Abstract
MicroRNAs (miRNAs) are small endogenous RNA molecules ∼22 nt in length. miRNAs are capable of posttranscriptional gene regulation by binding to their target messenger RNAs (mRNAs), leading to mRNA degradation or suppression of translation. miRNAs have recently been shown to play pivotal roles in skin development and are linked to various skin pathologies, cancer, and wound healing. This review focuses on the role of miRNAs in cutaneous biology, the various methods of miRNA modulation, and the therapeutic opportunities in treatment of skin diseases and wound healing.
Keywords: miRNA therapy, hypoxia
the skin is the largest organ of the body, accounting for ∼15% of total body weight in adult humans (60, 70). The skin protects the body against environmental hazards and prevents dehydration. The skin is made up of three distinct layers of tissue: epidermis, dermis, and hypodermis. The development of these three distinct layers as well as the normal physiological functions of the skin are highly orchestrated physiological processes involving numerous factors and a complex gene regulatory network that act in a temporally resolved integrated manner. The human genome consists of ∼20,000–25,000 protein coding genes (28, 88). Although it has been generally assumed that most genetic information is transacted by proteins, recent evidence suggests that the majority of the genomes of mammals and other complex organisms is in fact transcribed into noncoding RNAs, many of which are processed into smaller products. Little is known about these noncoding regions of the genome, which were once regarded as evolutionarily conserved “junk” DNA. MicroRNAs (miRNAs) are a major group of these noncoding RNAs and are now known to regulate almost a third of all the coding genes (104). They are small (∼22 nt long) endogenously formed repressors of gene expression. miRNAs usually bind to the 3′-untranslated region of the target messenger RNAs (mRNAs) and are capable of inducing posttranscriptional gene regulation by blocking translation, by degrading the target mRNA, or by doing both (5, 35). miRNAs can be classified into distinct families based on similarity in the 5′ seed sequences because target miRNA recognition depends on the 5′ seed sequence of the miRNA (90, 156). miRNA research in the field of dermatology is in its early phase, but the early finds are substantial, pointing toward a vast opportunity for developing effective therapies for treatment of skin diseases and wounds. The public health impact of chronic wounds is staggering. An estimated 1.3–3 million US individuals are believed to have pressure ulcers; and as many as 10–15% of the population with diabetes are at risk of developing diabetic ulcers, while many more have had venous ulcers or wounds that result from arterial disease (126). Treating these wounds is estimated to cost about $5–10 billion each year (79). miRNA-based therapeutics therefore provide opportunities for addressing chronic wounds as a major public health concern in the US and globally.
Mechanisms Underlying Biogenesis of MicroRNAs
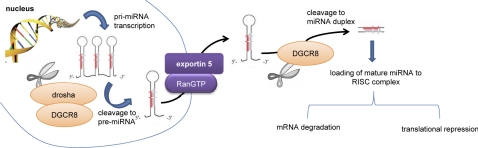
miRNA biogenesis, for the most part, occurs through the following sequential steps (50). miRNAs are encoded in the human genome as miRNA genes and are then processed to mature miRNAs. RNA polymerase II transcribes several-kilobyte-long fragments called primary miRNAs (pri-miRNAs), which are then capped and polyadenylated. The microprocessor complex, which is composed of the RNase III enzyme drosha and DGCR8, then cleaves the pri-miRNAs into ∼70-nt-long premature miRNAs (pre-miRNA). The resulting pre-miRNAs are then exported to the cytoplasm through the Ran-GTP-dependent nuclear export factor exportin-5. Another RNase III enzyme, dicer, then cleaves the pre-miRNAs into 18- to 24-nt double-stranded RNAs. The resulting RNA-duplex associates with the miRNA-induced silencing complex (RISC), where one of the strands is degraded while the other becomes the mature miRNA. The mature miRNAs interact with target mRNAs via complementarity binding with a particular region known as “seed sequence.” The resultant complex hinders assembly of ribosome, subsequently suppressing gene expression. An overview of the key processes involved in the biogenesis of miRNA is illustrated in Fig. 1.
Fig. 1.
MicroRNA (miRNA) biogenesis and posttranscriptional gene silencing mechanisms. pri-miRNA, primary miRNA; pre-miRNA, premature miRNA; RISC, miRNA-induced silencing complex.
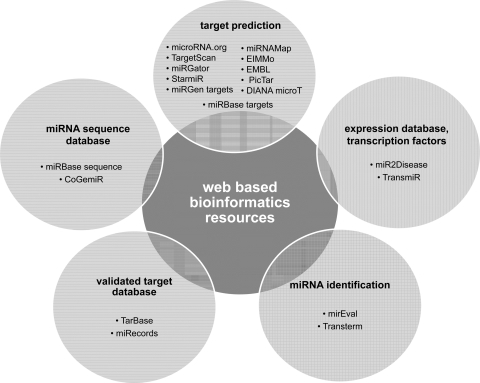
At present, a number of databases and online bioinformatics resources are available for miRNA target prediction in silico. Some of the most popular tools are listed in Fig. 2. On the basis of such resources, it is predicted that more than one-third of the mammalian mRNAs are potential targets of miRNAs (90).
Fig. 2.
Web-based bioinformatics resources used in miRNA research: PicTar (http://pictar.mdc-berlin.de/), microRNA.org (http://www.microrna.org/), TargetScan (http://www.targetscan.org/), miRGator (http://genome.ewha.ac.kr/miRGator/), StarmiR (http://sfold.wadsworth.org/starmir.pl), EMBL (http://www.russell.embl.de/miRNAs), miRGen targets (http://diana.pcbi.upenn.edu/cgi-bin/miRGen/v3/Targets.cgi), miRBase targets (http://www.mirbase.org/), EIMMo (http://www.mirz.unibas.ch/ElMMo2/), DIANA microT (http://diana.pcbi.upenn.edu/cgi-bin/micro_t.cgi), miRNAMap (http://mirnamap.mbc.nctu.edu/tw/index.php), mirEval (http://tagc.univ-mrs.fr/mireval/), Transterm (http://guinevere.otago.ac.nz/Transterm.html), miR2Disease (http://www.miR2Disease.org), TransmiR (http://202.38.126.151/hmdd/mirna/tf/), miRBase sequence (http://www.mirbase.org/), CoGemiR (http://cogemir.tigem.it/), TarBase (http://diana.cslab.ece.ntua.gr/tarbase/), miRecords (http://mirecords.biolead.org/).
MicroRNAs in Skin Morphogenesis
The skin is made of three distinct layers of tissue (130). The epidermis is populated mostly by keratinocytes along with dendritic cells, melanocytes, and Langerhans and Merkel cells. The dermis consists of collagenous and elastic fibers populated by fibroblasts, macrophages, mast cells, and lymphocytes. The dermis also consists of a glycosaminoglycan-proteoglycan fraction that functions as a supporting matrix or ground substance and makes up its base. It is composed of polysaccharides and protein that are linked to produce macromolecules and have important function in wound repair and tissue remodeling. Finally, the hypodermis is composed of adipocyte lobules. The skin contains hair follicles, which are epidermal outgrowths and have a reservoir of stem cells that may regenerate the epidermis (84, 89, 96, 139). The main functions of the skin include barrier defense, UV protection, thermoregulation, pigmentation, sensation of touch and pain, and regulation of water loss from the epidermis (130).
Dicer, which is the miRNA processing enzyme, is present both in the epidermis as well as in the outer root sheath of the hair follicles (1). Skin miRNAs can be classified into distinct groups based on analogy in the 5′ seed sequence of the miRNA (90, 156). Some of the most abundantly expressed skin miRNAs are listed in Table 1 (162). Among these most abundantly expressed miRNAs in the skin, the miRNA-200 and miRNA-19/20 families are heavily expressed in the epidermis while the miRNA-199 family is abundantly expressed highly in the hair follicles (162). This observation suggests that these miRNA families may have lineage-specific functions.
Table 1.
MicroRNAs most abundantly expressed in skin
|
Recent studies underscore the importance of miRNAs in skin development and epidermal differentiation. Dicer depletion in the epidermis results in failure of production of mature miRNAs. A striking difference in the morphogenesis of hair follicle was observed when dicer was depleted in embryonic skin progenitor cells. Mutant mice carrying floxed dicer gene were crossed to CD-1 transgenic mice expressing Cre recombinase under the control of the human keratin-14 promoter to obtain dicer1fl/fl, K14-Cre (conditional knockout) mice. Follicular epithelium progenitors evaginated toward the surface of the skin into the epidermis instead of normal invagination toward the dermis (122, 162). Loss of epithelial dicer affected both the epithelium and epithelial-mesenchymal signaling (1). In addition, absence of hair follicle stem cell marker expression and failure of dermal papilla and maintenance of the hair follicles were evidenced, resulting in stunted, hypoproliferative, and misoriented hair follicles (1). Hyperproliferation also was noted in the epidermis (1), which probably occurred because of arrest of physiological apoptosis, suggesting that the aging dicer-depleted skin might be susceptible to developing tumors. Dicer depletion also led to loss of expression of key signaling molecules such as sonic hedgehog (Shh) and Notch homolog 1 (Notch1) by postnatal day 7 (1), which may be responsible for the hyperproliferative epidermal phenotype (114) in the dicer-depleted epidermis. Inactivation of Notch1 has been also associated with hair loss followed by cyst formation (149), and thus Notch1 has emerged as a key miRNA-regulated protein in the skin that is silenced in response to dicer depletion, resulting in pathological conditions in the skin. Taken together, these data suggest that miRNAs are responsible for regulation of the genes involved in the development of the skin. Specific miRNAs required for the execution of key processes in skin morphogenesis have been identified.
In addition to its role in miRNA biogenesis, dicer has been implicated in the biogenesis of other small RNAs like endogenous small interfering RNAs (endo-siRNAs) and small nuclear (sn)/small nucleolar (sno) small RNAs. Thus whether the observations from the dicer knockout approach may therefore be suited to study the significance of miRNAs remains an open question. The role of DGCR8, on the other hand, is wholly dedicated to miRNA biogenesis (163). Phenotypically, no significant differences were observed between knockout of DGCR8 and dicer during embryonic skin development (163), thus establishing the fact that in the skin the primary function of both the proteins is miRNA biogenesis and that miRNAs are essential for skin development.
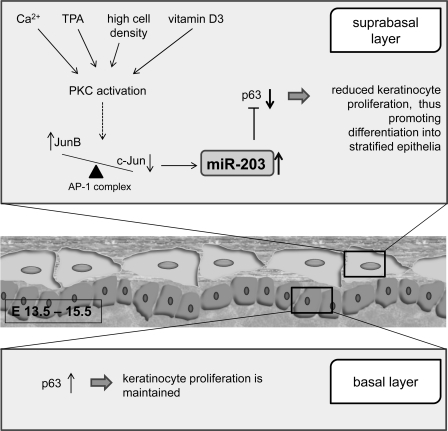
The specific role of miRNA-203 in skin morphogenesis has been tested. miRNA-203 posttranscriptionally represses p63, which is crucial in initiation of epithelial stratification and maintenance of the proliferative potential of mature keratinocytes in the basal layers (76). p63 is strongly expressed in the innermost basal layer, the home of epithelial cells with high clonogenic and proliferative capacity (127). Mice lacking all p63 isoforms have no epidermis, squamous epithelia, or epithelial appendages (103, 157, 158). Thus p63 plays a key role in the formation of epidermis and other stratified epithelia. Between embryonic days 13.5 and 15.5, the expression of miRNA-203 levels in mouse suprabasal cells increases compared with that in basal cells, leading to silencing of p63 expression and thus stalling the proliferation of the epidermis (76, 164). Although miRNA does not seem to completely silence p63 (87), it contributes as a switch between keratinocyte proliferation and differentiation in the adult epidermis. Recently, the mechanism for the induction of miRNA-203 has been reported. Ca2+, a protein kinase C (PKC) activator, is identified as an important signal in epidermal differentiation. Ca2+ regulates miRNA-203 expression in keratinocytes, and therefore miRNA-203 was expected to play an important role in development of the skin (134, 164). Specific inhibitors of PKC (GF109293X and Ro31–8220) were able to block such induction. Activator protein-1 (AP-1) proteins c-Jun and JunB were also found to be able to drive miRNA-203 expression in keratinocytes, thus suggesting that the upregulation of miRNA-203 is dependent on the activation of the PKC/AP-1 pathway (134) (Fig. 3).
Fig. 3.
miRNA-203 acts as a switch between keratinocyte proliferation and differentiation in adult epidermis and thus contributes to skin morphogenesis. TPA, 12-O-tetradecanoylphorbol-13-acetate: PKC, protein kinase C; AP-1, activator protein-1.
MicroRNA Functions in Regulation of Skin Physiology and Pathology
MicroRNAs in maintenance of skin function.
A key function of the skin is to serve as a first layer of defense against the outside environment. The epidermis is primarily responsible for this barrier function and also prevents loss of water from the organism. E-cadherin is an intercellular adhesion molecule that is specifically expressed in epithelial tissues and plays an important role in maintaining the epithelial architecture (142, 146). E-cadherin is required for the maintenance of proper localization of key tight junctional proteins, and its absence results in permeable tight junctions compromising epidermal barrier function of the skin (146). miRNA-200 and miRNA-205 are both highly expressed in normal skin and have been shown to specifically target ZEB1 and SIP1 (also known as ZEB2), the transcriptional repressors of E-cadherin (57, 74, 107). Thus miRNA-200 and miRNA-205 are expected to positively regulate E-cadherin and seem to be essential in maintaining epithelial stability. However, it must be noted that these studies were performed in cell lines and transformed cells. Therefore the significance of these results in normal skin biology remains to be elucidated.
Another important function of the skin is pigmentation. Human pigmentation involves production and dispersion of melanin by epidermal melanocytes to neighboring keratinocytes (75). Skin pigments are essential for absorbing the harmful ultraviolet radiations <310 nm and also regulate skin vitamin D production (106). miRNAs also have been reported to play a role in regulating skin pigmentation. miRNA profiling was done to compare expression in the skin of alpacas (domesticated species of South American camelids) with brown versus white coat color. Among the differentially expressed miRNAs, miRNA-25 repressed microphthalmia-associated transcription factor (MITF) in skin melanocytes, which regulates genes linked to coat color like tyrosinase (tyr) and tyrosinase-related protein 1 (172). Regulation of gene expression linked to skin color therefore has been identified as a novel functional role for miRNA-25. miRNA-434-5p is implicated in skin whitening and lightening by targeting tyr and hyaluronidase (hyal) genes (153a). Tyr plays an essential role in melanin production, and therefore Tyr repression by miRNA-434-5p resulted in significant loss of black color in murine skin as well as hair (153a).
MicroRNAs in skin cancer.
Skin cancer is classified into malignant melanoma, squamous cell carcinoma, and basal cell carcinoma (71). It is a common disease in all European-derived populations and has shown rapid increases in incidence in developed countries during the past several decades (10, 63, 108). High rates of incidence are found in Australia/New Zealand, North America, and northern Europe, and rapid increases in incidence and mortality are observed in both sexes in many countries, even where rates were formerly low (108).
Melanoma, a malignant tumor of melanocytes, causes the majority (75%) of skin cancer-related deaths (64, 161). A number of miRNAs have now been found to be associated with melanoma, while knowledge about the involvement of miRNAs in the other forms of skin cancer is still limited. The let-7 family has been implicated in the suppression of melanoma development. let-7a represses β3-integrin, while let-7b represses cyclin D1, D3, and A and cyclin-dependent kinase (Cdk)4, all of which play a role in melanoma development (105, 123). A significant upregulation of miRNA-221 and miRNA-222 has been observed in melanomas (49). This increased expression of the miRNAs activates cell proliferation and melanogenesis pathways by targeting p27Kip1 and c-Kit receptor regulation, respectively (49). Treatment with antagomiRs against miRNA-221 and miRNA-222 limited cell growth, invasion, chemotaxis, foci formation, and tumor progression (49), suggesting that approaches to antagonize miRNA-221 and miRNA-222 may have future therapeutic applications against melanoma. miRNA-196a, a miRNA that is strongly downregulated in melanoma cells, results in elevated levels of its target homeobox B7 (HOXB7) (16). HOXB7 acts as a transcription factor for basic fibroblast growth factor (bFGF), which subsequently results in elevated Ets-1 activity and BMP4 expression (20). The induction of BMP4 in turn acts as an inducer of migration of melanoma cells. miRNA-137 is also highly expressed in melanoma cell lines (8). Elevated expression of miRNA-137 represses MITF, which is known to be a master regulator of melanocyte development, survival, and function (8). Increased expression of miRNA-182 in melanoma stimulates the metastatic potential of melanocytes, whereas miRNA-182 downregulation impedes invasion and triggers apoptosis of these cancer cells (124). miRNA-182 targets MITF and FOXO3 (124), both of which antagonize invasion. miRNA-193b is downregulated in metastatic melanoma tissues (24), which represses cyclin D1 (CCND1) by directly binding to the 3′ untranslated region. Compromised expression of miRNA-193b results in increased cell proliferation (24) Several other miRNAs dysregulated in skin cancer have been validated to have oncogenic properties in other forms of cancer and may have similar function in skin. miRNA-21 is upregulated in melanoma (24) and is known to repress a number of tumor suppressors like phosphatase and tensin homolog (PTEN) (102), tromopyosin 1 (TPM1), programmed cell death 4 (PDCD4) (52, 169, 170), reversion-inducing cysteine-rich protein with Kazal motifs (RECK), and tissue inhibitor of matrix metalloproteinase 3 (TIMP3) (53). Members of the miRNA-17-92 cluster (miRNA-17–5p and miRNA-18a) and its paralog miRNA-106-25 cluster (miRNA-106b and miRNA-93) are upregulated in melanoma (24). They target E2Fs, cyclin-dependent kinase inhibitor CDKN1A, and proapoptotic protein BIM, thus regulating apoptosis and cell proliferation (101). Members of the miRNA-200 family (miRNA-200b, miRNA-200c, and miRNA-141), miRNA-205, let-7a, and let-7b are downregulated in melanomas and act as tumor suppressors (24). let-7a and let-7b are known to repress RAS, cyclins, and cyclin-dependent kinase 4 (CDK4). Cyclin D1 is a downstream effector of the RAS/MAPK signaling cascade and is an important regulator of the G1/S cell cycle transition, contributing to the phosphorylation of the retinoblastoma protein (pRB) by binding to CDK4 (113). These oncogenes mediate cellular responses to signals from growth factor receptors and thus regulate tumor progression (24, 68, 123). Downregulation of the miRNA-200 family and miRNA-205 also results in downregulation of E-cadherin, as explained above. Loss of E-cadherin results in loss of adhesion of the melanocytes to the keratinocytes and facilitates the invasion and metastasis of melanoma cells from primary tumors in the epidermis to surrounding tissues and distant organs (138).
MicroRNAs and psoriasis.
Psoriasis is the most prevalent immune-mediated chronic inflammatory disease of the skin (34), with an estimated prevalence of ∼2% of the US population (4). It is characterized by increase in differentiation and proliferation of keratinocytes and epidermal infiltration of inflammatory cells that leads to the formation of skin plaques (120), which are characterized by erythematous lesions covered with silvery scales (4).
To identify miRNAs associated with psoriasis, all human miRNAs registered in mirBase 8.0 in skin lesions of patients with psoriasis were compared with healthy human skin or to lesional skin from patients with a nonpsoriatic chronic inflammatory skin disease, atopic eczema (133). miRNA-203, miRNA-146a, and miRNA-21 were found to be upregulated in psoriasis, and miRNA-125b was found to be downregulated (15, 23). miRNA-146a represses IRAK1 and TRAF6, which mediate the signaling from the members of the tumor necrosis factor (TNF) receptor superfamily and also the members of the Toll/IL-1 family (133, 137). miRNA-125b, which is downregulated in psoriasis (132), is involved in posttranscriptional repression of TNF-α (141). TNF-α mediates leukocyte-keratinocyte interactions, and thus is involved in the pathogenesis of psoriasis (45, 94). Therefore, these two miRNAs have been implicated to play an important role in regulating the pathogenesis of psoriasis. miRNA-203 is another miRNA that is highly upregulated in psoriasis; however, the target gene for this miRNA in relevance to this disease remains to be elucidated. Targeting these specific miRNAs may therefore be of therapeutic value.
MicroRNAs and systemic sclerosis.
Systemic sclerosis (scleroderma) is a rare, chronic disorder characterized by scarring in the skin, joints, and internal organs and by blood vessel abnormalities. Systemic sclerosis can damage large areas of skin, whereby normal tissue is progressively replaced by collagen-rich extracellular matrix (ECM) (98), resulting in fibrosis of the skin and internal organs. This is believed to be caused by the transition of quiescent fibroblasts to activated myofibroblasts, which characteristically overproduce dermal fibrillar collagen (types I, III, V), collagen-modifying enzymes, and other ECM components (3). The pathogenesis of this disease is still unclear, although thickened dermis, because of uncontrolled excessive deposition of ECM, is considered the hallmark of this disease. The expression of various ECM proteins, mainly type I collagen, is upregulated in fibroblasts in systemic sclerosis patients (66). The skin on the face tightens, resulting in an inability to change facial expressions.
The miRNA-29 family has been identified as potential posttranscriptional regulators of collagen genes, of which miRNA-29a was predicted to have the best seed match to collagens and also broadly conserved among vertebrates (98). miRNA-29a targets type I and type III collagens and has been found to be strongly downregulated in skin biopsy and fibroblast samples from systemic sclerosis patients (98). miRNA-29a acts downstream of most of the profibrotic molecules identified previously such as transforming growth factor (TGF)-β, platelet-derived growth factor (PDGF)-B, and IL-4 (98), and therefore targeting miRNA-29 family members as posttranscriptional regulators could be a potent antifibrotic approach.
The miRNAs involved in the pathogenesis of the skin are summarized in Table 2.
Table 2.
MicroRNAs involved in skin diseases
| miRNAs | Levels | Gene Targets | Effect | References |
|---|---|---|---|---|
| Skin cancer | ||||
| miR-221/222 | Up | P27Kip1 | Increased cell proliferation | 49 |
| miR-221/222 | Up | cKit | Increased melanogenesis | 49 |
| miR-137 | Up | MITF | Dysregulated melanocyte development, survival, function | 8 |
| miR-182 | Up | MITF, FOXO3 | Increased metastasis | 124 |
| miR-21 | Up | PTEN, TPM1, PDCD4, RECK, TIMP3 | Increased metastasis | 24, 52, 53, 102, 169, 170 |
| miR-17-5p, miR-18a, miR-106b, miR-93 | Up | E2F, CDKN1, BIM | Increased cell proliferation, reduced apoptosis | |
| miR-196a | Down | HOXB7 | Increased migration | |
| miR-193b | Down | CCND1 | Increased cell proliferation | |
| miR-200b, miR-200c, miR-141, miR-205 | Down | E-cadherin | Increased invasion and metastasis | 138 |
| let-7a, let-7b | Down | ras, cyclin, CDK4 | Increased tumor progression | 24, 68, 113, 123 |
| Psoriasis | ||||
| miR-146a | Up | IRAK1/TRAF6 | 133, 137 | |
| miR-125b | Down | TNF-α | Constant activation of STAT3 and subsequent development of psoriatic plaques | 132, 141 |
| miR-203, miR-21 | Up | Not well defined | 15, 23 | |
| Systemic sclerosis | ||||
| miR-29a | Up | Type I and type III collagens | Increased collagen deposition and fibrosis | 98 |
MicroRNA Functions in Cutaneous Tissue Repair: Wound Healing
miRNAs are increasingly becoming important as players in wound healing (126). The repair of damaged human skin is a physiological process involving three overlapping phases (17, 18): the inflammatory phase, which is characterized by hemostasis and inflammation; the proliferation phase, which includes epithelialization, angiogenesis, granulation tissue formation, and collagen deposition; and the remodeling phase, in which organized deposition of collagen takes place over months even after the wound has closed. The cutaneous wound healing process involves changes in the expression of specific miRNAs at a specific phase of wound healing, and aberrant regulation of these specific miRNAs plays a key role in the abnormal healing of problem wounds (130).
Role of microRNAs in inflammatory phase and as regulators of angiogenesis.
The role of miRNAs in the inflammatory phase of wound healing is poorly understood. The inflammatory response to wounding starts with the passive leakage of neutrophils and other leukocytes through the damaged blood vessels into the wound (128). This is followed by a release of chemokine and cytokines like PDGF, platelet factor IV, TGF-β, and TNF-α by the immune cells in the wound (120). Neutrophils and monocytes are then recruited from nearby vessels. Neutrophils have a cleansing role and kill invading microorganisms. Extravasated monocytes mature into macrophages and demonstrate phagocytotic function. Monocytes may be either proinflammatory or anti-inflammatory and proangiogenic. The second category has been found to predominate in the later stages of wound repair (36).
One of the important roles of inflammatory cytokines at the wound site is to regulate angiogenesis (143). Endothelial cell migration and capillary formation represent the main aspects of this phase. Sprouting of capillaries into the wound bed is critical to support regenerating tissue, and therefore impairments in wound angiogenesis may lead to chronic problem wounds (22, 31, 54). The first evidence that indicated that the involvement of miRNAs in angiogenesis came from the observations that endothelial cell capillary sprouting, migration, and tubulogenesis were inhibited by knockdown of dicer and drosha (78, 129, 135). Dicer is also required for embryonic angiogenesis during mouse development (160), and the depletion of endothelial miRNAs by inactivation of dicer impairs postnatal angiogenic responses to a variety of stimuli, including exogenous vascular endothelial growth factor (VEGF), tumors, limb ischemia, and wound healing (136).
Current studies on miRNAs are mostly in vitro; however, they may provide important clues for further in vivo studies directly addressing the role of microRNAs in angiogenesis and wound healing.
Table 3 lists the key miRNAs implicated in regulating angiogenesis and predicted to have a role in angiogenesis and wound healing.
Table 3.
MicroRNAs involved in different phases of wound healing
| MicroRNAs | Targets | References |
|---|---|---|
| Inflammatory phase | ||
| miR-105 | TLR2 | 9 |
| miR-140 | PDGF receptor | 42 |
| miR-146a, miR-125b | TNF-α | 132 |
| Angiogenesis | ||
| Proangiogenic miRNAs | ||
| miR-17-92 | TSP-1, CTGF | 38, 135 |
| miR-126 | Spred1, PIK3R2 | 51, 80, 150 |
| miR-130a | GAX, HOXA5 | 25 |
| miR-210 | EFNA3 | 46, 115 |
| miR-296 | HGS | 154 |
| miR-378 | Fus-1, Sufu | 85 |
| Antiangiogenic miRNAs | ||
| miR-92a | Integrin-α5 | 13 |
| miR-17 | Janus Kinase 1 | 39 |
| miR-15b,miR-16, miR-20a, miR-20b | VEGF | 61 |
| miR-320 | IGF-1 | 152 |
| miR-221, miR-222 | c-kit | 112 |
| Proliferative phase | ||
| miR-184 | Akt | 51, 150, 166 |
| miR-205 | SHIP2, Rho-ROCK1 | 166 |
| miR-210 | E2F3, ISCU 1/2 | 11 |
| Remodeling phase | ||
| miR-29a | Type I and type II collagen | 98 |
| miR-29b,29c | Smads, β-catenin | 91, 148 |
| miR-192 | SIP1 | 72 |
Role of microRNAs in proliferative phase.
An essential aspect of the healing sequence in the proliferative phase is reepithelialization, which includes migration and proliferation of keratinocytes from the wound edge (116). Keratinocyte migration has been reported to be faster upon silencing of SH2-containing phosphoinositide 5-phosphatase 2 (SHIP2) and enhanced AKT signaling (165, 166). miRNA-205 has been found to repress SHIP2, which can interfere with the Akt signaling pathway (165, 166). miRNA-184, however, antagonizes the repression of SHIP2 by miRNA-205, and thus miRNA-184 indirectly represses AKT expression (165, 166). Furthermore, miRNA-205 is also required for downregulation of Rho-ROCK1 activity and thus repression of phospho-cofilin expression (165, 166). Dephosphorylated cofilin is active and severs actin filaments and regulates actin polymerization and depolymerization during migration (37). Furthermore, active cofilin increases cell motility (151). miRNA-205 upregulation thus decreases phospho-cofilin and increases cofilin expression and therefore modulates F-actin organization (165, 166) and enhances cell motility.
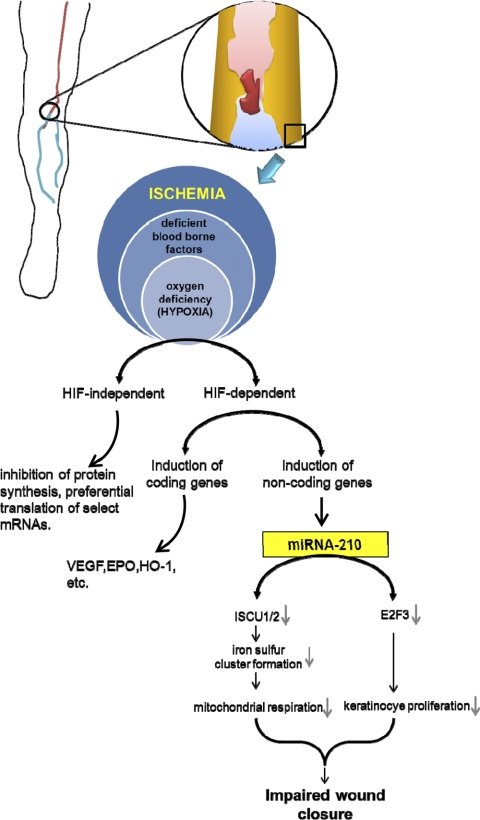
miRNA-210 is another miRNA that profoundly influences keratinocyte proliferation and thus wound closure (11, 48). miRNA-210 represents a class of miRNAs called “hypoxamiRs,” which are hypoxia sensitive (11, 48). This is of significance in the context of chronic wounds. Most chronic wounds are ischemic in nature. Ischemia is characterized by lack of perfusion, hypoxia (reduction in oxygen delivery below tissue demand), and insufficient nutrient supply. Chronic ischemic wounds are thus essentially hypoxic (125). Extreme near-anoxic hypoxia, as is commonly observed in these wounds, is not compatible with wound repair (125). Hypoxia sensing is generally mediated by a transcription factor called hypoxia-inducible factor (HIF). HIF binds to a hypoxia-responsive element (HRE) in the target genes. HIF, however, has two forms—HIF-1α (and its paralogs HIF-2α and HIF-3α) and HIF-1β. In normoxic conditions, molecular O2 targets HIF for degradation by posttranslational hydroxylation at specific prolyl residues (PHD domains) within these subunits, which increases the affinity for the ubiquitin ligases for proteolytic destruction by the ubiquitin/proteasome pathway. The O2-dependent hydroxylation process is, however, suppressed during hypoxia, resulting in stabilization of HIF-1α and subsequent binding to its constitutive partner HIF-1β to induce transactivation (125). Stabilization of HIF has been classically known to induce the transcription of coding genes like VEGF, erythropoietin, nitric oxide synthase-2, transferrin, and others (69). The transcriptional regulation of hypoxia-sensitive miRNAs by HIF has been recently reported (21, 81), and among these the most prominent is miRNA-210 (30, 81). miRNA-210 has been reported to be upregulated in hypoxia in almost all cell and tissue types tested (19, 30, 46, 56, 58, 61, 81, 110, 115, 171) and is hypoxia specific, since growth factor deprivation, osmotic stress, acidosis, and oxidative stress do not result in its induction (46).
Among the targets of miRNA-210, transcription factor E2F3, which is necessary for keratinocyte proliferation, has been implicated in wound healing (Fig. 4) (11). A number of E2F3-responsive genes like B-myb, cyclin A, cdc2, cdc6, and DHFR determine the timing of the G1/S transition, the rate of DNA synthesis, and thereby the rate of cellular proliferation (62). Because keratinocyte proliferation is an integral part of wound healing, induction of miRNA-210 and repression of E2F3 hinder closure of ischemic wounds (11).
Fig. 4.
Role of miRNA-210 in the impairment of healing in chronic ischemic wounds. HIF, hypoxia-inducible factor.
In another study, the iron sulfur cluster assembly proteins ISCU1 and ISCU2 have been identified as HIF-dependent genes and have been verified to be directly under the control of miRNA-210 (21). ISCU1/2 are essential for iron sulfur cluster biogenesis and are incorporated into wide variety of proteins, many of which, like Complex I and aconitase, are involved in mitochondrial metabolism. Chronic repression of mitochondrial function during hypoxia has been linked to various pathological consequences including ischemic diseases (12, 40, 109, 131). Therefore, this may be another possible mechanism for the observed impairment of closure upon HIF-dependent upregulation of miRNA-210.
MicroRNAs in remodeling: scarred and scarless healing.
Collagen deposition is an important aspect of the remodeling phase. As mentioned above, miRNA-29a directly regulates collagen expression at the posttranscriptional level (98). In normal skin fibroblasts, miRNA-29a is under the control of TGF-β, PDGF-B, and IL-4 (98). Mammalian fetal skin can heal without a scar (6, 7, 32, 59), whereas during the late gestational stage it transitions to a scarring phenotype (7, 29). Several miRNAs are differentially expressed between the two stages and probably contribute to this transition (26), and miRNA-29b, miRNA-29c, and miRNA-192 have been found to be the key mediators (26), with their levels being highly induced during the late gestation phase. miRNA-29b and miRNA-29c repress several ECM proteins, antifibrotic TGF-β, and proteins like Smads and β-catenin, which are involved in the signaling pathways important for scarless healing (91, 148). miRNA-192 enhances collagen 1α2 expression by targeting Smad-interacting protein 1 (SIP1) (72).
Clinical Applications of MicroRNAs and Therapeutic Strategies in Wound Healing
miRNA-based therapeutics provides a unique advantage, because by modulating a single miRNA a group of functionally related genes in a pathway can be targeted compared with modulating a single gene at a time as in conventional gene therapy. In addition, miRNAs can be efficiently inhibited both in vitro and in vivo. The levels of aberrantly expressed miRNAs can be reduced while the levels of the beneficial miRNAs are elevated to obtain desired results. The following are some of the clinical and therapeutic applications of miRNAs.
MicroRNAs as biomarkers.
miRNAs can serve as excellent biomarkers for prognosis of melanoma. In a recent study (86), ∼900 miRNA sequences were screened in blood samples of melanoma patients and healthy individuals: 21 statistically significant miRNAs were found to be downregulated and 30 miRNAs were upregulated in blood cells of melanoma patients, compared with blood cells of healthy control subjects. From these, 16 miRNAs were selected based on which the patients were classified as having melanoma or as being healthy. These miRNAs included hsa-miR-186, hsa-let-7d*, hsa-miR-18a*, hsa-miR-145, hsa-miR-99a, hsa-miR-664, hsa-miR-501-5p, hsa-miR-378*, hsa-miR-29c*, hsa-miR-1280, hsa-miR-365, hsa-miR-1249, hsa-miR-328, hsa-miR-422a, hsa-miR-30d, and hsa-miR-17*. This study highlights the outstanding potential of miRNA biomarker profiling from blood cells as a noninvasive biomarker test for melanoma and other forms of cancer. Preliminary reports also indicate that the miRNA biogenesis complex can act as a biomarker for prognosis of chronic ischemic wounds (47).
Upregulation of potentially beneficial microRNAs.
The overexpression of a miRNA can be achieved with the use of synthetic short double-stranded oligonucleotides (mimics). Mimics are double stranded, with one strand, called guide, whose sequence is same as the mature miRNA while the other, called passenger, is complementary to the mature sequence. Only the guide sequence is incorporated into the RISC complex (47). Another method of miRNA overexpression is the use of lentiviral vectors with built-in miRNA precursor constructs. Initial results from clinical trials using lentivirus have been positive (73, 159); however, further advancements are required before this technology can be used successfully in humans.
Downregulation of potentially harmful microRNAs.
As reported for ischemic wounds, miRNA-210 downregulation is expected to be beneficial for healing (11) and this can be achieved by complementary oligonucleotides. Anti-miRNAs essentially act as competitive inhibitors binding to the mature miRNA and also can affect miRNA maturation by binding to the pre-miRNA.
SPONGES.
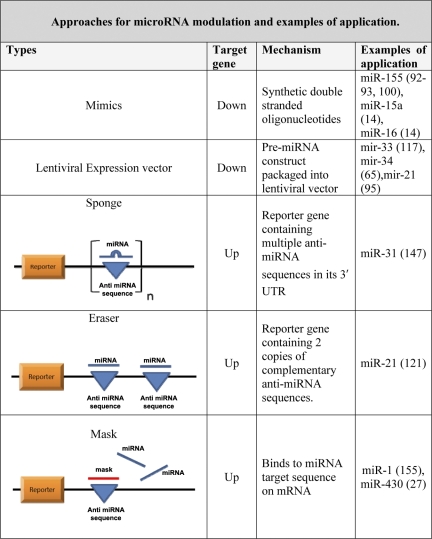
Sponges are essentially competitive inhibitors that contain multiple, tandem binding sites to a miRNA of interest. Sponges have a bulge at the position cleaved by the Ago2. They can thus stably interact with the miRNA target that cannot be sliced. Sponges can inhibit miRNA clusters with a complementary heptameric seed. Sponges therefore have the advantage of being able to block all the miRNAs that recognize the same sequence and thus inhibit all the miRNAs of the same family, resulting in a much more effective outcome (47).
ERASERS.
miRNA “erasers” are similar to sponges, except that they use only two copies of the perfectly complementary antisense sequence of the miRNA (47).
MASKS.
Since miRNAs regulate hundreds of genes, manipulating miRNA may repress other targets, many of which may be undesirable. To solve this problem, an oligonucleotide is made to bind to the miRNA target sequence of the specific mRNA of interest, thus preventing the miRNA/mRNA association (47). Thus, with this approach, the miRNA interaction with one specific target can be modulated (47). These approaches along with some examples of their application are summarized in Fig. 5.
Fig. 5.
Approaches for miRNA modulation and examples of application. UTR, untranslated region.
In vivo delivery systems.
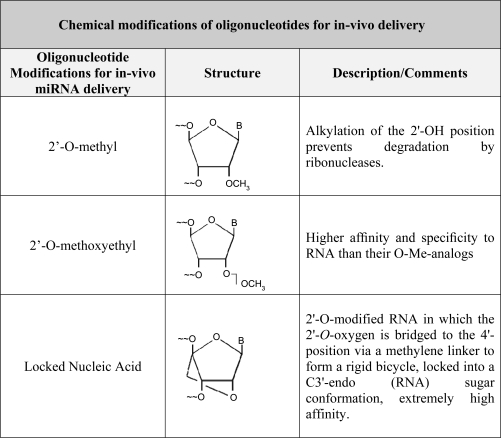
Successful delivery of the synthetic oligonucleotides listed in Fig. 5 will depend on their resistance to degradation in tissues, specificity, and high binding affinity to the specific miRNA in question. To achieve these goals chemical modifications of the oligonucleotides are often necessary. Delivery of anti-miRNAs to mammalian tissues is generally administered by either of these approaches (47): 1) intravenous injection of antagomiRs (chemically modified cholesterol-conjugated single-strand oligos) or 2) conjugation of RNA oligos with other lipophilic molecules, i.e., high-density lipoproteins. Three forms of chemically modified oligonucleotides that have been used are 1) 2′-O-methyl group (OMe)-modified oligonucleotides; 2) 2′-O-methoxyethyl-modified oligonucleotides, and 3) locked nucleic acid (LNA) (153) (Fig. 6).
Fig. 6.
Chemical modifications of oligonucleotides for in vivo delivery. OMe, 2′-O-methyl group.
Some of the noteworthy successes in the delivery of oligonucleotides to modulate miRNA levels include inhibition of miRNA-21 with a cholesterol-conjugated anti-miRNA in fibroblasts of the failing heart, which decreased interstitial fibrosis and cardiac hypertrophy (140). Exogenous delivery of synthetic let-7b miRNA by intratumoral injection or by intranasal delivery reduced tumor growth in non-small-cell lung cancer (NSCLC) patients, in the k-ras-dependent mouse model of NSCLC (44, 82), and in lung cancer xenograft mouse models (144). let-7b acts as a tumor suppressor, and therefore restoring let-7b level led to negative regulation of its target oncogenes such as RAS, MYC, and HMGA2 and cell cycle promoters such as CDCD25A, CDK6, and CCND2, thus reducing tumor growth (67, 68, 99, 119). Systemic delivery of miRNA-26a by adeno-associated virus (AAV) reduced tumor growth in a mouse model of hepatocellular carcinoma (77). Cholesterol-conjugated administration of anti-miRNA-126 suppressed allergic asthma response by airway hyperresponsiveness and inflammation (97). Antagonism of miRNA-122 by systemic administration of a miRNA-122 antisense oligonucleotide reduced plasma cholesterol levels and decreased hepatic fatty acid and cholesterol synthesis rates in mice (43). miRNA-122 is also an essential host factor for hepatitis C virus replication, and inhibition of miRNA-122 in liver cells caused reduction of replicating hepatitis C viral RNAs (83). pre-miRNA-1 cloned into suitable vectors and introduced into mice repressed its target Hand2 (168).
These results heighten interest in miRNA-based therapies.
Clinical Trials of MicroRNA-Mediated Therapy of Skin Diseases
Three clinical trials related to miRNA and skin have been completed but are yet to be reported. The purposes of these studies were (www.clinicaltrials.gov): 1) “MicroRNA Expression and Function in Cutaneous Malignant Melanoma” (completed 2007); 2) “Immunohistochemical Expression Patterns of MicroRNA Processing Enzyme Dicer in Cutaneous Malignant Melanoma, Benign and Dysplastic Melanocytic Naevi” (completed 2009); and 3) “Expression Levels of MicroRNA Processing Enzymes Dicer and Drosha in Epithelial Skin Cancer” (completed 2009). A clinical trial study on the “role of microRNA in the development of cutaneous squamous cell carcinoma” is under way and currently recruiting participants.
A phase I clinical trial in which an LNA-based anti-miRNA targeting miRNA-122 was developed as hepatitis C therapy has recently been completed. This further validates the viability of miRNAs as therapeutic targets and miRNA inhibitors and mimics as a new class of drugs (47).
Concluding Remarks
A majority of the human genome is composed of noncoding genes, and as of this date, very little knowledge is available about the functional significance of these noncoding regions. miRNAs are noncoding RNAs that recently have been reported to regulate skin development, pathogenesis of the skin, and wound healing. The discovery of miRNAs has opened up vast therapeutic opportunities. Chronic wounds present a major health burden and drain on resources, and developing newer and more effective treatments has therefore become a necessity. Knowledge of miRNA function in the regulation of wound healing and developing improved miRNA modulation techniques in the skin will help in translating this knowledge into more effective therapies. Of note, the same miRNAs often have been found to have different and contrasting function in different cell types. To solve this problem, a new technology employing laser capture microdissection can be used to perform cell type-specific miRNA studies in in vivo tissue samples. Recent publications demonstrate the feasibility of using this technique for analysis of genes captured from blood vessels from human tissues (118), prostate cancer epithelial and interstitial stromal cells, and epithelial cells from other regions.
GRANTS
This work was supported by National Institute of General Medical Sciences Grants GM-069589 and GM-077185 to C. K. Sen.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
REFERENCES
- 1. Andl T, Murchison EP, Liu F, Zhang Y, Yunta-Gonzalez M, Tobias JW, Andl CD, Seykora JT, Hannon GJ, Millar SE. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol 16: 1041–1049, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 27: 2128–2136, 2008 [DOI] [PubMed] [Google Scholar]
- 3. Asano Y. Future treatments in systemic sclerosis. J Dermatol 37: 54–70, 2010 [DOI] [PubMed] [Google Scholar]
- 4. Bagel J. Topical therapies for the treatment of plaque psoriasis. Cutis 84: 3–13, 2009 [PubMed] [Google Scholar]
- 5. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297, 2004 [DOI] [PubMed] [Google Scholar]
- 6. Beanes SR, Dang C, Soo C, Ting K. Skin repair and scar formation: the central role of TGF-beta. Expert Rev Mol Med 5: 1–22, 2003 [DOI] [PubMed] [Google Scholar]
- 7. Beanes SR, Hu FY, Soo C, Dang CM, Urata M, Ting K, Atkinson JB, Benhaim P, Hedrick MH, Lorenz HP. Confocal microscopic analysis of scarless repair in the fetal rat: defining the transition. Plast Reconstr Surg 109: 160–170, 2002 [DOI] [PubMed] [Google Scholar]
- 8. Bemis LT, Chen R, Amato CM, Classen EH, Robinson SE, Coffey DG, Erickson PF, Shellman YG, Robinson WA. MicroRNA-137 targets microphthalmia-associated transcription factor in melanoma cell lines. Cancer Res 68: 1362–1368, 2008 [DOI] [PubMed] [Google Scholar]
- 9. Benakanakere MR, Li Q, Eskan MA, Singh AV, Zhao J, Galicia JC, Stathopoulou P, Knudsen TB, Kinane DF. Modulation of TLR2 protein expression by miR-105 in human oral keratinocytes. J Biol Chem 284: 23107–23115, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Berwick M, Erdei E, Hay J. Melanoma epidemiology and public health. Dermatol Clin 27: 205–214, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Biswas S, Roy S, Banerjee J, Hussain SR, Khanna S, Meenakshisundaram G, Kuppusamy P, Friedman A, Sen CK. Hypoxia inducible microRNA 210 attenuates keratinocyte proliferation and impairs closure in a murine model of ischemic wounds. Proc Natl Acad Sci USA 107: 6976–6981, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Blomgren K, Zhu C, Hallin U, Hagberg H. Mitochondria and ischemic reperfusion damage in the adult and in the developing brain. Biochem Biophys Res Commun 304: 551–559, 2003 [DOI] [PubMed] [Google Scholar]
- 13. Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, Burchfield J, Fox H, Doebele C, Ohtani K, Chavakis E, Potente M, Tjwa M, Urbich C, Zeiher AM, Dimmeler S. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science 324: 1710–1713, 2009 [DOI] [PubMed] [Google Scholar]
- 14. Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D'Urso L, Pagliuca A, Biffoni M, Labbaye C, Bartucci M, Muto G, Peschle C, De Maria R. The miR-15a-miR-16–1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med 14: 1271–1277, 2008 [DOI] [PubMed] [Google Scholar]
- 15. Bostjancic E, Glavac D. Importance of microRNAs in skin morphogenesis and diseases. Acta Dermatovenerol Alp Panonica Adriat 17: 95–102, 2008 [PubMed] [Google Scholar]
- 16. Braig S, Mueller DW, Rothhammer T, Bosserhoff AK. MicroRNA miR-196a is a central regulator of HOX-B7 and BMP4 expression in malignant melanoma. Cell Mol Life Sci 67: 3535–3548, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Broughton G, 2nd, Janis JE, Attinger CE. The basic science of wound healing. Plast Reconstr Surg 117: 12S–34S, 2006 [DOI] [PubMed] [Google Scholar]
- 18. Broughton G, 2nd, Janis JE, Attinger CE. Wound healing: an overview. Plast Reconstr Surg 117: 1e-S–32e-S, 2006 [DOI] [PubMed] [Google Scholar]
- 19. Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H, Harris AL, Gleadle JM, Ragoussis J. hsa-miR-210 is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res 14: 1340–1348, 2008 [DOI] [PubMed] [Google Scholar]
- 20. Care A, Silvani A, Meccia E, Mattia G, Stoppacciaro A, Parmiani G, Peschle C, Colombo MP. HOXB7 constitutively activates basic fibroblast growth factor in melanomas. Mol Cell Biol 16: 4842–4851, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chan SY, Loscalzo J. MicroRNA-210: a unique and pleiotropic hypoxamir. Cell Cycle 9: 1072–1083, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chbinou N, Frenette J. Insulin-dependent diabetes impairs the inflammatory response and delays angiogenesis following Achilles tendon injury. Am J Physiol Regul Integr Comp Physiol 286: R952–R957, 2004 [DOI] [PubMed] [Google Scholar]
- 23. Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science 303: 83–86, 2004 [DOI] [PubMed] [Google Scholar]
- 24. Chen J, Feilotter HE, Pare GC, Zhang X, Pemberton JG, Garady C, Lai D, Yang X, Tron VA. MicroRNA-193b represses cell proliferation and regulates cyclin D1 in melanoma. Am J Pathol 176: 2520–2529, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen Y, Gorski DH. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood 111: 1217–1226, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cheng J, Yu H, Deng S, Shen G. MicroRNA profiling in mid- and late-gestational fetal skin: implication for scarless wound healing. Tohoku J Exp Med 221: 203–209, 2010 [DOI] [PubMed] [Google Scholar]
- 27. Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science 318: 271–274, 2007 [DOI] [PubMed] [Google Scholar]
- 28. Clamp M, Fry B, Kamal M, Xie X, Cuff J, Lin MF, Kellis M, Lindblad-Toh K, Lander ES. Distinguishing protein-coding and noncoding genes in the human genome. Proc Natl Acad Sci USA 104: 19428–19433, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Colwell AS, Krummel TM, Longaker MT, Lorenz HP. An in vivo mouse excisional wound model of scarless healing. Plast Reconstr Surg 117: 2292–2296, 2006 [DOI] [PubMed] [Google Scholar]
- 30. Crosby ME, Kulshreshtha R, Ivan M, Glazer PM. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res 69: 1221–1229, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. da Costa Pinto FA, Malucelli BE. Inflammatory infiltrate, VEGF and FGF-2 contents during corneal angiogenesis in STZ-diabetic rats. Angiogenesis 5: 67–74, 2002 [DOI] [PubMed] [Google Scholar]
- 32. Dang C, Ting K, Soo C, Longaker MT, Lorenz HP. Fetal wound healing current perspectives. Clin Plast Surg 30: 13–23, 2003 [DOI] [PubMed] [Google Scholar]
- 34. Davidovici BB, Sattar N, Jorg PC, Puig L, Emery P, Barker JN, van de Kerkhof P, Stahle M, Nestle FO, Girolomoni G, Krueger JG. Psoriasis and systemic inflammatory diseases: potential mechanistic links between skin disease and co-morbid conditions. J Invest Dermatol 130: 1785–1796 [DOI] [PubMed] [Google Scholar]
- 35. Dennis C. The brave new world of RNA. Nature 418: 122–124, 2002 [DOI] [PubMed] [Google Scholar]
- 36. Deonarine K, Panelli MC, Stashower ME, Jin P, Smith K, Slade HB, Norwood C, Wang E, Marincola FM, Stroncek DF. Gene expression profiling of cutaneous wound healing. J Transl Med 5: 11, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. DesMarais V, Ghosh M, Eddy R, Condeelis J. Cofilin takes the lead. J Cell Sci 118: 19–26, 2005 [DOI] [PubMed] [Google Scholar]
- 38. Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, Thomas-Tikhonenko A. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet 38: 1060–1065, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Doebele C, Bonauer A, Fischer A, Scholz A, Reiss Y, Urbich C, Hofmann WK, Zeiher AM, Dimmeler S. Members of the microRNA-17–92 cluster exhibit a cell-intrinsic antiangiogenic function in endothelial cells. Blood 115: 4944–4950, 2010 [DOI] [PubMed] [Google Scholar]
- 40. Duchen MR. Roles of mitochondria in health and disease. Diabetes 53, Suppl 1: S96–S102, 2004 [DOI] [PubMed] [Google Scholar]
- 41. Dziunycz P, Iotzova-Weiss G, Eloranta JJ, Lauchli S, Hafner J, French LE, Hofbauer GF. Squamous cell carcinoma of the skin shows a distinct microRNA profile modulated by UV radiation. J Invest Dermatol 130: 2686–2689, 2010 [DOI] [PubMed] [Google Scholar]
- 42. Eberhart JK, He X, Swartz ME, Yan YL, Song H, Boling TC, Kunerth AK, Walker MB, Kimmel CB, Postlethwait JH. MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat Genet 40: 290–298, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 3: 87–98, 2006 [DOI] [PubMed] [Google Scholar]
- 44. Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, Weidhaas JB, Brown D, Bader AG, Slack FJ. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle 7: 759–764, 2008 [DOI] [PubMed] [Google Scholar]
- 45. Ettehadi P, Greaves MW, Wallach D, Aderka D, Camp RD. Elevated tumour necrosis factor-alpha (TNF-alpha) biological activity in psoriatic skin lesions. Clin Exp Immunol 96: 146–151, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fasanaro P, D'Alessandra Y, Di Stefano V, Melchionna R, Romani S, Pompilio G, Capogrossi MC, Martelli F. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem 283: 15878–15883, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fasanaro P, Greco S, Ivan M, Capogrossi MC, Martelli F. microRNA: emerging therapeutic targets in acute ischemic diseases. Pharmacol Ther 125: 92–104, 2010 [DOI] [PubMed] [Google Scholar]
- 48. Fasanaro P, Greco S, Lorenzi M, Pescatori M, Brioschi M, Kulshreshtha R, Banfi C, Stubbs A, Calin GA, Ivan M, Capogrossi MC, Martelli F. An integrated approach for experimental target identification of hypoxia-induced miR-210. J Biol Chem 284: 35134–35143, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Felicetti F, Errico MC, Segnalini P, Mattia G, Care A. MicroRNA-221 and -222 pathway controls melanoma progression. Expert Rev Anticancer Ther 8: 1759–1765, 2008 [DOI] [PubMed] [Google Scholar]
- 50. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102–114, 2008 [DOI] [PubMed] [Google Scholar]
- 51. Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell 15: 272–284, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem 283: 1026–1033, 2008 [DOI] [PubMed] [Google Scholar]
- 53. Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, Krichevsky AM. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol 28: 5369–5380, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Galeano M, Deodato B, Altavilla D, Cucinotta D, Arsic N, Marini H, Torre V, Giacca M, Squadrito F. Adeno-associated viral vector-mediated human vascular endothelial growth factor gene transfer stimulates angiogenesis and wound healing in the genetically diabetic mouse. Diabetologia 46: 546–555, 2003 [DOI] [PubMed] [Google Scholar]
- 55. Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, Ambros VR, Israel MA. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res 67: 2456–2468, 2007 [DOI] [PubMed] [Google Scholar]
- 56. Giannakakis A, Sandaltzopoulos R, Greshock J, Liang S, Huang J, Hasegawa K, Li C, O'Brien-Jenkins A, Katsaros D, Weber BL, Simon C, Coukos G, Zhang L. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol Ther 7: 255–264, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10: 593–601, 2008 [DOI] [PubMed] [Google Scholar]
- 58. Guimbellot JS, Erickson SW, Mehta T, Wen H, Page GP, Sorscher EJ, Hong JS. Correlation of microRNA levels during hypoxia with predicted target mRNAs through genome-wide microarray analysis. BMC Med Genomics 2: 15, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hantash BM, Zhao L, Knowles JA, Lorenz HP. Adult and fetal wound healing. Front Biosci 13: 51–61, 2008 [DOI] [PubMed] [Google Scholar]
- 60. Healy B. Skin deep. As the body's largest organ, skin is a powerful yet unappreciated veneer. US News World Rep 139: 66–68, 2005 [PubMed] [Google Scholar]
- 61. Hua Z, Lv Q, Ye W, Wong CK, Cai G, Gu D, Ji Y, Zhao C, Wang J, Yang BB, Zhang Y. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One 1: e116, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Humbert PO, Verona R, Trimarchi JM, Rogers C, Dandapani S, Lees JA. E2f3 is critical for normal cellular proliferation. Genes Dev 14: 690–703, 2000 [PMC free article] [PubMed] [Google Scholar]
- 63. Jemal A, Devesa SS, Hartge P, Tucker MA. Recent trends in cutaneous melanoma incidence among whites in the United States. J Natl Cancer Inst 93: 678–683, 2001 [DOI] [PubMed] [Google Scholar]
- 64. Jerant AF, Johnson JT, Sheridan CD, Caffrey TJ. Early detection and treatment of skin cancer. Am Fam Physician 62: 357–368, 2000 [PubMed] [Google Scholar]
- 65. Ji Q, Hao X, Meng Y, Zhang M, Desano J, Fan D, Xu L. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer 8: 266, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jinnin M. Mechanisms of skin fibrosis in systemic sclerosis. J Dermatol 37: 11–25, 2010 [DOI] [PubMed] [Google Scholar]
- 67. Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, Chin L, Brown D, Slack FJ. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res 67: 7713–7722, 2007 [DOI] [PubMed] [Google Scholar]
- 68. Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell 120: 635–647, 2005 [DOI] [PubMed] [Google Scholar]
- 69. Kaluz S, Kaluzova M, Stanbridge EJ. Regulation of gene expression by hypoxia: integration of the HIF-transduced hypoxic signal at the hypoxia-responsive element. Clin Chim Acta 395: 6–13, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kanitakis J. Anatomy, histology and immunohistochemistry of normal human skin. Eur J Dermatol 12: 390–401, 2002 [PubMed] [Google Scholar]
- 71. Kasparian NA, McLoone JK, Meiser B. Skin cancer-related prevention and screening behaviors: a review of the literature. J Behav Med 32: 406–428, 2009 [DOI] [PubMed] [Google Scholar]
- 72. Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, Natarajan R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci USA 104: 3432–3437, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kohn DB. Lentiviral vectors ready for prime-time. Nat Biotechnol 25: 65–66, 2007 [DOI] [PubMed] [Google Scholar]
- 74. Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem 283: 14910–14914, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kosmadaki MG, Naif A, Hee-Young P. Recent progresses in understanding pigmentation. G Ital Dermatol Venereol 145: 47–55, 2010 [PubMed] [Google Scholar]
- 76. Koster MI, Kim S, Mills AA, DeMayo FJ, Roop DR. p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev 18: 126–131, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, Mendell JR, Mendell JT. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 137: 1005–1017, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res 101: 59–68, 2007 [DOI] [PubMed] [Google Scholar]
- 79. Kuehn BM. Chronic wound care guidelines issued. JAMA 297: 938–939, 2007 [DOI] [PubMed] [Google Scholar]
- 80. Kuhnert F, Mancuso MR, Hampton J, Stankunas K, Asano T, Chen CZ, Kuo CJ. Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126. Development 135: 3989–3993, 2008 [DOI] [PubMed] [Google Scholar]
- 81. Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM, Negrini M, Calin GA, Ivan M. A microRNA signature of hypoxia. Mol Cell Biol 27: 1859–1867, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA 105: 3903–3908, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327: 198–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lavker RM, Sun TT, Oshima H, Barrandon Y, Akiyama M, Ferraris C, Chevalier G, Favier B, Jahoda CA, Dhouailly D, Panteleyev AA, Christiano AM. Hair follicle stem cells. J Investig Dermatol Symp Proc 8: 28–38, 2003 [DOI] [PubMed] [Google Scholar]
- 85. Lee DY, Deng Z, Wang CH, Yang BB. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci USA 104: 20350–20355, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Leidinger P, Keller A, Borries A, Reichrath J, Rass K, Jager SU, Lenhof HP, Meese E. High-throughput miRNA profiling of human melanoma blood samples. BMC Cancer 10: 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lena AM, Shalom-Feuerstein R, Rivetti di Val Cervo P, Aberdam D, Knight RA, Melino G, Candi E. miR-203 represses “stemness” by repressing DeltaNp63. Cell Death Differ 15: 1187–1195, 2008 [DOI] [PubMed] [Google Scholar]
- 88. Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, Lin Y, MacDonald JR, Pang AW, Shago M, Stockwell TB, Tsiamouri A, Bafna V, Bansal V, Kravitz SA, Busam DA, Beeson KY, McIntosh TC, Remington KA, Abril JF, Gill J, Borman J, Rogers YH, Frazier ME, Scherer SW, Strausberg RL, Venter JC. The diploid genome sequence of an individual human. PLoS Biol 5: e254, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Levy V, Lindon C, Harfe BD, Morgan BA. Distinct stem cell populations regenerate the follicle and interfollicular epidermis. Dev Cell 9: 855–861, 2005 [DOI] [PubMed] [Google Scholar]
- 90. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20, 2005 [DOI] [PubMed] [Google Scholar]
- 91. Li Z, Hassan MQ, Jafferji M, Aqeilan RI, Garzon R, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem 284: 15676–15684, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Li Z, Zhan W, Wang Z, Zhu B, He Y, Peng J, Cai S, Ma J. Inhibition of PRL-3 gene expression in gastric cancer cell line SGC7901 via microRNA suppressed reduces peritoneal metastasis. Biochem Biophys Res Commun 348: 229–237, 2006 [DOI] [PubMed] [Google Scholar]
- 93. Liang Z, Wu H, Reddy S, Zhu A, Wang S, Blevins D, Yoon Y, Zhang Y, Shim H. Blockade of invasion and metastasis of breast cancer cells via targeting CXCR4 with an artificial microRNA. Biochem Biophys Res Commun 363: 542–546, 2007 [DOI] [PubMed] [Google Scholar]
- 94. Lowes MA, Bowcock AM, Krueger JG. Pathogenesis and therapy of psoriasis. Nature 445: 866–873, 2007 [DOI] [PubMed] [Google Scholar]
- 95. Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol 182: 4994–5002, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ma DR, Yang EN, Lee ST. A review: the location, molecular characterisation and multipotency of hair follicle epidermal stem cells. Ann Acad Med Singapore 33: 784–788, 2004 [PubMed] [Google Scholar]
- 97. Mattes J, Collison A, Plank M, Phipps S, Foster PS. Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc Natl Acad Sci USA 106: 18704–18709, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Maurer B, Stanczyk J, Jungel A, Akhmetshina A, Trenkmann M, Brock M, Kowal-Bielecka O, Gay RE, Michel BA, Distler JH, Gay S, Distler O. MicroRNA-29, a key regulator of collagen expression in systemic sclerosis. Arthritis Rheum 62: 1733–1743 [DOI] [PubMed] [Google Scholar]
- 99. Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 315: 1576–1579, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. McLaughlin J, Cheng D, Singer O, Lukacs RU, Radu CG, Verma IM, Witte ON. Sustained suppression of Bcr-Abl-driven lymphoid leukemia by microRNA mimics. Proc Natl Acad Sci USA 104: 20501–20506, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Mendell JT. miRiad roles for the miR-17–92 cluster in development and disease. Cell 133: 217–222, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133: 647–658, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398: 708–713, 1999 [DOI] [PubMed] [Google Scholar]
- 104. Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell 126: 1203–1217, 2006 [DOI] [PubMed] [Google Scholar]
- 105. Muller DW, Bosserhoff AK. Integrin beta3 expression is regulated by let-7a miRNA in malignant melanoma. Oncogene 27: 6698–6706, 2008 [DOI] [PubMed] [Google Scholar]
- 106. Neer RM. The evolutionary significance of vitamin D, skin pigment, and ultraviolet light. Am J Phys Anthropol 43: 409–416, 1975 [DOI] [PubMed] [Google Scholar]
- 107. Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 22: 894–907, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin 55: 74–108, 2005 [DOI] [PubMed] [Google Scholar]
- 109. Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N Engl J Med 350: 664–671, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Pineles BL, Romero R, Montenegro D, Tarca AL, Han YM, Kim YM, Draghici S, Espinoza J, Kusanovic JP, Mittal P, Hassan SS, Kim CJ. Distinct subsets of microRNAs are expressed differentially in the human placentas of patients with preeclampsia. Am J Obstet Gynecol 196: e261–e266, 2007 [DOI] [PubMed] [Google Scholar]
- 111. Pola R, Ling LE, Silver M, Corbley MJ, Kearney M, Blake Pepinsky R, Shapiro R, Taylor FR, Baker DP, Asahara T, Isner JM. The morphogen Sonic hedgehog is an indirect angiogenic agent upregulating two families of angiogenic growth factors. Nat Med 7: 706–711, 2001 [DOI] [PubMed] [Google Scholar]
- 112. Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, Mercatanti A, Hammond S, Rainaldi G. MicroRNAs modulate the angiogenic properties of HUVECs. Blood 108: 3068–3071, 2006 [DOI] [PubMed] [Google Scholar]
- 113. Polsky D, Cordon-Cardo C. Oncogenes in melanoma. Oncogene 22: 3087–3091, 2003 [DOI] [PubMed] [Google Scholar]
- 114. Proweller A, Tu L, Lepore JJ, Cheng L, Lu MM, Seykora J, Millar SE, Pear WS, Parmacek MS. Impaired notch signaling promotes de novo squamous cell carcinoma formation. Cancer Res 66: 7438–7444, 2006 [DOI] [PubMed] [Google Scholar]
- 115. Pulkkinen K, Malm T, Turunen M, Koistinaho J, Yla-Herttuala S. Hypoxia induces microRNA miR-210 in vitro and in vivo ephrin-A3 and neuronal pentraxin 1 are potentially regulated by miR-210. FEBS Lett 582: 2397–2401, 2008 [DOI] [PubMed] [Google Scholar]
- 116. Raja SK, Garcia MS, Isseroff RR. Wound re-epithelialization: modulating keratinocyte migration in wound healing. Front Biosci 12: 2849–2868, 2007 [DOI] [PubMed] [Google Scholar]
- 117. Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science 328: 1570–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Roy S, Patel D, Khanna S, Gordillo GM, Biswas S, Friedman A, Sen CK. Transcriptome-wide analysis of blood vessels laser captured from human skin and chronic wound-edge tissue. Proc Natl Acad Sci USA 104: 14472–14477, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res 67: 9762–9770, 2007 [DOI] [PubMed] [Google Scholar]
- 120. Sand M, Gambichler T, Sand D, Skrygan M, Altmeyer P, Bechara FG. MicroRNAs and the skin: tiny players in the body's largest organ. J Dermatol Sci 53: 169–175, 2009 [DOI] [PubMed] [Google Scholar]
- 121. Sayed D, Rane S, Lypowy J, He M, Chen IY, Vashistha H, Yan L, Malhotra A, Vatner D, Abdellatif M. MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Mol Biol Cell 19: 3272–3282, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Schultz HY, Goldsmith LA. Looking ahead by looking back. J Invest Dermatol 127: 1–2, 2007 [DOI] [PubMed] [Google Scholar]
- 123. Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res 18: 549–557, 2008 [DOI] [PubMed] [Google Scholar]
- 124. Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, Zakrzewski J, Blochin E, Rose A, Bogunovic D, Polsky D, Wei J, Lee P, Belitskaya-Levy I, Bhardwaj N, Osman I, Hernando E. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci USA 106: 1814–1819, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Sen CK. Wound healing essentials: let there be oxygen. Wound Repair Regen 17: 1–18, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Sen CK, Roy S. MicroRNA in cutaneous wound healing. In: Current Perspectives in MicroRNAs (miRNA), edited by Ying S-Y. New York: Springer Science+Business Media, 2008, p. 349–366 [Google Scholar]
- 127. Senoo M, Pinto F, Crum CP, McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell 129: 523–536, 2007 [DOI] [PubMed] [Google Scholar]
- 128. Shaw TJ, Martin P. Wound repair at a glance. J Cell Sci 122: 3209–3213, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Shilo S, Roy S, Khanna S, Sen CK. Evidence for the involvement of miRNA in redox regulated angiogenic response of human microvascular endothelial cells. Arterioscler Thromb Vasc Biol 28: 471–477, 2008 [DOI] [PubMed] [Google Scholar]
- 130. Shilo S, Roy S, Khanna S, Sen CK. MicroRNA in cutaneous wound healing: a new paradigm. DNA Cell Biol 26: 227–237, 2007 [DOI] [PubMed] [Google Scholar]
- 131. Sims NR, Anderson MF. Mitochondrial contributions to tissue damage in stroke. Neurochem Int 40: 511–526, 2002 [DOI] [PubMed] [Google Scholar]
- 132. Sonkoly E, Stahle M, Pivarcsi A. MicroRNAs: novel regulators in skin inflammation. Clin Exp Dermatol 33: 312–315, 2008 [DOI] [PubMed] [Google Scholar]
- 133. Sonkoly E, Wei T, Janson PC, Saaf A, Lundeberg L, Tengvall-Linder M, Norstedt G, Alenius H, Homey B, Scheynius A, Stahle M, Pivarcsi A. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS One 2: e610, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Sonkoly E, Wei T, Pavez Lorie E, Suzuki H, Kato M, Torma H, Stahle M, Pivarcsi A. Protein kinase C-dependent upregulation of miR-203 induces the differentiation of human keratinocytes. J Invest Dermatol 130: 124–134, 2009 [DOI] [PubMed] [Google Scholar]
- 135. Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res 100: 1164–1173, 2007 [DOI] [PubMed] [Google Scholar]
- 136. Suarez Y, Fernandez-Hernando C, Yu J, Gerber SA, Harrison KD, Pober JS, Iruela-Arispe ML, Merkenschlager M, Sessa WC. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci USA 105: 14082–14087, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA 103: 12481–12486, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Tang A, Eller MS, Hara M, Yaar M, Hirohashi S, Gilchrest BA. E-cadherin is the major mediator of human melanocyte adhesion to keratinocytes in vitro. J Cell Sci 107: 983–992, 1994 [DOI] [PubMed] [Google Scholar]
- 139. Taylor G, Lehrer MS, Jensen PJ, Sun TT, Lavker RM. Involvement of follicular stem cells in forming not only the follicle but also the epidermis. Cell 102: 451–461, 2000 [DOI] [PubMed] [Google Scholar]
- 140. Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, Koteliansky V, Rosenwald A, Basson MA, Licht JD, Pena JT, Rouhanifard SH, Muckenthaler MU, Tuschl T, Martin GR, Bauersachs J, Engelhardt S. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 456: 980–984, 2008 [DOI] [PubMed] [Google Scholar]
- 141. Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, Croce CM. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol 179: 5082–5089, 2007 [DOI] [PubMed] [Google Scholar]
- 142. Tinkle CL, Lechler T, Pasolli HA, Fuchs E. Conditional targeting of E-cadherin in skin: insights into hyperproliferative and degenerative responses. Proc Natl Acad Sci USA 101: 552–557, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Tonnesen MG, Feng X, Clark RA. Angiogenesis in wound healing. J Investig Dermatol Symp Proc 5: 40–46, 2000 [DOI] [PubMed] [Google Scholar]
- 144. Trang P, Medina PP, Wiggins JF, Ruffino L, Kelnar K, Omotola M, Homer R, Brown D, Bader AG, Weidhaas JB, Slack FJ. Regression of murine lung tumors by the let-7 microRNA: Oncogene 29: 1580–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Tsai YH, Wu MF, Wu YH, Chang SJ, Lin SF, Sharp TV, Wang HW. The M type K15 protein of Kaposi's sarcoma-associated herpesvirus regulates microRNA expression via its SH2-binding motif to induce cell migration and invasion. J Virol 83: 622–632, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Tunggal JA, Helfrich I, Schmitz A, Schwarz H, Gunzel D, Fromm M, Kemler R, Krieg T, Niessen CM. E-cadherin is essential for in vivo epidermal barrier function by regulating tight junctions. EMBO J 24: 1146–1156, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szasz AM, Wang ZC, Brock JE, Richardson AL, Weinberg RA. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell 137: 1032–1046, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 148. van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA 105: 13027–13032, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Vauclair S, Nicolas M, Barrandon Y, Radtke F. Notch1 is essential for postnatal hair follicle development and homeostasis. Dev Biol 284: 184–193, 2005 [DOI] [PubMed] [Google Scholar]
- 150. Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell 15: 261–271, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Wang W, Mouneimne G, Sidani M, Wyckoff J, Chen X, Makris A, Goswami S, Bresnick AR, Condeelis JS. The activity status of cofilin is directly related to invasion, intravasation, and metastasis of mammary tumors. J Cell Biol 173: 395–404, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Wang XH, Qian RZ, Zhang W, Chen SF, Jin HM, Hu RM. MicroRNA-320 expression in myocardial microvascular endothelial cells and its relationship with insulin-like growth factor-1 in type 2 diabetic rats. Clin Exp Pharmacol Physiol 36: 181–188, 2009 [DOI] [PubMed] [Google Scholar]
- 153. Weiler J, Hunziker J, Hall J. Anti-miRNA oligonucleotides (AMOs): ammunition to target miRNAs implicated in human disease? Gene Ther 13: 496–502, 2006 [DOI] [PubMed] [Google Scholar]
- 153a. Wu DTS, Chen JS, Chang DC, Lin SL. Mir-434-5p mediates skin whitening and lightening. Clin Cosmetic Invest Dermatol 1: 19–35, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Wurdinger T, Tannous BA, Saydam O, Skog J, Grau S, Soutschek J, Weissleder R, Breakefield XO, Krichevsky AM. miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell 14: 382–393, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Xiao J, Yang B, Lin H, Lu Y, Luo X, Wang Z. Novel approaches for gene-specific interference via manipulating actions of microRNAs: examination on the pacemaker channel genes HCN2 and HCN4. J Cell Physiol 212: 285–292, 2007 [DOI] [PubMed] [Google Scholar]
- 156. Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434: 338–345, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell 2: 305–316, 1998 [DOI] [PubMed] [Google Scholar]
- 158. Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, McKeon F. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398: 714–718, 1999 [DOI] [PubMed] [Google Scholar]
- 159. Yang S, Rosenberg SA, Morgan RA. Clinical-scale lentiviral vector transduction of PBL for TCR gene therapy and potential for expression in less-differentiated cells. J Immunother 31: 830–839, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Yang WJ, Yang DD, Na S, Sandusky GE, Zhang Q, Zhao G. Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem 280: 9330–9335, 2005 [DOI] [PubMed] [Google Scholar]
- 161. Yi R, Fuchs E. MicroRNA-mediated control in the skin. Cell Death Differ 17: 229–235, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Yi R, O'Carroll D, Pasolli HA, Zhang Z, Dietrich FS, Tarakhovsky A, Fuchs E. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet 38: 356–362, 2006 [DOI] [PubMed] [Google Scholar]
- 163. Yi R, Pasolli HA, Landthaler M, Hafner M, Ojo T, Sheridan R, Sander C, O'Carroll D, Stoffel M, Tuschl T, Fuchs E. DGCR8-dependent microRNA biogenesis is essential for skin development. Proc Natl Acad Sci USA 106: 498–502, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing “stemness.” Nature 452: 225–229, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Yu J, Peng H, Ruan Q, Fatima A, Getsios S, Lavker RM. MicroRNA-205 promotes keratinocyte migration via the lipid phosphatase SHIP2. FASEB J 24: 3950–3959, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Yu J, Ryan DG, Getsios S, Oliveira-Fernandes M, Fatima A, Lavker RM. MicroRNA-184 antagonizes microRNA-205 to maintain SHIP2 levels in epithelia. Proc Natl Acad Sci USA 105: 19300–19305, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, Yao G, Medina A, O'Brien-Jenkins A, Katsaros D, Hatzigeorgiou A, Gimotty PA, Weber BL, Coukos G. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci USA 103: 9136–9141, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 436: 214–220, 2005 [DOI] [PubMed] [Google Scholar]
- 169. Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol Chem 282: 14328–14336, 2007 [DOI] [PubMed] [Google Scholar]
- 170. Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res 18: 350–359, 2008 [DOI] [PubMed] [Google Scholar]
- 171. Zhu XM, Han T, Sargent IL, Yin GW, Yao YQ. Differential expression profile of microRNAs in human placentas from preeclamptic pregnancies vs normal pregnancies. Am J Obstet Gynecol 200: e661–e667, 2009 [DOI] [PubMed] [Google Scholar]
- 172. Zhu Z, He J, Jia X, Jiang J, Bai R, Yu X, Lv L, Fan R, He X, Geng J, You R, Dong Y, Qiao D, Lee KB, Smith GW, Dong C. MicroRNA-25 functions in regulation of pigmentation by targeting the transcription factor MITF in alpaca (Lama pacos) skin melanocytes. Domest Anim Endocrinol 38: 200–209, 2010 [DOI] [PubMed] [Google Scholar]