Abstract
The molecular mechanisms of lung injury and fibrosis are incompletely understood. MicroRNAs (miRNAs) are crucial biological regulators that act by suppressing their target genes and are involved in a variety of pathophysiological processes. To gain insight into miRNAs in the regulation of lung fibrosis, total RNA was isolated from mouse lungs harvested at different days after bleomycin treatment, and miRNA array with 1,810 miRNA probes was performed thereafter. MiRNAs expressed in lungs with bleomycin treatment at different time points were compared with miRNAs expressed in lungs without bleomycin treatment, resulting in 161 miRNAs differentially expressed. Furthermore, miRNA expression patterns regulated in initial and late periods after bleomycin were identified. Target genes were predicted in silico for differentially expressed miRNAs, including let-7f, let-7g, miR-196b, miR-16, miR-195, miR-25, miR-144, miR-351, miR-153, miR-468, miR-449b, miR-361, miR-700, miR-704, miR-717, miR-10a, miR-211, miR-34a, miR-367, and miR-21. Target genes were then cross-referenced to the molecular pathways, suggesting that the differentially expressed miRNAs regulate apoptosis, Wnt, Toll-like receptor, and TGF-β signaling. Our study demonstrated a relative abundance of miRNA levels in bleomycin-induced lung fibrosis. The miRNAs and their potential target genes identified may contribute to the understanding of the complex transcriptional program of lung fibrosis.
Keywords: lung fibrosis, microarray
progressive tissue fibrotic diseases affect a wide spectrum of organs and a large number of patients (49). The pulmonary fibrotic disorders include idiopathic pulmonary fibrosis (IPF), sarcoidosis, pneumoconioses, hypersensitivity pneumonitis, drug- and radiation-induced fibrosis, and the fibrosing alveolitis associated with collagen vascular diseases such as rheumatoid arthritis (7). IPF is a chronic, progressive, and usually lethal fibrotic lung disease with poor prognosis (7). The features of this disease are focal areas of alveolar epithelial cell injury and proliferation of mesenchymal cells in the interstitium, resulting in excessive deposition of extracellular matrix and distorted architecture leading to impaired gas exchange (42, 43). Prevailing hypotheses suggest that IPF is an epithelial-fibroblast cross-talk disorder that results from numerous microinjuries to the alveolar epithelia leading to a dysregulated fibrotic repair response (51, 52). Despite significant efforts, major gaps in understanding the genetic programs and signaling pathways that regulate severe pulmonary fibrosis remain.
The bleomycin-induced lung injury model is a widely used animal model in investigating the pathobiology of lung fibrosis (1, 2, 39). Intratracheal bleomycin instillation causes initial alveolar epithelial cell apoptosis, inflammatory cell infiltration, elevation of proinflammatory cytokines [interleukin (IL)-1, tumor necrosis factor (TNF)-α, IL-6, interferon-γ] by epithelial cells, inflammatory cells, and stromal cells, followed by increased expression of profibrotic markers [transforming growth factor (TGF)-β1, fibronectin, and procollagen-1] with a peak around day 14, and elevated till day 21 postinjury, during which fibroblasts are activated and release excessive extracellular matrix in the interstitium. Broadly, the pathological changes can be segregated into three stages: epithelial apoptosis, inflammation, and fibrosis (1, 2, 13, 23). Many experts in the field believe that the “switch” between inflammation and fibrosis appears to occur around day 9 after bleomycin (13).
MicroRNAs (miRNAs, also miRs) are a growing family of noncoding RNAs that have recently emerged as critical regulators of gene expression (5, 19). MiRNAs are short (∼22 bp in length), single-stranded, and noncoding RNAs that inhibit the production of target proteins or induce degradation of mRNAs by binding target mRNAs at complementary sites in 3′-untranslated regions (3′-UTRs) or coding sequences and thereby suppressing target gene expression (5, 19, 47). Many miRNAs exhibit temporal or tissue-specific expression patterns (33, 34) and are involved in a variety of crucial developmental, physiological, as well as disease processes (5, 19) including lung fibrosis. For instance, miR-21 regulates the activation of lung fibroblasts (36). MiR-155 regulates lung fibrosis by targeting keratinocyte growth factor (46). MiR-126 is involved in cystic fibrosis by regulating TOM1 in the Toll-like receptor (TLR) 2/4 signaling pathways (44). MiR-29 regulates collagen expression in systemic sclerosis (37). MiR-192 mediates renal fibrosis in a Smad3-dependent manner (15), while loss of miR-192 promotes fibrogenesis in diabetic nephropathy (30). These studies suggest that there are miRNAs involved in maintaining a fibrosis phenotype.
MiRNA microarray technology has been successfully exploited to generate miRNA gene expression profiles of lung cancer (32, 38), pulmonary hypertension in rats (10), and influenza virus H1N1-infected mice (35). Thus, miRNA gene expression profiling offers an effective means of acquiring novel and valuable information in gene regulation. Although recent reports have utilized miRNA array analysis to identify critical miRNAs in fibrogenesis in IPF patients (45), as well as bleomycin-treated mouse lungs (36), systematic analysis of miRNA expression in the diseases and in animal models is needed to enhance our understanding of the roles of miRNAs in the pathogenesis of progressive disease.
To elucidate the potential role of miRNA in the initiation and progression of pulmonary fibrosis, we investigated the time-dependent changes in miRNA expression in the bleomycin mouse model of lung fibrosis. Analysis of the target genes suggested that the differentially expressed miRNAs regulate critical pathways in apoptosis, inflammation, and fibrosis. These data may provide new insights into pathways contributing to the pathobiology of lung fibrosis and identify specific miRNAs that are targets for future interventional studies.
MATERIALS AND METHODS
Animals.
C57BL/6J mice were purchased from the Jackson Laboratory. All experiments were carried out using 8- to 12-wk-old female mice. All studies were conducted in accordance with National Institutes of Health guidelines for the care and use of animals and with approval from the Duke University Animal Care and Use Committee.
Bleomycin administration.
We administered 2.5 U/kg bleomycin (Blenoxane from Mayne Pharma, Paramus, NJ) dissolved in sterile PBS via trachea to mice under anesthesia, as previously described (20–22). Lung tissues were harvested at days 0, 3, 7, 14, and 21 postbleomycin challenges.
RNA isolation and quality assessment.
Total RNA from lung tissues was isolated using the mirVana miRNA isolation kit (Ambion, Austin, TX) according to the manufacturer's instructions. The quantity of the RNA was determined by optical density, measured at 260 nm by Nanodrop spectrophotometer. RNA quality was measured using the RNA Nano chip (Agilent Technologies, Santa Clara, CA) on the Agilent 2100 Bioanalyzer according to the manufacturer's instructions. RNA integrity number (RIN) of most samples are >7 except for one sample in the day 3 group (RIN = 6.7) according to the total RNA chip assay.
miRNA microarray hybridization.
All samples were profiled on the same miRNA-AI (Ambion and Invitrogen) array platform at Duke University microarray facility. This array combined probes from Ambion mirVana set2 and Invitrogen NCode multispecies miRNA probe set V2, which contains 1,810 probes covering 795 human, 487 mouse, and 282 rat miRNAs, respectively. RNA labeling and hybridization were completed according to the manufacturer's instructions. Briefly, RNA from each sample was labeled with Cy5 using miRCURY LNA miRNA Array labeling kit (Exiqon, Woburn, MA). After being labeled, the samples were loaded onto the microarray slides and incubated 16–18 h at 60°C. After hybridization, the slides were washed, dried by centrifugation, and scanned using the Axon GenePix 4000B Scanner (Axon, Sunnyvale, CA). Dye-swap designs were used as biological replicates rather than technical replicates to improve efficiency and simplify analysis (4).
miRNA microarray analysis.
The miRNA microarray output gpr files were loaded into GeneSpring GX 11.0 software (Agilent) for analysis. Average values of the replicate spots of each miRNA on the microarray were normalized by Lowess normalization. MiRNAs were filtered with raw value >10 to get 1,569 miRNAs above background. Volcano plot was used to do all the pair-wise comparisons to get the miRNA gene lists. The cutoff P value was <0.05 and fold change was at least 2. Profile plots and heat maps were subsequently produced to illustrate the splits. The miRNA array data are MIAME compliant and have been submitted to the National Center for Biotechnology Information Gene Expression Omnibus database (accession no.: GSE24323).
Gene target prediction for miRNAs and pathway analysis.
Online software packages were used to predict the genes targeted by miRNAs. In our study, gene target prediction was mainly performed by querying the miRNA Database miRanda (http://www.microrna.org/microrna/home.do) (24). Genes related to certain functions were inquired from Gene Ontology (http://www.geneontology.org/). Pathway analysis was performed by Gather KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis (http://gather.genome.duke.edu/) (12, 27). A Bayes factor was included in the consideration of KEGG pathway association (12).
RESULTS
miRNA expression patterns in lung tissues of bleomycin model.
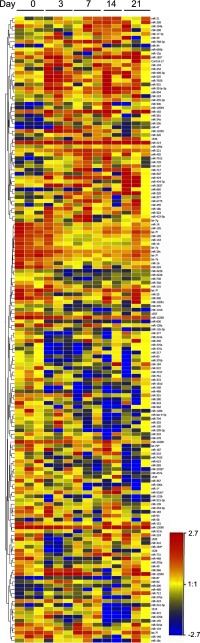
To gain an overview of changes in miRNA expression during bleomycin-induced lung injury and fibrosis, miRNA arrays were performed with total RNA isolated from mouse lung tissues harvested on days 0, 3, 7, 14, and 21 after bleomycin challenge. Samples were clustered according to their miRNA profiles using the hierarchical clustering algorithm of the GeneSpring software. There were 1,569 miRNAs expressed above background. There were 161 miRNAs that showed at least twofold changes (in either direction), significant at P < 0.05, in at least one time point after bleomycin when compared with control (day 0). Hierarchical clustering analysis of these 161 miRNAs is shown in Fig. 1.
Fig. 1.
Hierarchical clusters of differentially expressed microRNAs (miRNAs) in bleomycin-induced lung fibrosis. There were 161 miRNAs that found significantly differentially expressed (fold ≥2, P value < 0.05) for at least 1 of the 4 comparisons between bleomycin treatment and control (day 3 vs. day 0, day 7 vs. day 0, day 14 vs. day 0, and day 21 vs. day 0). Each row of the heat map represents an miRNA gene, and each column represents a sample in specific time point (n = 3 except for day 14 when nday14 = 2). Red indicates an increase in miRNA gene expression (relative to the other expression measurements in the same row), whereas blue indicates a decrease. Samples are identified at the top. MiRNA names are listed to the right.
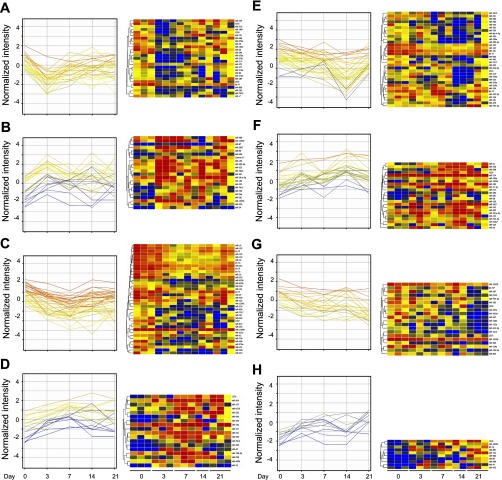
There are distinct temporal phases during bleomycin-induced lung injury and fibrosis. Hence, our analysis focused on these time points accordingly. Unsupervised clustering profiling was then performed to compare miRNA expression at each time point after bleomycin injury with control (day 0). Differentially expressed miRNAs in each comparison were identified. There were 27 downregulated (Fig. 2A) and 22 upregulated miRNAs (Fig. 2B) on day 3 vs. day 0. Comparing day 7 with day 0, 38 miRNAs were downregulated (Fig. 2C) and 18 miRNAs were upregulated (Fig. 2D). Thirty-three miRNAs were downregulated (Fig. 2E) and 23 miRNAs were upregulated on day 14 (Fig. 2F). There were 20 miRNAs downregulated (Fig. 2G) and 10 miRNAs upregulated on day 21 compared with day 0 (Fig. 2H), respectively. These data showed that the expression of miRNAs is differentially regulated during bleomycin-induced lung injury and fibrosis.
Fig. 2.
Supervised profiles and hierarchical clusters of miRNAs. Eight miRNA clusters obtained by comparisons between bleomycin treatment and control are shown. There were 27 miRNAs downregulated (A) and 22 miRNAs upregulated (B) between day 3 and day 0. There were 38 miRNAs downregulated (C) and 18 miRNAs upregulated (D) between day 7 and day 0. There were 33 miRNAs downregulated (E) and 23 miRNAs upregulated (F) on day 14 when compared with day 0. There were 20 miRNAs downregulated (G) and 10 miRNAs upregulated (H) on day 21 when compared with day 0. Blue indicates negative values, and red indicates positive values. MiRNA names are listed to the right, and sample identifications are listed at the bottom.
Differential miRNA expression during distinct pathologic phases of bleomycin-induced lung fibrosis.
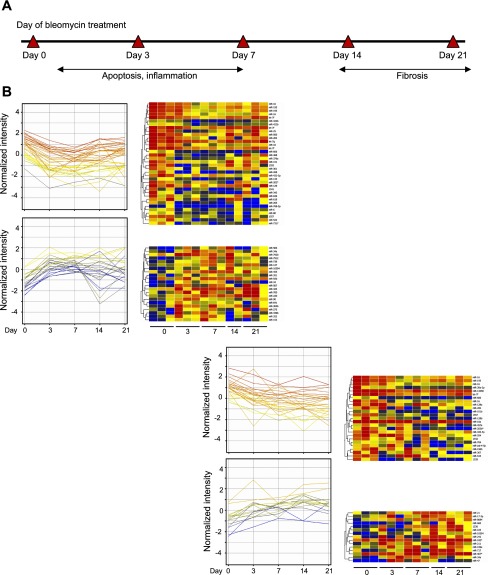
Characteristics of bleomycin-induced lung injury and fibrosis include epithelial apoptosis, inflammation, and fibrosis (1, 2, 13). To identify miRNA expressions related to lung injury and fibrosis, miRNAs differentially expressed at both day 3 and day 7 were separated from miRNAs regulated at day 14 and day 21. We reasoned that miRNAs differentially expressed at both day 3 and day 7 were mainly associated with epithelial cell apoptosis and the inflammatory process, whereas miRNAs differentially regulated at both day 14 and day 21 might play a pivotal role in regulating the fibrotic process (Fig. 3A). There were 58 miRNAs differentially regulated at both day 3 and day 7 when compared with control day 0. Among them, 36 miRNAs were downregulated at both day 3 and day 7, while 22 miRNAs were upregulated at both days (Fig. 3B). Also compared with day 0, a total of 40 miRNAs were differentially expressed at both day 14 and day 21. Among them, 25 miRNAs were upregulated and 15 miRNAs were downregulated. Furthermore, we analyzed the miRNAs regulated in the same direction for both the early and late phases postbleomycin lung injury. One miRNA (miR-34a) was upregulated and three miRNAs (miR-16, let-7f, miR-195) were downregulated in both the early and late phases. Thus, we have identified miRNAs associated with the pathological processes of bleomycin-induced lung injury and fibrosis.
Fig. 3.
MiRNAs in different pathological phases of bleomycin-induced lung fibrosis. A: graphic representation of the pathological stages of bleomycin-induced lung fibrosis. Two major pathological phases, including early epithelial apoptosis and inflammation phase (day 3 and day 7 postbleomycin), as well as late fibrosis phase (day 14 and day 21 postbleomycin). B: profiles and heat maps of differentially expressed miRNAs merged with each pathological phase. There were 36 miRNAs downregulated, and 22 miRNAs upregulated in early apoptosis and inflammation period of bleomycin treatment when compared with day 0. There were 25 miRNAs downregulated and 15 miRNAs upregulated in late fibrosis period of bleomycin treatment vs. day 0.
Predicted gene regulation by differentially expressed miRNA during bleomycin-induced lung fibrosis.
miRNAs regulate a large number of target genes and several databases based on various algorithms are available for predicting the targets of selected miRNAs. To investigate miRNA profiles associated with the molecular signaling pathways underlying lung injury and fibrosis, miRNA Database miRanda (http://www.microrna.org/microrna/home.do) (24) was used to predict mouse gene targets of the deregulated miRNAs identified. In brief, each differentially expressed miRNA target was extracted from miRanda. Overall, 7,509 target genes were predicted from miRNAs days 3 and 7 vs. day 0. The target genes were curated in gene ontology (GO) to identify the inflammation-, apoptosis-, and fibrosis-related genes. Analysis identified 386 genes related to apoptosis and 91 related to inflammation. Furthermore, there were 7,452 target genes for miRNAs at days 14 and 21 vs. day 0. GO analysis identified 154 genes related to fibrosis. Genes targeted by miRNAs deregulated in all time points were also identified by miRanda. There were 2,811 target genes predicted, including 133 genes related to apoptosis, 37 genes related to inflammation, and 51 genes related to fibrosis.
Pathway analysis of miRNA targets.
We further reasoned that miRNA regulation is the part of a biological network in a biological system. MiRNA-targeted genes may represent specific signaling pathways. KEGG is a project that classifies the current knowledge of molecular and cellular biology in terms of the information pathways that consist of interacting genes or molecules (26, 27). We next submitted the target genes of differentially regulated miRNAs during bleomycin-induced lung injury and repair to identify if these targets are involved in apoptosis, inflammation, and fibrosis. Alveolar epithelial cells overlying fibroblastic foci in IPF patients show evidence of apoptosis (54). In animal models, alveolar epithelial cell apoptosis was sufficient to induce fibrogenesis, whereas blockade of epithelial apoptosis attenuated fibrosis (31, 56). To investigate if miRNAs differentially expressed at both days 3 and 7 in our study play a role in apoptosis, we submitted the target genes enriched from GO to KEGG pathway analysis with Gather. There were 12 apoptosis-related pathways that were significantly enriched (P < 0.001) (Table 1) from the 386 apoptosis-related genes. Among them, the MAPK signaling pathway, apoptosis (Fig. 4A), and the Wnt signaling pathway (Fig. 4B) had the most Bayes factors, at 22, 8, and 7, respectively (Table 1). Other functional terms including cytokine-cytokine receptor interaction and TGF-β signaling pathway were also identified. These results suggest that the miRNAs regulated in early time points may play a role in apoptosis through these related pathways.
Table 1.
KEGG pathway identification of apoptosis genes targeted by microRNAs in early phase (day 3 and day 7) after bleomycin
| KEGG Pathway | Genes, n | P Value | Bayes Factor |
|---|---|---|---|
| MAPK signaling pathway | 40 | <0.0001 | 22 |
| Apoptosis | 17 | <0.0001 | 8 |
| Wnt signaling pathway | 21 | <0.0001 | 7 |
| Cytokine-cytokine receptor interaction | 27 | 0.0002 | 4 |
| TGF-β signaling pathway | 13 | 0.0004 | 4 |
| Toll-like receptor signaling pathway | 14 | 0.0005 | 3 |
| Adherens junction | 12 | 0.0006 | 3 |
| Jak-STAT signaling pathway | 18 | 0.001 | 3 |
| Alzheimer's disease | 6 | 0.001 | 2 |
| Focal adhesion | 23 | 0.001 | 2 |
KEGG, Kyoto Encyclopedia of Genes and Genomes.
Fig. 4.
Modeled miRNA mediated pathways in the pathogenesis of lung fibrosis. A: the mode of miRNAs that regulate apoptosis by modulating the apoptosis-signaling pathway is shown. B: an overview of miRNAs regulating the Wnt signaling pathway. C: miRNAs regulate the inflammation by mediating the Toll-like receptor pathway. D: miRNAs regulate the fibroblast activation and collagen deposition by modulating the TGF-β signaling pathway. Downregulated miRNAs are denoted in blue and upregulated in red.
Although inflammation is usually minimal by the time patients present with IPF, inflammation may still be involved in IPF (8). In the bleomycin model we hypothesized that miRNAs regulated at early time points play a role in inflammation. The 91 inflammation-related genes were submitted to KEGG pathway analysis through Gather and resulted in three inflammation-related pathways (P < 0.005) (Table 2). These were cytokine-cytokine receptor interaction, the TLR signaling pathway (Fig. 4C), and the Jak-STAT signaling pathway, with Bayes factors 15, 4, and 1, respectively. These data suggest that miRNAs may regulate inflammation through innate immune and adapted immune pathways.
Table 2.
KEGG pathway identification of inflammation genes targeted by microRNAs in early phase (day 3 and day 7) after bleomycin
| KEGG Pathway | Genes, n | P Value | Bayes Factor |
|---|---|---|---|
| Cytokine-cytokine receptor interaction | 20 | <0.0001 | 15 |
| Toll-like receptor signaling pathway | 8 | 0.0002 | 4 |
| Jak-STAT signaling pathway | 8 | 0.005 | 1 |
Fibrosis is a result of fibroblast activation and extracellular matrix deposition. This involves a series of molecular signaling pathways, which includes the TGF-β signaling pathway (9, 17, 18, 28). After pathway analysis of the 154 fibrosis-related genes, seven fibrosis-related pathways were identified (P < 0.01) (Table 3). They included cytokine-cytokine receptor interaction, focal adhesion, and the Jak-STAT signaling pathway. The Bayes factors for these are 28, 9, and 8, respectively. Other functional terms including the TGF-β signaling pathway (Fig. 4D) and adherens junction were also identified.
Table 3.
KEGG pathway identification of fibrosis genes targeted by micoRNAs in late phase (day 14 and day 21) after bleomycin
| KEGG Pathway | Genes, n | P Value | Bayes Factor |
|---|---|---|---|
| Cytokine-cytokine receptor interaction | 33 | <0.0001 | 28 |
| Focal adhesion | 21 | <0.0001 | 9 |
| Jak-STAT signaling pathway | 16 | <0.0001 | 8 |
| MAPK signaling pathway | 19 | <0.0001 | 6 |
| Adherens junction | 10 | <0.0001 | 6 |
| TGF-β signaling pathway | 9 | 0.0006 | 3 |
| Cell cycle | 8 | 0.006 | 1 |
Gene targets of the miRNAs deregulated by all time points after bleomycin were analyzed with KEGG. The analysis revealed that these target genes were also related to the pathways associated with apoptosis (Supplemental Table S1), inflammation (Supplemental Table S2), and fibrosis (Supplemental Table S3).1 For example, MAPK signaling pathway, cytokine-cytokine receptor interaction, and TGF-β signaling pathway were significantly enriched. In addition, complement and coagulation signaling were also identified (Supplemental Table S4). Complement signaling (3, 25) has been shown to play a role in regulating inflammation as well as TGF-β-mediated fibrosis during bleomycin-induced fibrosis (3).
In addition to the abovementioned pathways, we identified several pathways (such as oxidative response pathway and glutathione metabolism pathway) related to apoptosis and inflammation (Supplemental Table S4) and fibrosis (Supplemental Table S5). For example, reduced glutathione has been shown in the patients with IPF (29). Oxidative stress to epithelial cells has been suggested to play an important role in the pathogenesis of pulmonary fibrosis (8, 29). Furthermore, antioxidant therapy has some benefit in the treatment of IPF (8, 16).
Collectively, our study revealed that miRNAs were differentially expressed during bleomycin-induced lung injury and fibrosis. Further analysis demonstrated that differentially expressed miRNAs may regulate apoptosis, inflammation, and fibrosis by targeting the components in the apoptosis, Wnt signaling, TLR signaling, and TGF-β signaling pathways.
DISCUSSION
The molecular mechanisms that regulate lung tissue injury and repair are largely unknown. Signaling pathways regulating epithelial apoptosis, inflammatory cell infiltration/recruitment, cytokine release, and activation of fibroblasts/myofibroblasts have been investigated and implicated in the pathogenesis of progressive lung fibrosis. Many experts in the field believe that the regulation of fibrogenesis by the pathways and molecular processes is a well-orchestrated, tightly regulated process. Herein we provide novel information regarding profiling of miRNAs in different pathological phases of bleomycin-induced lung fibrosis in mice and demonstrate that miRNAs are differentially regulated during tissue injury and fibrosis. In light of the potential targets of these miRNAs, the differentially expressed miRNAs represent a new layer of regulatory machinery and an integrated part of a complex regulatory network to orchestrate tissue injury and fibrosis. In other systems, it has been shown that expression of miRNAs is varied in different pathological stages (40, 55).
Apoptosis and inflammation-related pathways were targeted by miRNAs in the initial period of bleomycin injury.
The bleomycin model of pulmonary fibrosis is characterized by initial Type II lung epithelial cell injury and apoptosis (17). Epithelial injury is followed by an initial influx of inflammatory cells that later instigates the fibrotic process. The magnitude of this initial inflammatory response is often correlated with enhanced fibrosis at late stages (53). The influx of inflammatory cells is between days 3 and 9, depending upon the dose of bleomycin used, in rats and mice (41). Concurrently, the inflammatory cells and stromal cells at the injury site produce chemokines, cytokines, and other inflammatory/immunoregulatory substances including the potent fibroblast growth factors TGF-β, TNF, endothelin 1, metalloproteinases, and the coagulation mediator tissue factor (11). We characterized miRNA expression in an initial period (day 3 and day 7) after bleomycin treatment and predicted their targets by using the miRanda algorithm. These miRNAs target important factors during apoptosis, including TNF-α, death activator FasL, effector caspases, which include caspases 3 and 7, proapoptotic members such as Bax, p53, and NF-κB, and antiapoptotic members such as IRAK-2 and BCL2 (Fig. 4A). The aforementioned pattern indicated that some differentially regulated miRNAs such as let-7f, let-7g, miR-144, miR-153, miR-16, miR-195, miR-25, miR-34a, miR-351, miR-361, miR-449b, miR-468, and miR-700 were in agreement with this prediction. In the present study, miR-153 was upregulated in both day 3 and day 7 and is also predicted to target Bcl-2, which is consistent with the previous report. It has been shown that overexpression of miR-153 in glioblastoma cells resulted in increased apoptosis by downregulating Bcl-2 (57).
Interestingly, we found that key factors in the Wnt signaling pathway (Fig. 4B), which have been shown to have a close relationship with lung fibrosis, were also targeted by these aforementioned miRNAs. The potential targets include the members of the Wnt family, such as Wnt5a, Wnt7a, Wnt7b, and Wnt9a, and the molecules in the Wnt pathway such as GSK3, APC, LEF, Rho, and Rock, and antagonists of Wnt signaling such as sFRP1 and 2. Therefore, miRNAs targeting the Wnt signaling pathway may play an important role in IPF. Aberrant canonical Wnt pathway was reported to exist in IPF along with the unregulated proliferation of Type II cells and mesenchymal fibroblasts (14).
The third set of genes targeted by miRNA deregulated in the initial period after bleomycin treatment is related to inflammatory pathways. These miRNAs target the key factors in the TLR signaling pathway (Fig. 4C), including members of the TLR family such as TLR4 and TLR5, and molecules in the pathway such as MAP2K3, MAPK8, IL-1β, IL-6, and TNF. We found that miR-34a was markedly upregulated on both day 3 and day 7 after bleomycin treatment. In silico analysis suggested that miR-34a was predicted to target IL-1β. MiRNA let-7g, which was markedly downregulated on day 3 and day 7, was predicted to target IL-6. Thus, our analysis is consistent with previous studies (48, 50). Expression of miR-34a was induced by proinflammatory cytokines IL-1β and TNF-α in vitro and in vivo (48). Let-7g was reported to be downregulated in LPS-induced acute inflammation in circulating leukocytes (50).
Fibrosis-related signaling pathway was targeted by miRNAs in the late period of bleomycin injury.
A large body of evidence has converged on a common theme: the central importance of the TGF-β pathway in the pathogenesis of pulmonary fibrosis (6, 17, 18, 28). In this study, miRNAs deregulated in the late period (days 14 and 21) indeed target key components in the TGF-β signaling pathway (Fig. 4D). These miRNAs include miR-196b, miR-704, miR-717, miR-16, miR-195, miR-10a, miR-211, miR-34a, miR-367, miR-21, and let-7f, which target TGF-β family members such as TGF-β2 and 3, TGF-β receptors such as TGF-β receptor I and II, Smad family members including Smad 3, 6, and 7, and procollagen type 1 alpha 2. A recent study showed that the miR-21 was upregulated in the lungs of mice with bleomycin-induced fibrosis and also in the lungs of patients with IPF (36). The study also showed that miR-21 may regulate lung fibrosis through targeting an inhibitory Smad, Smad7 (36).
In summary, the current work used microarray as a powerful tool to systematically and comprehensively examine global miRNA expression during bleomycin-induced lung injury and fibrosis. MiRNA target prediction and further functional characterization of differentially expressed miRNAs helped to elucidate the specific roles in lung injury and fibrosis. These findings demonstrated the cross talk between miRNAs and apoptosis, Wnt signaling, TLR signaling, and TGF-β signaling pathways, highlighting the role of miRNAs during apoptosis, inflammation, and fibrosis responses in lung fibrogenesis. The study points to a dynamic role of miRNAs in regulating critical components of complex regulatory network of the pathogenesis of tissue injury and fibrosis. The study also provides a framework in the multilayer regulatory machinery in orchestrating miRNA-molecular signaling-cellular process to control tissue injury and fibrosis. Collectively, this study may have important implications in our understanding of the molecular mechanisms underlying IPF. To further elucidate the role of specific miRNAs in lung fibrosis, additional studies are needed to investigate the function and targets of these miRNAs. Experiments along these lines are currently in progress in our laboratory.
GRANTS
This work was supported by National Heart, Lung, and Blood Institute R01-HL-060539 and R01-HL-077291. T. Xie was supported in part by a Ph.D. student fellowship from the China Scholarship Council.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
ACKNOWLEDGMENTS
We thank Zhengzheng Wei of Duke University for assistance with data analysis.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1. Adamson IY, Bowden DH.Bleomycin-induced injury and metaplasia of alveolar type 2 cells. Relationship of cellular responses to drug presence in the lung. Am J Pathol 96: 531–544, 1979. [PMC free article] [PubMed] [Google Scholar]
- 2. Adamson IY, Orr FW, Young L.Effects of injury and repair of the pulmonary endothelium on lung metastasis after bleomycin. J Pathol 150: 279–287, 1986. [DOI] [PubMed] [Google Scholar]
- 3. Addis-Lieser E, Kohl J, Chiaramonte MG.Opposing regulatory roles of complement factor 5 in the development of bleomycin-induced pulmonary fibrosis. J Immunol 175: 1894–1902, 2005. [DOI] [PubMed] [Google Scholar]
- 4. Altman N.Replication, variation and normalisation in microarray experiments. Appl Bioinformatics 4: 33–44, 2005. [DOI] [PubMed] [Google Scholar]
- 5. Ambros V.microRNAs: tiny regulators with great potential. Cell 107: 823–826, 2001. [DOI] [PubMed] [Google Scholar]
- 6. Araya J, Nishimura SL.Fibrogenic reactions in lung disease. Annu Rev Pathol 5: 77–98, 2010. [DOI] [PubMed] [Google Scholar]
- 7. ATS/ERS. Idiopathic pulmonary fibrosis: diagnosis and treatment International consensus statement American Thoracic Society (ATS), and the European Respiratory Society (ERS). Am J Respir Crit Care Med 161: 646–664, 2000. [DOI] [PubMed] [Google Scholar]
- 8. Bringardner BD, Baran CP, Eubank TD, Marsh CB.The role of inflammation in the pathogenesis of idiopathic pulmonary fibrosis. Antioxid Redox Signal 10: 287–301, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Broekelmann TJ, Limper AH, Colby TV, McDonald JA.Transforming growth factor beta 1 is present at sites of extracellular matrix gene expression in human pulmonary fibrosis. Proc Natl Acad Sci USA 88: 6642–6646, 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Caruso P, MacLean MR, Khanin R, McClure J, Soon E, Southgate M, MacDonald RA, Greig JA, Robertson KE, Masson R, Denby L, Dempsie Y, Long L, Morrell NW, Baker AH.Dynamic changes in lung microRNA profiles during the development of pulmonary hypertension due to chronic hypoxia and monocrotaline. Arterioscler Thromb Vasc Biol 30: 716–723, 2010. [DOI] [PubMed] [Google Scholar]
- 11. Chambers RC.Procoagulant signalling mechanisms in lung inflammation and fibrosis: novel opportunities for pharmacological intervention? Br J Pharmacol 153, Suppl 1: S367–S378, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chang JT, Nevins JR.GATHER: a systems approach to interpreting genomic signatures. Bioinformatics 22: 2926–2933, 2006. [DOI] [PubMed] [Google Scholar]
- 13. Chaudhary NI, Schnapp A, Park JE.Pharmacologic differentiation of inflammation and fibrosis in the rat bleomycin model. Am J Respir Crit Care Med 173: 769–776, 2006. [DOI] [PubMed] [Google Scholar]
- 14. Chilosi M, Poletti V, Zamo A, Lestani M, Montagna L, Piccoli P, Pedron S, Bertaso M, Scarpa A, Murer B, Cancellieri A, Maestro R, Semenzato G, Doglioni C.Aberrant Wnt/beta-catenin pathway activation in idiopathic pulmonary fibrosis. Am J Pathol 162: 1495–1502, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chung AC, Huang XR, Meng X, Lan HY.miR-192 Mediates TGF-beta/Smad3-Driven Renal Fibrosis. J Am Soc Nephrol 21: 1317–1325, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Demedts M, Behr J, Buhl R, Costabel U, Dekhuijzen R, Jansen HM, MacNee W, Thomeer M, Wallaert B, Laurent F, Nicholson AG, Verbeken EK, Verschakelen J, Flower CD, Capron F, Petruzzelli S, De Vuyst P, van den Bosch JM, Rodriguez-Becerra E, Corvasce G, Lankhorst I, Sardina M, Montanari M.High-dose acetylcysteine in idiopathic pulmonary fibrosis. N Engl J Med 353: 2229–2242, 2005. [DOI] [PubMed] [Google Scholar]
- 17. Gauldie J, Bonniaud P, Sime P, Ask K, Kolb M.TGF-beta, Smad3 and the process of progressive fibrosis. Biochem Soc Trans 35: 661–664, 2007. [DOI] [PubMed] [Google Scholar]
- 18. Goodwin A, Jenkins G.Role of integrin-mediated TGFbeta activation in the pathogenesis of pulmonary fibrosis. Biochem Soc Trans 37: 849–854, 2009. [DOI] [PubMed] [Google Scholar]
- 19. He L, Hannon GJ.MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5: 522–531, 2004. [DOI] [PubMed] [Google Scholar]
- 20. Jiang D, Liang J, Campanella GS, Guo R, Yu S, Xie T, Liu N, Jung Y, Homer R, Meltzer EB, Li Y, Tager AM, Goetinck PF, Luster AD, Noble PW.Inhibition of pulmonary fibrosis in mice by CXCL10 requires glycosaminoglycan binding and syndecan-4. J Clin Invest 120: 2049–2057, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jiang D, Liang J, Fan J, Yu S, Chen S, Luo Y, Prestwich GD, Mascarenhas MM, Garg HG, Quinn DA, Homer RJ, Goldstein DR, Bucala R, Lee PJ, Medzhitov R, Noble PW.Regulation of lung injury and repair by Toll-like receptors and hyaluronan. Nat Med 11: 1173–1179, 2005. [DOI] [PubMed] [Google Scholar]
- 22. Jiang D, Liang J, Hodge J, Lu B, Zhu Z, Yu S, Fan J, Gao Y, Yin Z, Homer R, Gerard C, Noble PW.Regulation of pulmonary fibrosis by chemokine receptor CXCR3. J Clin Invest 114: 291–299, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jiang D, Liang J, Noble PW.Hyaluronan in tissue injury and repair. Annu Rev Cell Dev Biol 23: 435–461, 2007. [DOI] [PubMed] [Google Scholar]
- 24. John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS.Human MicroRNA targets. PLoS Biol 2: e363, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jones HA, Schofield JB, Krausz T, Boobis AR, Haslett C.Pulmonary fibrosis correlates with duration of tissue neutrophil activation. Am J Respir Crit Care Med 158: 620–628, 1998. [DOI] [PubMed] [Google Scholar]
- 26. Kanehisa M.A database for post-genome analysis. Trends Genet 13: 375–376, 1997. [DOI] [PubMed] [Google Scholar]
- 27. Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M.KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res 38: D355–D360, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Khalil N, Greenberg AH.The role of TGF-beta in pulmonary fibrosis. Ciba Found Symp 157: 194–207, 1991. [DOI] [PubMed] [Google Scholar]
- 29. Kinnula VL, Fattman CL, Tan RJ, Oury TD.Oxidative stress in pulmonary fibrosis: a possible role for redox modulatory therapy. Am J Respir Crit Care Med 172: 417–422, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krupa A, Jenkins R, Luo DD, Lewis A, Phillips A, Fraser D.Loss of MicroRNA-192 promotes fibrogenesis in diabetic nephropathy. J Am Soc Nephrol 21: 438–447, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kuwano K, Hagimoto N, Kawasaki M, Yatomi T, Nakamura N, Nagata S, Suda T, Kunitake R, Maeyama T, Miyazaki H, Hara N.Essential roles of the Fas-Fas ligand pathway in the development of pulmonary fibrosis. J Clin Invest 104: 13–19, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Landi MT, Zhao Y, Rotunno M, Koshiol J, Liu H, Bergen AW, Rubagotti M, Goldstein AM, Linnoila I, Marincola FM, Tucker MA, Bertazzi PA, Pesatori AC, Caporaso NE, McShane LM, Wang E.MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res 16: 430–441, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lau NC, Lim LP, Weinstein EG, Bartel DP.An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862, 2001. [DOI] [PubMed] [Google Scholar]
- 34. Lee RC, Ambros V.An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862–864, 2001. [DOI] [PubMed] [Google Scholar]
- 35. Li Y, Chan EY, Li J, Ni C, Peng X, Rosenzweig E, Tumpey TM, Katze MG.MicroRNA expression and virulence in pandemic influenza virus-infected mice. J Virol 84: 3023–3032, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, Kaminski N, Abraham E.miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med 207: 1589–1597, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maurer B, Stanczyk J, Jungel A, Akhmetshina A, Trenkmann M, Brock M, Kowal-Bielecka O, Gay RE, Michel BA, Distler JH, Gay S, Distler O.MicroRNA-29, a key regulator of collagen expression in systemic sclerosis. Arthritis Rheum 62: 1733–1743, 2010. [DOI] [PubMed] [Google Scholar]
- 38. Miko E, Czimmerer Z, Csanky E, Boros G, Buslig J, Dezso B, Scholtz B.Differentially expressed microRNAs in small cell lung cancer. Exp Lung Res 35: 646–664, 2009. [DOI] [PubMed] [Google Scholar]
- 39. Moeller A, Ask K, Warburton D, Gauldie J, Kolb M.The bleomycin animal model: a useful tool to investigate treatment options for idiopathic pulmonary fibrosis? Int J Biochem Cell Biol 40: 362–382, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nana-Sinkam SP, Hunter MG, Nuovo GJ, Schmittgen TD, Gelinas R, Galas D, Marsh CB.Integrating the MicroRNome into the study of lung disease. Am J Respir Crit Care Med 179: 4–10, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nettelbladt O, Bergh J, Schenholm M, Tengblad A, Hallgren R.Accumulation of hyaluronic acid in the alveolar interstitial tissue in bleomycin-induced alveolitis. Am Rev Respir Dis 139: 759–762, 1989. [DOI] [PubMed] [Google Scholar]
- 42. Noble PW, Homer RJ.Back to the future: historical perspective on the pathogenesis of idiopathic pulmonary fibrosis. Am J Respir Cell Mol Biol 33: 113–120, 2005. [DOI] [PubMed] [Google Scholar]
- 43. Noble PW, Homer RJ.Idiopathic pulmonary fibrosis: new insights into pathogenesis. Clin Chest Med 25: 749–758, 2004. [DOI] [PubMed] [Google Scholar]
- 44. Oglesby IK, Bray IM, Chotirmall SH, Stallings RL, O'Neill SJ, McElvaney NG, Greene CM.miR-126 is downregulated in cystic fibrosis airway epithelial cells and regulates TOM1 expression. J Immunol 184: 1702–1709, 2010. [DOI] [PubMed] [Google Scholar]
- 45. Pandit KV, Corcoran D, Yousef H, Yarlagadda M, Tzouvelekis A, Gibson KF, Konishi K, Yousem SA, Singh M, Handley D, Richards T, Selman M, Watkins SC, Pardo A, Ben-Yehudah A, Bouros D, Eickelberg O, Ray P, Benos PV, Kaminski N.Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 182: 220–229, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pottier N, Maurin T, Chevalier B, Puissegur MP, Lebrigand K, Robbe-Sermesant K, Bertero T, Lino Cardenas CL, Courcot E, Rios G, Fourre S, Lo-Guidice JM, Marcet B, Cardinaud B, Barbry P, Mari B.Identification of keratinocyte growth factor as a target of microRNA-155 in lung fibroblasts: implication in epithelial-mesenchymal interactions. PLoS One 4: e6718, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rigoutsos I.New tricks for animal microRNAS: targeting of amino acid coding regions at conserved and nonconserved sites. Cancer Res 69: 3245–3248, 2009. [DOI] [PubMed] [Google Scholar]
- 48. Roggli E, Britan A, Gattesco S, Lin-Marq N, Abderrahmani A, Meda P, Regazzi R.Involvement of microRNAs in the cytotoxic effects exerted by proinflammatory cytokines on pancreatic beta-cells. Diabetes 59: 978–986, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rosenbloom J, Castro SV, Jimenez SA.Narrative review: fibrotic diseases: cellular and molecular mechanisms and novel therapies. Ann Intern Med 152: 159–166, 2010. [DOI] [PubMed] [Google Scholar]
- 50. Schmidt WM, Spiel AO, Jilma B, Wolzt M, Muller M.In vivo profile of the human leukocyte microRNA response to endotoxemia. Biochem Biophys Res Commun 380: 437–441, 2009. [DOI] [PubMed] [Google Scholar]
- 51. Selman M, Pardo A.The epithelial/fibroblastic pathway in the pathogenesis of idiopathic pulmonary fibrosis. Am J Respir Cell Mol Biol 29: S93–S97, 2003. [PubMed] [Google Scholar]
- 52. Selman M, Pardo A.Idiopathic pulmonary fibrosis: an epithelial/fibroblastic cross-talk disorder. Respir Res 3: 3, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shen AS, Haslett C, Feldsien DC, Henson PM, Cherniack RM.The intensity of chronic lung inflammation and fibrosis after bleomycin is directly related to the severity of acute injury. Am Rev Respir Dis 137: 564–571, 1988. [DOI] [PubMed] [Google Scholar]
- 54. Thannickal VJ, Horowitz JC.Evolving concepts of apoptosis in idiopathic pulmonary fibrosis. Proc Am Thorac Soc 3: 350–356, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, Yoshida K, Sasaki H, Nomura S, Seto Y, Kaminishi M, Calin GA, Croce CM.Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol 11: 136–146, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang R, Ibarra-Sunga O, Verlinski L, Pick R, Uhal BD.Abrogation of bleomycin-induced epithelial apoptosis and lung fibrosis by captopril or by a caspase inhibitor. Am J Physiol Lung Cell Mol Physiol 279: L143–L151, 2000. [DOI] [PubMed] [Google Scholar]
- 57. Xu J, Liao X, Wong C.Downregulations of B-cell lymphoma 2 and myeloid cell leukemia sequence 1 by microRNA 153 induce apoptosis in a glioblastoma cell line DBTRG-05MG. Int J Cancer 126: 1029–1035, 2010. [DOI] [PubMed] [Google Scholar]