Abstract
We performed a systematic BLAST analysis of 929 human disease gene entries associated with at least one mutant allele in the Online Mendelian Inheritance in Man (OMIM) database against the recently completed genome sequence of Drosophila melanogaster. The results of this search have been formatted as an updateable and searchable on-line database called Homophila. Our analysis identified 714 distinct human disease genes (77% of disease genes searched) matching 548 unique Drosophila sequences, which we have summarized by disease category. This breakdown into disease classes creates a picture of disease genes that are amenable to study using Drosophila as the model organism. Of the 548 Drosophila genes related to human disease genes, 153 are associated with known mutant alleles and 56 more are tagged by P-element insertions in or near the gene. Examples of how to use the database to identify Drosophila genes related to human disease genes are presented. We anticipate that cross-genomic analysis of human disease genes using the power of Drosophila second-site modifier screens will promote interaction between human and Drosophila research groups, accelerating the understanding of the pathogenesis of human genetic disease. The Homophila database is available at http://homophila.sdsc.edu.
Studies in the fruit fly Drosophila melanogaster have altered our estimate of the evolutionary relationship between vertebrate and invertebrate organisms. Key molecular pathways required for the development of a complex animal, such as patterning of the primary body axes, organogenesis, wiring of a complex nervous system, and control of cell proliferation have been highly conserved since the evolutionary divergence of flies and humans. When these pathways are disrupted in either vertebrates or invertebrates, similar defects are often observed. The utility of Drosophila as a model organism for the study of human genetic disease is now well documented. Developmental defects such as the mesenchymal malformations associated with Saethre-Chotzen syndrome (Howard et al. 1997), formation of intracellular inclusions in polyglutamine-tract repeat disorders such as spinocerebellar ataxia and Huntington disease (Fortini and Bonini 2000), and loss of cellular-growth control and malignancy resulting from mutations of tumor suppressor genes (Potter et al. 2000) have been analyzed effectively using Drosophila as the model genetic system. The many basic processes that are shared between Drosophila and humans, in conjunction with the recent completion of the Drosophila genomic sequence, provide the necessary ingredients for launching systematic analyses of human disease-causing genes in Drosophila. An important question that arises from the combination of this genomic information with the detailed mechanistic understanding of many Drosophila genes is, which human disease genes are most appropriate for study in Drosophila?
A survey of 289 Drosophila genes related to human disease genes has been presented in the context of the Drosophila genome sequence release (Rubin et al. 2000) and subsequently by Fortini et al. (2000). Additionally, more focused studies of Drosophila ion-channel genes (Littleton and Ganetzky 2000) and cancer-gene related sequences (Potter et al. 2000) have been published. Here, we report on results generated by a cross-genomic analysis of the 929 Locuslink entries of human disease genes known to have at least one mutant allele listed in the current version of the Online Mendelian Inheritance in Man (OMIM) (McKusick 2000) against the complete Drosophila genome sequence. We compiled this cross-genomic data into a database called Homophila, which presently contains a set of 714 clear-candidate human disease genes, their Drosophila counterparts (548 distinct genes), and any P-elements within 1 kb of these genes. This set of genes was categorized by human disease type and existing mutant alleles of these genes were identified. Analysis of this dataset and support material is currently available via the World Wide Web in the form of a searchable cross-genomic database (Homophila, http://homophila.sdsc.edu), which has been designed to be automatically updated as the number of disease-associated genes expands.
RESULTS
Development of Homophila as a Tool for Cross-Genomic Analysis
As a starting point for our analysis, we wished to determine which human disease genes have clearly related counterparts in Drosophila. To this end, we extracted the set of all known human-disease–associated genes from OMIM with Locuslink entries and compared this set of genes to the recently completed Drosophila genomic sequence (see Methods). By incorporating information about both the human disease gene and its Drosophila counterpart, it is possible to query the search results by key word, disease name, fly gene, and OMIM number. The outline of a typical query to Homophila is illustrated in Figure 1.
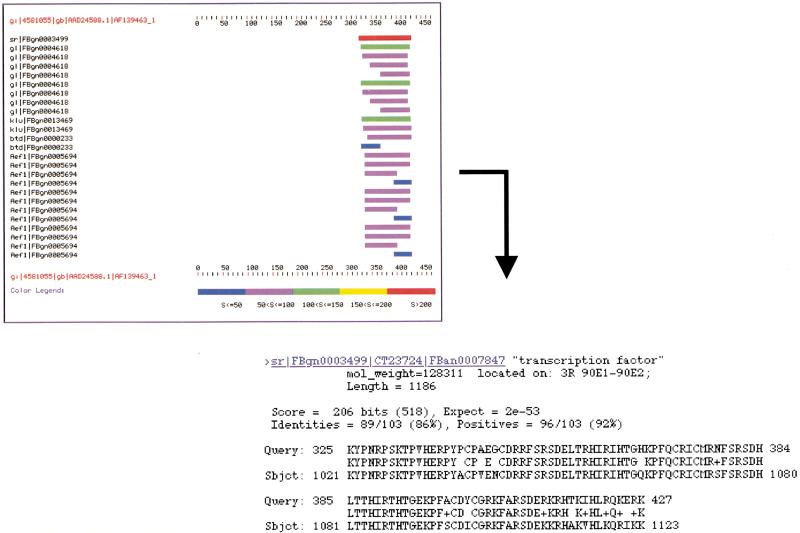
Figure 1.
How to query the Homophila database. (A) Schematic of a Homophila query. The user enters the text query in the form of human disease name, Online Mendelian Inheritance in Man (OMIM) number, fly gene name, or keyword search through the human disease entry box. The database then opens a window with information on the disease name, and human and fly genes that match the key word query. The user then can examine the details of an individual human to Drosophila BLAST comparison to get more information on the specific BLAST score, alignment, and other hits to this gene. In addition, P-element information is found at this level. (B) Example Homophila query using the keyword “neuropathy”. The user enters the key word and the database will return any human entry or Drosophila gene description that contains the key word. In this case, there are several human neuropathies listed, including a gene for peripheral neuropathy, which is a transcription factor (Krox20). By clicking the “details” button in this first window, one can examine the particular BLAST comparisons of the human genes to Drosophila genes as well as the P-element information. By scrolling down in this window, one can look at particular alignments between the query sequence and its Drosophila matches. In this case, the human Krox20 matches Drosophila stripe gene most strongly in the DNA-binding domains, but also retains some overall sequence similarity in other domains as can clearly be seen on the color graphical alignment of similar sequences.
Identification and Analysis of Drosophila Genes Related to Candidate Human Disease Genes
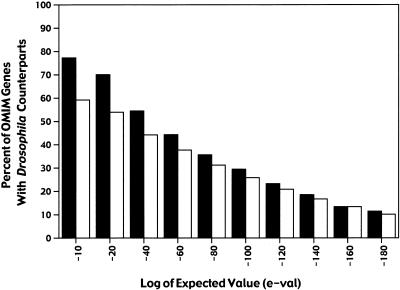
Using Homophila, we found that 714 of the 929 (77%) OMIM human disease gene entries have highly similar (E ≤10−10) cognates in Drosophila (Fig. 2), which we refer to as “related genes” hereafter. An E value of ≤10−10 indicates that the odds are <1 in 1010 that such a match would happen by chance alone given the sizes of the two compared databases (e.g., OMIM Locuslink entries and Flybase). We are aware that these Drosophila cognates may not be functional orthologs to the human disease genes and are using the less-stringent term “related genes” to describe these similar sequences. It is notable, however, that even at a higher E-value cut-off, a significant fraction of human disease genes have matches in Drosophila (Fig. 2, e.g., >54% with E ≤10−40 and 29% with E ≤10−100). A list of disease phenotypes resulting from mutations in genes that are highly related to Drosophila genes (E <10–10) is available as a separate table on the Homophila Web site (Reiter et al. 2000) as the clear-hit list, and has been categorized into various subclasses based on clinical phenotype (Table 1). Because some of the 714 distinct human disease genes match the same Drosophila-related sequences, the total number of different Drosophila counterparts of human clear-hit genes is 548 distinct Drosophila genes. We found a large number of human disease genes involved in nonmyelin-associated neurological disorders (74), cancer (79), skeletal disorders (26), and other developmental defects (35), as noted in previous studies. We also found a large number of metabolic and storage disorders (160), which were not highly sampled categories of genes in prior surveys. Consistent with the prevalence of disorders affecting metabolism and other general cellular functions, 409 of the clear-hit human genes (e.g., 57%) also have cognates in yeast (e.g., E ≤10−10). An interesting feature of this inclusive data set, which also was not evident from the earlier analyses of more selective sets of diseases, is the high-number of human genes affecting the visual (43), cardiovascular (26), auditory (13), skeletal (26), and endocrine (50) systems with Drosophila counterparts.
Figure 2.
Number of Drosophila sequences related to human disease genes as a function of E-value. A graph of the percent of human disease genes with similar sequences in Drosophila as a function of E-value. Black-filled bars indicate the percent of human Locuslink entries (929 total) with matches to Drosophila sequences. White-filled bars indicate the percent of unique Drosophila sequences that match one or more human disease gene sequences. Note that even at E-values of ≤10−40, 54% of human disease genes have matches to Drosophila sequences.
Table 1.
Classification of 714 Clear-Hit Drosophila Genes According to Human Disease Phenotypes
| Disorder | No. of genes |
|---|---|
| Neurological | 74 |
| Neuromuscular | 20 |
| Neuropsychiatric | 9 |
| CNS/Developmental | 8 |
| CNS/Ataxia | 9 |
| Mental retardation | 6 |
| Other | 22 |
| Endocrine | 50 |
| Diabetes | 10 |
| Other | 40 |
| Deafness | 13 |
| Syndromic | 7 |
| Nonsyndromic | 6 |
| Cardiovascular | 26 |
| Cardiomyopathy | 10 |
| Conduction defects | 4 |
| Hypertension | 7 |
| Atherosclerosis | 3 |
| Vascular malformations | 2 |
| Ophthalmologic | 43 |
| Anterior segment | (13) |
| Aniridia | 1 |
| Rieger syndrome | 1 |
| Mesenchymal dysgenisis | 2 |
| Iridogoniodysgenisis | 2 |
| Corneal dystrophy | 2 |
| Cataract | 3 |
| Glaucoma | 2 |
| Retina | (30) |
| Retinal dystrophy | 1 |
| Choroiderimea | 1 |
| Color vision defects | 4 |
| Cone dystrophy | 2 |
| Cone rod dystrophy | 1 |
| Night blindness | 8 |
| Leber congenital amaurosis | 2 |
| Macular dystrophy | 4 |
| Retinitis pigmentosa | 7 |
| Pulmonary | 4 |
| Gastrointestinal | 13 |
| Renal | 13 |
| Immunological | 33 |
| Complement mediated | 11 |
| Other | 22 |
| Hematologic | 42 |
| Erythrocyte, general | 29 |
| Porphyrias | 7 |
| Platelets | 6 |
| Coagulation abnormalities | 28 |
| Malignancies | 79 |
| Brain | 3 |
| Breast | 4 |
| Colon | 11 |
| Other gastrointestinal | 3 |
| Genitourinary | 5 |
| Gynocologic | 3 |
| Endocrine | 3 |
| Dermatologic | 3 |
| Xeroderma pigmentosa | 6 |
| Other/sarcomas | 9 |
| Hematologic malignancies | 29 |
| Skeletal development | 26 |
| Craniosynostosis | 5 |
| Skeletal dysplasia | 13 |
| Other | 8 |
| Soft tissue | 2 |
| Connective tissue | 18 |
| Dermatologic | 25 |
| Metabolic/mitochondrial | 123 |
| Pharmacologic | 12 |
| Peroxisomal | 9 |
| Storage | 37 |
| Glycogen storage | 11 |
| Lipid storage | 13 |
| Mucopolysaccaridosis | 10 |
| Other | 3 |
| Pleitropic developmental | 35 |
| Growth, immune, cancer | 7 |
| Apoptosis | 1 |
| Other | 27 |
| Complex other | 9 |
| Total | 714 |
Totals for categories of disease are in bold, subcategory totals are in parenthesis, and individual categories are in plain text.
To determine what fraction of Drosophila clear-hit genes already have been analyzed by loss-of-function genetics, we examined each entry in the list of 548 cognates of human disease genes in the clear-hit list and searched for alleles of each of these genes systematically using allele and gene tables available from Flybase (Flybase 1999) (see Methods). In this manner, 153 mutant alleles were identified (e.g., 28% of Drosophila clear-hit genes). These alleles and the human disease-related sequences can be found on our Web site in tabular form (Reiter et al. 2000).
A notable result of this allele analysis is that the great majority of Drosophila genes related to human disease genes (e.g., 395 of 548) have not yet been analyzed by loss-of-function genetics, which is consistent with the finding that only 14% of the genes identified by the Drosophila genome project had been identified previously by individual researchers working on specific hypothesis-driven projects (Rubin et al. 2000). We then determined what fraction of the 395 predicted Drosophila transcription units without known mutant alleles have P-elements inserted in or near them (e.g., within 1 kb of the gene-coding region). By aligning the map positions for 3442 known P-element insertions listed by the Berkeley genome project with the map positions of the Drosophila genes related to human clear-hit genes, we found 190 distinct P-element insertions that lie within or near disease-gene–related sequences. When corrected for multiple insertions, these 190 P-elements reduce to 102 distinct clear-hit Drosophila genes. Further analysis determined that 56 of these P-element insertions are the only known alleles of these genes. Using routine genetic methods in Drosophila, it should be possible to create null alleles of the 56 P-element tagged genes with relatively little difficulty by remobilizing the P-elements and screening for imprecise excisions that delete all or parts of the coding regions. Thus, loss-of-function analysis should be straightforward for 209 (153 + 56) of the 548 clear-hit genes, which represents a substantial proportion of these genes (e.g., 38%).
Defects at Multiple Tiers of Conserved Signal-Transduction Pathways Cause Human Disease
To explore the cross-genomic nature of the clear-hit gene dataset further, we subcategorized genes into one of several signal transduction pathways known to play important developmental functions in Drosophila and looked for trends in the resulting human phenotypes. Signal transduction pathways typically are activated by one or several ligands binding to one or more transmembrane receptors. Ligand binding activates the receptor and leads to modification of cytoplasmic transducers that enters the nucleus-altering gene expression. A feature common to many signaling pathways is that multiple ligands activate specific receptors, which converge upon one or a few common cytoplasmic transducer(s).
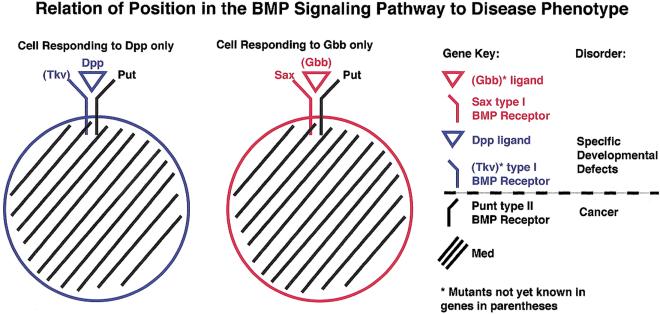
Among the disease genes on the clear-hit list, 56 (corresponding to 38 distinct Drosophila genes) encode components acting in well-characterized signaling pathways such as the bone morphogenic protein (BMP), receptor tyrosine kinase/reticular activating system (RTK/RAS), G-coupled receptor, JAK/STAT, Toll, Integrin, and axonal-guidance pathways (Table 2). Signaling components in these pathways have been ordered in Table 2 with respect to their position in known signaling cascades in Drosophila (e.g., ligand->receptor->cytoplasmic transducer->transcription-factor effector). A notable trend apparent in these tabulated data is that mutations affecting particular ligands or cell-type–specific receptors generally result in restricted developmental abnormalities in humans, whereas mutations in universally employed receptor subunits or downstream intracellular signal transducers tend to cause more global loss of cellular growth control or cancer in humans. For example, in the case of the BMP signaling pathway (Fig. 3), defects in specific BMP ligands result in human bone malformation (e.g., brachydactyly) and mutations of a selective type I BMP receptor subunit cause venous malformations (e.g., hereditary hemorrhagic telangiectasia). On the other hand, loss of the universal type II BMP receptor subunit, or the core signal transducer (e.g., vertebrate SMAD4 = Drosophila Medea) results in cancer in humans. This trend in which mutations in generally used signaling components often lead to loss of cellular growth control and cancer is consistent with many signaling pathways being directly or indirectly involved regulating cell proliferation.
Table 2.
Drosophila Genes From the Clear-Hit List That are in Known Signaling Pathways and the Human Phenotypes Associated with These Disease Genes
| Signaling pathway | Disease | OMIM# | Fly gene | Signaling component |
|---|---|---|---|---|
| BMP | Fibrodysplasia ossificans progressiva | 112262 | (dpp) | Ligand |
| Brachydactyly, type C | 113100 | (dpp) | Ligand | |
| Acromesomelic dysplasia, Hunter-Thompson type | 601146 | (dpp) | Ligand | |
| Hereditary hemorrhagic telangiectasia-2 | 601284 | (sax) | Specific type I receptor | |
| Persistent Mullerian duct syndrome, type II | 600956 | (wit) | Specific type II receptor | |
| Colorectal cancer, familial nonpolyposis, type 6 | 190182 | (put) | General type II receptor | |
| Polyposis, juvenile intestinal | 174900 | (med) | Cytoplasmic transducer | |
| Pancreatic cancer | 600993 | (med) | Cytoplasmic transducer | |
| Hedgehog | Holoprosencephaly-3 | 600725 | (hh) | Ligand |
| Basal cell nevus syndrome | 109400 | (ptc) | Co-receptor | |
| Basal cell carcinoma, sporadic | 601309 | (ptc) | Co-receptor | |
| Greig cephalopolysyndactyly syndrome | 165240 | (ci) | Transcription factor | |
| Wnt | Joubert syndrome | 213300 | (wg) | Ligand |
| Simpson dysmorphia syndrome | 300037 | (dally) | Proteoglycan (co-receptor?) | |
| Colorectal cancer | 116806 | (arm) | Cytoplasmic transducer | |
| Notch | Alagille syndrome | 601920 | (Ser) | Ligand |
| Cerebral ateriopathy with subcortical infarcts and leukoencephalopathy | 600276 | (N) | Receptor | |
| RTK | Obesity with impaired prohormone processing | 162150 | (Fur1) | Protease: Ligand activation? |
| Achondroplasia; Craniosynostosis; Crouzon syndrome | 134934 | (htl) | Receptor | |
| Pfeiffer syndrome | 136350 | (htl) | Receptor | |
| Venous malformations, multiple cutaneous and mucosal | 600221 | (htl) | Receptor | |
| Apert syndrome; Beare-Stevenson cutis gurata | 176943 | (htl) | Receptor | |
| Mast cell leukemia; Mastocytosis; Piebaldism | 164920 | (htl) | Receptor | |
| Diabetes mellitus, insulin-resistant; Leprechaunism; Rabson-Mendenhall syndrome | 147670 | (InR) | Receptor | |
| Renal cell carcinoma | 164860 | Receptor kinase-like gene | Receptor? | |
| Predisposition to myeloid malignancy | 164770 | Putative growth factor receptor | Receptor? | |
| Bladder cancer | 190020 | (Ras85D) | Cytoplasmic transducer | |
| Colorectal adenoma | 190070 | (Ras85D) | Cytoplasmic transducer | |
| Colorectal cancer | 164790 | (Ras85D) | Cytoplasmic transducer | |
| Colon cancer | 600679 | Tyrosine phosphatase 99A | Phosphatase | |
| Ehlers-Danlos syndrome, type X | 135600 | Tyrosine phosphatase 10D | Phosphatase | |
| Elliptocytosis-1 | 130500 | (cora) | Cytoskeletal scaffolding? | |
| Serpentine | Hypertension, salt-resistant | 108962 | guanylate cyclase receptor | Receptor |
| Night blindness, rhodopsin-related; Retinitis pigmentosa | 180380 | (ninaE) | Receptor (Rhodopsin 1) | |
| Colorblindness, deutan | 303800 | (ninaE) | Receptor | |
| Retinitis pigmentosa 4, included; rp4 | 180380 | (ninaE) | Receptor | |
| Night blindness, congenital stationary, rhodopsin-related | 190900 | (ninaE) | Receptor | |
| Autonomic nervous system dysfunction | 126452 | Dopamine receptor-like gene | Receptor | |
| Susceptibility to Schizophrenia? | 126451 | (DopR2) | Receptor | |
| Night blindness, congenital stationary, type 3 | 180072 | cGMP phosphodiesterase | Phosphodiesterase | |
| Retinitis pigmentosa, autosomal recessive | 180071 | cGMP phosphodiesterase | Phosphodiesterase | |
| Susceptibility to essential hypertension | 139130 | (Gbeta13F) | Cytoplasmic transducer | |
| Bleeding diathesis due to GNAQ deficiency | 600998 | (Galpha49B) | Cytoplasmic transducer | |
| JAK/STAT | SCID, autosomal recessive, T-negative/B-positive type | 600173 | (hop) | JAK kinase |
| Toll/NFkB | Leukemia/lymphoma, B-cell | 109560 | (cact) | Cytoplasmic transducer NFκI-like |
| Neuronal pathfinding | Propedrin deficiency | 312060 | Semaphorin family | Repulsive ligand |
| Polycystic kidney disease, type I | 601313 | Slit-like gene | Repulsive ligand | |
| Antithrombin III deficiency | 107300 | (sema-5c) | Ligand? | |
| Transcortin deficiency | 122500 | (sema-5c) | Ligand? | |
| Plasmin inhibitor deficiency | 262850 | (sema-5c) | Ligand? | |
| Hydrocephalus due to aqueductal stenosis, MASA syndrome, spastic paraplegia | 308840 | (Nrg) | Adhesion molecule (Neuroglian) | |
| Colorectal cancer | 120470 | (fra) | Receptor | |
| Integrin | Glazmann thrombasthenia, type A | 273800 | (if) | Integrin α-chain |
| Epidermolysis bullosa, junctional, with pyloric stenosis | 147556 | (mew) | Integrin α-chain | |
| Myopathy, congenital | 600536 | (mew) | Integrin α-chain | |
| Glycoprotein Ia deficiency | 192974 | (mew) | Integrin α-chain |
Figure 3.
Relationship of a components position in the bone morphogenetic protein (BMP) pathway to human disease phenotypes. In general, there is a relationship between the position of a component in signaling pathway to the disease phenotype resulting from inactivation of that component. An example of this trend is the BMP pathway. Mutations in components acting at the start of the BMP signal transduction cascade such as a particular BMP ligand (e.g., Drosophila Dpp = Human BMP4/BMP2) or a specialized BMP type I receptor (Drosophila Saxophone = type I receptor for the Screw and Glass Bottom Boat ligands) result in specific developmental defects (e.g., brachydactyly). Mutations acting on subsequent steps in the BMP pathway, which mediate the effects of several converging upstream inputs such as the universal type II BMP receptor (e.g., Drosophila Punt = type II receptor mediating all BMP signaling) or the cytoplasmic/nuclear SMAD transducer (e.g., Drosophila Medea = Human SMAD4) result in generalized misregulation of cellular growth control and cancer (e.g., colorectal or pancreatic cancer).
DISCUSSION
Our goal in conducting the analysis described in this study was to define a subset of human disease genes that would benefit most from molecular-genetic analysis in Drosophila. To this end we used Homophila, a searchable interactive database, to define a set of candidate human disease genes that have clearly related genes in Drosophila.
A strength of our current analysis with respect to previous studies is that it is inclusive and encompasses a much larger nonredundant set of human disease genes listed in OMIM with Locuslink entries. In contrast, previous studies have been more restrictive surveys focusing only on a subset of 289 genes selected a priori, which are known to be causally linked to a human disease (Fortini et al. 2000; Rubin et al. 2000) or those involved with a particular category of disease state (Littleton and Ganetzky 2000; Potter et al. 2000). This is a critical distinction because the current analysis reveals the relative proportions of different disease subclasses available for study in Drosophila. For example, in the previous two survey studies, genes affecting hearing and visual systems were relatively rare because of the more restrictive selection criteria used. Additionally, we identified 123 metabolic genes (17% of those analyzed) whereas the previous studies only included 17 metabolic genes in the dataset (6% of those analyzed).
Another problem with any type of cross-genomic analysis is that one must determine which sequence matches are significant enough to be considered similar in evolutionary origin. In addition, one must be able to distinguish domain-specific matches (e.g., a cross-species match of leucine zipper domains) versus matches that span the entire amino-acid sequence. For these reasons, we have provided a graph of the percent of human disease genes with Drosophila counterparts at a variety of E-values (Fig. 2). We also implemented a graphical interface for each match; this will provide the user with information about annotated domains of both the human and fly proteins (see Fig. 1).
Approximately Three-Quarters of the Candidate Human Disease Genes are Clearly Related to Genes in Drosophila
Analysis of the set of potential human disease genes related to Drosophila genes as defined in this study is informative in several respects. First, we find a high prevalence of neurological and neurodegenerative conditions (Table 1). This finding is not entirely unanticipated, because many of the components of neurogenesis (such as factors involving neural induction, guidance cues leading axons to their appropriate targets, the machinery for generating and propagating action potentials, and enzymes and molecular complexes involved in the synthesis and release of neurotransmitters) have been highly conserved during the course of evolution (Salzberg and Bellen 1996; Wu and Bellen 1997). Within the category of neurological diseases, the relatively large number of hearing conditions is noteworthy because these genes represent biologically analogous systems (e.g., the hairs in the inner ear versus the sensory bristles of Drosophila). Without the complete comparisons of the genomes in a database like Homophila, it would not be immediately obvious that genes responsible for human deafness could be functionally analyzed in an organism like Drosophila, which has no external auditory specializations analogous to ears. Second, we find that components of signal transduction pathways are frequent targets of human disease. An interesting relationship regarding this category of disease genes is that mutations in different components of various signaling pathways can result in very different disease phenotypes in humans. Components acting at early stages in a given pathway, such as genes encoding extracellular ligands, tend to have more specific and limited phenotypes, while genes acting in more downstream capacities, such as those encoding ligand receptors and downstream intracellular signaling molecules exhibit broader sets of defects resulting from disruption of several converging upstream signals.
Loss-of-Function Genetics to Study Human Disease Genes In Drosophila
Another striking feature of the list of potential human disease genes with related genes in Drosophila (i.e., the clear-hit list) is that only a minority of these genes already have been studied by classical loss-of-function genetics (i.e., 28% of the 548 Drosophila genes related to human disease genes on the clear-hit list). This number highlights the substantial number of yet-unstudied Drosophila cognates of human disease genes, which could be analyzed using the molecular genetic tools of Drosophila. It should be possible to study the majority of these genes using various previously-established methods. For example, wild-type or disease-causing mutant variants of any candidate human disease gene can be misexpressed in Drosophila using routine methods and the resulting gain-of-function phenotypes assayed either during development or in the adult. Because developmental pathways have been extensively studied in Drosophila, observation of gain-of-function phenotypes often will immediately implicate particular candidate pathways. For example, in the Drosophila wing it is possible to distinguish phenotypes resulting from disruption of components in the EGF-Receptor (RTK), Notch, Wingless, Hedgehog, and Drosphila decapentaplegic (Dpp) signaling pathways based on wing shape, integrity of the wing border, and the position and number of wing veins. Isolation of a new mutant with phenotypes resembling those of mutants in one of these known pathways would suggest obvious follow-up experiments to verify that the new gene was indeed involved in the suspected pathway. It also is possible to assay neurobehavioral phenotypes in Drosophila such as defects in vision, chemosensation, touch, hearing, and rudimentary learning. The few studies of this kind that have been carried out to date are very encouraging in that misexpression of disease alleles of human genes often results in visible morphological defects or behavioral deficits. A particularly promising aspect of several of these studies is that the function of normal versus mutant alleles of the human disease gene can be distinguished. A likely mechanistic basis for the different activity of wild-type versus mutant forms of candidate human-disease genes is that the mutant may act as a dominant negative in Drosophila as a result of a nonproductive interaction with a conserved component shared between flies and humans.
The function of the endogenous Drosophila counterparts of candidate human disease genes also can be analyzed by gain-of-function studies. More critically, however, loss-of-function analyses can be initiated to determine the consequence of removing the activity of these genes in Drosophila. Such loss-of-function analysis can be carried out for any of the 56 yet-to-be-analyzed P-element tagged genes. Additionally, because it is now practical to make targeted mutants in Drosophila (Rong and Golic 2000), it should soon be feasible to generate loss-of-function mutants in any of these genes. If misexpression of a human disease gene (normal or altered) or mutation of its Drosophila counterpart leads to scorable phenotypes in flies, second-site modifier screens typically can be designed to identify further genetic components acting in the same pathway as the gene of interest. The use of Drosophila to identify second-site modifier loci, which can then be tested for potential contribution to human disease (or modification of disease phenotypes), is likely to emerge as the most valuable application of Drosophila as model system for analysis of human disease genes because similar screens cannot be carried out on a significant scale in vertebrate systems.
Which Candidate Human Disease Genes are Best Suited for Analysis in Drosophila?
The motivation for conducting the above analysis was the practical issue of identifying Drosophila genes related to candidate human disease genes that are likely to be productively studied in Drosophila. It is evident that not all genes on the clear-hit list are necessarily best suited for study in Drosophila. For example, the great majority of the clear-hit human disease genes involved in metabolic and mitochondrial disorders (123) also have direct counterparts in yeast. Because many of these genes control similar basic cellular processes in yeast, flies, and humans, they may be more effectively analyzed in yeast rather than in Drosophila. It is also the case that some genes common to Drosophila and humans may not be performing equivalent functions in these two organisms. For example, it is likely that some of the genes involved in human-blood diseases affecting specific cell types may have other functions in Drosophila, which has a relatively simpler hemolymph system compared to the complexity of vertebrate blood.
These general guiding principles should not be adhered to dogmatically, however. For example, the gene for the metabolic disorder acute porphyria (OMIM #176000), a defect in the gene for porphobilinogen deaminase (PBG), has a clearly related gene in Drosophila (E = 10−78). Although there are gene sequences related to this uroporphyrinogen synthetase in many lower organisms, including yeast, the phenotype in humans involves paralysis and seizures as a result of secondary neurotoxicity from the buildup of excess porphyrin precursors. The study of such secondary effects of biochemical defects and the suppressors of these effects is more suited to an organism like Drosophila, which has a complex nervous system. As it happens, the fly gene most related to PBG (CG9165) contains a P-element insertion EP(3)0419. Thus, this gene seems to be an excellent candidate gene for study in Drosophila.
Another limitation of the cross-genomic comparison of human disease genes is that some of the Drosophila genes related to human disease genes may not be functionally equivalent (or orthologous) to the human disease gene in question, but rather may be more related by sequence and/or activity to another human gene that has a different function than the human disease gene. It is therefore to be anticipated that the clear-hit list contains matches between human and Drosophila genes that are members of a related but functionally diverged gene family. True orthologs may be identified through functional studies of individual genes in Drosophila. Thus, an important first step in analyzing any human disease gene in flies will be to demonstrate that the wild-type form of the human disease gene can substitute for (or rescue) loss-of-function mutants in the Drosophila gene. It is worth noting in this regard that a significant number of human disease genes have very strong matches to Drosophila counterparts (e.g., 274 disease genes = 29% match with E ≤10−100), suggesting that this stringent criterion of functional equivalence will be satisfied in many cases. Our group is in the process of studying several human disease genes using misexpression in Drosophila. Our initial findings indicate that at least for the human CYP2D6 gene, the Drosophila cyp18 gene is an ortholog and that regulation of the Drosophila gene can be disrupted via misexpression of the human gene (L. Reiter, pers. comm.).
With the above considerations and qualifications in mind, we believe that there are broad categories of candidate disease genes that are likely to be particularly amenable to study in Drosophila. Thus, among the human disease, clear-hit genes, 74 result in neurological disorders. Given the substantial existing evidence indicating that basic neuronal systems have been conserved between flies and humans, this set of disease genes is likely to be effectively analyzed in Drosophila. As mentioned above, analysis of Drosophila mutants involved in synaptic transmission and action potential propagation has proven to be directly relevant to these processes in vertebrates (Salzberg and Bellen 1996; Wu and Bellen 1997). Also, the 296 genes that represent developmental, neurological, cardiovascular, ophthalmologic, and hearing disorders as well as cancers, appear to be good candidates for study using Drosophila because there is reason to believe that the underlying molecular mechanisms controlling these organismic processes also are highly similar in flies and humans.
For the individual researcher, we suggest that the best approach to using our dataset is to query Homophila for a particular disease or key word representing a class of disorders. From these results, one can judge the degree of similarity between the human and fly genes as well as the domains that are similar (for example, the HOX genes show homology only in the DNA-binding domain). The domain graphic below the best match enables the user to determine if the best match is in a known domain and not across the entire gene. One should be mindful when using this information not to discard hits with only localized domains of homology. For example, in the case of the HOX genes, it has been well established that functionally orthologous genes in highly diverged species share high-sequence similarity only within the DNA-binding homeobox domain. Yet this relatively small portion of the molecule seems to carry key developmental information. There are links to both OMIM for the human disease information as well as to Flybase to determine allele and P-element information. In addition to determining if the human and fly genes are likely to perform similar functions, reasonable criteria for a good-candidate disease gene for study in Drosophila would include the following: (1) There is at least good circumstantial evidence that the gene is involved in the disease condition, (2) the mechanism by which the human gene functions is poorly understood (e.g., has not been placed in the context of a known pathway), and, pragmatically, (3) there is at least one mutant allele in that gene (153 genes) or a P-element insertion in or near that gene (56 genes).
Future Development of Homophila as an Interactive Tool for Cross-Genomic Analysis
We will continue to develop the Homophila database to bridge the gap between the human disease (OMIM) and Drosophila (Flybase) databases, which were not originally designed to facilitate cross-genomic browsing. Given that ∼4000 human disease phenotypes may have a genetic basis (Scriver 1995), it seems likely that the number of genes currently listed in OMIM will continue to grow at a rapid pace and that the frequent updates to the Homophila database will provide researchers with state-of-knowledge links to Drosophila counterparts of these genes. In addition, we currently are creating software to facilitate discovery of secondary associations among human and fly genes, which is now the focus of our next phase in development of the database. In particular, we plan to implement a version of the database that will allow for phenotype key word searches in both human and fly databases. This modified alignment technology would create key word strings for each disease-gene entry in OMIM and each fly-gene entry (e.g., by distilling key words from OMIM, Flybase, Interactive Fly, or Medline review abstracts) and then allow researchers to search these unordered strings against one another for statistically significant similarities (e.g., neurological diseases with cell loss in the cerebellum). The idea would be to then examine the fly cognates of genes responsible for similar diseases identified by such key word searches, and ask if these fly genes have some interesting feature or function in common (e.g., they are part of a common signaling pathway or molecular machine of some kind). It also would be possible to do this in reverse (e.g., cluster fly genes and ask if the corresponding human diseases share any common disease phenotypes).
Another addition to Homophila we are planning is software to identify potential candidate disease genes based on predicted phenotypes. This idea is based on the fact that while there are many examples in which several human disease genes belong to a common signaling pathway or functional module, there typically are not known diseases associated with all components in these pathways as defined by studies in Drosophila or other systems. In principle, one could guess the types of disease phenotypes that might arise from mutations in human orthologs of these other components (based on the disease phenotypes of mutations in existing components, the phenotypes of mutations of these other components in Drosophila, and the expression pattern of these components in mice or other vertebrates). The software we are currently developing will be used to identify human counterparts of fly genes in a systematic fashion and to ask if any diseases matching the predicted phenotypes have been mapped to regions of the human genome containing those genes. We anticipate that with the input of both the human and Drosophila genetics communities, Homophila will become a valuable cross-genomics tool in the post-genome-sequence era.
METHODS
Identification of Drosophila Genes Related to Human Disease Genes
This work reflects version 3.01 of the Homophila database (released Feb. 1, 2001). Our analysis began with the OMIM morbid map, a catalog of genetic diseases and their cytogenetic map locations, which is available electronically at ftp://ncbi.nlm.nih.gov/repository/OMIM/morbidmap. It was not possible to simply download the sequences related to each disease in the on-line version of OMIM because the protein and nucleic sequences associated with each OMIM entry often include unrelated genes mentioned in the text. Thus, a more involved procedure, relying on the NCBI Locuslink database, was required. Beginning with each of the 1792 genetic diseases specified in the OMIM morbid map, each disease was identified in the Locuslink mim2loc table, which relates OMIM entries to NCBI locus records. Each locus record then was used to locate the correct protein and nucleic-acid sequence records using the Locuslink loc2UG, loc2acc, and loc2ref tables, which specify entries in the NCBI Unigene, protein, nucleic acid, and RefSeq databases, respectively. This process was simplified by downloading the Locuslink tables (mim2loc, loc2ref, loc2acc, and loc2UG) and importing them directly into the Homophila database. The result of this procedure was a list of 4104 protein-sequence entries associated with 929 OMIM disease loci, and 4643 nucleic-acid sequence entries associated with 941 OMIM disease loci. Each of the protein-sequence entries was compared to the complete Drosophila genome sequence (Adams et al. 2000) using the BLASTP and TBLASTX programs (Altschul et al. 1997). BLAST comparisons were performed using BLAST v2.09 and the standard BLOSUM 62 and E = 10 settings. Many OMIM disease entries have multiple protein sequences linked to the disease through Locuslink. The BLAST search results for each of the probe sequences are merged, and the most significant hit (smallest E value) taken to construct the table of clear hits (Drosophila cognates of human disease genes, Table 1).
A relational database has been implemented to allow queries on these results and is available on-line (http://homophila.sdsc.edu) using the MySQL relational database management system (Dubois 2000). PERL scripts using the DBI package are used to convert queries entered on the Homophila Web pages to SQL queries to the actual RDBMS. A complete list of P-element locations in the Drosophila genomic sequence was kindly provided by FlyBase (Flybase 1999).
Acknowledgments
The authors thank the members of the human genetics community for their suggestions and comments during the preparation of this manuscript. We also thank Dr. Victor McKusick, creator of the OMIM database that made our study possible, for his insightful comments. This work was supported in part by grants to E.B. from the NIH (NS29870 and GM60585) and NSF (IBN-9604048) and from NIH P41 RR08605, National Biomedical Computation Resource which provides the server and also provides support to M.G. and S.C. L.T.R. was supported in part by a grant from the Glaucoma Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ebier@ucsd.edu; FAX (858) 822-2044.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.169101.
REFERENCES
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois P. MySQL. Indianapolis, IN: New Riders Publishing; 2000. [Google Scholar]
- Flybase. 1999. The FlyBase database of the Drosophila Genome Projects and community literature. The FlyBase Consortium. (http://flybase.bio.indiana.edu/). [DOI] [PMC free article] [PubMed]
- Fortini ME, Bonini NM. Modeling human neurodegenerative diseases in Drosophila: On a wing and a prayer. Trends Genet. 2000;16:161–167. doi: 10.1016/s0168-9525(99)01939-3. [DOI] [PubMed] [Google Scholar]
- Fortini ME, Skupski MP, Boguski MS, Hariharan IK. A survey of human disease gene counterparts in the Drosophila genome. J Cell Biol. 2000;150:F23–30. doi: 10.1083/jcb.150.2.f23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard TD, Paznekas WA, Green ED, Chiang LC, Ma N, Ortiz de Luna RI, Garcia Delgado C, Gonzalez-Ramos M, Kline AD, Jabs EW. Mutations in TWIST, a basic helix-loop-helix transcription factor, in Saethre-Chotzen syndrome. Nat Genet. 1997;15:36–41. doi: 10.1038/ng0197-36. [DOI] [PubMed] [Google Scholar]
- Littleton JT, Ganetzky B. Ion channels and synaptic organization: Analysis of the Drosophila genome. Neuron. 2000;26:35–43. doi: 10.1016/s0896-6273(00)81135-6. [DOI] [PubMed] [Google Scholar]
- McKusick, V.A. 2000. Online Mendelian Inheritance in Man, OMIM. (http://www.ncbi.nlm.nih.gov/omim/). [DOI] [PubMed]
- Potter CJ, Turenchalk GS, Xu T. Drosophila in cancer research: An expanding role. Trends Genet. 2000;16:33–39. doi: 10.1016/s0168-9525(99)01878-8. [DOI] [PubMed] [Google Scholar]
- Reiter, L., Beir, E., and Gribskov, M. 2000. Homophila. (http://homophila.sdsc.edu).
- Rong YS, Golic KG. Gene targeting by homologous recombination in Drosophila. Science. 2000;288:2013–2018. doi: 10.1126/science.288.5473.2013. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Yandell MD, Wortman JR, Gabor Miklos GL, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W, et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg A, Bellen HJ. Invertebrate versus vertebrate neurogenesis: Variations on the same theme? Dev Genet. 1996;18:1–10. doi: 10.1002/(SICI)1520-6408(1996)18:1<1::AID-DVG1>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Scriver CR. The metabolic and molecular bases of inherited disease. 7th ed. New York, NY: McGraw-Hill Health Professions Division; 1995. [Google Scholar]
- Wu MN, Bellen HJ. Genetic dissection of synaptic transmission in Drosophila. Curr Opin Neurobiol. 1997;7:624–630. doi: 10.1016/s0959-4388(97)80081-5. [DOI] [PubMed] [Google Scholar]