Abstract
The rice prolamins consist of cysteine-rich 10 kDa (CysR10), 14 kDa (CysR14) and 16 kDa (CysR16) molecular species and a cysteine-poor 13 kDa (CysP13) polypeptide. These storage proteins form protein bodies (PBs) composed of single spherical intracisternal inclusions assembled within the lumen of the rough endoplasmic reticulum. Immunofluorescence and immunoelectron microscopy demonstrated that CysR10 and CysP13 were asymmetrically distributed within the PBs, with the former concentrated at the electron-dense center core region and the latter distributed mainly to the electron-lucent peripheral region. These results together with temporal expression data showed that the formation of prolamin-containing PB-I in the wild-type endosperm was initiated by the accumulation of CysR10 to form the center core. In mutants deficient for cysteine-rich prolamins, the typical PB-I structures containing the electron-dense center core were not observed, and instead were replaced by irregularly shaped, electron-lucent, hypertrophied PBs. Similar, deformed PBs were observed in a CysR10 RNA interference plant line. These results suggest that CysR10, through its formation of the central core and its possible interaction with other cysteine-rich prolamins, is required for tight packaging of the proteins into a compact spherical structure.
Keywords: Cysteine-rich prolamin, Endoplasmic reticulum, Mutant, Protein body, Rice, RNAi
Introduction
The cereal prolamins are broadly classified into cysteine (Cys)-poor and Cys-rich species (Shewry and Tatham 1999). In wheat and barley, most prolamins are Cys-rich species (gliadins and glutenins in wheat and hordeins in barley) and are deposited in protein storage vacuoles (PSVs) in the form of very large aggregates (Cameron-Mills and von Wettstein 1980, Levanony et al. 1992). In contrast, maize endosperm contains relatively large amounts of Cys-poor prolamins, α-zeins, which form single spherical inclusion granules surrounded by the endoplasmic reticulum (ER) membrane (Lending and Larkins 1989). These two major cereals exemplify the two modes of intracellular deposition of seed storage proteins.
The rice prolamins account for about 18–20% of the total seed protein content (Ogawa et al. 1987, Li and Okita 1993). They consist of 60% Cys-rich prolamins and 40% Cys-poor prolamins (Ogawa et al. 1987, Kim and Okita 1993). The prolamin fraction of the japonica rice variety Kinmaze consists of 10, 13 (indicated as 13b in Ogawa et al., 1987), 14 (indicated as 13a in Ogawa et al., 1987) and 16 kDa molecular species. Ogawa et al. (1987) demonstrated that the 10, 14 and 16 kDa prolamins are Cys-rich species, while the 13 kDa prolamin is a Cys-poor species.
Based on the primary sequences derived from cDNA sequences, the four prolamins are encoded by three distinct classes of genes (Kim and Okita 1988a, Kim and Okita 1988b, Masumura et al. 1989, Masumura et al. 1990, Shyur and Chen 1990, Shyur et al. 1992, Shyur and Chen 1993, Shyur et al. 1994). The Cys-poor 13 kDa (CysP13) and Cys-rich 14 kDa (CysR14) and 16 kDa prolamins (CysR16) share considerable homology (∼70%) and differ only in that the former species lack cysteine residues. The 10 kDa prolamins (CysR10) share minimal sequence homology with the other two classes and are characterized by their high content of methionine (20%) and cysteine (10%) residues (Masumura et al. 1989). Both Cys-rich prolamin classes contain the three A, B and C cysteine motifs which are typically observed in cereal Cys-rich prolamins (Shewry et al. 1995).
Two types of protein bodies (PBs), called PB-I and PB-II, are observed in rice (Bechtel and Juliano 1980, Tanaka et al. 1980). Prolamins are accumulated in PB-Is as intracisternal protein granules, while glutelins are accumulated in PB-IIs derived from the PSV (Tanaka et al. 1980, Ogawa et al. 1987). PB-I is spherical with a diameter of 1–2 μm and surrounded by rough ER membranes with attached polysomes (Bechtel and Juliano 1980, Tanaka et al. 1980, Muench and Okita 1997, Muench et al. 1999). When viewed by electron microscopy, the structure of PB-I consists of an electron-dense center core surrounded by electron-lucent layers, which are interspersed with concentric rings of varying electron density (Bechtel and Juliano 1980, Tanaka et al. 1980, Krishnan et al. 1986, Ogawa et al. 1987). Similar PB structures are also observed in Sorghum (Shull et al. 1992) and Setaria (Rost 1972). It is not known how the electron-dense core structure is formed and how prolamin polypeptides assemble to form a tightly compact, spherical intracisternal inclusion granule within the ER.
As first observed for the maize zeins, the rice prolamins are synthesized on rough ER membranes and are co-translationally translocated into the ER lumen (Yamagata and Tanaka 1986). In maize, the various zein classes are not randomly distributed within the PBs; the Cys-rich β-zeins and γ-zeins are localized to the PB periphery, which surrounds the centrally located Cys-poor α-zeins and Cys-rich δ-zeins (Lending and Larkins 1989, Esen and Stetler 1992). PB formation is initiated by the accumulation of Cys-rich γ-zeins and β-zeins to give a small electron-dense granule, whereupon accumulation of Cys-poor α-zeins displaces the β- and γ-zeins from the center to the periphery (Lending and Larkins 1989). These cytochemical results suggest that Cys-rich β- and γ-zeins play an important role for initiation of PB formation and the sequestration of α-zeins within the PBs in maize endosperm.
Kumamaru et al. (1987, 1988) characterized rice mutants for storage protein and isolated three prolamin mutant classes. The varied prolamin polypeptide composition was reflected in the morphology of their prolamin PBs (Ogawa et al. 1989). Endosperm storage protein mutants esp1 and Esp4 are characterized by low levels of CysP13, with the latter also containing elevated levels of Cys-rich prolamins. On the other hands, the esp3 mutant contains low levels of CysR10, CysR14 and CysR16 (Kumamaru et al. 1987, Kumamaru et al. 1988, Ogawa et al. 1989).
In order to identify the processes responsible for PB-I formation, we evaluated the temporal accumulation of CysR10 and CysP13 during rice seed development and their spatial distributions within the PB using immunocytochemical and immunofluorescence analysis of the wild type, genetic mutants for prolamin accumulation and CysR10 RNA interference (RNAi) lines. Our results indicate that these prolamin species are asymmetrically distributed within PB-I and that CysR10 is essential for formation of the tightly compact, spherical shaped structure of PB-I.
Results
The distribution of CysR10 and CysP13 in PB-I
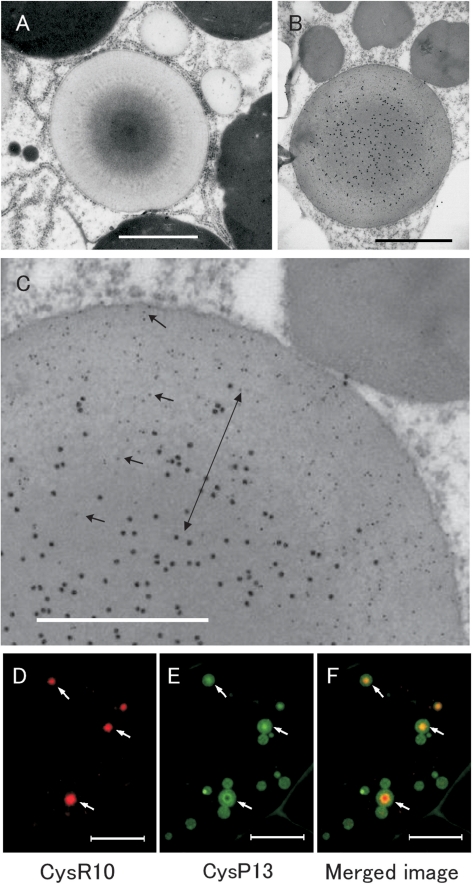
PB-I exhibits a non-uniform pattern of electron density when stained with lead acetate and uranyl acetate (Tanaka et al. 1980). The intracisternal inclusion granule contains a dark staining center core surrounded by one or more layers of increasing lighter density extending from the core to the periphery (Fig. 1A, C). In many instances the electron-lucent peripheral region is interspersed with non-continuous, electron-dense concentric rings.
Fig. 1.
Distribution of CysR10 and CysP13 in PB-Is of developing rice endosperm at 3 WAF. (A) Electron micrographs of PB-Is. (B, C) Immunoelectron microscopy of PB-Is which were labelled with anti-CysR10 antibody (15 nm gold particles) and anti-CysP13 antibody (5 nm gold particles). (C) Enlarged image of (B). Arrows in C show the 5 nm gold particles (CysP13). The double arrow in C denotes the middle layer containing CysR10 and CysP13. Bars in (A, B): 1 μm, bars in (C): 0.5 μm. (D–F) Immunofluorescence microscopy of PB-Is from developing rice endosperm at 3 WAF. (D) The distribution of CysR10 as visualized using anti-CysR10 and rhodamine-conjugated secondary antibodies. (E) The localization of CysP13 using anti-CysP13 and FITC-conjugated secondary antibodies. (F) Merged image of D and E. Bars in (D–F): 10 μm.
To investigate the spatial relationship of prolamin polypeptides within PB-I, immunoelectron microscopy was carried out using thin sections taken from developing rice endosperm at 3 weeks after flowering (WAF). Fig. 1B and C show the distribution of CysR10 labeled with secondary antibodies conjugated with 15 nm gold particles and CysP13 visualized with 5 nm gold particles. The CysR10 was concentrated at the interior while the CysP13 was distributed to the outer electron-lucent peripheral layer (Fig. 1B, C). Additionally, a middle layer containing both CysR10 and the CysP13 was evident (Fig. 1C, double arrow).
The spatial distribution of these prolamins within the PBs was confirmed by immunofluorescence light microscopy. CysR10 was visualized as small particles when labeled by anti-CysR10 followed by rhodamine-conjugated secondary antibodies (Fig. 1D). The distribution of CysR10 co-localized with CysP13, which was visualized with fluorescein isothiocyanate (FITC)-conjugated secondary antibodies (Fig. 1E, F arrows). CysR10 was observed at the center of the PBs, whereas the distribution of CysP13 was more varied. In some PBs, CysP13 appeared to be distributed uniformly throughout the PB, while in others, CysP13 was found either at lower levels or was entirely absent from the center core region (Fig. 1E, F).
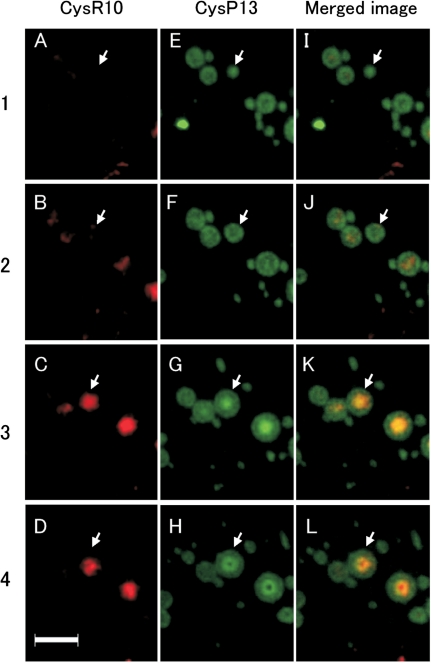
To examine the spatial relationship between the prolamins in more detail, serial thin sections from developing endosperm at 3 WAF were obtained and analyzed by immunofluorescence microscopy. In Fig. 2 image 1, a PB sectioned near its periphery is labeled with the CysP13 antibody but not by the CysR10 antibody (arrow). The succeeding series of images 2–4 show cross-sections of this same PB taken from the periphery towards the center. In image 2, the CysP13 was distributed mainly around the periphery with the center of the PB showing a trace of the CysR10. The next image (Fig. 2 image 3), which depicts a section near the center of the PB, shows a non-uniformed distribution of CysP13 with lower levels at the center region, which coincided with the location of CysR10, and higher amounts at the peripheral areas. Image 4 shows a section at the center of the PB where the center contains an ‘apparent cavity’ when viewed for CysP13 but an area filled with CysR10. These observations are consistent with the distribution of these prolamin polypeptides in PB-I as viewed by immunocytochemistry at the electron microscope level. PB-I is composed of a center core of CysR10, surrounded by a middle layer of a mixture of CysR10 and CysP13, and then followed by a peripheral layer containing CysP13 devoid of CysR10.
Fig. 2.
Immunofluorescence microscopy of serial sections of PB-Is obtained from developing rice endosperm at 3 WAF. The numbers at the left of the image denote the order of the sections. The PBs in each section were labeled with antibodies against CysR10 and CysP13. The CysR10–antibody interactions were visualized with rhodamine-conjugated secondary antibodies (A–D), while those for CysP13s were visualized with FITC-conjugated secondary antibodies (E–H). (I–L) Merged images. The arrow indicates the location of the same PB in the serial sections. Bars: 4 μm.
Developmental changes in the prolamin composition of the interior PB-I
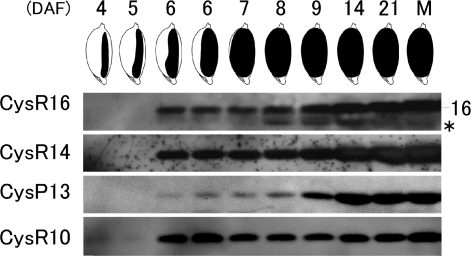
The temporal accumulation patterns of the various prolamin classes were examined by SDS–PAGE followed by immunoblot analysis. The accumulation of the CysR10 polypeptide was first detected at 4 days after flowering (DAF) and attained maximum steady-state levels (on a seed mg basis) at 6 DAF (Fig. 3, Supplementary Fig. S1). Significant levels of CysR14 and CysR16 were initially detected at 6 DAF and steadily increased as the seed developed (Fig. 3, Supplementary Fig. S1). Only small amounts of CysP13 were detected in developing seed at 6–8 DAF although their levels increased prominently from 9 to 14 DAF. These results show that the rice prolamins are differentially accumulated during seed development. CysR10 was initially detected as early as 4 DAF, followed closely by the accumulation of CysR14 and CysR16 polypeptides. In turn, CysP13 was the last prolamin to be accumulated during seed development. The results of shorter and longer exposures of the membranes in all the immunoblotting analyses (Supplementary Fig. S1) were essentially the same as those shown in Fig. 3.
Fig. 3.
Immunoblot analysis of the four prolamin species during rice seed development. DAF are shown in the top of the figure, and the illustrations depict the shape of the developing seeds at the corresponding DAF. Asterisk shows a non-specific band in CysR16 prolamin. M, mature seed.
The steady-state RNA levels for CysR10 and CysR16 as assessed by reverse transcription–PCR (RT–PCR) (Supplementary Fig. S2) are consistent with their respective polypeptide levels during seed development. CysR10 RNAs were readily detected at 2 DAF and attain their maximum levels at 3 DAF, while significant levels of CysR16 were evident at 4 DAF and reach their highest levels at 8 DAF. Interestingly, steady-state RNA levels for CysR14 and CysP13 were very similar during early seed development, a pattern which contrasts with the differential temporal accumulation patterns of their polypeptides (Fig. 3). As CysR14 was readily observed at 6 DAF whereas a significantly high level of CysP13 was only detected 3 days later (Fig. 3), CysR14 RNAs appear to be preferentially translated compared with CysP13 transcripts during early seed development.
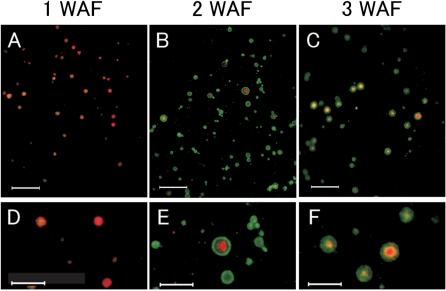
To examine further the development and maturation of PB-I formation, double indirect immunofluorescence studies using anti-CysR10 and anti-CysP13 antibodies were carried out using developing seeds harvested at 1, 2 and 3 WAF. In developing seeds at 1 WAF, PB-Is were observed as small reddish particles with a diameter of 0.5–2.1 μm containing mainly CysR10 (Fig. 4A). Small amounts of CysP13 were also seen as small locules embedded in the matrix of larger PBs composed of CysR10 (Fig. 4D). As the developing seed matures (2 WAF), CysP13 was detected in all of the PBs (Fig. 4B), while CysR10 was detected at the center in some PBs with a diameter of 0.7–3.0 μm (Fig. 4B, E). Interestingly in the very large PB depicted in Fig. 4E, CysP13 was not uniformly distributed in the region surrounding the CysR10 central core. Instead a narrow layer devoid of CysR10 and CysP13 was evident. The basis for this layer is probably due to the presence of other prolamins (see Discussion). At 3 WAF, PBs with a diameter of 1.0–3.4 μm contained the CysR10 core (reddish center) surrounded by a mixture of CysR10 and CysP13 (yellowish layer) which, in turn, was surrounded by CysP13 (greenish layer) (Fig. 4C, F).
Fig. 4.
Double indirect immunofluorescence microscopy of PB-Is in rice endosperm during seed development. Endosperm sections were labeled with the antibodies against CysR10 and CysP13 and visualized by rhodamine- and FITC-conjugated antibodies, respectively. (D–F) Enlarged images of (A–C), respectively. Bars in (A–C): 10 μm. Bars in (D–F): 5 μm.
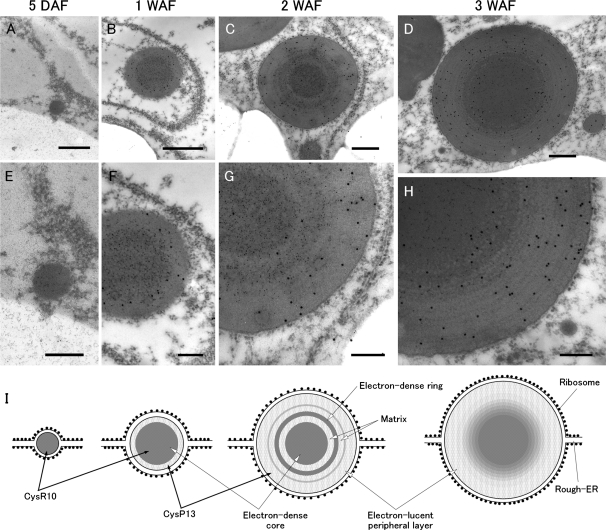
To obtain a more detailed structure of PB-I, we analyzed its ultrastructure during different stages of seed development by immunoelectron microscopy (Fig. 5). In developing seeds at 5 DAF (see seed illustration in Fig. 3), PB-I was seen as a small particle with an average diameter of about 0.3 μm with attached polysomes (Fig. 5A). These small particles were readily labeled with antibodies to CysR10 but not with those to CysP13 (Fig. 5A, E). At 1 WAF, the PB was spherical with an average diameter of about 0.9 μm (Fig. 5B). CysR10 was located in the electron-dense center region which accounted for the bulk of the PB, with a small amount of the CysP13 localized to the peripheral region (Fig. 5B, F). These results are consistent with those observed by immunofluorescence microscopy (Fig. 3). At 2 WAF, the observed spherical PB was about 1.3 μm in average diameter (Fig. 5C). Several electron-dense ring structures, which surrounded the electron-dense core structure, were observed (Fig. 5C). These ring structures reacted with the CysR10 antibody, indicating that CysR10 is one of the polypeptides that comprise this structure (Fig. 5G). CysP13 was evident in the electron-lucent peripheral layer and the electron-lucent matrix between the electron-dense ring and core structures in PB-I (Fig. 5G). At 3 WAF, the PBs, which increased to an average size of about 1.5 μm in diameter, were devoid of the middle ring structures observed in PBs at 2 WAF (Fig. 5D). The middle concentric electron-dense matrix appeared to merge with the center region, with its outer layer gradually being more electron lucent towards the periphery (Fig. 5H). The distribution of CysR10 gradually decreased in this middle region, while that for CysP13 increased toward the periphery (Fig. 5H).
Fig. 5.
Immunoelectron microscopy of PB-I structure during rice endosperm development. PB-I was labeled with antibodies against CysR10 (5 nm gold particles) and CysP13 antibodies (15 nm gold particles). (E–H) Enlarged images of (A–D), respectively. (I) Schematic images of A–D. Bars in (A–D): 400 nm. Bars in (E–H): 200 nm.
Changes in the structure of PB-I in rice mutants
Various seed mutants which show altered levels of rice prolamins were previously identified and studied (Kumamaru et al. 1987, Kumamaru et al. 1988, Ogawa et al. 1989). Mature esp3 seeds contain low amounts of CysR10 as well as reduced levels of CysR14 and CysR16. The esp1 mutant showed low levels of CysP13 while the Esp4 mutant showed low levels of CysP13 as well as high levels of Cys-rich prolamins (Ogawa et al. 1989). To determine how these changes in the rice prolamin levels affect PB formation, the distribution of CysR10 and CysP13 in the PBs from these rice prolamin mutant endosperms was analyzed.
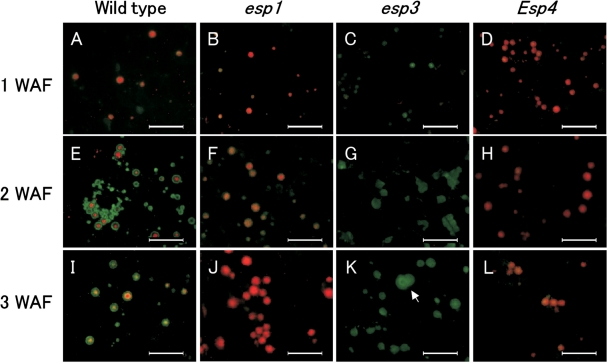
Immunofluorescence microscopy of esp1 at 1 WAF indicated that there were many small PBs up to 1.8 μm in diameter containing CysR10 (red) and very little, if any, CysP13 (green) (Fig. 6B). Similarly, Esp4 endosperm at 1 WAF contained many small PBs up to 1.5 μm in diameter containing CysR10 and very little, if any, CysP13 (Fig. 6D). The structure of PBs in both mutants was similar to that of the wild-type PB at 1 WAF (Fig. 6A). At 2 WAF, PBs of esp1 and Esp4 were labeled almost entirely with the CysR10 antibody and had an average size similar to that of the PBs in wild-type endosperm (Fig. 6E, F, H). The peripheral layer containing CysP13 typically observed in wild-type PBs was not observed in Esp4 (Fig. 6H). At 3 WAF, the PBs of esp1 increased in size, with an average size of about 2.6 μm (Fig. 6J), and the PBs of Esp4 increased only slightly in size, with an average size of about 1.6 μm (Fig. 6L). While the level of CysP13 was lower in Esp4 endosperm, this was compensated by increases in levels of all of the Cys-rich prolamins (Kumamaru et al. 1988, Ogawa et al. 1989). This result, together with the cytological findings reported here, suggests that the reduction in size of Esp4 PB-Is is due to the tighter packaging of Cys-rich prolamins and loss of CysP13, which accumulates in the peripheral region of PBs in the wild type.
Fig. 6.
The structure of PB-I in the wild type, esp1, esp3 and Esp4 as viewed by double indirect immunofluorescence microscopy. Endosperm sections from the wild type and the three mutant lines were labeled with antibodies against CysR10 and CysP13, followed by labeling with secondary antibodies conjugated with rhodamine and FITC, respectively. Note that the small pinpoint red dots in (A–E) are non-specific background, which is observed occasionally in immunofluorescence microscopy. Bars: 10 μm.
Immunofluorescence microscopy of esp3 showed that only PBs labeled by the CysP13 antibody were observed at all stages of seed development (Fig. 6). At 1 WAF, PBs labeled by the CysP13 antibody were about 1.6 μm in diameter (Fig. 6C), similar in size to those from the wild type (Figs. 6A, 4D). At 2 WAF, the PBs, which were wholly recognized by anti-CysP13 antibody, were significantly larger (about 1.8–2.8 μm in diameter) than those from the wild type (Figs. 6G, 4B). At 3 WAF, larger PBs about 2.8–4.2 μm in diameter were observed (Fig. 6K). Though individual PBs were spherical in shape, many were clustered and appeared to be fused together (Fig. 6K, arrow).
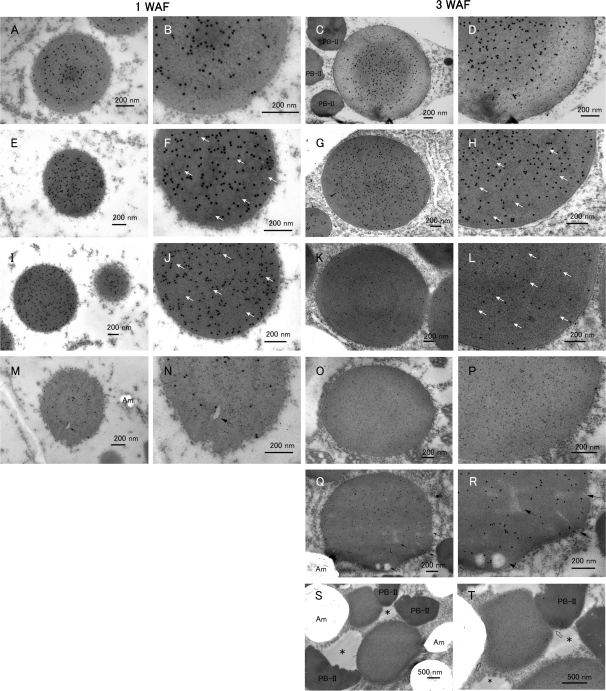
To analyze the PB structures in these mutants in more detail, immunoelectron microscopy was performed. At 1 WAF, the esp1 and Esp4 PBs exhibited a homogeneous electron density, with CysR10 (15 nm gold particles) distributed nearly uniformly over the whole PB (Fig. 7F, J). A few gold particles (5 nm) denoting the presence of CysP13 polypeptide were evident in several PBs (Fig. 7F, J, arrows). At 3 WAF, the localization of both prolamins was randomly distributed within clearly spherical PB-Is, and the size was slightly larger than at 1 WAF (Fig. 7H, L).
Fig. 7.
The structure of PB-I in the wild type, esp1, esp3 and Esp4 as viewed by immunocytochemistry at the electron microscopy level. Endosperm sections, except for (K, L and Q, R), were incubated with anti-CysR10 and anti-CysP13 antibodies followed by treatment with secondary antibodies which were conjugated with 15 and 5 nm gold particles, respectively. The sections in (K, L and Q, R) were labeled with anti-CysR10 antibody (5 nm gold particles) and anti-CysP13 antibody (15 nm gold particles). (A–D) The wild type (Kinmaze). (E–H) esp1. (I–L) Esp4. (M–T) esp3. White arrows in (F, H, J and L) show the 5 nm gold particles (CysP13). The left two tiers show the PB-I at 1 WAF and the right two tiers show the PB-I at 3 WAF. (B, D, F, H, J, L, N, P, R and T) Enlarged images of each left panel. Arrows in (M, N, Q and R) show the cracks in the PBs. Open arrows in T show the boundary of PB-I compressed by PB-II. Asterisks in (S, T) show the matrix in protein storage vacuoles (PSVs). Am, amyloplast. Bars in (A–R): 200 nm, Bars in (S, T): 500 nm.
The morphology of the PBs in the esp3 endosperm at 1 WAF was significantly different from that seen in wild-type endosperm (Fig. 7A, B). The PBs were oval in shape, were electron lucent, and displayed a wrinkled surface (Fig. 7M, N). At 1 WAF, CysP13 (5 nm gold particles) was uniformly distributed throughout the whole PB (Fig. 7M, N). In some instances, small amounts of CysR10 (15 nm gold particles) were evident and distributed as a ring (Fig. 7M, N). At 3 WAF, the PBs were labeled with only the CysP13 antibody (Fig. 7O, P). The electron-dense core and ring structures observed in the wild type (Fig. 7C, D) were not observed in esp3 PBs, which were electron lucent and of uniform electron density (Fig. 7O, P and Q, R). The PBs were elongated (Fig. 7Q) or irregularly shaped, and appeared to be less rigid as their shape was compressed by other organelles (Fig. 7S, and arrowheads in Fig. 7T). These irregular-shaped PBs contained cracks and cavities, spaces presumably free of prolamins (Fig. 7Q, R, arrows). These cavities were detected in PBs at 1 WAF (Fig. 7M, N, arrow). The larger PB size but lower levels of the Cys-rich prolamins (Kumamaru et al. 1988, Ogawa et al. 1989) indicate that prolamins are packaged much less compactly in esp3 compared with normal conditions. These results indicate that the decrease in the levels of the Cys-rich prolamins results in the formation of abnormal PB-Is. The degeneration of the electron-dense core containing CysR10 is especially remarkable. Although the levels of the other Cys-rich prolamins were reduced in esp3, we hypothesized that the reduction of CysR10 and, in turn, the absence of the core were responsible for the deformation of PB-I.
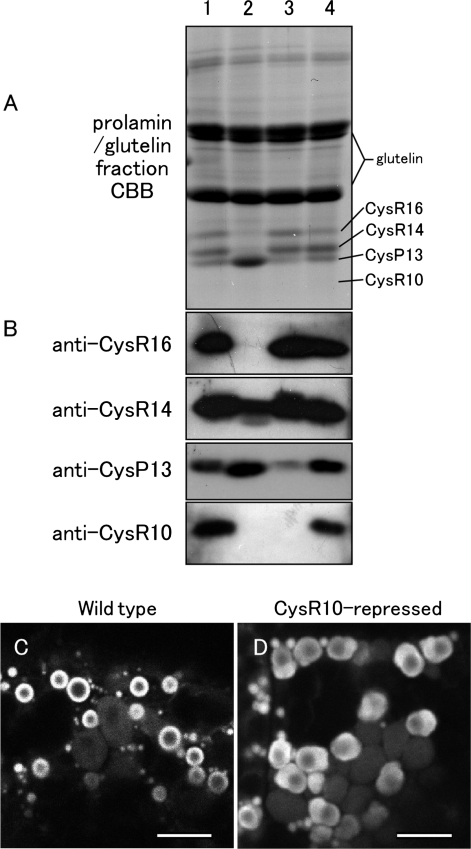
To determine whether CysR10 is required for normal PB-I formation, an RNAi cassette for CysR10 was transferred into rice by Agrobacterium-mediated transformation, resulting in the generation of 10 independent transformant lines. The strongest silenced line, which showed significantly reduced amounts of CysR10 but normal levels of the other Cys-rich prolamins, was selected for further study (Fig. 8A, B). This RNAi line also contained lower amounts of CysP13 (Fig. 8A, B, and Supplementary Fig. S3). The PB-Is in seeds at 2 WAF from the CysR10-repressed transformant and wild type were stained with rhodamine and examined by confocal laser microscopy. PB-Is from the wild type are seen as ring-like structures as the hydrophobic character of the prolamins causes them to bind to this vital stain (Choi et al. 2000, Onda et al. 2009) (Fig. 8C). Although the PB-I organelles could also be detected by rhodamine staining in developing endosperm of the CysR10 RNAi plant, their structures were significantly altered. The PB-Is in the RNAi line were non-spherical (Fig. 8D). The PB-I in the CysR10 RNAi plant was observed by immunocytochemistry at the electron microscope level (Fig. 9). The ribosome-attached membrane was observed around the non-spherical PBs (Fig. 9C, D), indicating that the PBs were derived from the ER. Except for a change in size of the PBs, the deformed PBs from the CysR10 RNAi plant were similar to the irregular-shaped PBs in esp3 which contained reduced levels of CysR10, CysR14 and CysR16 (Figs. 6K, 7M, Q). These results support the view that the compact size and spherical morphology of PB-I is dependent on the presence of CysR10.
Fig. 8.
Down-regulation of CysR10 by RNAi. (A) SDS–PAGE analysis of the prolamin/glutelin fraction extracted from mature seed. Lane 1, wild-type, Kinmaze; lane 2, esp3 mutant derived from Kinmaze; lane 3, CysR10-repressed transformant derived from Yukihikari; lane 4, wild-type, Yukihikari. (B) Immunoblot analysis of the prolamin/glutelin fraction with anti-prolamin antibodies. (C) Confocal laser scanning microscopy of rhodamine-stained wild-type endosperm cells. (D) Confocal laser scanning microscopy of rhodamine-stained endosperm cells from the CysR10-repressed transformant. Bars in (C, D): 5 μm.
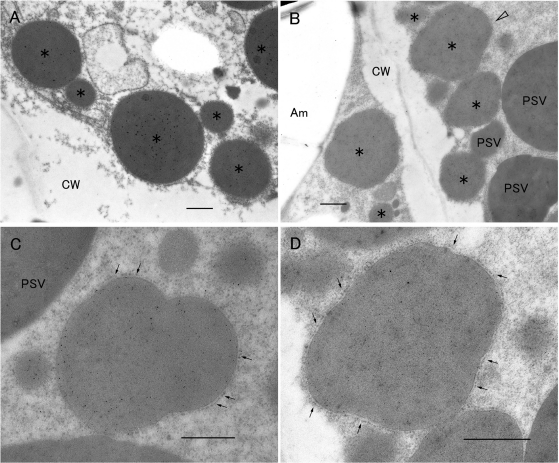
Fig. 9.
The effect of reduced levels of CysR10 on PB-I formation as viewed by immunoelectron microscopy of the CysR10-RNAi transformant. PB-I was labeled with anti-CysR10 (15 nm gold particles) and anti-CysP13 (5 nm gold particles) antibodies (A–D). (A) Wild-type PB-I at 3 WAF. (B–D) CysR10-repressed transformants at 3 WAF. (D) Enlarged image of the PB-I (open arrowhead) shown in B. Asterisks in (A) and (B) denote PB-I. Arrows in (C) and (D) indicate the rough ER. Am, amyloplast; CW, cell wall; PSV, protein storage vacuole. Bars: 500 nm.
Discussion
The results in this study demonstrate that the formation of the rice prolamin-containing PB (PB-I) is initiated by the accumulation of CysR10. We suggest that CysR10 is needed for the initial assembly of prolamins to form a small focus which ultimately becomes the core of PB-I and that CysP13 is synthesized and layered over the core in two separate concentric rings. We found that the inner ring immediately surrounded the central core and contained both CysR10 and CysP13, whereas the outer ring contained only the latter species (Supplementary Figs. S4, S5). Interestingly, in the very large PB (Fig. 4E) at 2 WAF, there was a thin ring between the CysP13 peripheral layer and the CysR10 core which was not recognized by either antibody (Fig. 4E). This clear ring most probably contains other prolamin classes such as CysR14 and CysR16. We indeed found that the distribution of CysR16 within PB-I was very defined and appeared as two concentric rings (Supplementary Fig. S4B, panel c). It is thus possible that the inner CysR16 ring may correspond to the thin ring lacking both CysR10 and CysP13 in the PB depicted in Fig. 4E. The distribution of CysR14 in PB-I was also non-uniform. Immunofluorescence studies showed that CysR14 was distributed in a donut pattern where it was excluded from the central core region (Supplementary Fig. S5). Such an arrangement was verified by immunoelectron microscopy where CysR14 was located around the central core and distributed in a pattern similar to that seen for CysP13 (Supplementary Fig. S5E–H).
The prolamin-containing PB-Is were non-spherical in the CysR10-repressed endosperm and esp3 mutant. Additionally, the PBs in esp3 were significantly larger. Unlike the case in esp3 where the levels of CysR14 and CysR16 were also reduced, their amounts are not significantly different in the CysR10-repressed line. These observations strongly suggest that CysR10 is required for forming PB-Is into the spherical shape and for the compact size, and that it not only forms the central core but also interacts with other Cys-rich prolamins to assemble the concentric ring structure within the PB-I.
Although the PBs in the CysR10-repressed line and esp3 mutant appear similar in being non-spherical, there are notable differences. PB-Is in esp3 appear to be more elastic as their irregular shape could be further misshapen by other organelles including starch-containing amyloplasts and glutelin-containing PSVs (Fig. 7T). In contrast, the irregular shape of PB-Is in the CysR10-repressed line is independent of other organelles and appears to be due to the less ordered arrangement of prolamin polypeptides (Fig. 9C, D and Supplementary Fig. S6). It is inferred from the structural difference in PB-I between CysR10-RNAi transformant and esp3 that the deformed shape of PB-I in the CysR10-RNAi transformant is induced by the presence of CysR14 and CysR16.
Interestingly, RNAi suppression of CysR10 also slightly reduced the expression level of CysP13. The molecular basis for this reduction in the levels of CysP13 is unclear, but the interaction of CysR10 with the other Cys-rich prolamins around the core region may be required to stabilize the accumulation of CysP13. It should be pointed out that the decline in CysP13 is not the reason for the irregular shape of PB-Is in the CysR10-repressed line as PB-Is in CysP13-deficient esp1 and Esp4 are spherical (Fig. 7F, J; Kumamaru et al. 1988, Ogawa et al. 1989).
The smaller size of PB-I in esp1 and Esp4 mutant lines is consistent with the results of a recent study (Kawakatsu et al. 2010), in which the expression of 13 kDa prolamins is repressed. However, unlike this study where large, abnormally shaped PB-I structures were seen in our CysR10-deficient RNAi line, smaller spherical PB-Is were observed when CysR10 was down-regulated (Kawakatsu et al. 2010). The reason for the discrepancy is not clear, but we note that the CysR10 RNAi plants in this study and that of Kawakatsu et al. (2010) had significant differences in the quantities of the other prolamin species. Seeds of our CysR10 RNAi line contained normal or slightly higher amounts of CysR16 and CysR14 and lower amounts of CysP13 (Fig. 8A). Although lower amounts of CysP13 (RM2 and RM4) prolamins were also observed by Kawakatsu et al. (2010), their 10 kDa prolamin-suppressed plant accumulated substantially higher levels (about three times compared with the wild type) of CysR14 (RM1 and RM9) and CysR16 (RP16) prolamins than normal. Hence, the elevated levels of CysR14 and CysR16 may compensate for the loss of CysR10 and contribute to form a rigid normal sized PB-I. This view is supported by esp3 which contains reduced levels of CysR16, CysR14 and CysR10 and displays abnormal large PBIs.
Alternatively, differences in the methods for inducing RNAi may, in part, account for the different PB-I structures. In the study of Kawakatsu et al. (2010), the target protein genes are down-regulated by expressing a modified human Glucagon-like peptide-1 (GLP-1). Although highly effective in repressing target genes, the mechanism remains unclear (Yasuda et al. 2005). In the current study, we employed a conventional RNAi method, in which a DNA fragment containing a part of the CysR10 open reading frame in an inverted manner is designed to be transcribed under the control of an endosperm-specific promoter, as reported previously (Onda et al. 2009, Onda et al. 2011). The two approaches are likely to be responsible for the differences in the expression of CysR14 and CysR16 between the two CysR10 RNAi lines.
The stratified distribution of Cys-rich (CysR10) and Cys-poor (CysP13) prolamins in PB-I differs substantially from that observed for the maize PBs. Cys-rich β- and γ-zeins in maize initially form a small PB, but they are subsequently displaced to the peripheral region by the accumulation of the Cys-poor δ-zein and Cys-rich δ-zein in the core (Lending and Larkins 1989, Esen and Stetler 1992). The exact reason for the different arrangements of prolamin species in the PBs of rice and maize is not known, but it has been suggested that the overall differences in hydrophobicity of maize prolamins may play a role in the formation of the respective PBs (Kim et al. 2002). Analysis of the zein primary amino acid sequences indicates that the hydrophobicity of δ- and α-zein is substantially higher than that of the β- and γ-zeins, with δ-zein being the most hydrophobic (Kim et al. 2002; Supplementary Fig. S7). Segregation of the maize prolamins inside the PB according to their hydrophobicity profiles may establish a more thermodynamically stable arrangement of the prolamins. Interestingly, however, a similar hydropathy analysis of rice prolamins revealed that differences in the overall hydrophobicity profiles between CysR10 (RP10) and CysP13 are not as readily apparent as those seen for the maize prolamins (Supplementary Fig. S8). In view of the differences in spatial arrangement of the maize and rice Cys-rich and Cys-poor prolamins and in their overall hydrophobicity, the mechanisms responsible for PB formation are likely to be different in these cereals.
Since neither rice nor maize prolamins contain the typical C-terminal KDEL retrieval signal (Masumura et al. 1989, Kumamaru et al. 2007), ER retention of both prolamins is likely to be dependent on their capacity to assemble into an intracisternal granule at a rate greater than their capacity to be exported from the ER. The localization of these RNAs to the PB ER would facilitate the assembly of these proteins to form an inclusion granule by concentrating their proteins within the PB lumen (Crofts et al. 2005, Washida et al. 2009). The CysR10 polypeptides are the first prolamin species to be synthesized, followed soon after by the other species. Immunofluorescence microscopy using anti-CysR14 indicated that CysR14 surrounded the center core containing CysR10 (Supplementary Fig. S4). This region also contained CysR10 (Figs. 4, 5) and as well as CysR16 and CysP13, as the synthesis of these proteins began a few days later. The CysR16, CysR14 and CysR10 species could interact with each other in the layer surrounding the center core by non-covalent as well as covalent bonds via disulfide linkages. Interactions by the latter are likely to be responsible for the electron-dense rings evident in PBs in seeds at mid-development (Fig. 5). Although CysP13, which lacks a cysteine residue, is unable to bind to other prolamins through disulfide bonds, the prolamin shares considerable sequence identity with CysR14 and CysR16 (Kim and Okita 1988a, Kim and Okita 1988b, Masumura et al. 1989, Masumura et al. 1990, Shyur and Chen 1990, Shyur et al. 1992, Shyur and Chen 1993, Shyur et al. 1994), suggesting possible hydrophobic interactions between CysP13 and Cys-rich prolamins. Although direct evidence for protein–protein interactions for formation of the PB-I in the rice endosperm has not yet been obtained, the available evidence supports such a role. The Cys-rich prolamins contain the three conserved A, B and C motifs containing cysteine residues (Shewry et al. 1995), which have been demonstrated to be involved in interchain disulfide bonding (Kawagoe et al. 2005). In a recent study, we have shown that protein disulfide isomerase-like protein (PDIL) 2;3 is localized mainly on the surface of PB-I in the ER lumen, and that PDIL1;1 (Esp2) and PDIL2;3 play distinct roles in the accumulation of CysR10 and CysP13 in PB-I (Onda et al. 2011). It is possible that intermolecular disulfide bond formation between CysR molecules facilitated by PDILs may be important for the spatial arrangement of prolamins in PB-I.
Materials and Methods
Plant materials
The esp1, Esp4 and esp3 mutant lines, CM21, PM164 and PM163, respectively, induced by N-methyl-N-nitrosourea (MNU) treatment, and the wild-type rice cultivar, Kinmaze, were used in this study. esp1 shows low levels of CysP13, Esp4 shows low levels of CysP13 and high levels of CysR10, CysR14 and CysR16, while esp3 shows low levels of the Cys-rich prolamins and high levels of CysP13 (Kumamaru et al. 1987, Kumamaru et al. 1988, Kumamaru et al. 1990). esp1, Esp4 and esp3 mutant lines are registered as CM21, CM1834 and CM1675, respectively, in the Rice National Bio Resources Project database, Oryzabase (http://www.shigen.nig.ac.jp/rice/oryzabase/top/top.jsp). These rice plants were grown in field plots at the university farm of Kyushu University and the Agricultural Experimental Station of Yamaguchi Prefecture. The RNAi plant was cultivated in the greenhouse at the National Institute of Agrobiological Science. Developing seeds were tagged on the day after flowering and collected at various developmental stages. The seeds for chemical analysis were immediately frozen with liquid nitrogen then store at −80°C before use, while developing seeds for cytochemical analysis were immediately chemically fixed.
SDS–PAGE and immunoblot analysis
SDS–PAGE of the total rice seed protein extract and immunoblot analysis were performed as described previously (Takemoto et al. 2002, Kumamaru et al. 2010). The total rice protein was extracted in 20% (v/v) glycerol, 4% (w/v) SDS, 88 M urea, 5% 2-mercaptethanol, 500 mM Tris–HCl, pH 6.8 (30 μl of extraction buffer per 11 mg of developing seeds), and 7 μl of the extracted sample was subjected to SDS–PAGE. The proteins separated by SDS–PAGE were transferred to a polyvinylidene fluoride (PVDF) membrane. Antisera against CysR10, CysP13, CysR14 and CysR16 were diluted 1/4,000, 1/2,000, 1/8,000 and 1/4,000, respectively.
Antibodies
The CysR10, CysR14 and CysR16 proteins were purified by elimination of Cys-poor prolamins from matured rice seed powder by 60% (v/v) n-propanol solution and then extraction of Cys-rich prolamins by 60% (v/v) n-propanol solution containing 5% (v/v) 2-mercaptoethanol. Proteins from the latter extraction were separated by SDS–PAGE. Each prolamin band was excised and separated again by SDS–PAGE. The purified CysP13 was produced by extraction of the Cys-poor prolamin from matured rice seed powder in 60% (v/v) n-propanol solution. The extracted proteins were then separated by isoelectric focusing and the major CysP13 band (pI 6.65) was excised and then re-purified by preparative SDS–PAGE. Antibodies against CysR10 and CysR14 were prepared in rabbits, while antibodies against CysP13 and CysR16 were raised in mice. Antibodies against CysR10 and CysR14 are very specific because prolonged exposures showed no cross-reaction with other prolamins (Fig. 3, Supplementary Fig. S1). Antibody against CysP13 showed a weak cross-reaction with unidentified proteins in the total protein containing the albumin or globulin fraction (Fig. 3), but the antibody reacted only with CysP13 in the prolamin fraction (Fig. 8B). Antibodies to CysR16 occasionally reacted faintly with a polypeptide of small size (∼15 kDa) (Fig. 3), but the identity of the polypeptide was not investigated further because the reaction was sporadic (Fig. 8B).
Microscopy analysis
For immunoelectron and immunofluorescence microscopic observations, the samples were fixed as described previously (Takemoto et al. 2002, Satoh-Cruz et al. 2010). Sections were prepared using a microtome (Leica) and then immunolabeled with antisera toward each prolamin. Incubation of the sections with conjugated secondary antibodies for each microscopic technique was performed as described previously (Takemoto et al. 2002, Satoh-Cruz et al. 2010). Images obtained by fluorescence microscopy were analyzed by Image-Pro plus (Planetron). Immunolabeled sections for electron microscopy were sequentially stained with 0.25% KMnO4 and 1% uranyl acetate. Samples for electron microscopy were embedded in epon and sections were stained in Reynolds's lead staining solution consisting of 3.44% Pb(NO3)2, 4.63% Na2(H6H5O7)·2H2O, 0.21 N NaOH in CO2 free distilled water. The diameters (minimum, maximum and average) of PBs at various developing stages of seed development were measured from multiple micrographs using a graphic software package (Canvas X, ACD Systems of America Inc).
Plasmid construction and rice transformation
A full-length cDNA clone for CysR10 (RP10, AK108254) was obtained from the National Institute of Agrobiological Sciences (Tsukuba, Japan). For the CysR10-specific RNAi construction, 4,299 bp (Asp41 to stop codon) of the cDNA clone for CysR10 was used to make inverted repeats. The RNAi cassette was transferred to the binary vector containing the α-globulin promoter and the nopaline synthase (Nos) terminator described previously (Wu et al. 1998, Kawagoe et al. 2005), and the hygromycin phosphotransferase gene. The CysR10 RNAi plasmid was transformed into the rice variety Yukihikari as described previously (Goto et al. 1999, Onda et al. 2009).
Extraction of prolamin/glutelin fraction
The powder of an individual rice seed was suspended in 200 mM Tris–HCl (pH 6.8) containing 0.55 M NaCl (60 μl per 11 mg of mature seeds). The suspension was sonicated for 55 min on ice. After centrifugation, the supernatant was discarded. The precipitate was suspended with 0.55 ml of 0.1255 M Tris–HCl (pH 6.8) containing 4% (w/v) SDS, 88 M urea and 5% (v/v) 2-mercaptoethanol. After centrifugation, the supernatant was collected as the prolamin/glutelin fraction. The protein composition of the fraction was analyzed by SDS–PAGE.
Confocal laser scanning microscopy
Rhodamine labeling of PBs in developing rice seed at 2 WAF was performed as described previously (Onda et al. 2009). The fluorescence images of the subaleurone cells were analyzed with a confocal laser scanning microscope with a laser beam of 5,433 nm (TCS SP2 AOBS; Leica).
Supplementary data
Supplementary data are available at PCP online.
Funding
This work was supported by the Japan Society for the Promotion of Science [grant-in-aid for Scientific Research to T.K. (16380009 and 21380008]; Bio-oriented Technology Research Advanced Institution (BRAIN) [the program for Promotion of Basic Research Activities for Innovative Biosciences]; the Ministry of Agriculture, Forestry and Fisheries of Japan [grant to Y.K. (Genomics for Agricultural Innovation, IPG-0023); United States National Science Foundation [grants IOS-1021699 and DBI-0605016 to T.W.O.]. United States Department of Agriculture-Cooperative State Research, Education and Extension-National Research Initiative [grant 2006-35301-17043 to T.W.O.].
Supplementary Material
Acknowledgments
We thank Mr. Kazuhiko Kaneko, Mr. Hiroshi Kajiwara and Mr. Masayasu Hajima for enabling us to use the transmission electron microscope at the Yamaguchi Prefectural Agricultural Experimental Station.
Glossary
Abbreviations
- DAF
days after flowering
- ER
endoplasmic reticulum
- esp
endosperm storage protein mutant
- FITC
fluorescein isothiocyanate
- PB
protein body
- PDIL
protein disulfide isomerase-like protein
- PSV
protein storage vacuole
- RNAi
RNA interference
- WAF
weeks after flowering.
References
- Bechtel DB, Juliano BO. Formation of protein bodies in the starchy endosperm of rice (Oryza sativa L.): a re-investigation. Ann. Bot. 1980;45:503–509. [Google Scholar]
- Cameron-Mills V, von Wettstein D. Protein body formation in the developing barley endosperm. Carlsberg Res. Commun. 1980;45:577–594. [Google Scholar]
- Choi SB, Wang C, Muench DG, Ozawa K, Franceschi VR, Wu Y, et al. Messenger RNA targeting of rice seed storage proteins to specific ER subdomains. Nature. 2000;407:765–767. doi: 10.1038/35037633. [DOI] [PubMed] [Google Scholar]
- Crofts AJ, Washida H, Okita TW, Satoh M, Ogawa M, Kumamaru T, et al. The role of mRNA and protein sorting in seed storage protein synthesis, transport, and deposition. Biochem. Cell Biol. 2005;83:728–737. doi: 10.1139/o05-156. [DOI] [PubMed] [Google Scholar]
- Esen A, Stetler DA. Immunocytochemical localization of δ-zein in the protein bodies of maize endosperm cells. Amer. J. Bot. 1992;79:243–248. [Google Scholar]
- Goto F, Yoshihara T, Shigemoto N, Toki S, Takaiwa F. Iron fortification of rice seed by the soybean ferritin gene. Nat. Biotechnol. 1999;17:282–286. doi: 10.1038/7029. [DOI] [PubMed] [Google Scholar]
- Kawagoe Y, Suzuki K, Tasaki M, Yasuda H, Akagi K, Katoh E, et al. The critical role of disulfide bond formation in protein sorting in the endosperm of rice. Plant Cell. 2005;17:1141–1153. doi: 10.1105/tpc.105.030668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, Hirose S, Yasuda H, Takaiwa F. Reducing rice seed storage protein accumulation leads to changes in nutrient quality and storage organelle formation. Plant Physiol. 2010;154:1842–1854. doi: 10.1104/pp.110.164343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CS, Woo Ym YM, Clore AM, Burnett RJ, Carneiro NP, Larkins BA. Zein protein interactions, rather than the asymmetric distribution of zein mRNAs on endoplasmic reticulum membranes, influence protein body formation in maize endosperm. Plant Cell. 2002;14:655–672. doi: 10.1105/tpc.010431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WT, Okita TW. Nucleotide and primary sequence of a major rice prolamine. FEBS Lett. 1988a;231:308–310. doi: 10.1016/0014-5793(88)80839-1. [DOI] [PubMed] [Google Scholar]
- Kim WT, Okita TW. Structure, expression, and heterogeneity of the rice seed prolamines. Plant Physiol. 1988b;88:649–655. doi: 10.1104/pp.88.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WT, Okita TW. Expression of storage protein multigene families in developing rice endosperm. Plant Cell Physiol. 1993;34:595–603. [Google Scholar]
- Krishnan HB, Franceschi VR, Okita TW. Immunochemical studies on the role of the Golgi complex in protein-body formation in rice seeds. Planta. 1986;169:471–480. doi: 10.1007/BF00392095. [DOI] [PubMed] [Google Scholar]
- Kumamaru T, Ogawa M, Satoh H, Okita TW. Protein body biogenesis in cereal endosperms. In: Olsen OA, editor. Endosperm—Development and Molecular Biology. Berlin: Springer-Verlag; 2007. pp. 141–158. [Google Scholar]
- Kumamaru T, Satoh H, Iwata N, Omura T, Ogawa M. Mutants for rice storage proteins. III. Genetic analysis of mutants for storage proteins of protein bodies in the starchy endosperm. Jpn. J. Genet. 1987;62:333–339. doi: 10.1007/BF00288825. [DOI] [PubMed] [Google Scholar]
- Kumamaru T, Satoh H, Iwata N, Omura T, Ogawa M, Tanaka K. Mutants for rice storage proteins. 1. Screening of mutants for rice storage proteins of protein bodies in the starchy endosperm. Theor. Appl. Genet. 1988;76:11–16. doi: 10.1007/BF00288825. [DOI] [PubMed] [Google Scholar]
- Kumamaru T, Satoh H, Omura T, Ogawa M. Mutants for rice storage proteins. IV. Maternally inherited mutants for storage proteins of protein bodies in the starchy endosperm. Heredity. 1990;64:9–15. [Google Scholar]
- Kumamaru T, Uemura Y, Inoue Y, Takemoto Y, Siddiqui SU, Ogawa M, et al. Vacuolar processing enzyme plays an essential role in the crystalline structure of glutelin in rice seed. Plant Cell Physiol. 2010;51:38–46. doi: 10.1093/pcp/pcp165. [DOI] [PubMed] [Google Scholar]
- Lending CR, Larkins BA. Changes in the zein composition of protein bodies during maize endosperm development. Plant Cell. 1989;1:1011–1023. doi: 10.1105/tpc.1.10.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanony H, Rubin R, Altschuler Y, Galili G. Evidence for a novel route of wheat storage proteins to vacuoles. J. Cell Biol. 1992;119:1117–1128. doi: 10.1083/jcb.119.5.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Okita TW. Accumulation of prolamines and glutelins during rice seed development: a quantitative evaluation. Plant Cell Physiol. 1993;34:385–390. [Google Scholar]
- Masumura T, Hibino T, Kidzu K, Mitsukawa N, Tanaka K, Fujii S. Cloning and characterization of a cDNA encoding a rice 13 kDa prolamin. Mol. Gen. Genet. 1990;221:1–7. doi: 10.1007/BF00280360. [DOI] [PubMed] [Google Scholar]
- Masumura T, Shibata D, Hibino T, Kato T, Kawabe K, Takeba G, et al. cDNA cloning of an mRNA encoding a sulfur-rich 10 kDa prolamin polypeptide in rice seeds. Plant Mol. Biol. 1989;12:123–130. doi: 10.1007/BF00020497. [DOI] [PubMed] [Google Scholar]
- Muench DG, Ogawa M, Okita TW. The prolamins of rice. In: Shewry PR, Casey R, editors. Seed Protein. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. pp. 93–108. [Google Scholar]
- Muench DG, Okita TW. The storage proteins of rice and oat. In: Larkins BA, Vasil IK, editors. Cellular and Molecular Biology of Plant Seed Development. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. pp. 404–412. [Google Scholar]
- Ogawa M, Kumamaru T, Satoh H, Iwata N, Omura T, Kasai Z, et al. Purification of protein body-I of rice seed and its polypeptide composition. Plant Cell Physiol. 1987;28:1517–1527. [Google Scholar]
- Ogawa M, Kumamaru T, Satoh H, Omura T, Park T, Shintaku K, et al. Mutants for rice storage proteins. 2. Isolation and characterization of protein bodies from rice mutants. Theor. Appl. Genet. 1989;78:305–310. doi: 10.1007/BF00265288. [DOI] [PubMed] [Google Scholar]
- Onda Y, Kumamaru T, Kawagoe Y. ER membrane-localized oxidoreductase Ero1 is required for disulfide bond formation in the rice endosperm. Proc. Natl Acad. Sci. USA. 2009;106:14156–14161. doi: 10.1073/pnas.0904429106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onda Y, Nagamine A, Sakurai M, Kumamaru T, Ogawa M, Kawagoe Y. Distinct roles of protein disulfide isomerase and p5 sulfhydryl oxidoreductases in multiple pathways for oxidation of structurally diverse storage proteins in rice. Plant Cell. 2011;23:210–223. doi: 10.1105/tpc.110.079509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost TL. The ultrastructure and physiology of protein bodies and lipids from hydrated dormant and nondormant embryos of Setaria lutescens (Gramineae) Amer. J. Bot. 1972;59:607–616. [Google Scholar]
- Satoh-Cruz M, Crofts AJ, Takemoto-Kuno Y, Sugino A, Washida H, Crofts N, et al. Protein disulfide isomerase like 1-1 participates in the maturation of proglutelin within endoplasmic reticulum in rice endosperm. Plant Cell Physiol. 2010;51:1581–1593. doi: 10.1093/pcp/pcq098. [DOI] [PubMed] [Google Scholar]
- Shewry PR, Napier JA, Tatham AS. Seed storage proteins: structures and biosynthesis. Plant Cell. 1995;7:945–956. doi: 10.1105/tpc.7.7.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewry PR, Tatham AS. The characteristics, structure and evolutionary relationships of prolamins. In: Shewry PR, Casey R, editors. Seed Protein. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. pp. 11–33. [Google Scholar]
- Shull JM, Watterson JJ, Kirleis AW. Purification and immunocytochemcial localization of kafirins in Sorghum bicolor (L. Moench) endosperm. Protoplasma. 1992;171:64–74. [Google Scholar]
- Shyur LF, Chen CS. Nucleotide sequence of two rice prolamin cDNAs. Nucleic Acids Res. 1990;18:6683. doi: 10.1093/nar/18.22.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyur LF, Chen CS. Rice prolamins: heterogeneity of cDNAs and synthesis of precursors. Bot. Bull. Acad. Sin. 1993;34:143–154. [Google Scholar]
- Shyur LF, Wen TN, Chen CS. cDNA cloning and gene expression of the major prolamins of rice. Plant Mol. Biol. 1992;20:323–326. doi: 10.1007/BF00014501. [DOI] [PubMed] [Google Scholar]
- Shyur LF, Wen TN, Chen CS. Purification and characterization of rice prolamins. Bot. Bull. Acad. Sin. 1994;35:65–71. [Google Scholar]
- Takemoto Y, Coughlan SJ, Okita TW, Satoh H, Ogawa M, Kumamaru T. The rice mutant esp2 greatly accumulates the glutelin precursor and deletes the protein disulfide isomerase. Plant Physiol. 2002;128:1212–1222. doi: 10.1104/pp.010624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Sugimoto T, Ogawa M, Kasai Z. Isolation and characterization of two types of protein bodies in the rice endosperm. Agric. Biol. Chem. 1980;44:1633–1639. [Google Scholar]
- Washida H, Sugino A, Kaneko S, Crofts N, Sakulsingharoj C, Kim D, et al. Identification of cis-localization elements of the maize 10-kDa delta-zein and their use in targeting RNAs to specific cortical endoplasmic reticulum subdomains. Plant J. 2009;60:146–155. doi: 10.1111/j.1365-313X.2009.03944.x. [DOI] [PubMed] [Google Scholar]
- Woo YM, Hu DW, Larkins BA, Jung R. Genomics analysis of genes expressed in maize endosperm identifies novel seed proteins and clarifies patterns of zein gene expression. Plant Cell. 2001;13:2297–2317. doi: 10.1105/tpc.010240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CY, Adachi T, Hatano T, Washida H, Suzuki A, Takaiwa F. Promoters of rice seed storage protein genes direct endosperm-specific gene expression in transgenic rice. Plant Cell Physiol. 1998;39:885–889. [Google Scholar]
- Yamagata H, Tanaka K. The site of synthesis and accumulation of storage proteins. Plant Cell Physiol. 1986;27:135–145. [Google Scholar]
- Yasuda H, Tada Y, Hayashi Y, Jomori T, Takaiwa F. Expression of the small peptide GLP-1 in transgenic plants. Transgenic Res. 2005;14:677–684. doi: 10.1007/s11248-005-6631-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.