Abstract
Chronic wounds represent a major and rising socioeconomic threat affecting over 6.5 million people in the United States costing in excess of US $25 billion annually. Wound healing is a physiological response to injury that is conserved across tissue systems. In humans, wounding is followed by instant response aimed at hemostasis, which in turn provides the foundation for inflammatory processes that closely follow. Inflammation is helpful and a prerequisite for healing as long as it is mounted and resolved in a timely manner. Chronic inflammation derails the healing cascade resulting in impaired wound closure. Disruption of Dicer, the RNase III enzyme that generates functional miRNAs, has a major impact on the overall immune system. Emerging studies indicate that miRNAs, especially miR-21, miR-146a/b, and miR-155, play a key role in regulating several hubs that orchestrate the inflammatory process. Direct evidence from studies addressing wound inflammation being limited, the current work represents a digest of the relevant literature that is aimed at unveiling the potential significance of miRNAs in the regulation of wound inflammation. Such treatment would help establish new paradigms highlighting a central role of miRs in the understanding and management of dysregulated inflammation as noted in conjunction with chronic wounds.
Keywords: wound healing
BACKGROUND
The process of wound healing is well regulated and for the ease of understanding is divided into specific functional phases: hemostasis, inflammation, proliferation, and remodeling (43, 49, 73, 80). Chronic wounds fail to progress through the normal phases of healing and therefore enter a state of prolonged pathologic inflammation (49). An improved understanding of the molecular mechanisms that regulate and co-ordinate the inflammatory response in wounds has potential for improved therapeutic intervention of chronic wounds. We first hypothesized in 2007 and reported in 2008 that the understanding of microRNA (miRNA or miR) biology is critically important to develop a comprehensive understanding of the molecular mechanisms that govern wound healing (6, 74, 75, 78, 79). Emerging studies suggest that miRNAs play a significant role in the immune responses (3). Likewise, the endonuclease enzyme Dicer, the RNase III enzyme that generates functional miRNAs, plays a key role in the regulation of the immune system (13). While the role of miRNA in inflammatory response associated with cancer has been extensively studied and reviewed (31, 59, 82, 89), how miRNA may govern wound inflammation remains unexplored. Such information will help understand persistent and unresolved inflammation as seen in chronic wounds. The role of miRNA in processes that drive wound healing such as epithelialization and angiogenesis has been discussed elsewhere (6, 74, 75, 78, 79). In this work, emphasis is directed toward addressing the significance of miRNA in regulating the component biological processes underlying wound inflammation. We first discuss the major miRNAs that are known to regulate the innate immune responses. Next, we address the regulation of key cytokines, chemokines, and growth factors that regulate the inflammatory response following wounding (Fig. 1). Direct evidence from studies addressing wound inflammation being limited, the current work represents a digest of the relevant literature that is aimed at unveiling the potential significance of miRNAs in the regulation of wound inflammation. Such treatment would help establish new paradigms highlighting a central role of miRs in the understanding and management of dysregulated inflammation as noted in conjunction with chronic wounds.
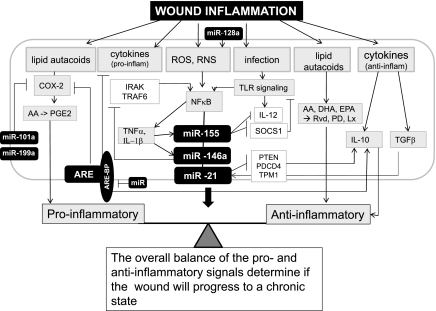
Fig. 1.
Potential role of microRNA (miRNA, also miR) in regulation of wound inflammatory response. The inflammation response to wound is tightly regulated by proinflammatory signals as well as signals that are anti-inflammatory to resolve inflammation. An imbalance between these signals results in chronic inflammation and derails the healing cascade. miRNA have been shown responsive to as well as regulate some of the key mediators of inflammatory response in the course of wound healing. The details of the miRNA or mediators have been discussed in the review. miRNA regulate the expression of the components of the ARE-BPs that are known to regulate cytokine and Cox-2 gene expression. The miRNA and the targets of miR have been presented as filled and open boxes, respectively. AA, arachidonic acid; ARE-BP, AU-rich element binding protein; COX-2, cyclooxygenase-2; DHA, docosahexaenoic acid, EPA, eicosapentaenoic; Lx, lipoxin; PGE2, prostaglandin E2; Pd, protectin; RvD, resolvin; ROS, reactive oxygen species; RNS, reactive nitrogen species; TLR, Toll-like receptor.
miRNA IN REGULATION OF INNATE IMMUNE RESPONSE
The innate immune response provides primary defense against infection by external pathogens such as bacteria, fungi, and viruses. The presence of invading pathogens is commonly detected by tissue macrophages using receptors known as PAMP (pathogen-associated molecular pattern) receptors (95). Multiple families of PAMP receptors have been identified, including the Toll-like receptors (TLRs), nucleotide-binding domain (NOD)-like receptors (NLRs), and the retinoic acid-inducible gene (RIG)-like receptors (RLRs) families (15). The first report linking miRNA with immune responses came from miRNA expression profiling performed in a monocytic cell line treated with the lipopolysaccharide (LPS), a TLR4 ligand (86). The expression of miR-146a, miR-155, and miR-132 were induced in response to LPS stimulation (Fig. 2)(86). miRNAs are known to influence the fate of immune cells (e.g., miR-223) as well as to regulate adaptive immune responses such as antigen presentation (e.g., miR-155) and T-cell receptor signaling (miR-181a) (81). miR-146, miR-155, and miR-21 have been of particular interest for investigations associated with inflammatory and immune responses. These miRNAs are induced by proinflammatory stimuli such as IL-1β, TNF-α, and TLRs (76). In the following section we discuss the expression, regulation well as the targets of miR-146, miR155, and miR-21.
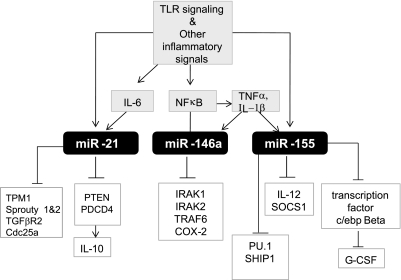
Fig. 2.
Three major miRNA induced as a response to inflammation. Known key activation signal and the targets of miR-146, miR-21, and miR-155 in macrophages. G-CSF, granulocyte colony-stimulating factor; SHIP-1, src homology 2-containing inositol 5-phosphatase 1; SOCS1, suppressor of cytokine signaling; TGFβR2, transforming growth factor-β receptor 2; TPM1, tropomyosin1.
miR-146
The miR-146 family is composed of two members, miR-146a and miR-146b, that are located on chromosomes 5 and 10, respectively (95). miR-146 is induced as a general response in myeloid cells through TLR-2, -4, or -5 ligands (e.g., bacterial and fungal components) or following exposure to the pro-inflammatory cytokines (such as TNF-α or IL-1β) (56, 86, 90). This miRNA, however, is not responsive to TLR -3, -7, or -9 activation (e.g., single- or double-stranded RNA and CpG motifs) (95). A detailed list of activators and conditions known to change expression of miR-146 is displayed in Table 1. The miRNA-146 family (miR-146a/b) regulates TLR4 through a negative feedback loop mechanism (35). IRAK1 and TRAF6 represent two prominent targets of miR-146a that help it to negatively regulate the release of IL-8 and RANTES (95). In addition to TRAF6 and IRAK-1, IRAK2 has been identified as another target of miR-146a, which regulates IFN-γ production (29). Furthermore, miR-146a silences cyclooxygenase-2 (Cox-2) mRNA in fibroblasts from patients suffering fromchronic obstructive pulmonary disease (COPD). Thus, lowering of miR-146a expression may prolong the half-life of Cox-2 in COPD patients (69).Differential expression and/or binding of miR-146a and miR-146b have been implicated in a number of pathological conditions displayed in Table 2. Dysregulated expression of this miR has been associated with metastatic and proliferative response observed in a number of neoplastic disorders such as papillary thyroid carcinoma, cervical, ovarian and breast cancers (61). Elevated expression of miR-146a in tissues is also associated with chronic inflammatory diseases, such as psoriasis (81), rheumatoid arthritis (32, 53) and COPD (69).
Table 1.
miR-146, miR-155, and miR-21: activators and targets
| miR | Expression | Activators/Conditions | Cell/Tissue | Ref. No. |
|---|---|---|---|---|
| miR-146 | up | A TLR4 ligands (e.g., LPS) | Thp-1, monocytic cell line | 86 |
| up | TLR-2, -4, or -5 ligands (e.g., bacterial and fungal components) and proinflammatory cytokines TNF-α or IL-1b #28 | myeloid cells | 56, 86, 90 | |
| up | IL-1 receptor signaling & inflammatory cytokines associated with senescence-associated secretory phenotype (SASP) | senescencent cells | 5, 14 | |
| up | 12-O-tetradecanoylphorbol-13-acetate (TPA) treated in vitro-differentiating HL-60 cells | myeloid cell differentiation | 12 | |
| down | high glucose (20 mM, 24 h) compared with low (5.6 mM) glucose | cultured human islets | 22 | |
| up | IL-1β | A549 cells, BEAS2B cells and primary human airway epithelial cells | 60 | |
| miR-155 | up | inflammatory mediators [e.g., TNF-α, LPS, polyriboinosinic:polyribocytidylic (PI:PC) acid and IFNβ] | macrophages | 56 |
| up | LPS stimulation | mouse Raw 264.7 macrophages | 90 | |
| up | ip injection with LPS | C57 BL6 mice | 90 | |
| up (>100-fold) | maturation of DCs | DCs | 44 | |
| up | innate and adaptive immune responses | macrophages and B- and T-cells | 87 | |
| miR-21 | up | administration of aerosolized LPS to mouse lung | mouse lung | 51 |
| up | IL-6 induces the expression of miR-21 in a STAT3-dependent manner | multiple myeloma cells | 39 |
LPS, lipopolysaccharide; DC, dendritic cell.
Table 2.
miR-146, miR-155, and miR-21: dysregulation and health disorders
| Expression | Disease/Pathology | Cells/Tissues | Ref. No. | |
|---|---|---|---|---|
| miR-146 | up | acute coronary syndrome (ACS) patients | peripheral blood mononuclear cells (PBMC) | 27 |
| up | vesicular stomatitis virus (VSV) infection | macrophages | 29 | |
| up | human memory T-cells compared to human naive T cells | T-cells | 16 | |
| up | T-cell receptor (TCR) stimulation | human primary T-lymphocytes | 16 | |
| up | human rheumatoid arthritis | synovial fibroblasts | 53 | |
| up | Helicobacter pylori infection | gastric mucosa | 37 | |
| miR-155 | LPS/d-galactosamine-induced septic shock and endotoxin shock | mice | 90 | |
| up | lymphomas of activated-B-cell origin, including Hodgkin's lymphoma and diffuse large cell B-cell lymphoma | B-cells | 87 | |
| miR-21 | up | overexpressed in multiple cancer forms and tightly associated with cancerogenesis | cancer cells | 30 |
| up | infarcted region of the ischemia-reperfused heart | cardiac fibroblasts | 66 | |
| up | allergic airway inflammation | lung | 40 |
1Ref, reference.
miR-155
microRNA-155 (miR-155) represents a common target of a broad range of inflammatory mediators including TNF-α, LPS, polyriboinosinic:polyribocytidylic (PI:PC) acid and IFN-β (56) (Table 1). The transcription factor c/ebp-β is a direct target of miR-155 (97). Silencing of miR-155 in murine macrophages, human monocytic cells, as well as in LPS-treated mice causes marked derepression of the c/ebp-β isoforms and downregulation of granulocyte colony-stimulating factor (G-CSF). Overexpression of miR-155 in the THP1 monocytic cell line decreases PU.1 protein levels at both mRNA as well as protein levels (44). Using gain- and loss-of-function approaches, miR-155 was observed to repress src homology-2 domain-containing inositol 5-phosphatase 1 (SHIP1) through direct 3′-untranslated region (UTR) interactions. Repression of endogenous SHIP1 by miR-155 occurred following sustained overexpression of miR-155 in hematopoietic cells both in vitro as well as in vivo. Primary macrophages from miR-155 knockout mice also demonstrated repressed SHIP1 expression, unveiling a molecular link between miR-155 and SHIP1 (55). miR-155 directly targets transcript coding for several proteins involved in LPS signaling such as the Fas-associated death domain protein (FADD), IκB kinase epsilon, and the receptor (TNFR superfamily)-interacting serine-threonine kinase 1 (Ripk1) (90).
miR-155 has emerged as a central regulator of the immune system (81, 86). A potential role of miRNA-155 in the adaptive immune response was provided from studies using knockout mice (63, 88, 91). These miRNA-deficient animals displayed severe immunodeficiencies, particularly impaired B-cell responses and skewed Th2-helper T-cell responses (63, 88, 91). In addition to the deficiency in adaptive immunity, autoimmune phenotype in the lungs of miR-155 null mice was also observed (63). Increased airway remodeling and leukocyte invasion suggested that miR-155 plays a key role in regulating the response of the immune system to self-antigens (63). miR-155 is encoded within an exon of the noncoding RNA known as bic (B-cell integration cluster). To uncover the significance of miR-155 in regulating immune function in vivo, bic/miR-155 (bic) null mice were developed. These mice fail to generate high levels of class-switched antibody upon immunization with thymus-dependent and thymus-independent antigens. The bic-deficient T-cells show skewed differentiation into the Th2 lineage under a variety of in vitro conditions. Thus, miR-155 emerged as a central regulator of lymphocyte differentiation (91). Thai et al. (88) generated two mutant mouse strains with miR-155 deficiency. In the first strain, a major portion of the bic second exon, including miR-155, was replaced by a β-galactosidase (lacZ) reporter, generating a loss-of-function allele designated bic/miR-155−/−. The second mutant strain was generated using an established knock-in strategy. In these mice, miR-155, together with an enhanced green fluorescent protein (EGFP) reporter, was conditionally expressed in mature B-cells in a Cre-dependent manner. These approaches, using a combined genetic loss- and gain-of-function approach, demonstrated that miR-155 regulates specific differentiation processes in the immune response and that it exerts its functions at least partly by influencing cytokine production (88).
miR-21
miR-21 is likely one of the most studied miRs to date. miR-21 initially was described as an “oncomir” overexpressed in multiple cancer forms and tightly associated with cancerogenesis (Table 2). It has been commonly noted that inflammatory stimuli induce miR-21 (Table 1). A single primary transcript containing miR-21 (pri-miR-21) is transcribed from an evolutionarily conserved promoter that resides in an intron of an overlapping coding gene, TMEM49 (23). The putative miR-21 promoter region contains three AP1 and one PU.1 binding sites (23). Computational analyses predicted transcription repressor NFIB mRNA as a target for miR-21 and the miR-21 promoter itself contains a conserved binding site for the NFIB protein (23, 30). In silico analyses combined with experimental biology approaches have identified numerous target proteins whose expression is regulated by miR-21. Phosphatase and tensin homolog (PTEN) is a phosphatidylinositol-3,4,5-trisphosphate 3 (PIP3)-phosphatase that inhibits phosphoinositide-3-kinase (PI3K) pathway while dephosphorylating (PIP3) and thus prevents Akt activation (85). The first evidence that PTEN is a valid miR-21 target was reported in cholangiocarcinoma cell line Mz-ChA-1 (48). Using laser-capture microdissection technique we demonstrated that miR-21 signal was localized to cardiac fibroblasts of the infarcted region of the ischemia-reperfused heart. PTEN was identified as a direct target of miR-21 in cardiac fibroblasts (66). The tumor suppressor PDCD4 is a proinflammatory protein that promotes activation of the transcription factor NF-κB and suppresses interleukin 10 (IL-10). Transfection of cells with a miR-21 precursor blocked NF-κB activity and promoted the production of anti-inflammatory IL-10 in response to LPS. Transfection with antisense oligonucleotides to miR-21 or targeted protection of the miR-21 site in PCDC4 mRNA displayed the opposite effect (77). This study demonstrated that miR-21 regulates PDCD4 expression following LPS stimulation (77). In addition to PTEN and PDCD4, a number of other targets of miR-21 including tropomyosin (TPM1), sprouty1 and 2, TGF-β receptor (TGFBR2) Cdc25a have been validated (30).
AU-RICH ELEMENTS AND ARE-BINDING PROTEINS
The AU-rich elements (AREs), located in 3′-UTR of transcripts, are well-established determinants of RNA stability (19). AREs are prerequisite for regulating the half-life of many cytokines and are central to achieving temporal and spatial regulation of these genes (65). The ARE-binding proteins (ARE-BPs) regulate this process either by decay promotion/destabilization (e.g., tristetraprolin, TTP; AU-rich binding factor 1, AUF1) or by stabilization (e.g., Hu protein R, HuR) (4). Both TTP and HuR are predicted to be miRNA targets (4). TTP destabilizes the expression of class II AREs that are present in many proinflammatory growth factors and cytokines such as TNF-α and GM-CSF (4, 8).
WOUND INFLAMMATORY RESPONSE
Wound-induced inflammatory response constitutes one of the early events that determine the fate and quality of healing (18). Infiltrating leukocytes represent the principal cellular components of the inflammatory response. These cells not only fight infection but also deliver cytokines, chemokines, and growth factors, laying the foundation for tissue repair. Controlled influx of specific populations of blood-borne cells, including leukocytes, marks the early phase of healing (58). Understanding the mechanisms that regulate the inflammatory response in wound repair is the foundation that will help design innovative strategies to address dysregulatedinflammation as noted in say chronic ulcers where resolution of inflammation is often impaired. In the following section, we describe how miRNA may regulate wound inflammation.
Chemokines, Cytokines, and Growth Factors
MCP-1.
Following wounding, the CC chemokine macrophage chemoattractant protein (MCP-1/CCL2) is one of the major chemo-attractants for monocytes/macrophages. It also helps recruit a subset of T-cells and on mast cells carrying the CCR3 receptor (94). The expression of MCP-1 was highly upregulated (∼70 fold) 12h following wounding (67). A putative consensus site for miR-124a binding in the 3′-UTR of MCP-1 mRNA has been identified. miR-124a specifically suppresses the reporter activity driven by the 3′-UTR of MCP-1 mRNA suggesting that miR-124a is directly implicated in the posttranscriptional silencing of MCP-1 expression (52).
TNF-α.
TNF-α is a potent proinflammatory cytokine produced by activated macrophages known to drive the inflammatory response to wounding. Depending on the concentration, length of exposure, and presence of other cytokines, the effect of TNF-α can be beneficial or deleterious. TNF-α is involved in tissue remodeling, mounting and sustenance of inflammation, cachexia, shock, and cell death (68). Lowering of the functionally available levels of TNF-α, using anti-TNF-α therapy directed at managing activated macrophages, restores diabetic wound healing in ob/ob mice (25). miR-125b targets the 3′-UTR of TNF-α transcripts. Thus, LPS-induced downregulation of miR-125b may be instrumental in bolstering the production of TNF-α. In cells, TLR4 activation downregulates miR-125b expression (90). Also, TNF-α-mediated induction of endothelial adhesion molecules can be regulated by miRNAs that are induced by TNF-α. Specifically, E-selectin and ICAM-1 are targets of TNF-induced miRNAs, miR-31 and miR-17–3p, respectively (84). The posttranscriptional mechanisms gradually and variably impose a series of flexible rate-limiting controls to modify the abundance of the TNF-α mRNA and the rate of its translation in response to inflammatory signals (83). These posttranscriptional controls consist of signaling networks converging on RNA-binding proteins and miRNAs, which in turn target a code of secondary or tertiary ribonucleotide structures located on the TNF mRNA (83).
IL-10.
IL-10 is recognized as a major suppressor of the inflammatory response (50). IL-10 downregulates the expression of proinflammatory genes. In fetal wounds, IL-10 plays a major role in subduing the expression of proinflammatory cytokines resulting in minimized matrix deposition and scar-free healing (36). An imbalance featuring increased levels of the proinflammatory cytokines TNF-α and IL-6 and a decreased level of IL-10 is noted in diabetic wounds. These observations point towards IL-10 insufficiency as being a key factor underlying the exaggerated and sustained inflammatory response commonly noted in diabetic wounds (33). Of note, IL-10 inhibits the LPS-inducible expression of miR-155 (47), while miR-21 or miR-146a remain unaffected. IL-10 inhibits the transcription of miR-155 from the BIC gene in a STAT3-dependent manner. Such inhibition of miR-155 upregulates the abundance of miR-155 target protein such as SHIP1. Through its inhibitory effect on miR-155, IL-10 promotes the expression of anti-inflammatory genes (47). In macrophages stimulated with TLR ligand, miR-466l can upregulate both mRNA and protein expression of IL-10 via competitive binding to the 3′-UTR contain AREs. The RNA-binding protein TTP mediates rapid degradation of IL-10 mRNA. Thus, the net effect of the binding of miR-466l to IL-10 AREs is to prevent IL-10 mRNA degradation mediated by TTP, resulting in extended half-life of IL-10 mRNA, which in turn elevates IL-10 expression (41). The regulation of IL-10 by miR-21 via PDCD4 has been discussed in an earlier section.
TGF-β1 Signaling
As a physiological response to wounding, TGF-β1 is released in large amounts from platelets. It serves as a chemoattractant for neutrophils, macrophages, and fibroblasts. These cell types further augment TGF-b1 levels in the wound environment via a feed-forward loop (94). In addition to the functionally active form, latent TGF-βs are also produced and sequestered within the wound matrix, allowing sustained release upon action of proteolytic enzymes. The TGF-β superfamily encompasses a diverse range of proteins, many of which play important roles during development, tissue homeostasis, disease processes, and repair (94). Signaling via active TGF-βs is mediated by a heterotetrameric complex of two trans-membrane receptor serine/threonine kinases, consisting of a type II ligand binding receptor (TβRII) and a type I signaling receptor (TβRI) (93). Transcriptional regulators Smad2 and Smad3 are direct substrates of TGFβRI. Upon activation, the phosphorylated Smads2 and 3 undergo conformational change, which allows them to bind to cytoplasmic Smad4, a shuttle to the nucleus, resulting in induction of TGF-β gene expression (93). Full-thickness incisional wounding in Smad3-null mice show accelerated closure, characterized by an increased rate of re-epithelialization and reduced inflammation (2). miRNA biogenesis by ligand-specific SMAD proteins is now known to control vascular smooth muscle cell phenotype (17). Smads play a regulatory role in the processing of miRNA into the nucleus (28). Receptor-activated Smads regulate the processing of a subset of miRNAs, particularly miR-21 (37). In breast cancer, miR-155 play an important role in TGF-β-induced epithelial-mesenchymal transition (EMT), cell migration, and invasion by targeting RhoA(34). Among miRNAs that directly target molecules in TGF-β signaling pathway, miR-128a negatively target TGFβRI protein expression by binding to the 3′-UTR region of the gene (45).
TLR Signaling
Inflammatory cells, including macrophages and neutrophils, recognize invading microbial pathogens primarily through TLRs (1). Based on the adaptor molecules recruited to the TLR intracellular domain after ligand engagement, the TLR-activated signaling events are largely defined as myeloid differentiation primary response gene 88 (MyD88)-dependent or TIR-domain-containing adapter-inducing IFN-β (TRIF)-dependent (57). MyD88-deficient mice exhibit severely impaired wound healing phenotype characterized by delayed granulation tissue formation and compromised blood vessel formation (42). The impairment of wound healing in these animals is not associated with increased complication caused by infection. Thus, in addition to the host pathogen response, signaling through the MyD88-dependent pathway regulates infection-independent processes in wound healing (42). MyD88 signaling involves recruitment and activation of IRAK4, IRAK1, and TRAF6, leading to the activation of the NF-κB, AP1, and MAP kinase pathways (57). Expression profiling of miRNAs in human monocyte cell line THP1 treated with LPS (TLR4 agonist) identified miR-146, miR-155, and miR-132 as LPS responsive (86). miR-146a negatively regulates TLR signaling via targeting TRAF6 and IRAK-1, IRAK2 (86). TLR-signaling responsive miRs-146a and 155 have been discussed elsewhere in this work.
NADPH Oxidase and Reactive Oxygen Species Production
In its molecular form, oxygen is required for oxidative metabolism-derived energy synthesis, protein synthesis, and the maturation (hydroxylation) of extracellular matrices such as collagen (73). In a wound setting, large amounts of molecular oxygen are partially reduced to form reactive oxygen species (ROS). Oxygen free radicals such as superoxide anion as well its nonradical derivative hydrogen peroxide (H2O2) are released during the inflammatory response (73). Among driving other key processes, ROS drives endothelial cell signaling required for successful angiogenesis. We provided first evidence demonstrating that miRNA may regulate ROS-driven redox signaling (78). Arrest of the miRNA biogenesis enzyme dicer led to lower inducible production of ROS in human endothelial cells when activated with phorbol ester, TNF-α, or vascular endothelial growth factor (78). NADPH oxidases represent a major source of superoxide anion radicals at the wound site. NADPH oxidases in phagocytic cells help fight infection. The p47phox component of NADPH oxidase complex has been identified as one of the targets of miRNA that regulates the ROS production and the angiogenic properties of endothelial cells (78). Microarray analysis of hormone refractory cell lines identified hsa-miR-128a as being hormone-responsive (45). Regulation of ROS by miR-128a via the specific inhibition of the Bmi-1 oncogene has been recently demonstrated (92).
Lipid Mediators
Lipid mediators such as eicosanoids consist of a family of biologically active metabolites, including prostaglandins (PG), prostacyclin (PC), thromboxanes (TX), leukotrienes (LT), and lipoxins (LX) (26). Free arachidonic acid may be metabolized through the COX pathway, involving COX-1 and COX-2, along with terminal synthases, to generate PG, PC, and TX. Eicosanoids are well known to initiate, amplify, and perpetuate inflammation in both acute as well as chronic wounds (7). The ω-3 polyunsaturated fatty acids eicosapentaenoic (EPA; i.e., ω-3, C20:5) and docosahexaenoic acid (DHA; i.e., ω-3, C22:6) are transformed, in a manner equivalent to arachidonic acid metabolism, by COX-2 and LOX enzymes to generate novel classes of endogenous lipid autacoids with anti-inflammatory and protective activities (26). Induction of COX-2 represents one of the earliest responses following cutaneous injury (58). Conserved ARE in the promoter of COX-2 gene has been identified (98). Rapid degradation of COX-2 mRNA has been attributed to AREs at 3′-UTR of COX-2 (46). ARE-BP HuR has been shown to bind the COX-2 3′-UTR and stabilize the transcript (46). miR-101a and miR-199a appear to control COX-2 expression in the mouse uterus during embryo implantation (9).
Differentiation of the Inflammatory Leukocytes
miRNAs play important roles in normal or malignant hematopoiesis, including the differentiation of hematopoietic stem cell (HSC), their self-renewal, and the function of immune cells (24). Early expression profiling studies demonstrated that the expression of specific miRNA such as miR-181, miR-223, and miR-142s were limited to cells of the bone marrow, spleen, and thymus (Fig. 3). Of these, miR-181 was limited to B-cells, whereas miR-223 expression was isolated to myeloid cells (11, 81). Despite its preferential expression in myeloid cells, ectopic expression of miR-223 in hematopoietic progenitor cells did not have a profound effect on myeloid differentiation (81). Later studies proved miR-223 to be an essential modulator of myeloid differentiation in human. Overexpression of miR-223 doubled the cells committed to the granulocyte-specific lineage in a granulocyte differentiation model, whereas knock-down of miR-223 had the opposite effect (20). miR-223 is involved in an autoregulatory feedback loop to control its own expression and enhance granulocytic differentiation (81). The relative levels of PU.1 and C/EBPα determine cell fate between monocyte and granulocyte as end-products (62, 72). PU.1 activates the transcription of miR-424 stimulating monocyte differentiation through miR-424-dependent translational repression of NFIA transcription factor. Ectopic expression of miR-424 in precursor cells enhances monocytic differentiation. These data point underscore the significance of miR-424 in controlling the monocyte/macrophage differentiation program (64). Downregulation of miRNAs 17-5p-20a-106a regulate monocytopoiesis through targeting of AML1 and upregulation of M-CSF receptor (21). miRNAs, specifically miR-21, miR-155, miR-424, and miR-17–92, and their transcriptional regulatory control are directly implicated in monocytic differentiation (71).
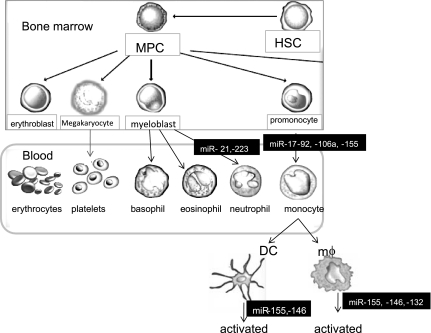
Fig. 3.
Overview of major miRNAs in myeloid cell differentiation. This illustration summarizes the known changes in miRNA expression during differentiation of granulocytic and monocytic cells. DC, dendritic cell; HSC, hematopoietic stem cell; mϕ, macrophage; MPC, myeloid progenitor cell.
miRNA AS BIOMARKERS AND THERAPEUTIC TARGET IN INFLAMMATION
Circulating miRNAs as Prognostics and Therapeutic Biomarkers
Numerous biological molecules from tissues of the body can be found in human serum. The simplicity of obtaining a blood sample and ease of testing makes the research of finding specific biomarkers for prognostic/therapeutics purposes very tempting. A small number of serum-based biomarkers for a specific disease, such as alpha-fetoprotein (AFP) for liver cancer or C-reactive protein (CRP) for inflammation, have been used for diagnosis for several years (96). The potential of using circulating miRNA levels as diagnostic markers has been reported for cancers (70, 96). Studies have compared the expression levels of miRNAs in plasma or serum from cancer cases to healthy control populations. For example, miR-17-3p and miR-92 expression levels in plasma were found to be elevated in colorectal cancer patients (54, 70). Likewise, in the sera of lung cancer patients, miRNA profile was significantly different compared with healthy subjects, with 28 miRNAs missing and 63 new miRNA species detected. Two highly expressed miRNAs in lung cancer, miR-25 and miR-223, were analyzed by qRT-PCR and confirmed for their ability to serve as blood-based biomarkers for lung cancer in an independent trial of 75 healthy donors and 152 cancer patients (12). miRNAs have been suggested to have high value as biomarkers because of highly sensitive PCR detection methods and low complexity compared with protein biomarkers (96).
miRNA in Therapeutics
The potential to therapeutically regulate miRNA levels at the accessible skin wound site make miRNA-based therapies attractive in wound care. miRNA-based therapies may be also used to address chronic inflammation. To date, the main RNA inhibition agents used in preclinical and clinical studies include antisense oligonucleotides, ribozymes, and the DNAzymes, small interfering RNAs and short hairpin RNAs, and anti-miRNA agents such as antisense oligonucleotides, locked nucleic acids, and antagomirs(99). At present several clinical trials are under way to test the therapeutic efficacy of miRNA-based therapies. In 2011, several such trials evaluating efficacy for cancer treatments are expected to be launched (70). Within the next several years, it is anticipated that miRNA-based therapeutics, alone or in combination with other modalities, will be clinically useful treatments for various cancers and inflammatory disorders (70).
GRANTS
Wound healing research in the authors' laboratory is funded by National Institutes of Health Grants DK-076566 (S. Roy) and GM-069589, GM-077185, and NS-42617 (C. K. Sen).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
REFERENCES
- 1. Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol 4: 499–511, 2004 [DOI] [PubMed] [Google Scholar]
- 2. Ashcroft GS, Roberts AB. Loss of Smad3 modulates wound healing. Cytokine Growth Factor Rev 11: 125–131, 2000 [DOI] [PubMed] [Google Scholar]
- 3. Asirvatham AJ, Gregorie CJ, Hu Z, Magner WJ, Tomasi TB. MicroRNA targets in immune genes and the Dicer/Argonaute and ARE machinery components. Mol Immunol 45: 1995–2006, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Asirvatham AJ, Magner WJ, Tomasi TB. miRNA regulation of cytokine genes. Cytokine 45: 58–69, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bhaumik D, Scott GK, Schokrpur S, Patil CK, Orjalo AV, Rodier F, Lithgow GJ, Campisi J. MicroRNAs miR-146a/b negatively modulate the senescence-associated inflammatory mediators IL-6 and IL-8. Aging (Albany NY) 1: 402–411, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Biswas S, Roy S, Banerjee J, Hussain SR, Khanna S, Meenakshisundaram G, Kuppusamy P, Friedman A, Sen CK. Hypoxia inducible microRNA 210 attenuates keratinocyte proliferation and impairs closure in a murine model of ischemic wounds. Proc Natl Acad Sci USA 107: 6976–6981, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Broughton G, 2nd, Janis JE, Attinger CE. The basic science of wound healing. Plast Reconstr Surg 117: 12S–34S, 2006 [DOI] [PubMed] [Google Scholar]
- 8. Carballo E, Lai WS, Blackshear PJ. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood 95: 1891–1899, 2000 [PubMed] [Google Scholar]
- 9. Chakrabarty A, Tranguch S, Daikoku T, Jensen K, Furneaux H, Dey SK. MicroRNA regulation of cyclooxygenase-2 during embryo implantation. Proc Natl Acad Sci USA 104: 15144–15149, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen A, Luo M, Yuan G, Yu J, Deng T, Zhang L, Zhou Y, Mitchelson K, Cheng J. Complementary analysis of microRNA and mRNA expression during phorbol 12-myristate 13-acetate (TPA)-induced differentiation of HL-60 cells. Biotechnol Lett 30: 2045–2052, 2008 [DOI] [PubMed] [Google Scholar]
- 11. Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science 303: 83–86, 2004 [DOI] [PubMed] [Google Scholar]
- 12. Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Zen K, Zhang CY. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 18: 997–1006, 2008 [DOI] [PubMed] [Google Scholar]
- 13. Cobb BS, Hertweck A, Smith J, O'Connor E, Graf D, Cook T, Smale ST, Sakaguchi S, Livesey FJ, Fisher AG, Merkenschlager M. A role for Dicer in immune regulation. J Exp Med 203: 2519–2527, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, Nelson PS, Desprez PY, Campisi J. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol 6: 2853–2868, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Creagh EM, O'Neill LA. TLRs, NLRs and RLRs: a trinity of pathogen sensors that co-operate in innate immunity. Trends Immunol 27: 352–357, 2006 [DOI] [PubMed] [Google Scholar]
- 16. Curtale G, Citarella F, Carissimi C, Goldoni M, Carucci N, Fulci V, Franceschini D, Meloni F, Barnaba V, Macino G. An emerging player in the adaptive immune response: microRNA-146a is a modulator of IL-2 expression and activation-induced cell death in T lymphocytes. Blood 115: 265–273, 2010 [DOI] [PubMed] [Google Scholar]
- 17. Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature 454: 56–61, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eming SA, Krieg T, Davidson JM. Inflammation in wound repair: molecular and cellular mechanisms. J Invest Dermatol 127: 514–525, 2007 [DOI] [PubMed] [Google Scholar]
- 19. Espel E. The role of the AU-rich elements of mRNAs in controlling translation. Semin Cell Dev Biol 16: 59–67, 2005 [DOI] [PubMed] [Google Scholar]
- 20. Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell 123: 819–831, 2005 [DOI] [PubMed] [Google Scholar]
- 21. Fontana L, Pelosi E, Greco P, Racanicchi S, Testa U, Liuzzi F, Croce CM, Brunetti E, Grignani F, Peschle C. MicroRNAs 17–5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat Cell Biol 9: 775–787, 2007 [DOI] [PubMed] [Google Scholar]
- 22. Fred RG, Bang-Berthelsen CH, Mandrup-Poulsen T, Grunnet LG, Welsh N. High glucose suppresses human islet insulin biosynthesis by inducing miR-133a leading to decreased polypyrimidine tract binding protein-expression. PLoS One 5: e10843, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K, Iba H. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol 378: 492–504, 2008 [DOI] [PubMed] [Google Scholar]
- 24. Garzon R, Croce CM. MicroRNAs in normal and malignant hematopoiesis. Curr Opin Hematol 15: 352–358, 2008 [DOI] [PubMed] [Google Scholar]
- 25. Goren I, Muller E, Schiefelbein D, Christen U, Pfeilschifter J, Muhl H, Frank S. Systemic anti-TNFalpha treatment restores diabetes-impaired skin repair in ob/ob mice by inactivation of macrophages. J Invest Dermatol 127: 2259–2267, 2007 [DOI] [PubMed] [Google Scholar]
- 26. Gronert K. Lipid autacoids in inflammation and injury responses: a matter of privilege. Mol Interv 8: 28–35, 2008 [DOI] [PubMed] [Google Scholar]
- 27. Guo M, Mao X, Ji Q, Lang M, Li S, Peng Y, Zhou W, Xiong B, Zeng Q. miR-146a in PBMCs modulates Th1 function in patients with acute coronary syndrome. Immunol Cell Biol 88: 555–564, 2010 [DOI] [PubMed] [Google Scholar]
- 28. Hata A, Davis BN. Control of microRNA biogenesis by TGFbeta signaling pathway-A novel role of Smads in the nucleus. Cytokine Growth Factor Rev 20: 517–521, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hou J, Wang P, Lin L, Liu X, Ma F, An H, Wang Z, Cao X. MicroRNA-146a feedback inhibits RIG-I-dependent Type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J Immunol 183: 2150–2158, 2009 [DOI] [PubMed] [Google Scholar]
- 30. Jazbutyte V, Thum T. MicroRNA-21: from cancer to cardiovascular disease. Curr Drug Targets 11: 926–935, 2010 [DOI] [PubMed] [Google Scholar]
- 31. Jiang S, Zhang HW, Lu MH, He XH, Li Y, Gu H, Liu MF, Wang ED. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res 70: 3119–3127, 2010 [DOI] [PubMed] [Google Scholar]
- 32. Jones SW, Watkins G, Le Good N, Roberts S, Murphy CL, Brockbank SM, Needham MR, Read SJ, Newham P. The identification of differentially expressed microRNA in osteoarthritic tissue that modulate the production of TNF-alpha and MMP13. Osteoarthritis Cartilage 17: 464–472, 2009 [DOI] [PubMed] [Google Scholar]
- 33. Khanna S, Biswas S, Shang Y, Collard E, Azad A, Kauh C, Bhasker V, Gordillo GM, Sen CK, Roy S. Macrophage dysfunction impairs resolution of inflammation in the wounds of diabetic mice. PLoS One 5: e9539, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kong W, Yang H, He L, Zhao JJ, Coppola D, Dalton WS, Cheng JQ. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol 28: 6773–6784, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lederhuber H, Baer K, Altiok I, Sadeghi K, Herkner KR, Kasper DC. MicroRNA-146: tiny player in neonatal innate immunity? Neonatology 99: 51–56, 2010 [DOI] [PubMed] [Google Scholar]
- 36. Liechty KW, Kim HB, Adzick NS, Crombleholme TM. Fetal wound repair results in scar formation in interleukin-10-deficient mice in a syngeneic murine model of scarless fetal wound repair. J Pediatr Surg 35: 866–872, 2000 [DOI] [PubMed] [Google Scholar]
- 37. Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, Kaminski N, Abraham E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med 207: 1589–1597, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu Z, Xiao B, Tang B, Li B, Li N, Zhu E, Guo G, Gu J, Zhuang Y, Liu X, Ding H, Zhao X, Guo H, Mao X, Zou Q. Up-regulated microRNA-146a negatively modulate Helicobacter pylori-induced inflammatory response in human gastric epithelial cells. Microbes Infect 12: 854–863, 2010. [DOI] [PubMed] [Google Scholar]
- 39. Loffler D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermuller J, Kretzschmar AK, Burger R, Gramatzki M, Blumert C, Bauer K, Cvijic H, Ullmann AK, Stadler PF, Horn F. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood 110: 1330–1333, 2007 [DOI] [PubMed] [Google Scholar]
- 40. Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol 182: 4994–5002, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ma F, Liu X, Li D, Wang P, Li N, Lu L, Cao X. MicroRNA-466l upregulates IL-10 expression in TLR-triggered macrophages by antagonizing RNA-binding protein tristetraprolin-mediated IL-10 mRNA degradation. J Immunol 184: 6053–6059, 2010 [DOI] [PubMed] [Google Scholar]
- 42. Macedo L, Pinhal-Enfield G, Alshits V, Elson G, Cronstein BN, Leibovich SJ. Wound healing is impaired in MyD88-deficient mice: a role for MyD88 in the regulation of wound healing by adenosine A2A receptors. Am J Pathol 171: 1774–1788, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Martin P. Wound healing–aiming for perfect skin regeneration. Science 276: 75–81, 1997 [DOI] [PubMed] [Google Scholar]
- 44. Martinez-Nunez RT, Louafi F, Friedmann PS, Sanchez-Elsner T. MicroRNA-155 modulates the pathogen binding ability of dendritic cells (DCs) by down-regulation of DC-specific intercellular adhesion molecule-3 grabbing non-integrin (DC-SIGN). J Biol Chem 284: 16334–16342, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Masri S, Liu Z, Phung S, Wang E, Yuan YC, Chen S. The role of microRNA-128a in regulating TGFbeta signaling in letrozole-resistant breast cancer cells. Breast Cancer Res Treat 124: 89–99, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mbonye UR, Song I. Posttranscriptional and posttranslational determinants of cyclooxygenase expression. BMB Rep 42: 552–560, 2009 [DOI] [PubMed] [Google Scholar]
- 47. McCoy CE, Sheedy FJ, Qualls JE, Doyle SL, Quinn SR, Murray PJ, O'Neill LA. IL-10 inhibits miR-155 induction by toll-like receptors. J Biol Chem 285: 20492–20498, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, Jiang J, Schmittgen TD, Patel T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology 130: 2113–2129, 2006 [DOI] [PubMed] [Google Scholar]
- 49. Menke NB, Ward KR, Witten TM, Bonchev DG, Diegelmann RF. Impaired wound healing. Clin Dermatol 25: 19–25, 2007 [DOI] [PubMed] [Google Scholar]
- 50. Moore KW, de Waal Malefyt R, Coffman RL, O'Garra A. Interleukin-10 and the interleukin-10 receptor. Ann Rev Immunol 19: 683–765, 2001 [DOI] [PubMed] [Google Scholar]
- 51. Moschos SA, Williams AE, Perry MM, Birrell MA, Belvisi MG, Lindsay MA. Expression profiling in vivo demonstrates rapid changes in lung microRNA levels following lipopolysaccharide-induced inflammation but not in the anti-inflammatory action of glucocorticoids. BMC Genomics 8: 240, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nakamachi Y, Kawano S, Takenokuchi M, Nishimura K, Sakai Y, Chin T, Saura R, Kurosaka M, Kumagai S. MicroRNA-124a is a key regulator of proliferation and monocyte chemoattractant protein 1 secretion in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Rheum 60: 1294–1304, 2009 [DOI] [PubMed] [Google Scholar]
- 53. Nakasa T, Miyaki S, Okubo A, Hashimoto M, Nishida K, Ochi M, Asahara H. Expression of microRNA-146 in rheumatoid arthritis synovial tissue. Arthritis Rheum 58: 1284–1292, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut 58: 1375–1381, 2009 [DOI] [PubMed] [Google Scholar]
- 55. O'Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci USA 106: 7113–7118, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA 104: 1604–1609, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. O'Neill LA. How Toll-like receptors signal: what we know and what we don't know. Curr Opin Immunol 18: 3–9, 2006 [DOI] [PubMed] [Google Scholar]
- 58. Oberyszyn TM. Inflammation and wound healing. Front Biosci 12: 2993–2999, 2007 [DOI] [PubMed] [Google Scholar]
- 59. Okada H, Kohanbash G, Lotze MT. MicroRNAs in immune regulation–opportunities for cancer immunotherapy. Int J Biochem Cell Biol 42: 1256–1261, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Perry MM, Moschos SA, Williams AE, Shepherd NJ, Larner-Svensson HM, Lindsay MA. Rapid changes in microRNA-146a expression negatively regulate the IL-1beta-induced inflammatory response in human lung alveolar epithelial cells. J Immunol 180: 5689–5698, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Perry MM, Williams AE, Tsitsiou E, Larner-Svensson HM, Lindsay MA. Divergent intracellular pathways regulate interleukin-1beta-induced miR-146a and miR-146b expression and chemokine release in human alveolar epithelial cells. FEBS Lett 583: 3349–3355, 2009 [DOI] [PubMed] [Google Scholar]
- 62. Radomska HS, Huettner CS, Zhang P, Cheng T, Scadden DT, Tenen DG. CCAAT/enhancer binding protein alpha is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol Cell Biol 18: 4301–4314, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, Vetrie D, Okkenhaug K, Enright AJ, Dougan G, Turner M, Bradley A. Requirement of bic/microRNA-155 for normal immune function. Science 316: 608–611, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rosa A, Ballarino M, Sorrentino A, Sthandier O, De Angelis FG, Marchioni M, Masella B, Guarini A, Fatica A, Peschle C, Bozzoni I. The interplay between the master transcription factor PU.1 and miR-424 regulates human monocyte/macrophage differentiation. Proc Natl Acad Sci USA 104: 19849–19854, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ross J. mRNA stability in mammalian cells. Microbiol Rev 59: 423–450, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Roy S, Khanna S, Hussain SR, Biswas S, Azad A, Rink C, Gnyawali S, Shilo S, Nuovo GJ, Sen CK. MicroRNA expression in response to murine myocardial infarction: miR-21 regulates fibroblast metalloprotease-2 via phosphatase and tensin homologue. Cardiovasc Res 82: 21–29, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Roy S, Khanna S, Rink C, Biswas S, Sen CK. Characterization of the acute temporal changes in excisional murine cutaneous wound inflammation by screening of the wound-edge transcriptome. Physiol Genomics 34: 162–184, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sander AL, Henrich D, Muth CM, Marzi I, Barker JH, Frank JM. In vivo effect of hyperbaric oxygen on wound angiogenesis and epithelialization. Wound Repair Regen 17: 179–184, 2009 [DOI] [PubMed] [Google Scholar]
- 69. Sato T, Liu X, Nelson A, Nakanishi M, Kanaji N, Wang X, Kim M, Li Y, Sun J, Michalski J, Patil A, Basma H, Holz O, Magnussen H, Rennard SI. Reduced MiR-146a increases prostaglandin E2 in chronic obstructive pulmonary disease fibroblasts. Am J Respir Crit Care Med 182: 1020–1029, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis 31: 37–49, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schmeier S, MacPherson CR, Essack M, Kaur M, Schaefer U, Suzuki H, Hayashizaki Y, Bajic VB. Deciphering the transcriptional circuitry of microRNA genes expressed during human monocytic differentiation. BMC Genomics 10: 595, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science 265: 1573–1577, 1994 [DOI] [PubMed] [Google Scholar]
- 73. Sen CK. Wound healing essentials: let there be oxygen. Wound Repair Regen 17: 1–18, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sen CK, Roy S. miRNA: licensed to kill the messenger. DNA Cell Biol 26: 193–194, 2007 [DOI] [PubMed] [Google Scholar]
- 75. Sen CK, Roy S. Redox signals in wound healing. Biochim Biophys Acta 1780: 1348–1361, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sheedy FJ, O'Neill LA. Adding fuel to fire: microRNAs as a new class of mediators of inflammation. Ann Rheum Dis 67, Suppl 3: iii50–iii55, 2008 [DOI] [PubMed] [Google Scholar]
- 77. Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O'Leary JJ, Ruan Q, Johnson DS, Chen Y, O'Neill LA. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol 11: 141–147, 2010 [DOI] [PubMed] [Google Scholar]
- 78. Shilo S, Roy S, Khanna S, Sen CK. Evidence for the involvement of miRNA in redox regulated angiogenic response of human microvascular endothelial cells. Arterioscler Thromb Vasc Biol 28: 471–477, 2008 [DOI] [PubMed] [Google Scholar]
- 79. Shilo S, Roy S, Khanna S, Sen CK. MicroRNA in cutaneous wound healing: a new paradigm. DNA Cell Biol 26: 227–237, 2007 [DOI] [PubMed] [Google Scholar]
- 80. Singer AJ, Clark RA. Cutaneous wound healing. N Engl J Med 341: 738–746, 1999 [DOI] [PubMed] [Google Scholar]
- 81. Sonkoly E, Stahle M, Pivarcsi A. MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin Cancer Biol 18: 131–140, 2008 [DOI] [PubMed] [Google Scholar]
- 82. Sorrentino A, Liu CG, Addario A, Peschle C, Scambia G, Ferlini C. Role of microRNAs in drug-resistant ovarian cancer cells. Gynecol Oncol 111: 478–486, 2008 [DOI] [PubMed] [Google Scholar]
- 83. Stamou P, Kontoyiannis DL. Posttranscriptional regulation of TNF mRNA: a paradigm of signal-dependent mRNA utilization and its relevance to pathology. Curr Dir Autoimmun 11: 61–79, 2010 [DOI] [PubMed] [Google Scholar]
- 84. Suarez Y, Wang C, Manes TD, Pober JS. Cutting edge: TNF-induced microRNAs regulate TNF-induced expression of E-selectin and intercellular adhesion molecule-1 on human endothelial cells: feedback control of inflammation. J Immunol 184: 21–25, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Suzuki A, Nakano T, Mak TW, Sasaki T. Portrait of PTEN: messages from mutant mice. Cancer Sci 99: 209–213, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA 103: 12481–12486, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Teng G, Papavasiliou FN. Shhh! Silencing by microRNA-155. Philos Trans R Soc Lond B Biol Sci 364: 631–637, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. Regulation of the germinal center response by microRNA-155. Science 316: 604–608, 2007 [DOI] [PubMed] [Google Scholar]
- 89. Tili E, Croce CM, Michaille JJ. miR-155: on the crosstalk between inflammation and cancer. Int Rev Immunol 28: 264–284, 2009 [DOI] [PubMed] [Google Scholar]
- 90. Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, Croce CM. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol 179: 5082–5089, 2007 [DOI] [PubMed] [Google Scholar]
- 91. Turner M, Vigorito E. Regulation of B- and T-cell differentiation by a single microRNA. Biochem Soc Trans 36: 531–533, 2008 [DOI] [PubMed] [Google Scholar]
- 92. Venkataraman S, Alimova I, Fan R, Harris P, Foreman N, Vibhakar R. MicroRNA 128a increases intracellular ROS level by targeting Bmi-1 and inhibits medulloblastoma cancer cell growth by promoting senescence. PLoS One 5: e10748, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wakefield LM, Roberts AB. TGF-beta signaling: positive and negative effects on tumorigenesis. Curr Opin Genet Dev 12: 22–29, 2002 [DOI] [PubMed] [Google Scholar]
- 94. Werner S, Grose R. Regulation of wound healing by growth factors and cytokines. Physiol Rev 83: 835–870, 2003 [DOI] [PubMed] [Google Scholar]
- 95. Williams AE, Perry MM, Moschos SA, Larner-Svensson HM, Lindsay MA. Role of miRNA-146a in the regulation of the innate immune response and cancer. Biochem Soc Trans 36: 1211–1215, 2008 [DOI] [PubMed] [Google Scholar]
- 96. Wittmann J, Jack HM. Serum microRNAs as powerful cancer biomarkers. Biochim Biophys Acta 2010 [DOI] [PubMed] [Google Scholar]
- 97. Worm J, Stenvang J, Petri A, Frederiksen KS, Obad S, Elmen J, Hedtjarn M, Straarup EM, Hansen JB, Kauppinen S. Silencing of microRNA-155 in mice during acute inflammatory response leads to derepression of c/ebp Beta and down-regulation of G-CSF. Nucleic Acids Res 37: 5784–5792, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Young LE, Sanduja S, Bemis-Standoli K, Pena EA, Price RL, Dixon DA. The mRNA binding proteins HuR and tristetraprolin regulate cyclooxygenase 2 expression during colon carcinogenesis. Gastroenterology 136: 1669–1679, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zhang S, Chen L, Jung EJ, Calin GA. Targeting microRNAs with small molecules: from dream to reality. Clin Pharmacol Ther 87: 754–758, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]