations of various microarray technologies (Richmond et al. 1999). Our experience with nylon microarrays (GeneFilters; Research Genetics) is that certain clones are detected consistently at higher levels than others and that certain experiments have greater sensitivity than others (G. García-Cardeña and J. Comander, unpubl.). The causes of these patterns are diverse and ultimately comprehensible, but in any case averaging reliable and unreliable readings (as performed in other analysis packages) is unlikely to produce an optimal result. In essence, the sources and distribution of variance in a given data set, on which all statistical techniques should be based, must be characterized before an optimal statistical model can be used.
Because such a characterization has not been performed yet on all common sources of microarray data, Argus is intentionally written so that only a single measurement has to meet the search criteria for a clone to be returned in the results page. The user should verify that those results were acceptably replicated in the rest of the data set. The Argus details page conveniently provides all data, replicate and otherwise, for a particular clone and reference condition. Data from all clones belonging to a certain gene also can be presented on a single page, as described above. As the statistical models are fine-tuned for particular combinations of biological experiments and implementations of array technologies, we plan to incorporate those models into Argus.
Future Directions
Argus creates hyperlinks to various Internet databases of biological information, providing an easy way for the user to efficiently investigate a clone or gene of interest. We encourage publishers of such databases to contact us for hyperlinks to be included in future Argus versions. We also would like to incorporate more clone and gene information into the Argus database itself, such as additional means of assigning gene names, providing consensus sequences for expressed sequence tag clusters, and assigning systematic functional categories to all genes. Such integration would allow, for example, automatic testing of regulated genes for enrichment by certain functional categories. We also envision a tighter integration between Argus and higher-order analyses, such as clustering tools, classification algorithms, and analysis of upstream promoter regions of regulated genes.
Argus is freely available to academic institutions at http://vessels.bwh.harvard.edu/software/Argus.
METHODS
Argus was written in Microsoft Visual Basic 6.0, and the Web site it produces uses VBScript and Javascript.
For the sample data set, 33P-labeled cDNA from four conditions was hybridized to a series of three nylon GeneFilter arrays (Research Genetics). Microarray spots were located and quantified using Pathways 3.0 (Research Genetics). The UniGene database was downloaded from National Center for Biotechnology Information at ftp://ftp.ncbi.nlm.nih.gov/repository/UniGene. GeneFilter spot identification information was downloaded from ftp://ftp.resgen.com/pub/genefilters. Each three-array set used in the sample data set contained a total of 15,244 spots to quantify and contained cDNA clones from 13,325 unique accession numbers. Excluding spots with no accession numbers, the entire data set of 12 arrays contains 59,344 data points, and the same number of spot images. Gene names of the 13,325 cDNA fragments were obtained from the UniGene database, downloaded June 20, 2000. Clones that showed obvious artifacts (such as miscentered spots or contamination from a bright neighboring spot) were removed by setting their quality values to zero.
Search Algorithm
The large size of microarray databases creates a massive computational problem. Without optimization, the computational time required for direct queries on sample data sets is often several hours, too slow by at least four orders of magnitude for Argus to be a useful tool. To improve the performance of the Web site, we stored the data files in an efficient relational database system.
Argus also creates lookup tables to further improve performance of queries for regulated genes. Briefly, the lookup tables contain several precalculated intermediate values, including the maximum intensity and ratio for every pair of spots in the database with the same array location and experiment number. Because these values are required for each Web query, performing the calculations once and storing them in the lookup tables greatly improves performance. The computation time to produce the lookup tables will increase with the number of clones on the arrays, the number of experiments in the database, and the number of conditions in each experiment. (See Argus Web site for more detail: http://vessels.bwh.harvard.edu/software/Argus).
When a user submits a query, the lookup tables are used to rapidly identify and sort accession numbers of all spot pairs meeting the search criteria. These accession numbers are merged with other database tables that contain all of the related information that is presented in the results page (see Fig. 4). This strategy produces results quickly enough to allow real-time, interactive searches (see Benchmarks above).
Figure 4.
Query processing using lookup tables. When a user submits a query from a Web browser, precalculated lookup tables are used to identify a list of accession numbers that match the search criteria. These accession numbers are sorted and merged with the additional information shown in italics, and the results are displayed in a Web browser window.
Acknowledgments
We thank Chaz Yoon for programming assistance, Lucila Ohno-Machado for helpful discussions and comments on the manuscript, and John Aach, Jeanne Kiely, and Yvonne Ou for critical reading of manuscript. This study was supported by grants from the National Heart Lung and Blood Institute, NIH (P50-HL56985; R37-HL511509). J.C. also was supported by NRSA grants T32-GM07753 and T32-HLI07623. G.M.W. also was supported by NLM Grants R01LM06538-01 and T15LM07092-07.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ggarcia-cardena@rics.bwh.harvard.edu; FAX (617) 732-5933.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.186601.
REFERENCES
- Aach J, Rindone W, Church GM. Systematic management and analysis of yeast gene expression data. Genome Res. 2000;10:431–445. doi: 10.1101/gr.10.4.431. [DOI] [PubMed] [Google Scholar]
- Aach J, Bulyk ML, Church GM, Comander J, Derti A, Shendure J. Computational comparison of two draft sequences of the human genome. Nature. 2001;409:856–859. doi: 10.1038/35057055. [DOI] [PubMed] [Google Scholar]
- Alter O, Brown PO, Botstein D. Singular value decomposition for genome-wide expression data processing and modeling. Proc Natl Acad Sci. 2000;97:10101–10106. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguski MS, Schuler GD. ESTablishing a human transcript map. Nat Genet. 1995;10:369–371. doi: 10.1038/ng0895-369. [DOI] [PubMed] [Google Scholar]
- Bowtell DD. Options available—from start to finish—for obtaining expression data by microarray. Nat Genet. 1999;21:25–32. doi: 10.1038/4455. [DOI] [PubMed] [Google Scholar]
- Claverie JM. Computational methods for the identification of differential and coordinated gene expression. Hum Mol Genet. 1999;8:1821–1832. doi: 10.1093/hmg/8.10.1821. [DOI] [PubMed] [Google Scholar]
- Dudoit S, Yang YH, Callow M, Speed T. Technical report 578: Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Stanford, CA: Standford University School of Medicine; 2000. [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolaeva O, Rastogi M, Pruitt KD, Schuler GD, Bittner ML, Chen Y, Simon R, Meltzer P, Trent JM, Boguski MS. Data management and analysis for gene expression arrays. Nat Genet. 1998;20:19–23. doi: 10.1038/1670. [DOI] [PubMed] [Google Scholar]
- Furey TS, Cristianini N, Duffy N, Bednarski DW, Schummer M, Haussler D. Support vector machine classification and validation of cancer tissue samples using microarray expression data. Bioinformatics. 2000;16:906–914. doi: 10.1093/bioinformatics/16.10.906. [DOI] [PubMed] [Google Scholar]
- García-Cardeña G, Comander J, Anderson KR, Blackman BR, Gimbrone MA., Jr Biomechanical activation of vascular endothelium as a determinant of its functional phenotype. Proc Natl Acad Sci. 2001;98:4478–4485. doi: 10.1073/pnas.071052598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Grant GR, Manduchi E, Stoeckert CJ., Jr . Critical Assessment of Techniques for Microarray Data Analysis (CAMDA ‘00) Proceedings. 2000. Using non-parametric methods in the context of multiple testing to determine differentially expressed genes.http://www.cbil.upenn.edu/PaGE/camda.pdf http://www.cbil.upenn.edu/PaGE/camda.pdf. [Google Scholar]
- Holter NS, Mitra M, Maritan A, Cieplak M, Banavar JR, Fedoroff NV. Fundamental patterns underlying gene expression profiles: Simplicity from complexity. Proc Natl Acad Sci. 2000;97:8409–8414. doi: 10.1073/pnas.150242097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JD, Estep PW, Tavazoie S, Church GM. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J Mol Biol. 2000;296:1205–1214. doi: 10.1006/jmbi.2000.3519. [DOI] [PubMed] [Google Scholar]
- Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JC, Trent JM, Staudt LM, Hudson J, Boguski MS, et al. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- Kadota K, Miki R, Bono H, Shimizu K, Okazaki Y, Hayashizaki Y. Preprocessing implementation for microarray (PRIM): An efficient method for processing cDNA microarray data. Physiol Genomics. 2001;4:183–188. doi: 10.1152/physiolgenomics.2001.4.3.183. [DOI] [PubMed] [Google Scholar]
- Livesey FJ, Furukawa T, Steffen MA, Church GM, Cepko CL. Microarray analysis of the transcriptional network controlled by the photoreceptor homeobox gene Crx. Curr Biol. 2000;10:301–310. doi: 10.1016/s0960-9822(00)00379-1. [DOI] [PubMed] [Google Scholar]
- Manduchi E, Grant GR, McKenzie SE, Overton GC, Surrey S, Stoeckert CJ., Jr Generation of patterns from gene expression data by assigning confidence to differentially expressed genes. Bioinformatics. 2000;16:685–698. doi: 10.1093/bioinformatics/16.8.685. [DOI] [PubMed] [Google Scholar]
- McKusick, V.A. 2000. Online Mendelian Inheritance in Man, OMIM (TM). McKusick-Nathans Institute for Genetic Medicine, Johns Hopkins University (Baltimore, MD) and National Center for Biotechnology Information, National Library of Medicine (Bethesda, MD). http://www.ncbi.nlm.nih.gov/omim/
- Newton MA, Kendziorski CM, Richmond CS, Blattner FR, Tsui KW. On differential variability of expression ratios: Improving statistical inference about gene expression changes from microarray data. J Computat Biol. 2001;8:37–52. doi: 10.1089/106652701300099074. [DOI] [PubMed] [Google Scholar]
- Richmond CS, Glasner JD, Mau R, Jin H, Blattner FR. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 1999;27:3821–3835. doi: 10.1093/nar/27.19.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth FP, Hughes JD, Estep PW, Church GM. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat Biotechnol. 1998;16:939–945. doi: 10.1038/nbt1098-939. [DOI] [PubMed] [Google Scholar]
- Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR. Interpreting patterns of gene expression with self-organizing maps: Methods and application to hematopoietic differentiation. Proc Natl Acad Sci. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM. Systematic determination of genetic network architecture. Nat Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]