Abstract
Debranching enzymes, which hydrolyze α-1 and 6-glucosidic linkages in α-polyglucans, play a dual role in the synthesis and degradation of starch in plants. A transposon-inserted rice mutant of isoamylase3 (isa3) contained an increased amount of starch in the leaf blade at the end of the night, indicating that ISA3 plays a role in the degradation of transitory starch during the night. An epitope-tagged ISA3 expressed in Escherichia coli exhibited hydrolytic activity on β-limit dextrin and amylopectin. We investigated whether ISA3 plays a role in amyloplast development and starch metabolism in the developing endosperm. ISA3–green fluorescent protein (GFP) fusion protein expressed under the control of the rice ISA3 promoter was targeted to the amyloplast stroma in the endosperm. Overexpression of ISA3 in the sugary1 mutant, which is deficient in ISA1 activity, did not convert water-soluble phytoglycogen to starch granules, indicating that ISA1 and ISA3 are not functionally redundant. Both overexpression and loss of function of ISA3 in the endosperm generated pleomorphic amyloplasts and starch granules. Furthermore, chloroplasts in the leaf blade of isa3 seedlings were large and pleomorphic. These results suggest that ISA3 facilitates starch metabolism and affects morphological characteristics of plastids in rice.
Keywords: Amyloplast, Chloroplast, Plastid division, Starch degradation, Sugary, Transitory starch
Introduction
In cereals, storage starch is synthesized in the endosperm as the primary storage substance. Starch consists of two classes of α-polyglucans: amylose and amylopectin. Amylose is an essentially linear α-1, 4-linked glucose polymer, and amylopectin is a branched glucose polymer connected by α-1, 4 and α-1, 6 linkages. Amylose is synthesized by granule-bound starch synthase (GBSS), whereas amylopectin is synthesized by the coordinated actions of soluble starch synthases (SSs), branching enzymes (BEs) and debranching enzymes (DBEs) (for reviews, see James et al. 2003, Tetlow 2006). A number of mutations have been identified that affect starch synthesis and degradation (Fujita et al. 2006, Lu and Sharkey 2006, Tetlow 2006, Fujita et al. 2007). In addition, comprehensive genome-wide expression analyses have greatly enhanced our knowledge of how storage and transitory starch is metabolized in different tissues at different developmental stages (Hirose and Terao 2004, Smith et al. 2004, Ohdan et al. 2005).
Some mutations affecting starch metabolism in Arabidopsis leaves result in decreased plant growth (Zeeman et al. 2007, Zeeman et al. 2010). Transitory starch in Arabidopsis leaves is degraded by the coordinated actions of several different classes of enzymes, including glucan, water dikinase (GWD), phosphoglucan, water dikinase (PWD), chloroplastic β-amylase (BAM) and DBE (Smith et al. 2005, Delatte et al. 2006, Edner et al. 2007, Zeeman et al. 2007). The ISA3 (isoamylase3) mutants of Arabidopsis grow more slowly than the wild type (Delatte et al. 2005, Delatte et al. 2006), and mutants deficient in BAMs have elevated starch levels in their leaves (Fulton et al. 2008). In contrast, starch metabolism in the amy1/amy2/amy3 triple α-amylase mutant of Arabidopsis leaves appears to be normal (Yu et al. 2005). The rice genome encodes at least 10 α-amylase genes (Asatsuma et al. 2005) and seven β-amylase genes (Fulton et al. 2008). Asatsuma et al. (2005) reported that the N-glycosylated α-amylaseI-1 in Nipponbare plays a significant role in the degradation of transitory starch in leaves.
DBEs are divided into two groups according to their substrate specificities: isoamylases (ISA1, ISA2 and ISA3) and pullulanase, which is also called limit dextrinase (abbreviated hereinafter to PUL/LD). Both groups of DBEs hydrolyze α-1, 6 branches in α-polyglucans such as amylopectin, whereas PUL/LD, but not ISAs, readily hydrolyzes α-1, 6 linkages of pullulan, yielding maltotriose. Recombinant potato ISA3 has a 70-fold higher activity on β-limit dextrin than does recombinant ISA1; this is consistent with the notion that ISA3 and BAM degrade cooperatively transitory starch (Hussain et al. 2003). In Arabidopsis protoplasts expressing ISA3 fused with green fluorescent protein (ISA3–GFP), the fusion protein is localized in the stroma of chloroplasts and concentrated on the surface of granule-like structures (Delatte et al. 2006). In addition to their role in starch turnover in the leaf chloroplast at night, DBEs are also essential factors in the synthesis of starch in different tissues, including the endosperm of maize (Dinges et al. 2001), barley (Burton et al. 2002) and rice (Kawagoe et al. 2005, Kubo et al. 2005), and Arabidopsis leaf (Delatte et al. 2005, Delatte et al. 2006). In the sugary1 endosperm of rice, in which ISA1 activity is very low, the inner endosperm cells produce water-soluble α-glucan, also called phytoglycogen, in the amyloplast, whereas the outer endosperm cells synthesize sugary-amylopectin in starch granules that are much smaller than those in the wild type. In the developing rice endosperm, ISA1 forms homopentamers or heterohexamers with ISA2, and the latter form is heat resistant in vitro (Utsumi and Nakamura 2006). PUL/LD is also involved in the synthesis of storage starch in cereal endosperm. It is noted that the maize zpu1 mutation enhances a sugary phenotype (Dinges et al. 2003). In rice, PUL/LD activity is significantly reduced in sugary mutants (Kubo et al. 1999). A recent study has demonstrated that the variation of the sugary1 phenotype is not significantly dependent on PUL activities (Fujita et al. 2009).
In the rice endosperm, amyloplasts divide simultaneously at multiple constriction sites, and small amyloplasts bud from the surface (Yun and Kawagoe 2009). When the unicellular alga Ostreococcus tauri divides, a single starch granule is divided into two granules by the degradation of a starch granule at the equatorial plane of the dividing chloroplast (Ral et al. 2004). However, such a mode of amyloplast division has not been observed in the rice endosperm. Rice endosperm produces characteristic compound granules in each amyloplast. We recently showed that a septum-like structure containing inner envelope membrane divides granules in the amyloplast and that plastid division proteins including FtsZ, Min, ARC5 and PDV2 proteins play roles not only in amyloplast division but also in compound granule synthesis (Yun and Kawagoe 2009, Yun and Kawagoe 2010). These results strongly suggest that amyloplast division and compound granule synthesis are closely related processes. In this study, we characterized in detail an isa3 mutant of rice, screened from transposon insertion mutant lines (Miyao et al. 2003). Overexpression of ISA3 in the endosperm of the sugary1 mutant of rice did not convert water-soluble phytoglycogen into starch granules in the amyloplast, indicating that ISA1 and ISA3 are not functionally redundant. Comparative analyses of a null isa3 mutant and endosperm-specific ISA3-knockdown (KD) transgenic plants have revealed that glucan debranching by ISA3 facilitates the formation of spherical amyloplasts containing compound granules in the endosperm. In addition, we have revealed that the leaf blade of the isa3 seedling contains pleomorphic chloroplasts. These results suggest that ISA3 plays roles in starch metabolism and affects plastid morphogenesis in rice.
Results
Characterization of ISA3 activity
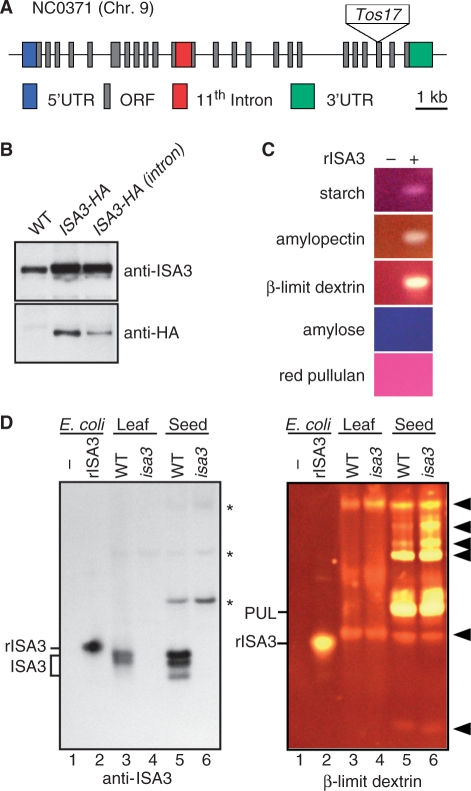
Rice ISA3 consists of 24 exons on chromosome 9 and is predicted to encode a polypeptide of 782 amino acid residues with a putative transit peptide from Met1 to Arg68. A full-length cDNA clone J033050A06 (AK101554) contains all the predicted 24 exons and the putative 11th intron (Fig. 1A). To clarify whether the 11th intron in the full-length cDNA is spliced out when expressed in rice plants, we tagged the intron-containing ISA3 and intron-removed ISA3 with the hemagglutinin (HA) epitope at the C-termini. The HA epitope-tagged ISA3 with or without the 11th intron was expressed in the endosperm under the control of the ADP-glucose pyrophosphorylase (AGPS2b) promoter (Ohdan et al. 2005). Immunoblot analysis revealed that antibodies for ISA3 and the HA epitope both recognized a single band of apparent molecular mass of 79 kDa, indicating that the 11th intron was efficiently spliced out in the endosperm (Fig. 1B).
Fig. 1.
Gene structure of ISA3 and substrate specificity of ISA3. (A) Gene structure of ISA3 on chromosome 9. The transposon Tos17 is inserted at the 3′ splice site of the 21st intron in the isa3 mutant line NC0371. Exons are indicated by boxes and the unspliced 11th intron in the full-length cDNA (AK101554) is indicated by a red box. (B) ISA3 protein levels in seed extracts of wild-type control (WT) and transformants expressing HA epitope-tagged ISA3 without the 11th intron (ISA3-HA) and with the intron [ISA3-HA (intron)] were compared by immunoblotting with anti-ISA3 and anti-HA epitope antibodies. (C) Zymogram analysis with recombinant ISA3 (rISA3) expressed in E. coli with different α-glucans. Vector control (−); HA epitope-tagged rISA3 (+). An equal amount (2 μl) of samples was loaded on native polyacrylamide gels for electrophoresis and then transferred to gels containing the indicated substrates. (D) Zymogram analysis with leaf and seed extracts. Crude extracts of the leaf (10 μl) and developing seed (5 μl) of wild-type Nipponbare control (WT) and isa3 were separated on a native polyacrylamide gel. Proteins were transferred to a PVDF membrane for immunoblotting with anti-ISA3 antibody (left panel) or a polyacrylamide gel containing β-limit dextrin for zymogram analysis (right panel). Cell extracts (2 μl) of E. coli transformed with a control vector (lane 1) and the vector containing HA epitope-tagged ISA3 (lane 2) were used as controls. The E. coli cell extracts for the immunoblotting analysis were diluted 5,000-fold relative to those used for the zymogram analysis. Note that rISA3 containing the HA epitope and His-tag at the C-terminus migrated more slowly than endogenous ISA3 bands extracted from the leaf and seed. Although specific fragments are recognized by the anti-ISA3 antibody (left panel), debranching activity of the endogenous ISA3 at the corresponding positions is under the limit of detection by zymogram analysis (right panel). Arrowheads indicate unidentified enzymes or complexes that modify the substrate in the gel but do not contain ISA3. Asterisks show non-specific fragments recognized by the ISA3 antibody.
The debranching enzyme activity of rice ISA3 was examined with a recombinant protein expressed in Escherichia coli. The DNA sequences encoding a transit peptide (Met1–Arg68) and the 11th intron were removed from the full-length cDNA (AK101554) and the HA epitope was inserted at the C-terminus. A semi-quantitative native PAGE analysis, also known as zymogram analysis, was conducted to investigate the substrate specificity of the recombinant ISA3 (rISA3). Crude cell extracts containing rISA3 were separated by native gel electrophoresis and electrotransferred overnight to gels containing starch, amylopectin, β-limit dextrin, amylose or red pullulan. Iodine staining of the gels revealed that rISA3 exhibited hydrolytic activities on β-limit dextrin, amylopectin and starch, but no hydrolytic activity was detectable with amylose (Fig. 1C), indicating that rISA3 primarily hydrolyzes the α-1, 6 linkages. Because rISA3 did not readily hydrolyze red pullulan, the zymogram analyses have revealed that rISA3 and PUL/LD have different substrate specificities.
We then conducted zymogram analysis with crude extracts of mature leaf blades and developing seeds of rice with β-limit dextrin. Note that the extracts of E. coli cells, but not those of the leaf and seed, used for the immunoblot analysis were diluted 5,000-fold relative to those used for the zymogram analysis (Fig. 1D). Immunoblot analysis detected two and three bands of the endogenous ISA3 in the extracts of the leaf and seed, respectively. The reason for the detection of multiple ISA3 bands by immunoblot analysis is not clear, but it is possible that ISA3 proteins are in different conformations or have a different phosphorylation status. Although the zymogram analysis readily detected substrate-modifying activities, including the debranching activity of PUL/LD, the debranching activity of endogenous ISA3 in the leaf and seed extracts was undetectable at the corresponding positions of ISA3 identified by the immunoblot analysis.
Role of ISA3 in starch metabolism in leaf blades
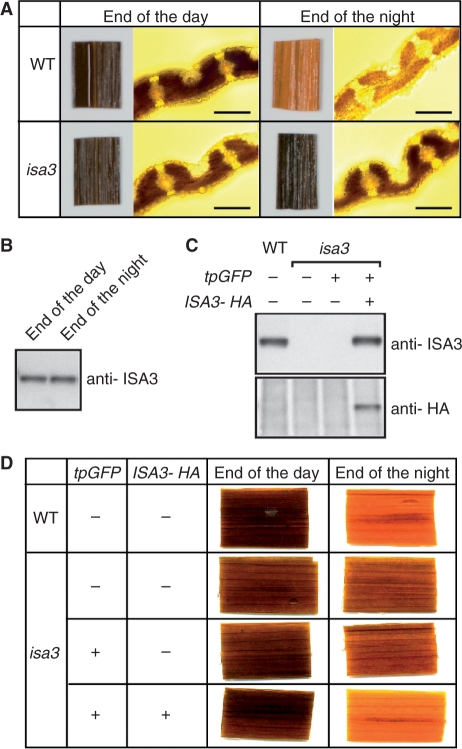
The transposon Tos17-tagged mutant line NC0371 contains the transposon at the 3′ splice site of the 21st intron of ISA3, and is thus presumably a null mutant (Fig. 1A). We screened for homozygous isa3 mutant plants by PCR with appropriate primers. The plant height of isa3 was not significantly different from that of the wild type when grown in the field. We examined the starch metabolism in the leaf blades of isa3 by staining the leaf with iodine at the end of the day and at the end of the night (Fig. 2A). The color and intensity of the iodine staining were similar for the leaves of isa3 and the wild type at the end of the day. In contrast, at the end of the night, the wild-type leaf was light brown, whereas the isa3 leaf remained deep brown, suggesting that starch degradation in the isa3 leaf is inefficient during the night. We then quantified the starch content in mature leaf blades of the wild type and isa3 at the beginning and end of the night, and found that the starch content in the isa3 leaves was markedly higher than that in the wild type at the end of the night (Table 1). The chain length profile of amylopectin in the starch purified from isa3 leaf blades at the end of the day showed that the relative amounts of chains of degree of polymerization (DP) 10–22 were reduced compared with those of the wild type (Supplementary Fig. S1). The chain length profiles of the amylopectin of isa3 isolated at the end of the day and at the end of the subsequent night were similar (Supplementary Fig. S1). The protein levels of ISA3 in the leaf blades extracted at the end of the day and at the end of the night were also similar (Fig. 2B). Quantitative measurements of the activities of α- and β-amylases in the leaf blades revealed that both activities were higher in isa3 than in the wild type (Table 1). These results suggest that the activities of α- and β-amylases, even at the increased levels, are insufficient for the efficient degradation of transitory starch during the night, and that ISA3 plays an important role in starch metabolism in the leaf blades.
Fig. 2.
Starch metabolism in the leaf blade. (A) The leaf blades of the wild type (upper panels) and isa3 (lower panels) were harvested at the end of the day and at the end of the night, and stained with iodine solution. The leaves were scanned (left panels) and the cross-sections of the leaf were viewed under a microscope (right panels). Bars = 0.1 mm. (B) Immunoblotting with anti-ISA3 antibody against leaf proteins of wild-type Nipponbare extracted at the end of the day and at the end of the night. (C, D) Complementation assays. ISA3 protein levels in leaf blades were compared by immunoblotting with anti-ISA3 antibody (upper) and anti-HA epitope antibody (lower) (C). The isa3 mutant was transformed with either tpGFP or tpGFP and the 11th intron-containing ISA3 under the control of the ISA3 promoter (ISA3-HA). Leaf blades were harvested at the end of the day and at the end of the night, and stained with iodine solution (D).
Table 1.
Starch content and amylase activities in the leaf blades
| Plant | Starch content (% FW) |
α-Amylase activity (U FW g−1)a | β-Amylase activity (U FW g−1)b | |
|---|---|---|---|---|
| End of the day | End of the night | |||
| Wild type | 1.98 ± 0.04 | 0.05 ± 0.01 | 0.96 ± 0.04 | 1.02 ± 0.01 |
| isa3 | 2.09 ± 0.06 | 1.23 ± 0.03* | 1.10 ± 0.01* | 1.16 ± 0.01* |
The values are the average for three replications (mean ± SE).
Significant differences from the wild type are marked with an asterisk (P < 0.01, t-test).
a One unit of α-amylase activity, Ceralpha Unit (Megazyme), is defined as the amount of enzyme, in the presence of excess α-glucosidase and glucoamylase, required to release 1 μmol of p-nitrophenol from BPNPG7 in 1 min under the defined assay conditions.
b One unit of β-amylase activity, Betamyl Unit (Megazyme), is defined as the amount of enzyme required, in the presence of excess β-glucosidase, to release 1 μmol of p-nitrophenol from PNPG5 in 1 min under the defined assay conditions.
We performed complementation analysis to confirm that the observed phenotypes were the consequence of the loss of ISA3 function. The HA epitope-tagged ISA3 containing the 11th intron was placed under the control of the rice ISA3 promoter (Fig. 2C). This ISA3-HA expression cassette and/or another expression cassette for a plastid-targeted GFP (tpGFP) were introduced into isa3 calli using an Agrobacterium-mediated transformation method. Immunoblot analysis with anti-ISA3 and anti-HA epitope antibodies showed that ISA3-HA was produced in the leaf blades of transgenic rice, whereas the endogenous ISA3 was not detectable in isa3 and the transformant expressing tpGFP alone (Fig. 2C). Starch in the leaf blades of the transformant expressing tpGFP had a substantial amount of starch in the leaf blade at the end of the night, whereas the starch in the leaf of transformants expressing both ISA3-HA and tpGFP was efficiently degraded during the night (Fig. 2D). Based on these results, we conclude that ISA3 activity is necessary for the efficient breakdown of transitory starch in the leaves during the night.
Role of ISA3 in developing seed
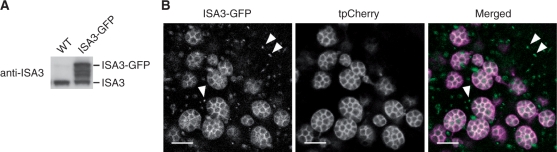
Although isa3 plants showed a reduction in fertility, the seeds were viable and the average seed weight was comparable with that of the control plants (Table 2). The starch content in polished grains of isa3 was lower than that of the wild type, whereas the starch content in the bran of isa3 was significantly higher than that of the wild type (Table 2). Because the aleurone cells of isa3 were not noticeably different from the wild-type cells, it is probable that ISA3 facilitates the degradation of transitory starch in the pericarp (Hirose and Terao 2004). The amylose contents of the isa3 and wild-type seeds were not significantly different (Table 2). Interestingly, the activity of α-amylase was up-regulated in the developing isa3 seed (Table 2). To determine whether ISA3 is targeted to amyloplasts in the developing endosperm, we expressed a GFP-fused ISA3 (ISA3–GFP) under the control of the ISA3 promoter together with an amyloplast-targeted tpCherry, red fluorescent protein (Fig. 3A). Semi-crystalline starch granules did not take up Cherry, and only the stroma surrounding the starch granules was visible (Fig. 3B). ISA3–GFP was similarly localized in the stroma and in small particles, whose identity was not further investigated (Fig. 3B).
Table 2.
Starch content in endosperm and bran, and amylase activities
| Plant | Weight of 100 grains (g) | Endosperm |
Bran | |||||
|---|---|---|---|---|---|---|---|---|
| Starch content (% FW)a | Amylose content (% total starch) | α-Amylase activity (U FW g−1)b |
β-Amylase activity (U FW g−1)c |
Starch content (% FW)a | ||||
| 5 DAF | 10 DAF | 5 DAF | 10 DPA | |||||
| Wild type | 2.89 ± 0.01 | 80.9 ± 0.1 | 23.0 ± 0.1 | 0.40 ± 0.01 | 0.28 ± 0.01 | 0.28 ± 0.02 | 0.24 ± 0.01 | 6.6 ± 0.1 |
| isa3 | 2.89 ± 0.02 | 79.1 ± 0.3* | 22.9 ± 0.1 | 0.52 ± 0.01* | 0.36 ± 0.01* | 0.31 ± 0.01 | 0.32 ± 0.02 | 19.5 ± 0.2* |
The values are the average for three replications (mean ± SE).
Significant differences from the wild type are marked with an asterisk (P < 0.01, t-test).
a Mature isa3 and wild-type grains were polished with a rice polisher (Kett Electric Laboratory) until 10% polishing was achieved. The starch contents of the polished endosperm and the bran were measured.
b One unit of α-amylase activity, Ceralpha Unit (Megazyme), is as in Table 1.
c One unit of β-amylase activity, Betamyl Unit (Megazyme), is as in Table 1.
Fig. 3.
ISA3–GFP is localized in the amyloplast stroma in the rice endosperm. (A) Immunoblotting with anti-ISA3 antibody on seed proteins extracted from developing seeds of wild-type and transgenic plants expressing ISA3-GFP under the control of the rice ISA3 promoter. (B) Co-expression of ISA3-GFP under the control of the ISA3 promoter and tpCherry under the control of the α-globulin promoter in the developing endosperm. ISA3–GFP is localized in the amyloplast stroma and small particles indicated by arrowheads. The fluorescence signals of ISA3–GFP (left panel) and tpCherry (middle panel) are converted to green and magenta, respectively, and the images are merged (right panel). Bars = 5 μm.
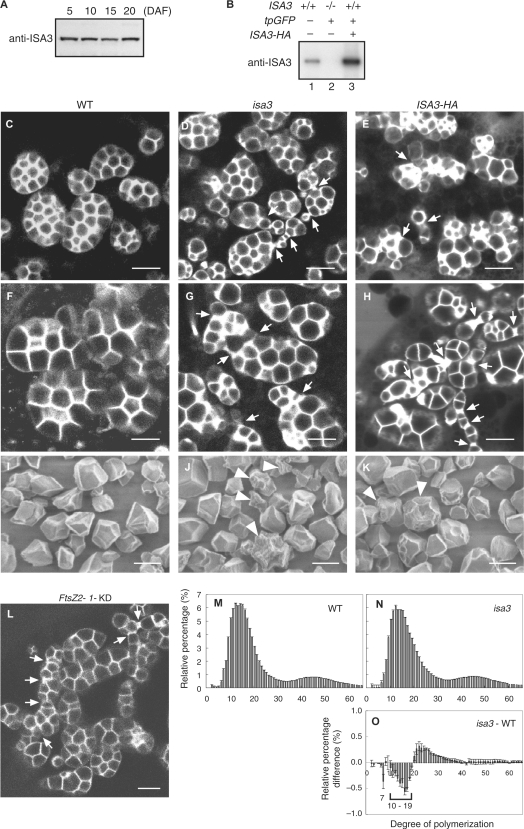
The ISA3 protein level was relatively constant up to 20 days after flowering (DAF) (Fig. 4A). ISA3 was undetectable in the endosperm of isa3 (Fig. 4B). As a control, overexpression of HA epitope-tagged ISA3 was induced with the seed-specific AGPS2b promoter, and a high accumulation of ISA3-HA was detected in the endosperm (Fig. 4B, lane 3). We compared the morphology of amyloplasts in the outer endosperm cells of the wild type, isa3 and ISA3-HA using tpGFP as a molecular probe. The size and shape of the starch granules in each amyloplast of the wild type were mostly uniform (Fig. 4C, F). In contrast, isa3 amyloplasts often had starch granules with different sizes and shapes (Fig. 4D, G). Many amyloplasts of isa3 were pleomorphic with irregular or long-necked shapes (Fig. 4D, G, arrows). Transformants of ISA3-HA also showed irregularly shaped amyloplasts, in which heterogeneous starch granules were formed (Fig. 4E, H). We note that aberrant amyloplasts in the isa3 endosperm are similar to some of those produced in transformants deficient in the function of plastid division proteins such as ARC5, FtsZ1 and FtsZ2-1 (Yun and Kawagoe 2009, Yun and Kawagoe 2010). In particular, the aberrant amyloplasts of isa3 are similar to those of FtsZ2-1-KD, in which amyloplasts develop multiple division septa that are not fully constricted, thereby generating pleomorphic amyloplasts (Fig. 4L).
Fig. 4.
Amyloplast and starch granule morphology in the endosperm cells. (A) ISA3 protein levels in developing seeds. Proteins were extracted from developing seeds at the indicated days after flowering (DAF) and were analyzed by immunoblotting with anti-ISA3 antibody. (B) ISA3 is not detectable in isa3 seed. Proteins were extracted from mature seeds of Nipponbare wild type (lane 1), isa3 transformed with tpGFP (lane 2) and wild-type Nipponbare transformed with tpGFP and ISA3-HA under the control of the AGPS2b promoter (lane 3). (C–H) Amyloplasts in the outer endosperm cells at 8 DAF visualized with stroma-localized GFP. Seed sections of the wild type (C, F), isa3 (D, G) and transformant expressing ISA3-HA (E, H). The starchy endosperm cells shown in C–E and F–H are those in cell layers 1–3 and 4–7 from the outermost aleurone cell layer, respectively. Note that the amyloplasts and granules in the inner cells (F–H) are larger than those of outer cells (C–E). Some amyloplasts in isa3 and ISA3-HA contain starch granules that are heterogeneous in size. Arrows point to constricted or long-necked sites. Bars = 5 μm. (I–K) Scanning electron micrographs of starch granules. Purified starch of WT (I), isa3 (J) and ISA3-HA (K) were directly viewed using an SEM without surface coating. Arrowheads indicate granules that are irregularly shaped and rough on the surface. Bars = 5 μm. (L) Amyloplasts in the outer endosperm cells of FtsZ2-1-KD at 8 DAF visualized with stroma-localized GFP (Yun and Kawagoe 2010). Arrows indicate putative division sites, where constriction of the division septa is not complete. Bar = 5 μm. (M, N) Chain length distribution of wild-type and isa3 amylopectin. Purified starch from wild-type Nipponbare (M) and isa3 (N) seeds was debranched with a bacterial isoamylase, labeled with APTS (8-aminopyrene-1,3,6-trisulfonic acid), and analyzed using capillary electrophoresis. Peak areas were summed, and the areas of individual peaks were expressed as a percentage of the total. The mean ± SE of three independent isoamylase digests is shown. (O) Difference plots derived by subtracting the relative percentage values of the wild type from those of isa3.
We purified starch granules from the mature seeds of the wild type, isa3 and ISA3-HA, and directly observed starch granules with a scanning electron microscope (SEM) under low-voltage conditions (Fig. 4I–K). The average and SD of granule sizes (n = 1458 for each) were 5.03 ± 1.43, 5.19 ± 1.54 and 4.96 ± 1.54 μm for the wild type, isa3 and ISA3-HA, respectively. Consistent with the differences in SD, granules <2 μm or >8 μm were 2.7, 5.3 and 4.8% for the wild type, isa3 and ISA3-HA, respectively, indicating that the granules produced in isa3 and ISA3-HA are more heterogeneous.
We next compared the chain length profiles of amylopectin in the starch purified from isa3 and wild-type endosperm (Fig. 4M, N). The proportion of chains of DP 7 and DP 10–19 in isa3 was lower than that in the wild type (Fig. 4O). In contrast, the chain length profile of amylopectin purified from ISA3-HA was not significantly different from that of the wild type (Supplementary Fig. S2). We then compared the gelatinization properties of starch purified from isa3 and wild-type seeds by differential scanning calorimetry (DSC) and found that the peak temperature of the isa3 starch was lower than that of the wild type (Table 3). The viscosity profile as revealed by a rapid visco-analyzer (RVA) showed the viscosity values of isa3 flour to be lower than those of the wild type (Table 4). These results together provide evidence that ISA3 affects starch properties in the endosperm.
Table 3.
Thermal properties of starch of wild-type and isa3 endosperm
| Plant | To (°C) | Tp (°C) | Tc (°C) | ΔH (J g−1) |
|---|---|---|---|---|
| Wild type | 47.9 ± 0.4 | 63.4 ± 0.3 | 74.0 ± 0.5 | 5.9 ± 0.2 |
| isa3 | 46.9 ± 0.3 | 61.1 ± 0.2* | 72.6 ± 0.2 | 6.0 ± 0.1 |
To, onset temperature; Tp, peak temperature; Tc, conclusion temperature; ΔH, enthalpy change of gelatinization and melting.
The values are the average for three replications (mean ± SE).
Significant differences from the wild type are marked with an asterisk (P < 0.01, t-test).
Table 4.
Viscosity analysis of rice flour of the wild type and isa3
| Plant | Maximum viscosity (RVU) | Minimum viscosity (RVU) | Final viscosity (RVU) | Breakdown (RVU) | Setback (RVU) | Peak time (min) | Pasting temperature (°C) |
|---|---|---|---|---|---|---|---|
| Wild type | 560.8 ± 0.3 | 226.0 ± 1.8 | 334.8 ± 2.1 | 375.4 ± 1.3 | 149.4 ± 0.5 | 6.0 ± 0.0 | 66.5 ± 0.0 |
| isa3 | 512.5 ± 0.1* | 212.3 ± 4.1 | 300.3 ± 4.1* | 344.1 ± 2.7* | 131.9 ± 1.9* | 6.0 ± 0.0 | 66.5 ± 0.0 |
RVU, rapid visco-analyzer unit.
The values are the average for three replications (mean ± SE).
Significant differences from the wild type are marked with an asterisk (P < 0.01, t-test).
It is possible that the altered starch metabolism in the leaf of isa3 affects sucrose transport to the endosperm, which may indirectly affect starch properties and morphological characteristics of amyloplasts in the endosperm. It is also possible that the lack of ISA3 function has pleiotropic effects on the function of plastid division proteins in the endosperm. To examine these possibilities, we generated ISA3-KD transformants, in which ISA3 expression is repressed in an endosperm-specific manner by RNA interference (RNAi) using the seed storage protein α-globulin promoter. We extracted proteins from the seed of the wild type, isa3 arc5, a null mutant of ARC5 (Yun and Kawagoe, 2009) and transformants expressing tpGFP under the background of wild-type (tpGFP) or RNAi for ISA3 (ISA3-KD) or FtsZ2-1 (FtsZ2-1-KD) (Yun and Kawagoe 2010). Immunoblot analyses were conducted with antibodies against plastid division proteins FtsZ1, FtsZ2-1 and ARC5. The expression level of soluble starch synthase IVb (SSIVb) was also compared because, in Arabidopsis, SSIV plays a major role in the initiation of starch granule synthesis in the leaf mesophyll cells (Szydlowski et al. 2009). The protein levels of FtsZ1 and FtsZ2-1 were not significantly altered in isa3, ISA3-KD and arc5 (Fig. 5A). Similarly, the SSIVb level was quite similar in all the samples analyzed. As we reported previously (Yun and Kawagoe 2009), the level of ARC5 in the mature seed of the wild type was below the limit of detection, and ARC5 in the isa3 mature seed was not elevated to a detectable level (data not shown).
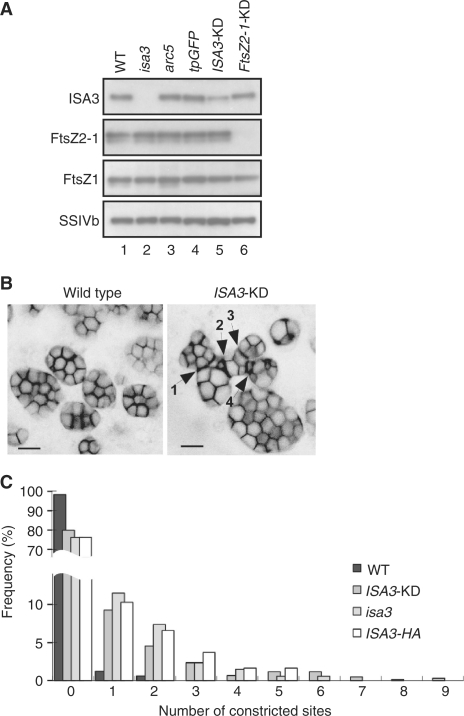
Fig. 5.
Endosperm-specific repression of ISA3 alters amyloplast morphology. (A) Immunoblot analysis of seed proteins with antibodies against ISA3, FtsZ2-1, FtsZ1 and SSIVb. Total proteins were extracted from mature seeds of the wild type (WT, lane 1), isa3 (lane 2), arc5 (lane 3), transformants expressing tpGFP in the background of the WT (lane 4), tpGFP in the background of ISA3-KD induced with the endosperm-specific α-globulin promoter (lane 5), tpCherry and BT1-GFP in the background of FtsZ2-1-KD (lane 6, Yun and Kawagoe, 2010). As reported previously (Yun and Kawagoe 2009), ARC5 was not detectable in the proteins extracted from mature seeds by immunoblot analysis (see also Fig. 7C). (B) Confocal image of stroma-targeted tpGFP in the developing seed of the wild type (left panel) and ISA3-KD (right panel). The image of fluorescence signals of tpGFP is converted to the negative image to show clearly the shape of amyloplasts and the stroma between granules. Arrows show constricted sites. Bars = 5 μm. (C) Frequencies of aberrant amyloplasts. The amyloplast shown in the right panel of (B) has four constricted sites. Up to nine such constricted sites are identified. Amyloplasts in the outer endosperm cells of the wild type (965 amyloplasts in total), ISA3-KD (917), isa3 (340) and ISA3-HA (244) were analyzed.
We analyzed the morphology of amyloplasts produced in the ISA3-KD endosperm. Interestingly, pleomorphic amyloplasts were indeed observed in the ISA3-KD endosperm (Fig. 5B). To analyze these phenotypes more quantitatively, we counted the irregularly shaped amyloplasts in the seeds of the wild type, ISA3-KD, isa3 and ISA3-HA. About 20% of the amyloplasts in the endosperm of ISA3-KD, isa3 and ISA3-HA were pleomorphic with irregular or long-necked shapes, whereas such aberrant amyloplasts accounted for only 2% in the wild type (Fig. 5C).
Distinct roles of ISA1 and ISA3 in developing seeds
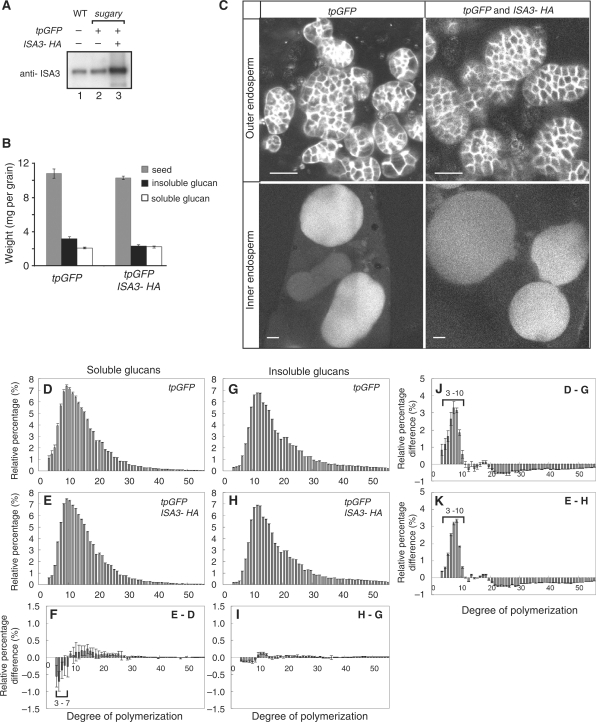
To clarify whether the activities of ISA1 and ISA3 are redundant in the synthesis of starch in the rice endosperm, we overexpressed ISA3-HA in the ISA1-deficient sugary1 mutant (Kawagoe et al. 2005). Immunoblot analysis with anti-ISA3 antibody showed a markedly enhanced level of ISA3 in the transformed endosperm of sugary1 (Fig. 6A). The overexpression of ISA3-HA in the sugary1 endosperm did not convert water-soluble glucans, phytoglycogen, to insoluble glucans (Fig. 6B). As reported previously (Kawagoe et al. 2005), the amyloplasts in the inner endosperm of sugary1 contained no detectable starch granules, whereas the amyloplasts in the outer endosperm contained a markedly increased number of small granules (Fig. 6C, left panels). Amyloplasts in the inner and outer endosperm of sugary1 transformed with tpGFP and ISA3-HA were essentially indistinguishable from those of sugary1 transformed with tpGFP alone (Fig. 6C, right panels). These results indicate that ISA3 and ISA1 are not functionally redundant in the rice endosperm. The phytoglycogen produced in the sugary1 mutant contained an increased amount of short chains of DP 3–10 compared with those in insoluble glucans (Fig. 6D, G, J). When ISA3-HA was overexpressed in the sugary1 mutant, the relative percentage of chains of DP 3–7 in phytoglycogen was reduced (Fig. 6D–F). In contrast, the chain length profiles of insoluble glucans were essentially unchanged by ISA3 overexpression in the sugary1 endosperm (Fig. 6G–I).
Fig. 6.
ISA3 expression in the sugary1 endosperm. (A) Immunoblotting with anti-ISA3 antibody. Seed proteins were isolated from mature seeds of the wild type (lane 1), and sugary1 expressing either tpGFP (lane 2) or tpGFP and ISA3-HA (lane 3). (B) Contents of soluble and insoluble glucans extracted from mature seeds of sugary1 transformed with either tpGFP or tpGFP and ISA3-HA. The means and SE of three replicates are shown. (C) Amyloplasts in the outer and inner endosperm cells of sugary1 expressing tpGFP (left panels), and tpGFP and ISA3-HA (right panels) at 14 DAF. Note that the amyloplasts in the inner endosperm cells do not contain visible starch granules. Bars = 5 μm. (D, E) Chain length profiles of the soluble glucans extracted individually from three seeds of sugary1 expressing tpGFP (D) or tpGFP and ISA3-HA (E). (F) Difference plots derived by subtracting the relative percentage values of the soluble glucans extracted from sugary1 expressing tpGFP (D) from those of sugary1 expressing tpGFP and ISA3-HA (E). (G, H) Chain length profiles of the insoluble glucans extracted individually from three seeds of sugary1 expressing tpGFP (G) or tpGFP and ISA3-HA (H). (I) Difference plots derived by subtracting the relative percentage values of the insoluble glucans extracted from sugary1 expressing tpGFP (G) from those of sugary1 expressing tpGFP and ISA3-HA (H). (J, K) Difference plots derived by subtracting the relative percentage values of the insoluble glucans from those of soluble glucans.
Role of ISA3 in chloroplast development
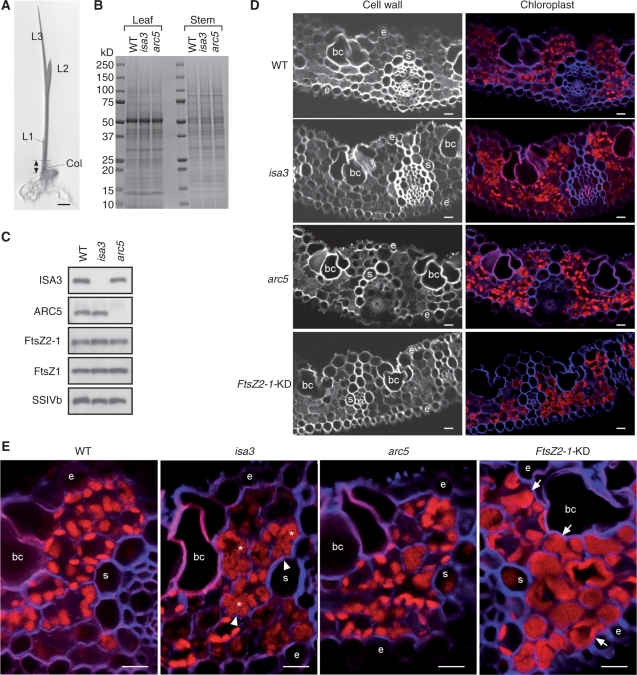
The finding of the peculiar role of ISA3 in amyloplast morphogenesis in the endosperm prompted us to investigate possible roles of ISA3 in chloroplast development in the leaf. We germinated seeds of the wild type, isa3 and arc5 under continuous light for 2 weeks. The growth rate of the seedlings was very similar between the wild type and the mutants under these growth conditions. Proteins were extracted from the leaf blade of the second leaf (L2) and the basal stem region, which contains leaf primordia (Itoh et al. 2005), and subjected to SDS–PAGE (Fig. 7B) and immunoblot analyses with antibodies against the plastid division proteins and SSIVb (Fig. 7C). Protein levels of FtsZ1, FtsZ2-1, ARC5 and SSIVb in the basal stem region were quite similar between the wild type and the two mutants, indicating that the loss of ISA3 function does not significantly induce pleiotropic effects on the expression of the plastid division proteins and SSIVb. We similarly conducted immunoblot analyses with the proteins extracted from the leaf blade of L2 of the wild type, isa3 and arc5, but failed to detect FtsZ1, FtsZ2-1 and ARC5 proteins in all the samples (data not shown), suggesting that chloroplast division is no longer active in the expanded leaf blade of L2 and that the division proteins are degraded during the development of L2 blade. We next analyzed the morphology of chloroplasts in the leaf blade of L2 of the 2-week-old seedling. The cross-sections of the leaf blade were stained with Calcofluor White, which binds β-glucans in the cell wall, and the chloroplasts were visualized by taking advantage of Chls that readily autofluoresce. Most chloroplasts in the wild type were oval in shape, well separated and distributed against the cell wall (Fig. 7D, E). Large starch granules were not readily identified in the chloroplasts of mesophyll cells. Interestingly, a significant fraction of chloroplasts in isa3 were large and pleomorphic (Fig. 7D, E). As we reported previously (Yun and Kawagoe 2009), some, but not all, chloroplasts in arc5 were irregularly shaped. Finally, chloroplasts in FtsZ2-1-KD were markedly larger than those of the wild type; a single chloroplast occupied almost all part of the cell. Most of the large chloroplasts in FtsZ2-1-KD exhibited a round or smooth outline, whereas the large, pleomorphic chloroplasts of isa3 were characterized by deep grooves (Fig. 7E, arrowheads). These results have strongly suggested that ISA3 affects morphological characteristics of chloroplast in the leaf.
Fig. 7.
Role of ISA3 in chloroplast development in the seedling. (A) Seedling of Nipponbare grown for 2 weeks. L2 and the double-headed arrow show the leaf blade of the second leaf and basal stem region, from which proteins were extracted for SDS–PAGE (B) and immunoblot analyses (C). Col, coleoptile; L1, first leaf; L2, second leaf; L3, third leaf. Bar = 5 mm. (B) SDS–PAGE analysis of proteins extracted from the leaf blade of L2 (Leaf) and the basal stem region (Stem) of the seedling of wild-type (Nipponbare), isa3 and arc5. Total proteins were separated on a 10–20% pre-cast gradient gel (ATTO), and stained with CBB. The amounts of samples were normalized by adjusting the volume of homogenization buffer (15 μl mg FW−1) and 2 μl of supernatant was applied on the gel. (C) Immunoblot analysis of the proteins extracted from the basal stem region of the seedling with antibodies against ISA3, FtsZ2-1, ARC5, FstZ1 and SSIVb. The samples (5 μl of the supernatant) were separated on a 10–20% pre-cast gradient gel. (D, E) Cross-sections of the leaf blade of L2 of 2-week-old seedlings. The sections were stained with 1% (v/v) Calcofluor White in PBS. (D) The left panels show the fluorescence signals of the β-glucans in the cell wall. The fluorescence images of the cell wall were converted to blue and merged with autofluorescence signals (red) including those of Chls (right panels). (E) Merged images of the fluorescence signals of the cell wall (blue) and Chls (red). Asterisks indicate pleomorphic chloroplasts containing deep grooves (indicated by arrowheads). Arrows point to exceptionally large chloroplasts. bc, bulliform cell; s, bundle sheath cell; e, epidermis Bars = 10 μm.
Discussion
ISA3 facilitates degradation of transitory starch in the leaf blade
In Arabidopsis leaf, ISA3 and BAMs play a central role in the breakdown of transitory starch (Delatte et al. 2005, Fulton et al. 2008). It is likely that BAMs convert amylopectin to β-limit dextrin in the chloroplast during the night. The debranching enzyme ISA3 has a high hydrolytic activity on β-limit dextrin (Hussain et al. 2003) and is necessary for efficient starch degradation in Arabidopsis leaves (Delatte et al. 2005, Delatte et al. 2006, Wattebled et al. 2008). Rice rISA3 also showed a high debranching activity on β-limit dextrin (Fig. 1C). In the leaf blade of Nipponbare rice, the transcript level of ISA3 is ∼6 and 14 times higher than that of PUL/LD and ISA1, respectively (Ohdan et al. 2005). Consistent with the expression profile, starch degradation in the leaf blade of rice isa3 during the night was severely inhibited (Fig. 2A), although the activities of α-amylase and β-amylase were significantly up-regulated in the isa3 leaf blade (Table 1). These results indicate that ISA3 plays an essential role in the degradation of transitory starch in the leaf blade during the night. We noted that the starch content in the leaf of rice isa3 was comparable with that of the wild type at the end of the day (Table 1). In Arabidopsis, however, the starch content in the isa3 leaf was 2.5 times higher than that of the wild type at the end of the light period (Wattebled et al. 2005). It is possible that the field conditions for rice were close to being optimal for the synthesis of transitory starch in the leaf blade, whereas the growth conditions for Arabidopsis may be inadequate for efficient synthesis of transitory starch in the leaf.
Role of isoamylase in starch synthesis
Mutation on ISA1 causes the accumulation of water-soluble polysaccharides (WSPs) called phytoglycogen in storage tissues such as the endosperm of the sugary1 mutant (Kawagoe et al. 2005). The trimming model, which describes a possible role for ISA1/ISA2 in starch synthesis, predicts that the enzyme complex selectively removes branches that are superfluous or inappropriately positioned for the formation of semi-crystalline structures (Ball et al. 1996, Myers et al. 2000, Nakamura 2002). Another model, called the WSP-clearing model, predicts that the ISA1–ISA2 complex prevents the diversion of synthetic enzymes from their actions at the granule surface to WSPs (Zeeman et al. 1998). Studies on the function of the four DBEs in the synthesis of transitory starch in the Arabidopsis leaf have indeed revealed that DBEs promote amylopectin crystallization (Streb et al. 2008). However, the functions of the four DBEs are not mandatory for starch synthesis because additional loss of the chloroplastic α-amylase AMY3 restores starch granule synthesis (Streb et al. 2008). Here we have shown that the seeds of isa3 do not flatten as they mature and that the overexpression of ISA3-HA in sugary1 endosperm was unable to convert phytoglycogen to starch (Fig. 6). These results clearly indicate that ISA1 and ISA3 are not functionally redundant in the synthesis of storage starch. The chain length profile of soluble glucans in the sugary1 endosperm expressing ISA3-HA exhibited a reduction in the short chains of DP 3–7, whereas the chain length profile of insoluble glucans in the sugary1 endosperm was hardly affected (Fig. 6D–I). The overexpression of ISA3-HA in wild-type endosperm did not significantly change the chain length profile of amylopectin (Supplementary Fig. S2). Although the properties of starch purified from isa3 were different from those of wild-type starch (Tables 3, 4), relatively small changes in the chain length profiles of amylopectins suggest that ISA3 does not play a major role in amylopectin synthesis in the endosperm.
Role of ISA3 in plastid morphogenesis
Significantly increased numbers of amyloplasts and granules were pleomorphic in the endosperms of ISA3-knockout (isa3) and ISA3-KD plants (Figs. 4D, G, 5B). Furthermore, many chloroplasts in the leaf blade of isa3 seedling were irregularly enlarged (Fig. 7E). ISA3–GFP was targeted to developing amyloplasts when expressed under the control of the ISA3 promoter (Fig. 3), and overexpression of ISA3-HA in the endosperm induced aberrant morphology of amyloplasts and granules (Fig. 4E, H). It is important to note that aberrant amyloplasts in ISA3-KD plants showed structural similarities to those in FtsZ2-1-KD plants, which contained multiple division sites where constriction processes are apparently inhibited (Figs. 4L, 5B; Yun and Kawagoe 2010). Although ISA3 has not been considered as a plastid division protein (Yang et al. 2008, Miyagishima and Kabeya 2010), these characteristic phenotypes led us to speculate that ISA3 plays a role in plastid division or development in rice. The results of current and previous studies (Yun and Kawagoe 2009, Yun and Kawagoe 2010) support the notion that starch metabolism and plastid division are interdependent processes regulated by common factors including plastid division proteins, and that ISA3 is involved in both processes. However, the mechanism by which ISA3 plays a role in plastid division remains unclear. One possibility is that ISA3 may contribute to the degradation of soluble and insoluble glucans in the stroma at the division site. During cell division of the unicellular alga O. tauri, a single starch granule is divided into two granules at the equatorial plane of the dividing chloroplast (Ral et al. 2004). It is possible that soluble or insoluble glucans containing abnormal α-1, 6 branches in isa3 and ISA3-HA plastids may physically interfere with the assembly and constriction of the division machinery at the division site, which may eventually lead to the formation of pleomorphic plastid. In this context, it is important to note that a fraction of secretory α-amylase is targeted to plastids via the Golgi body (Kitajima et al. 2009) and the loss of ISA3 function affects the activity of other glucan-metabolizing enzymes including α-amylase and β-amylase (Tables 1, 2). In addition, PUL/LD affects starch synthesis in the rice endosperm (Fujita et al. 2009). Further studies are needed to elucidate whether ISA3, PUL/LD and other amylases have distinct or overlapping roles in the processes of plastid division in rice. Another possibility is that ISA3 may participate in the degradation or constriction of nanofilaments consisting of polyglucans at the plastid-dividing ring, which has been characterized in the unicellular alga Cyanidioschyzon merolae (Yoshida et al. 2010), although it remains to be investigated whether the polyglucans at the plastid-dividing ring are α-1, 6 branched. The polyglucan synthesis at the plastid-dividing ring depends on glycogenin-like protein encoded by plastid-dividing ring 1 (PDR1), and PDR1 homologs have been identified in land plants (Yoshida et al. 2010). We are currently studying the possible role of PDR homologs in plastid division in rice.
Materials and Methods
Materials
Seeds of the Tos17-inserted line of ISA3 (NC0371) and sugary1 (EM5) were obtained from the National Institute of Agrobiological Sciences (NIAS, Tsukuba, Japan). Both isa3 and the wild type (Oryza sativa L. Nipponbare) were grown during the summer months under natural environmental conditions in an experimental paddy field at Tsukuba. Full-length cDNA clones of ISA3 (AK101554) and SSIVb (AK067577) were obtained from NIAS. The antibodies against recombinant ISA3, ARC5, FtsZ1 and FtsZ2-1 have been described previously (Yun and Kawagoe 2009, Yun and Kawagoe 2010). The antibody against SSIVb was produced against a recombinant SSIVb protein. The DNA fragment encoding A630-V908 of SSIVb was PCR-amplified using primers 5′-ATACCATGGCAATTGTGTATTCCAAC-3′ and 5′-TATCTCGAGCACTGCTCTTTGATAGAT-3′, and the full-length cDNA as template, and cloned in pET23d (Novagen). The polypeptide was expressed in Rosetta2 (DE3) (Novagen), purified from inclusion bodies, eluted from an Ni column and injected into rat for antibody production.
Gene construction and rice transformation
Promoters of AGPS2b (endosperm-specific, Ohdan et al. 2005) and the terminator of Gt1 (seed storage protein glutelin) were used to make expression cassettes in an entry vector of the Gateway system (Invitrogen) (Onda et al. 2009). A 2 kb promoter fragment of ISA3 was amplified with primers 5′-ATAGTCGACGCTGCTACATGGGCCTATAAATTTG-3′ and 5′-GGGGACACGGGTGCCTAGTTATAGG-3′, and genomic DNA isolated from a seedling of O. sativa Nipponbare. The 11th intron of ISA3 in the cDNA (AK101554) was removed and the HA epitope was inserted at the C-terminus by the following two-step PCRs. First, the N- and C-terminal fragments separated by the 11th intron were individually amplified with 5′-ATAACCGGTCTGCAGTTTTGGATGGATTCCATT-3′ and 5′-GTTGCAGTTCAGTGTATTCCCGCAGCCTGAGAAGTTCA-3′ for the N-terminal fragment, and with 5′-GGAATACACTGAACTGCAAC-3′ and 5′-TATCTCGAGCTAAGCGTAGTCTGGAACGTCGTATGGGTAAGGCTTTGCCTTGAGCAG-3′ for the C-terminal fragment. The two PCR products were purified and mixed with the 5′ end primer used in the N-terminal PCR and the 3′ end primer used in the C-terminal PCR, followed by another round of PCR. As a control, the full-length ISA3 cDNA containing the 11th intron and HA epitope was also amplified with the same pair of primers. These ISA3 cDNAs with or without the 11th intron were then cloned in appropriate entry vectors. As for the construction of an ISA3 RNAi-inducing vector (ISA3-KD), the cDNA clone (AK101554) was digested with NcoI and XhoI, generating a 960 bp fragment (base pairs 2,311–3,270 of the cDNA clone), which was cloned in inverted repeats. This ISA3-RNAi cassette was inserted between the rice α-globulin promoter and the nos terminator (Kawagoe et al. 2005). The binary vector containing a hygromycin-resistant hpt cassette and tpGFP (Kawagoe et al. 2005) was converted to a destination vector of the Gateway system by the Gateway Vector Conversion system according to the manufacturer's instructions (Yun and Kawagoe 2009). Entry vectors containing the HA epitope-tagged ISA3 expression cassettes or the ISA3 RNAi-inducing cassette were transferred to the destination vector by an LR reaction, yielding binary vectors. The transformation of calli derived from Nipponbare rice seeds and the regeneration of plants were conducted as described previously (Kawagoe et al. 2005).
Starch content of leaves
For measuring the leaf starch content, starch was isolated from the leaf blade of the fifth and sixth leaves at 3 weeks after planting by following the method described previously (Dinges et al. 2003). Leaf tissues (250 mg) were harvested at the beginning and end of the day, and immediately boiled in 10 ml of 80% (v/v) ethanol for 10 min three times. The decolorized leaves were then homogenized in 80% (v/v) ethanol with a pestle and mortar. The homogenates were centrifuged at 3,000 × g for 10 min at 4°C, and the pellet was washed twice with 80% (v/v) ethanol. The total starch content was quantified by measuring the amount of glucose released by treatment with α-amylase and amyloglucosidase (Total Starch Assay Kit, Megazyme) according to the manufacturer's instructions.
Protein extraction and immunoblot analysis
Seed proteins were extracted from a fine flour of mature hulled seeds in extraction buffer consisting of 50 mM Tris–HCl (pH 6.8), 8 M urea, 4% (w/v) SDS, 20% (v/v) glycerol and 5% (v/v) β-mercaptoethanol (35 μl mg−1) overnight at room temperature. For protein extractions from leaf blades and immature hulled seeds, the samples were homogenized in the extraction buffer (10 μl mg−1) with a plastic homogenizer. As for the protein extraction from the leaf blade of L2 and basal stem region of seedling, the seeds were soaked in water and grown on wet paper towels for 2 weeks in a plastic box under continuous fluorescent light at 27°C. Twenty leaf blades and 10 basal stem sections, which were cross-sectioned through the basal stem nodes and 5 mm above, of the seedlings were pooled and homogenized with a pestle and mortar in the extraction buffer (15 μl mg−1) containing protease inhibitors (Complete Mini, Roche, 1 tablet ml−1). After centrifugation at 12,000 × g for 5 min, proteins in the supernatant were separated by SDS–PAGE on a pre-cast 10–20% polyacrylamide gel (ATTO). Equal amounts (5 μl) of seed and leaf extracts were loaded on gels, unless otherwise indicated. The gels were either stained with Coomassie Brilliant Blue (CBB) or blotted onto a polyvinylidene fluoride (PVDF) membrane for immunoblot analyses with antibodies and an ECL kit (GE Healthcare).
Zymogram analysis and amylase assays
As for protein extractions from leaf blades and seeds for zymogram analysis, the samples were homogenized in the extraction buffer (10 μl mg−1) consisting of 50 mM HEPES (pH 7.8), 4 mM MgCl2, 50 mM β-mercaptoethanol and 12.5% (v/v) glycerol with a plastic homogenizer. To express HA epitope-tagged ISA3 in E. coli, we used the entry vector containing the HA epitope-tagged ISA3 without the 11th intron described above as a PCR template and conducted PCR with primers 5′-ATACCATGGCTAGCACCACGGCGAGAGCTC-3′ and 5′-TATCTCGAGAGCGTAGTCTGGAACGTCGTA-3′, yielding a fragment encoding ISA3 (S69–P782) without the transit peptide. This HA epitope-tagged ISA3 fragment was cloned in pET-23d (Novagen), yielding ISA3 with an HA epitope and His-tag at the C-terminus. To produce ISA3 in an active form in E. coli XL1-Blue, cells were cultured in 80 ml of LB at 200 r.p.m. at 25°C for 24 h without isopropyl-β-d-thiogalactopyranoside (IPTG) induction, and collected by centrifugation at 8,000 × g for 10 min at 4°C. The pellet was suspended in 4 ml of lysis buffer consisting of 50 mM Tris–HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA and lysozyme (1 mg ml−1) for 1 h on ice. After brief sonication, the disrupted cells were centrifuged at 12,000 × g for 10 min at 4°C. The supernatant was saved and mixed with the same volume of buffer consisting of 50 mM Tris–HCl (pH 8.0) and 20% (v/v) glycerol, and stored at −20°C until use. The proteins were separated on a 7.5% pre-cast gel (ATTO) at 10 mA for 150 min at 4°C, and transferred at 10 V overnight at 25°C to a 7% polyacrylamide gel containing one of the following α-glucans: 0.3% amylose (Sigma, Type III, A0512), 0.3% starch (Sigma, S2004), 0.3% amylopectin (Sigma, 10118), 0.2% β-limit dextrin (Megazyme) or 1% red pullulan (Megazyme) in a transfer buffer consisting of 25 mM Tris, 192 mM glycine and 50 mM β-mercaptoethanol. The gels, except for the red pullulan-containing gel, were stained with iodine solution (1% KI and 0.1% I2). The α-amylase and β-amylase activities in the leaf blade and endosperm were measured using the Ceralpha and Betamyl assay kits (Megazyme), respectively.
Laser scanning confocal microscopy
Seeds without husks were embedded in 5% agarose and cross-sectioned through the middle portion in 100 μm thick sections with a PR07 linear slicer (D.S.K.). The sections were incubated in phosphate-buffered saline (PBS), and the samples were examined using a TCS SP2 AOBS laser scanning confocal microscope (Leica Microsystems) as described previously (Yun and Kawagoe 2009). The leaf blade of L2 of 2-week-old seedling was similarly embedded in 5% agarose and cross-sectioned in 80 μm thick sections with a PR07 linear slicer. The sections were stained with 1% (v/v) Calcofluor White in PBS. The sections were illuminated with a laser beam at 405 nm and the fluorescence signals of β-glucans in the cell wall (420–480 nm) and the autofluorescence signals of Chl (660–710 nm) were obtained simultaneously using laser scanning confocal microscopy.
Seed starch preparation and scanning electron microscopy
Starch was purified from seeds of isa3, ISA3-HA and wild-type plants. Mature seeds were reduced to powder with a hammer, and the resulting powder was suspended in 1 ml of starch extraction buffer consisting of 55 mM Tris–HCl (pH 6.8), 2.3% (w/v) SDS, 10% (v/v) glycerol and 5% (v/v) β-mercaptoethanol for 20 min. The sample was centrifuged at 2,500 × g for 5 min and the supernatant was discarded. The starch pellet was washed twice with the same buffer, three times with distilled water and twice with methanol. The starch, which was resuspended in methanol, was then filtered through a layer of 30 μm mesh and dried. The resulting starch granules were directly observed under a VE-8800 SEM (Keyence) at 1 kV. Soluble and insoluble glucans were extracted from the seeds of sugary1 expressing tpGFP or tpGFP and ISA3-HA by a modified method described by Zeeman et al. (1998). Briefly, individual mature grain was ground to fine powder and the powder was suspended in 300 μl of extraction buffer consisting of 100 mM MOPS (pH 7.0) and 5 mM EDTA. The samples were centrifuged at 3,000 × g for 5 min at 4°C, and the supernatant was saved. An equal volume of extraction buffer was added to the pellet and mixed by pipeting. The samples were centrifuged again as described and the supernatant was saved and pooled. This process was repeated once more. Proteins in the pooled supernatant were removed and then phytoglycogen was collected by the method described by Zeeman et al. (1988). The amount of soluble and insoluble glucans was quantified by using a Total Starch Assay Kit (Megazyme) according to the manufacturer's instructions.
Amylose content and chain length distributions of amylopectins
Mature isa3 and wild-type grains were polished with a rice polisher (Kett Electric Laboratory) until 10% polishing was achieved. The polished grains were powdered with a high-speed vibrating mill (TI-100, Fujiwara Co.) for 1 min and the starch was purified as described above. The amylose and amylopectin contents of the purified starch were determined by using an Amylose/Amylopectin Assay Procedure kit (Megazyme) following the manufacturer's instructions. For measurement of the amount of transitory starch in the flag leaves, flag leaves at 3 weeks after flowering were harvested at the end of the day and the end of the night, and reduced to powder in liquid nitrogen in the starch extraction buffer (1 g FW per 5 ml of starch extraction buffer). The homogenate was filtered through two layers of Miracloth and centrifuged at 4,000 × g for 15 min at 4°C. The pellet was resuspended in the starch extraction buffer and centrifuged at 10,000 × g for 2 min at 4°C, and the supernatant was discarded. The pellet was similarly washed once with 90% Percoll, twice with distilled water and four times with 80% ethanol, and the pellet was air-dried. The chain length distributions of amylopectin after debranching with isoamylase were determined as previously described (Fujita et al. 2001, Umemoto et al. 2004). β-Limit dextrin and maltotriose were used as controls for short chains (i.e. DP 2 and 3).
Starch gelatinization
Starch thermographs were recorded using DSC (DSC6200S, Seiko Instruments Inc.) as described previously (Umemoto et al. 2004) with some modifications. Purified starch (5 mg) from polished grains was mixed with 45 μl of distilled water. The heating rate was 3°C min−1 over a 10–120°C temperature range. The pasting property of rice flour was measured using an RVA (RVA-4, Newport Scientific) as previously described (Umemoto et al. 2004). Flour (3.5 g) was gelatinized in 25 ml of distilled water. All analyses were carried out in triplicate.
Supplementary data
Supplementary data are available at PCP online.
Funding
This work was supported by the Bio-Oriented Technology Research Advanced Institution [Research and Development Program for New Bio-Industry Initiatives]; the Ministry of Agriculture, Forestry and Fisheries of Japan [Genomics for Agricultural Innovation, grant No. IPG-0023].
Supplementary Material
Acknowledgments
We thank Dr. Tamao Hatta at the Japan International Research Center for Agricultural Sciences for his help with DSC, and Dr. Noriaki Aoki at the National Institute of Crop Science for his help with RVA. We are grateful to Mayu Hiratsuka at the National Institute of Crop Sciences for assistance with capillary electrophoresis.
Glossary
Abbreviations
- BAM
β-amylase
- CBB
Coomassie Brilliant Blue
- DAF
days after flowering
- DBE
debranching enzyme
- DP
degree of polymerization
- DSC
differential scanning calorimetry
- GFP
green fluorescent protein
- HA
hemagglutinin
- ISA
isoamylase
- KD
knockdown
- PBS
phosphate-buffered saline
- PUL/LD
pullulanase/limit dextrinase
- RNAi
RNA interference
- RVA
rapid visco-analyzer
- SEM
scanning electron microscope
- SSIVb
soluble starch synthase IVb
- tpGFP
plastid-targeted green fluorescent protein
- WSP
water-soluble polysaccharide.
References
- Asatsuma S, Sawada C, Itoh K, Okito M, Kitajima A, Mitsui T. Involvement of alpha-amylase I-1 in starch degradation in rice chloroplasts. Plant Cell Physiol. 2005;46:858–869. doi: 10.1093/pcp/pci091. [DOI] [PubMed] [Google Scholar]
- Ball S, Guan HP, James M, Myers A, Keeling P, Mouille G, et al. From glycogen to amylopectin: a model for the biogenesis of the plant starch granule. Cell. 1996;86:349–352. doi: 10.1016/s0092-8674(00)80107-5. [DOI] [PubMed] [Google Scholar]
- Burton RA, Jenner H, Carrangis L, Fahy B, Fincher GB, Hylton C, et al. Starch granule initiation and growth are altered in barley mutants that lack isoamylase activity. Plant J. 2002;31:97–112. doi: 10.1046/j.1365-313x.2002.01339.x. [DOI] [PubMed] [Google Scholar]
- Delatte T, Trevisan M, Parker ML, Zeeman SC. Arabidopsis mutants Atisa1 and Atisa2 have identical phenotypes and lack the same multimeric isoamylase, which influences the branch point distribution of amylopectin during starch synthesis. Plant J. 2005;41:815–830. doi: 10.1111/j.1365-313X.2005.02348.x. [DOI] [PubMed] [Google Scholar]
- Delatte T, Umhang M, Trevisan M, Eicke S, Thorneycroft D, Smith SM, et al. Evidence for distinct mechanisms of starch granule breakdown in plants. J. Biol. Chem. 2006;281:12050–12059. doi: 10.1074/jbc.M513661200. [DOI] [PubMed] [Google Scholar]
- Dinges JR, Colleoni C, Myers AM, James MG. Molecular structure of three mutations at the maize sugary1 locus and their allele-specific phenotypic effects. Plant Physiol. 2001;125:1406–1418. doi: 10.1104/pp.125.3.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinges JR, Colleoni C, James MG, Myers AM. Mutational analysis of the pullulanase-type debranching enzyme of maize indicates multiple functions in starch metabolism. Plant Cell. 2003;15:666–680. doi: 10.1105/tpc.007575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edner C, Li J, Albrecht T, Mahlow S, Hejazi M, Hussain H, et al. Glucan, water dikinase activity stimulates breakdown of starch granules by plastidial beta-amylases. Plant Physiol. 2007;145:17–28. doi: 10.1104/pp.107.104224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Hasegawa H, Taira T. The isolation and characterization of a waxy mutant of diploid wheat (Triticum monococcum L.) Plant Sci. 2001;160:595–602. doi: 10.1016/s0168-9452(00)00408-8. [DOI] [PubMed] [Google Scholar]
- Fujita N, Toyosawa Y, Utumi Y, Higuchi T, Hanashiro I, Ikegami A, et al. Characterization of pullulanase (PUL)-deficient mutants of rice (Oryza sativa L.) and the function of PUL on starch biosynthesis in the developing rice endosperm. J. Exp. Bot. 2009;60:1009–1023. doi: 10.1093/jxb/ern349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Yoshida M, Asakura N, Ohdan T, Miyao A, Hirochika H, et al. Function and characterization of starch synthase I using mutants in rice. Plant Physiol. 2006;140:1070–1084. doi: 10.1104/pp.105.071845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Yoshida M, Kondo T, Saito K, Utsumi Y, Tokunaga T, et al. Characterization of SSIIIa-deficient mutants of rice (Oryza sativa L.); the function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol. 2007;144:2009–2023. doi: 10.1104/pp.107.102533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton DC, Stettler M, Mettler T, Vaughan CK, Li J, Francisco P, et al. beta-AMYLASE4, a noncatalytic protein required for starch breakdown, acts upstream of three active beta-amylases in Arabidopsis chloroplasts. Plant Cell. 2008;20:1040–1058. doi: 10.1105/tpc.107.056507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T, Terao T. A comprehensive expression analysis of the starch synthase gene family in rice (Oryza sativa L.) Planta. 2004;220:9–16. doi: 10.1007/s00425-004-1314-6. [DOI] [PubMed] [Google Scholar]
- Hussain H, Mant A, Seale R, Zeeman S, Hinchliffe E, Edwards A, et al. Three isoforms of isoamylase contribute different catalytic properties for the debranching of potato glucans. Plant Cell. 2003;15:133–149. doi: 10.1105/tpc.006635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh J-I, Nonomura K-I, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, et al. Rice plant development: from zygote to spikelet. Plant Cell Physiol. 2005;46:23–47. doi: 10.1093/pcp/pci501. [DOI] [PubMed] [Google Scholar]
- James MG, Denyer K, Myers AM. Starch synthesis in the cereal endosperm. Curr. Opin. Plant Biol. 2003;6:215–222. doi: 10.1016/s1369-5266(03)00042-6. [DOI] [PubMed] [Google Scholar]
- Kawagoe Y, Kubo A, Satoh H, Takaiwa F, Nakamura Y. Roles of isoamylase and ADP-glucose pyrophosphorylase in starch granule synthesis in rice endosperm. Plant J. 2005;42:164–174. doi: 10.1111/j.1365-313X.2005.02367.x. [DOI] [PubMed] [Google Scholar]
- Kitajima A, Asatsuma S, Okada H, Hamada Y, Kaneko K, Nanjo Y, et al. Plastid targeting of α-amylase glycoprotein from the Golgi apparatus through the secretory pathway. Plant Cell. 2009;21:2844–2858. doi: 10.1105/tpc.109.068288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Fujita N, Harada K, Matsuda T, Satoh H, Nakamura Y. The starch-debranching enzymes isoamylase and pullulanase are both involved in amylopectin biosynthesis in rice endosperm. Plant Physiol. 1999;121:399–409. doi: 10.1104/pp.121.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Rahman S, Utsumi Y, Li ZY, Mukai Y, Yamamoto M, et al. Complementation of sugary-1 phenotype in rice endosperm with the wheat isoamylase1 gene supports a direct role for isoamylase1 amylopectin biosynthesis. Plant Physiol. 2005;137:43–56. doi: 10.1104/pp.104.051359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Sharkey TD. The importance of maltose in transitory starch breakdown. Plant Cell Environ. 2006;29:353–366. doi: 10.1111/j.1365-3040.2005.01480.x. [DOI] [PubMed] [Google Scholar]
- Miyagishima S-Y, Kabeya Y. Chloroplast division: squeezing the photosynthetic captive. Curr. Opin. Microbiol. 2010;13:738–746. doi: 10.1016/j.mib.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Miyao A, Tanaka K, Murata K, Sawaki H, Takeda S, Abe K, et al. Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell. 2003;15:1771–1780. doi: 10.1105/tpc.012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AM, Morell MK, James MG, Ball SG. Recent progress toward understanding biosynthesis of the amylopectin crystal. Plant Physiol. 2000;122:989–998. doi: 10.1104/pp.122.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y. Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: rice endosperm as a model tissue. Plant Cell Physiol. 2002;43:718–725. doi: 10.1093/pcp/pcf091. [DOI] [PubMed] [Google Scholar]
- Ohdan T, Francisco PB, Sawada T, Hirose T, Terao T, Satoh H, et al. Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J. Exp. Bot. 2005;56:3229–3244. doi: 10.1093/jxb/eri292. [DOI] [PubMed] [Google Scholar]
- Onda Y, Kumamaru T, Kawagoe Y. ER membrane-localized oxidoreductase Ero1 is required for disulfide bond formation in the rice endosperm. Proc. Natl Acad. Sci. USA. 2009;106:14156–14161. doi: 10.1073/pnas.0904429106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ral JP, Derelle E, Ferraz C, Wattebled F, Farinas B, Corellou F, et al. Starch division and partitioning. A mechanism for granule propagation and maintenance in the picophytoplanktonic green alga Ostreococcus tauri. Plant Physiol. 2004;136:3333–3340. doi: 10.1104/pp.104.044131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Zeeman SC, Smith SM. Starch degradation. Annu. Rev. Plant Biol. 2005;56:73–98. doi: 10.1146/annurev.arplant.56.032604.144257. [DOI] [PubMed] [Google Scholar]
- Smith SM, Fulton DC, Chia T, Thorneycroft D, Chapple A, Dunstan H, et al. Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and posttranscriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol. 2004;136:2687–2699. doi: 10.1104/pp.104.044347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streb S, Delatte T, Umhang M, Eicke S, Schorderet M, Reinhardt D, et al. Starch granule biosynthesis in Arabidopsis is abolished by removal of all debranching enzymes but restored by the subsequent removal of an endoamylase. Plant Cell. 2008;20:3448–3466. doi: 10.1105/tpc.108.063487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szydlowski N, Ragel P, Raynaud S, Lucas MM, Roldán I, Montero M, et al. Starch granule initiation in Arabidopsis requires the presence of either Class IV or Class III strach synthase. Plant Cell. 2009;21:2443–2457. doi: 10.1105/tpc.109.066522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetlow IJ. Understanding storage starch biosynthesis in plants: a means to quality improvement. Can. J. Bot.-Rev. Can. Bot. 2006;84:1167–1185. [Google Scholar]
- Umemoto T, Aoki N, Lin HX, Nakamura Y, Inouchi N, Sato Y, et al. Natural variation in rice starch synthase IIa affects enzyme and starch properties. Funct. Plant Biol. 2004;31:671–684. doi: 10.1071/FP04009. [DOI] [PubMed] [Google Scholar]
- Utsumi Y, Nakamura Y. Structural and enzymatic characterization of the isoamylase1 homo-oligomer and the isoamylase1–isoamylase2 hetero-oligomer from rice endosperm. Planta. 2006;225:75–87. doi: 10.1007/s00425-006-0331-z. [DOI] [PubMed] [Google Scholar]
- Wattebled F, Dong Y, Dumez S, Delvalle D, Planchot R, Berbezy P, et al. Mutants of Arabidopsis lacking a chloroplastic isoamylase accumulate phytoglycogen and an abnormal form of amylopectin. Plant Physiol. 2005;138:184–195. doi: 10.1104/pp.105.059295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattebled F, Planchot V, Dong Y, Szydlowski N, Pontoire B, Devin A, et al. Further evidence for the mandatory nature of polysaccharide debranching for the aggregation of semicrystalline starch and for overlapping functions of debranching enzymes in Arabidopsis leaves. Plant Physiol. 2008;148:1309–1323. doi: 10.1104/pp.108.129379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Glynn JM, Olson B, Schmitz AJ, Osteryoung KW. Plastid division: across time and space. Curr. Opin. Plant Biol. 2008;11:577–584. doi: 10.1016/j.pbi.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Kuroiwa H, Misumi O, Yoshida M, Ohnuma M, Fujiwara T, et al. Chloroplasts divide by contraction of a bundle of nanofilaments consisting of polyglucan. Science. 2010;329:949–953. doi: 10.1126/science.1190791. [DOI] [PubMed] [Google Scholar]
- Yu TS, Zeeman SC, Thorneycroft D, Fulton DC, Dunstan H, Lue WL, et al. alpha-Amylase is not required for breakdown of transitory starch in Arabidopsis leaves. J. Biol. Chem. 2005;280:9773–9779. doi: 10.1074/jbc.M413638200. [DOI] [PubMed] [Google Scholar]
- Yun M-S, Kawagoe Y. Amyloplast division progresses simultaneously at multiple sites in the endosperm of rice. Plant Cell Physiol. 2009;50:1617–1626. doi: 10.1093/pcp/pcp104. [DOI] [PubMed] [Google Scholar]
- Yun M-S, Kawagoe Y. Septum formation in amyloplasts produces compound granules in the rice endosperm and is regulated by plastid division proteins. Plant Cell Physiol. 2010;51:1469–1479. doi: 10.1093/pcp/pcq116. [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Kossmann J, Smith AM. Starch: its metabolism, evolution, and biotechnological modification in plants. Annu. Rev. Plant Biol. 2010;61:209–234. doi: 10.1146/annurev-arplant-042809-112301. [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Smith SM, Smith AM. The diurnal metabolism of leaf starch. Biochem. J. 2007;401:13–28. doi: 10.1042/BJ20061393. [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Umemoto T, Lue WL, Au-Yeung P, Martin C, Smith AM, et al. A mutant of Arabidopsis lacking a chloroplastic isoamylase accumulates both starch and phytoglycogen. Plant Cell. 1998;10:1699–1711. doi: 10.1105/tpc.10.10.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.