Abstract
The common polymorphism SCN5A-S1103Y (∼13% allelic frequency in African Americans) is a risk factor for arrhythmia, sudden unexplained death (SUD), and sudden infant death syndrome. Prompted by a case of autopsy-negative SUD in a 23-year-old African American man who collapsed while playing football, we hypothesized that S1103Y interacted with other SCN5A variants to pathologically modify sodium current (INa). Mutational analysis of arrhythmia-associated genes in the victim revealed the variants SCN5A-R680H and SCN5A-S1103Y. These variants were made both separately and in the same cDNA construct of the alternative splice variant backgrounds (SCN5A-Q1077del and Q1077) and expressed in HEK293 cells. In the most abundant SCN5A-Q1077del, late INa for S1103Y alone was not significantly different from wild type (WT). However, late INa for R680H, R680H+S1103Y (coexpressed), and R680H/S1103Y (on the same cDNA) was increased 2.1-, 3.4-, and 3.6-fold, respectively, compared with WT. Intracellular acidosis (pH 6.7) increased late INa for S1103Y, R680H, R680H+S1103Y, and R680H/S1103Y by 2.2-, 2.4-, 5.0-, and 5.5-fold, respectively, compared with WT at pH 6.7. Expression in the less abundant SCN5A-Q1077 showed no increased late INa. This is the initial report of a functional interaction for the common polymorphism S1103Y with another mutation in the major transcript Q1077del of SCN5A. The “double hit” and environmental factor of acidosis may have converged to cause arrhythmic sudden death in this case.
Keywords: sudden death, ion channels, sodium current, genetics
mutations in the SCN5A-encoded α-subunit of the voltage-dependent sodium channel NaV1.5 can alter properties of the cardiac sodium current (INa) to cause inherited arrhythmia syndromes such as long-QT syndrome (LQTS) type 3 (LQT3) and Brugada syndrome (BrS) type 1 (BrS1). S1103Y, one of the eight common (allelic frequency >0.5%) SCN5A polymorphisms with an allelic frequency of ∼13% in African Americans (1), has been associated with a small risk of acquired arrhythmia, particularly in the setting of medications and hypokalemia (18), and with familial syncope, ventricular fibrillation, and sudden death (6). Postmortem molecular analysis in cohorts of both sudden unexplained cardiac death (SUD) (5) and sudden infant death syndrome (SIDS) (16, 21) identified S1103Y as a risk factor for increased sudden death. Electrophysiological cellular studies indicated that Y1103-containing channels had small shifts in kinetics under baseline conditions (19), but when they were exposed to cellular acidosis late INa was increased markedly (16).
Messenger RNA for two splice variants of SCN5A, one lacking a glutamine at position 1077 (Q1077del) and one containing the glutamine at position 1077 (Q1077), exist in the human heart at a ratio of ∼2:1 (Q1077del:Q1077) (12), and the function of INa for both polymorphisms (19) and channelopathic mutations (20) may depend on which splice variant background the mutation is studied. We encountered and genotyped a 23-year-old African American male SUD victim and identified two SCN5A single amino acid substitutions: the common S1103Y polymorphism and a rare missense mutation, R680H, which was reported previously in a deceased Norwegian infant. Like S1103Y, R680H had a latent dysfunctional biophysical phenotype that depended on acidosis to produce late INa (22). Hypothesizing a possible synergistic interaction, we investigated the interactions of both R680H and S1103Y in both the Q1077del and Q1077 SCN5A backgrounds as well as the pH dependence of late INa. We found that the “double hit” in combination with the environmental factor of acidosis caused a more severe pathological biophysical phenotype of late INa that critically depended on the sodium channel transcript's status at residue 1077.
MATERIALS AND METHODS
This study was approved by Mayo Clinic's Institutional Review Board as a consented study for postmortem genetic testing.
Mutational analysis.
Genomic DNA was extracted from frozen myocardial necropsy tissue with the Qiagen DNeasy Tissue Kit (Qiagen, Valencia, CA). Comprehensive open reading frame/splice site mutational analysis of the three most common susceptibility genes for LQTS and the most common susceptibility gene for BrS (KCNQ1, KCNH2, and SCN5A) was performed with polymerase chain reaction (PCR), denaturing high-performance liquid chromatography (DHPLC), and direct DNA sequencing as previously described (2). Primer sequences, PCR conditions, and DHPLC conditions are available on request.
Plasmid constructions.
R680H and S1103Y were created separately and also together in the two SCN5A splice variant backgrounds designated Q1077del (GenBank accession no. AY148488) and Q1077 (GenBank accession no. AC1377587) with a site-directed mutagenesis kit (Stratagene, La Jolla, CA). In all, eight SCN5A constructs were made and tested. The pcDNA3 plasmid vector (Invitrogen, Carlsbad, CA) was used as previously reported (12, 19). All clones were sequenced to confirm the presence of the introduced mutations and the absence of Taq polymerase-induced substitutions that may occur during PCR.
Mammalian cell transfection.
The wild-type (WT) and mutant channels in these two alternatively spliced transcripts of SCN5A were transiently transfected into HEK293 cells with FuGENE6 reagent (Roche Diagnostics, Indianapolis, IN) according to manufacturer's instructions.
Electrophysiological measurements.
Macroscopic voltage-gated INa was measured 24 h after transfection with the standard whole cell patch-clamp method at 21–23°C in HEK293 cells. The extracellular (bath) solution contained the following (in mM): 140 NaCl, 4 KCl, 1.8 CaCl2, 0.75 MgCl2, and 5 HEPES and was adjusted to pH 7.4 with NaOH. The intracellular (pipette) solution contained the following (in mM): 120 CsF, 20 CsCl2, 2 EGTA, 5 NaCl, and 5 HEPES and was adjusted to pH 7.4 or 6.7 with CsOH. Microelectrodes were manufactured from borosilicate glass with a puller (P-87, Sutter Instrument, Novato, CA) and were heat polished with a microforge (MF-83, Narishige, Tokyo, Japan). The resistances of microelectrodes ranged from 1.0 to 2.0 MΩ. Voltage clamp data were generated with pClampex 10.2 and analyzed with Clampfit 10.2 (Molecular Devices, Sunnyvale, CA). Membrane current data were digitalized at 100 kHz, low-pass filtered at 5 kHz, and then normalized to membrane capacitance. Standard voltage clamp protocols are presented with the data, and data were measured and analyzed as described previously (7, 12, 19) and with additional details provided in Figs. 2–4.
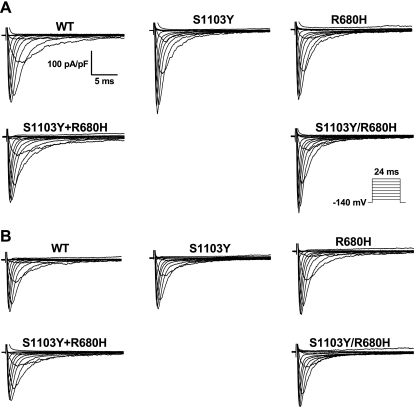
Fig. 2.
Representative whole cell current traces of wild-type (WT) and variant sodium channels. Currents were recorded at membrane potentials between −120 and +60 mV in 10-mV increments from a holding potential of −140 mV as depicted in the protocol inset. A: representative whole cell current traces under baseline pH. B: representative whole cell current traces under internal acidosis condition (pH 6.7).
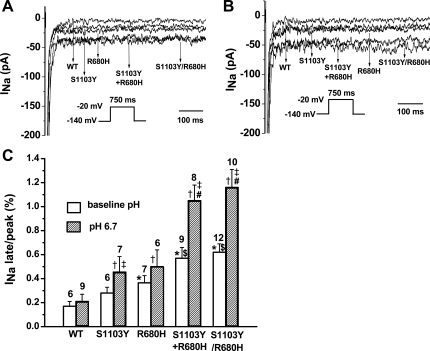
Fig. 3.
pH dependence of late current in WT and mutant constructs in Q1077del background. A: representative traces showing increased late sodium current (INa) associated with variants compared with WT under baseline pH condition. The corresponding peak INa were WT = −3,044 pA, S1103Y = −4,936 pA, R680H = −5,346 pA, S1103Y+R680H = −5,263 pA, and S1103Y/R680H = −5,209 pA. B: representative traces showing increased late INa associated with S1103Y, R680H, S1103Y+R680H, and S1103Y/R680H compared with WT under pH 6.7. The corresponding peak INa were WT = −3,809 pA, S1103Y = −4,170 pA, R680H = −4,663 pA, S1103Y+R680H = −4,285 pA, and S1103Y/R680H = −4,655 pA. C: summary data of late INa normalized to peak INa after leak subtraction. Late INa was measured as the mean current between 600 and 700 ms after the initiation of the depolarization from −140 mV to −20 mV for 750 ms (see protocol insets) after passive leak subtraction. Numbers of experiments are indicated above bars. Data were analyzed with 1-way ANOVA followed by a Tukey test. *P < 0.05 vs. WT at baseline pH; $P < 0.05 vs. S1103Y or R680H at baseline pH; †P < 0.05 vs. WT at pH 6.7; #P < 0.05 vs. S1103Y or R680H at pH 6.7. ‡P < 0.05 vs. the same variant at baseline pH (Student's t-test).
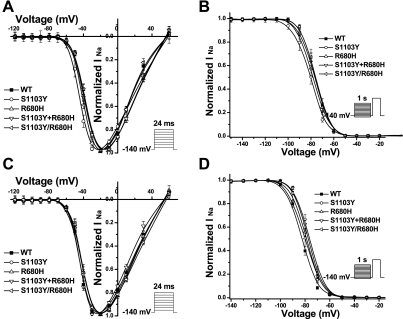
Fig. 4.
pH effects on gating kinetics of WT and mutant constructs in Q1077del background. A: voltage dependence of activation for WT and variants under baseline pH condition. Activation was measured with the protocol shown in the inset. The curves were fit with a Boltzmann function in which GNa = [1 + exp(V1/2 − V)/K]−1, where G, V1/2, and K are conductance, midpoint, and slope factor, respectively. G/GNa = INa(norm)/(V − Vrev) where Vrev is the reversal potential and V is the membrane potential. B: steady-state inactivation for WT and variants under baseline pH condition. Steady-state availability from inactivation was measured with the protocol shown in the inset and was determined by fitting the data to the Boltzmann function INa = INa-max [1 + exp(Vc − V1/2)/K]−1, where V1/2 and K are midpoint and slope factor, respectively, and Vc is the membrane potential. C: voltage dependence of activation for WT and variants under pH 6.7. None of the variants altered steady-state activation parameters significantly. D: steady-state inactivation for WT and variants at pH 6.7. S1103Y, S1103Y+R680H, and S1103Y/R680H caused a statistically significant depolarizing shift in inactivation by 4–6 mV. See Tables 1 and 2 for summary of parameter fits and n values.
Statistical analysis.
All data points are reported as means and SE. Determinations of statistical significance were performed with a Student's t-test for comparisons of two means or with analysis of variance (ANOVA) for comparisons of multiple groups. Statistical significance was determined by a value of P < 0.05.
RESULTS
Case report of sudden cardiac death.
A 23-year-old African American man collapsed while playing football. He was transported immediately to a hospital, where he was pronounced dead. Postmortem examination demonstrated no obvious pathological changes to explain the SUD of this apparently healthy athlete. Further histopathological examination revealed no serious structural changes in vital organs. Tissue samples were submitted for a cardiac channel molecular autopsy by the medical examiner.
Mutational analysis.
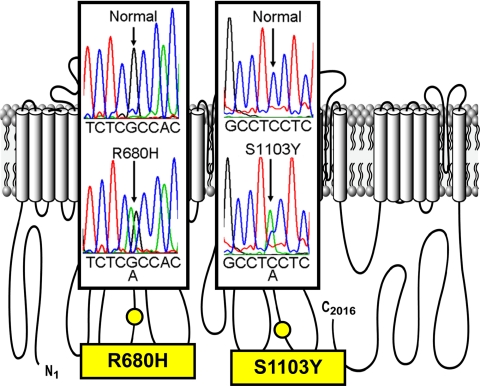
Comprehensive open reading frame/splice site mutational analysis for both LQTS- and BrS-associated genes (KCNQ1, KCNH2, and SCN5A) revealed two SCN5A variants (2039 G→A, R680H and 3308 C→A, S1103Y; Fig. 1). However, the cis- or trans-status could not be determined in the victim's heart because the quality of the RNA isolated from the submitted autopsy sample was not adequate for long-range PCR; therefore we tested all combinations. R680H was absent in >1,300 reference alleles and demonstrated various levels of conservation across species. Regrettably, no other information could be obtained from family members.
Fig. 1.
Postmortem genetic testing identifies SCN5A mutations in sudden unexplained death (SUD). DNA sequence chromatograms superimposed on locations in the amino acid channel topology are shown. The cis- or trans-status could not be determined in the victim's heart because the quality of the RNA isolated from the submitted autopsy sample was not adequate for long-range PCR.
Electrophysiology.
We studied WT, the single mutations R680H and S1103Y expressed separately, the two single mutations coexpressed together (R680H + S1103Y) but on separate plasmid, and the double mutation R680H/S1103Y in the same plasmid, using the Q1077del cDNA that reflects the most abundant alternatively spliced SCN5A transcript (∼65%) in human hearts (12). Representative families of current traces at normal pH (Fig. 2A) and with acidosis (Fig. 2B) showed no obvious differences, and summary data for peak INa density (Tables 1 and 2) showed no significant differences. Representative traces on an expanded amplitude scale to depict late INa (Fig. 3, A and B) showed an obvious increase in late INa under acidic pH (Fig. 3B) compared with normal pH (Fig. 3A) as summarized in Fig. 3C. At baseline pH the late INa for S1103Y by itself showed no significant difference from WT, but R680H, R680H+S1103Y, and R680H/S1103Y showed significant 2.1-, 3.4-, and 3.6-fold increases, respectively (Fig. 3C; Table 1). The degree of accentuated late INa for the combined mutants is comparable to that observed in patients established to have LQT3-associated mutations in SCN5A (3). Moreover, late INa for both R680H+S1103Y and R680H/S1103Y was significantly greater than that of R680H alone (P < 0.05). The double variants, whether on the same allele or on separate alleles that were coexpressed, had an interactive effect to produce increased late INa. Lowering the intracellular pH to 6.7 significantly increased late INa compared with baseline for each group tested except for WT (Fig. 3C; Tables 1 and 2), but the effect was especially marked for both R680H+S1103Y and R680H/S1103Y, where the already increased (relative to WT) late INa was doubled.
Table 1.
Biophysical properties of WT or variant sodium channels in Q1077del background in HEK293 cells under baseline pH
| Peak INa |
Activation |
Inactivation |
Late INa |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Samples | pA/pF | n | V1/2, mV | K | n | V1/2, mV | K | n | % | n |
| WT | −278 ± 40 | 8 | −37.6 ± 1.2 | 4.4 | 8 | −76.1 ± 1.7 | 5.0 | 7 | 0.17 ± 0.04 | 6 |
| S1103Y | −367 ± 49 | 11 | −43.7 ± 1.3* | 4.1 | 10 | −80.7 ± 1.9 | 5.1 | 9 | 0.28 ± 0.05 | 6 |
| R680H | −288 ± 42 | 12 | −38.8 ± 1.1 | 4.5 | 12 | −77.0 ± 1.6 | 5.0 | 10 | 0.36 ± 0.06* | 7 |
| S1103Y+R680H | −256 ± 51 | 15 | −37.8 ± 0.9 | 4.5 | 12 | −79.0 ± 1.5 | 5.1 | 12 | 0.57 ± 0.09† | 9 |
| S1103Y/R680H | −305 ± 57 | 17 | −39.6 ± 0.8 | 4.3 | 13 | −76.3 ± 0.9 | 5.0 | 14 | 0.62 ± 0.07† | 12 |
Values are means ± SE for n experiments. INa, sodium current; pA/pF, current density; V1/2, voltage of half-maximal activation/inactivation; K, slope factor. The late INa level is described as % of peak INa. All parameters were analyzed with 1-way ANOVA followed by a Tukey test.
P < 0.05 vs. wild type (WT);
P < 0.01 vs. WT, P < 0.05 vs. S1103Y or R680H.
Table 2.
Biophysical properties of WT or variant sodium channels in Q1077del background in HEK293 cells under internal acidosis condition (pH 6.7)
| Peak INa |
Activation |
Inactivation |
Late INa |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Samples | pA/pF | n | V1/2, mV | K | n | V1/2, mV | K | n | % | n |
| WT | −223 ± 34 | 18 | −42.9 ± 1.0 | 4.5 | 17 | −82.7 ± 0.8 | 5.0 | 16 | 0.21 ± 0.06 | 9 |
| S1103Y | −215 ± 40 | 12 | −41.1 ± 1.4 | 4.0 | 8 | −76.4 ± 1.0* | 4.8 | 11 | 0.45 ± 0.13* | 7 |
| R680H | −264 ± 38 | 11 | −41.8 ± 1.8 | 5.1 | 10 | −80.3 ± 1.8 | 5.0 | 10 | 0.50 ± 0.14* | 6 |
| S1103Y+R680H | −205 ± 23 | 16 | −44.0 ± 0.6 | 4.0 | 11 | −77.7 ± 0.8* | 4.9 | 15 | 1.05 ± 0.13† | 8 |
| S1103Y/R680H | −234 ± 32 | 16 | −41.2 ± 0.9 | 4.4 | 14 | −78.7 ± 0.7* | 5.1 | 14 | 1.16 ± 0.15† | 10 |
Values are means ± SE for n experiments. All parameters were analyzed with 1-way ANOVA followed by a Tukey test.
P < 0.05 vs. WT;
P < 0.01 vs. WT, P < 0.05 vs. S1103Y or R680H.
We analyzed the kinetic parameters of activation (Fig. 4, A and C) and inactivation (Fig. 4, B and D) at baseline pH (Fig. 4, A and B) and pH 6.7 (Fig. 4, C and D) for all variants and compared these data with WT. Parameter values for the fits of activation and inactivation fits at baseline pH (Table 1; Fig. 4, A and B) showed a 6-mV negative shift in activation midpoint for S1103Y, but otherwise no significant differences in activation, inactivation, and recovery from inactivation parameters were noted. At pH 6.7, compared with WT, none of the variants showed a significant difference in activation (Fig. 4C; Table 2) while S1103Y, R680H+S1103Y, and R680H/S1103Y caused a statistically significant depolarizing shift (4–6 mV) in channel inactivation (Fig. 4D; Table 2). Fast inactivation properties of all variants were analyzed by two-exponential fits of the decay phase of macroscopic sodium current measured at various test potentials. Only R680H/S1103Y had significantly larger fast time constant (τf) values across a wide range of test potentials than WT at pH 6.7 (data not shown). For recovery from inactivation, S1103Y, R680H+S1103Y, and R680H/S1103Y exhibited faster recovery from inactivation and had significantly smaller time constant [τf and slow time constant (τs)] values compared with WT channels under low-pH conditions (Table 3).
Table 3.
Recovery of WT and variant sodium channels in Q1077del background under both baseline pH and internal acidosis (pH 6.7) conditions
| Recovery at Baseline pH |
Recovery at pH 6.7 |
|||||||
|---|---|---|---|---|---|---|---|---|
| Samples | τf, ms | τs, ms | As, % | n | τf, ms | τs, ms | As, % | n |
| WT | 1.8 ± 0.3 | 45.1 ± 6.2 | 25.3 ± 1.7 | 7 | 3.2 ± 0.2 | 71.2 ± 6.2 | 24.4 ± 1.4 | 18 |
| S1103Y | 2.2 ± 0.3 | 45.0 ± 8.8 | 22.0 ± 1.7 | 8 | 2.4 ± 0.2* | 56.1 ± 5.2* | 24.4 ± 1.5 | 12 |
| R680H | 1.7 ± 0.2 | 34.2 ± 3.0 | 24.7 ± 2.4 | 9 | 3.7 ± 0.7 | 79.6 ± 9.8 | 23.2 ± 1.9 | 8 |
| S1103Y+R680H | 1.8 ± 0.2 | 42.1 ± 4.1 | 23.6 ± 1.7 | 10 | 2.5 ± 0.2* | 50.4 ± 3.8* | 25.7 ± 1.6 | 16 |
| S1103Y/R680H | 1.7 ± 0.2 | 37.7 ± 3.0 | 23.8 ± 1.1 | 14 | 2.5 ± 0.1* | 53.2 ± 3.3* | 25.5 ± 1.9 | 16 |
Values are means ± SE for n experiments. Time course of recovery from inactivation was analyzed by fitting data with a 2-exponential (exp) function: normalized INa (t) = Af[1 − exp(−t/τf)] + As[1 − exp(−t/τS)], where t is time, Af and As are fractional amplitudes of fast and slow components, respectively, and τf and τs are fast and slow time constants, respectively.
P < 0.05 vs. WT (1-way ANOVA followed by a Tukey test).
We previously reported (12, 19, 20) that some SCN5A variants had different biophysical properties in the minor splice variant Q1077 background compared with Q1077del background. Wang et al. (22)also found that, in the Q1077 background, some SCN5A mutations including R680H exhibited no significant increase of late INa that had been seen in the Q1077del background. To determine whether the minor alternatively spliced transcript exerts an effect on these case variants, we also tested R680H, S1103Y, R680H+S1103Y, and R680H/S1103Y in the Q1077-containing background. Compared with SCN5A-Q1077 WT, all variants showed no significant difference in both peak and late INa, activation, and inactivation (Table 4).
Table 4.
Biophysical properties of WT or variant sodium channels in Q1077 background
| Peak INa |
Activation |
Inactivation |
Late INa |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Samples | pA/pF | n | V1/2, mV | K | n | V1/2, mV | K | n | % | n |
| WT | −267 ± 54 | 7 | −39.6 ± 2.5 | 4.0 | 5 | −74.6 ± 1.6 | 5.0 | 7 | 0.19 ± 0.06 | 6 |
| S1103Y | −246 ± 56 | 5 | −41.2 ± 1.2 | 4.0 | 5 | −77.2 ± 1.4 | 5.0 | 5 | 0.31 ± 0.05 | 5 |
| R680H | −213 ± 43 | 5 | −42.2 ± 0.6 | 4.0 | 5 | −76.1 ± 1.3 | 5.0 | 5 | 0.27 ± 0.11 | 5 |
| S1103Y+R680H | −228 ± 32 | 11 | −40.1 ± 0.7 | 4.0 | 7 | −74.9 ± 1.2 | 5.1 | 11 | 0.22 ± 0.04 | 8 |
| S1103Y/R680H | −235 ± 69 | 7 | −40.4 ± 1.3 | 4.0 | 6 | −75.5 ± 0.7 | 5.0 | 7 | 0.26 ± 0.07 | 6 |
Values are means ± SE for n experiments. All parameters were analyzed by 1-way ANOVA followed by a Tukey test. Compared with WT, all P > 0.05.
DISCUSSION
The common polymorphism SCN5A-S1103Y is a proarrhythmic, sudden cardiac death-predisposing risk factor in African Americans. Clinical, molecular (5, 6, 16, 18, 21), and functional (16, 18) investigations indicate that S1103Y may increase risk for arrhythmia susceptibility in the presence of environmental risk factors such as acidosis, medications, hypokalemia, and structural heart disease.
In the present study, we showed that Y1103-containing channels exhibit a statistically significant negative shift in activation and that the rare R680H mutation causes a mild increase in late INa as previously reported (18, 22). The new findings in this study are that S1103Y interacts with R680H to significantly increase late INa and that this marked accentuation occurs whether the two mutations reside on the same allele or on separate alleles and then coexpressed together. Loss-of-function mutations in SCN5A have been “rescued” by the interaction with another common polymorphism, H558R (17, 24), and with R1193Q (15). Two rare mutations in SCN5A (R34fs/60 and R1195H) also interact to augment late INa (13). The present study shows for the first time that the common polymorphism S1103Y might also act as an interacting genetic variant as a “double hit” or compound heterozygosity (4) to cause disease. This has particular interest because of its frequent prevalence in the African American population (1) and its implication that intragenic interaction is an additional mechanism for S1103Y as a proarrhythmic risk factor. It has been previously suggested based on cooperative gating that NaV1.5 α-subunits, despite their ability to form a complete/self-sufficient pore as a monomeric structure, coassemble and influence each monomer's properties (20). An interaction between different alleles has been noted previously for the common SCN5A polymorphism H558R on current density in BrS (17), and we now show such an interaction for late current.
Plant et al. (16) and Wang et al. (22) previously showed that intracellular acidosis (pH 6.7) increased late INa for S1103Y and R680H, respectively, in SIDS and suggested this as an etiologic factor. Internal acidification markedly increased late INa by more than fivefold WT levels with both mutations (both R680H+S1103Y and R680H/S1103Y). We speculate that acidosis associated with extremely intense exercise may have been an environmental trigger for a fatal arrhythmia in this case. An additional trigger that may have played a role is exercise-induced tachycardia. Many LQT3 mutations tend to be bradycardia dependent because accumulation of inactivation at rapid stimulation rates decreases late INa (14). However, the increased late INa in this case did not show a decay (Fig. 3), suggesting that it is not subject to accumulation of inactivation and therefore late INa is maintained at higher rates.
As previously reported (22) and confirmed in the present study, R680H did not show a significant increase in late INa in the minor transcript (SCN5A-Q1077) compared with SCN5A-Q1077del (WT). Even the double mutation (both R680H+S1103Y and R680H/S1103Y) exhibited WT-like late INa in the Q1077 background. In the Q1077del background, however, these mutations showed marked increases in late INa. We have shown previously that the expression levels and kinetics of eight common SCN5A polymorphisms depend upon the Q1077 or Q1077del splice variant background (19) and that plasma membrane protein expression levels of the BrS G1406R mutation was influenced by the alternatively spliced SCN5A transcript (20). Wang et al. (22) have identified two SCN5A variants in SIDS patients (delAL586–587 and V1951L) that required expression in Q1077del to exhibit late INa. These results emphasize the importance of studying putative arrhythmia mutations in both backgrounds to assess the plausibility of pathogenicity. Notably, it is being increasingly recognized that the function of voltage-gated cardiac ion channels can be regulated by splice variants in their subunits such as potassium channel KV11.1 (23), L-type calcium channel CaV1.2 (11), type 2 ryanodine receptor (RyR2) (10), and calcium channel β2-subunit (8). Thus whether the dependence of functional expression on the Q1077del/Q1077 backgrounds implicates a potential splice variant-based regulatory mechanism for normal function in cardiac sodium channel deserves further investigation.
This study has several limitations. Heterologous systems do not faithfully recapitulate the actual cellular phenotypes because they lack subunits and interacting proteins found in myocytes. Despite this limitation we have described current dysfunction that is plausibly pathogenic. Also, the exercise phenotype of this case is different from conventional LQT3 late current-induced arrhythmia mechanisms, but we did not study the possible effect of increased sympathetic stimulation on late INa in this case, because of the limitations of heterologous systems, or of rate dependence on late INa.
In summary, the SCN5A biophysical phenotype associated with this SUD case suggests that pathogenicity may involve 1) a “double hit” involving a rare mutation, R680H, and a common polymorphism, S1103Y, 2) in the most common alternatively-spliced SCN5A transcript, with 3) acidosis as a contributing environmental trigger. Although the voltage clamp data provide plausible mechanisms for the presumed arrhythmia in this SUD case, the association is conjectural as it was not supported by a linkage study or other corroborative evidence of causality. This report, however, does lend further support to the proarrhythmic, sudden death-predisposing potential of the S1103Y common polymorphism in African Americans and to our knowledge is the first report of an intragenic interaction for S1103Y. This may have implications for screening and prevention of arrhythmia as well as postmortem genotyping of SUD cases in African Americans. In addition, the data show that separate SCN5A α-subunits interact to affect function and suggest they may assemble together as part of the NaV1.5 macromolecular complex (9).
GRANTS
This work was supported by the University of Wisconsin Cellular and Molecular Arrhythmia Research Program (J. C. Makielski), the Mayo Clinic Windland Smith Rice Comprehensive Sudden Cardiac Death Program (M. J. Ackerman), the established Investigator Award from the American Heart Association (to M. J. Ackerman), National Heart, Lung, and Blood Institute Grants HL-42569 (to M. J. Ackerman), HL-60723 (to C. T. January), and HL-71092 (to J. C. Makielski), and Grant 30973367 (to J. Cheng) from the National Natural Science Foundation of China.
DISCLOSURES
M. J. Ackerman is a consultant for PGxHealth and chairs their FAMILION Medical/Scientific Advisory Board (approved by Mayo Clinic's Medical-Industry Relations Office and Conflict of Interests Review Board). In addition, “cardiac channel gene screen” and “know-how relating to long QT genetic testing” license agreements, resulting in consideration and royalty payments, were established between Genaissance Pharmaceuticals (now PGxHealth) and Mayo Medical Ventures (now Mayo Clinic Health Solutions) in 2004. C. T. January is a cofounder of Cellular Dynamics International.
ACKNOWLEDGMENTS
We thank Jessie Ou, Neal Haas, and Qing Zhou for technical assistance.
REFERENCES
- 1. Ackerman MJ, Splawski I, Makielski JC, Tester DJ, Will ML, Timothy KW, Keating MT, Jones G, Chadha M, Burrow CR, Stephens JC, Xu C, Judson R, Curran ME.Spectrum and prevalence of cardiac sodium channel variants among black, white, Asian, and Hispanic individuals: implication for arrhythmogenic susceptibility and Brugada/long QT syndrome genetic testing. Heart Rhythm 1: 600–607, 2004 [DOI] [PubMed] [Google Scholar]
- 2. Ackerman MJ, Tester DJ, Jones G, Will MK, Burrow CR, Curran M.Ethnic differences in cardiac potassium channel variants: implications for genetic susceptibility to sudden cardiac death and genetic testing for congenital long QT syndrome. Mayo Clin Proc 78: 1479–1487, 2003 [DOI] [PubMed] [Google Scholar]
- 3. Bennett PB, Yazawa K, Makita N, George AL., JrMolecular mechanism for an inherited cardiac arrhythmia. Nature 376: 683–685, 1995 [DOI] [PubMed] [Google Scholar]
- 4. Benson DW.Compound heterozygous SCN5A mutations: does the sum of the parts equal the whole? Heart Rhythm 6: 1176–1177, 2009 [DOI] [PubMed] [Google Scholar]
- 5. Burke A, Creighton W, Mont E, Li L, Hogan S, Kutys R, Fowler D, Virmani R.Role of SCN5A Y1102 polymorphism in sudden cardiac death in blacks. Circulation 112: 798–802, 2005 [DOI] [PubMed] [Google Scholar]
- 6. Chen S, Chung MK, Martin D, Rozich R, Tchou PJ, Wang Q.SNP S1103Y in the cardiac sodium channel gene SCN5A is associated with cardiac arrhythmias and sudden death in a white family. J Med Genet 39: 913–915, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheng J, Van Norstrand DW, Medeiros-Domingo A, Valdivia C, Tan BH, Ye B, Kroboth S, Vatta M, Tester DJ, January CT, Makielski JC, Ackerman MJ.Alpha1-syntrophin mutations identified in sudden infant death syndrome cause an increase in late cardiac sodium current. Circ Arrhythm Electrophysiol 2: 667–676, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chu PJ, Larsen JK, Chen CC, Best PM.Distribution and relative expression levels of calcium channel beta subunits within the chambers of the rat heart. J Mol Cell Cardiol 36: 423–434, 2004 [DOI] [PubMed] [Google Scholar]
- 9. Gavillet B, Rougier JS, Domenighetti AA, Behar R, Boixel C, Ruchat P, Lehr HA, Pedrazzini T, Abriel H.Cardiac sodium channel Nav1.5 is regulated by a multiprotein complex composed of syntrophins and dystrophin. Circ Res 99: 407–414, 2006 [DOI] [PubMed] [Google Scholar]
- 10. George CH, Rogers SA, Bertrand BM, Tunwell RE, Thomas NL, Steele DS, Cox EV, Pepper C, Hazeel CJ, Claycomb WC, Lai FA.Alternative splicing of ryanodine receptors modulates cardiomyocyte Ca2+ signaling and susceptibility to apoptosis. Circ Res 100: 874–883, 2007 [DOI] [PubMed] [Google Scholar]
- 11. Liao P, Yu D, Li G, Yong TF, Soon JL, Chua YL, Soong TW.A smooth muscle Cav1.2 calcium channel splice variant underlies hyperpolarized window current and enhanced state-dependent inhibition by nifedipine. J Biol Chem 282: 35133–35142, 2007 [DOI] [PubMed] [Google Scholar]
- 12. Makielski JC, Ye B, Valdivia CR, Pagel MD, Pu J, Tester DJ, Ackerman MJ.A ubiquitous splice variant and a common polymorphism affect heterologous expression of recombinant human SCN5A heart sodium channels. Circ Res 93: 821–828, 2003 [DOI] [PubMed] [Google Scholar]
- 13. Medeiros-Domingo A, Tan BH, Iturralde-Torres P, Tester DJ, Tusié-Luna T, Makielski JC, Ackerman MJ.Unique mixed phenotype and unexpected functional effect revealed by novel compound heterozygosity mutations involving SCN5A. Heart Rhythm 6: 1170–1175, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nagatomo T, January CT, Ye B, Abe H, Nakashima Y, Makielski JC.Rate-dependent QT shortening mechanism for the LQT3 DeltaKPQ mutant. Cardiovasc Res 54: 624–629, 2002 [DOI] [PubMed] [Google Scholar]
- 15. Niu DM, Hwang B, Hwang HW, Wang NH, Wu JY, Lee PC, Chien JC, Shieh RC, Chen YT.A common SCN5A polymorphism attenuates a severe cardiac phenotype caused by a nonsense SCN5A mutation in a Chinese family with an inherited cardiac conduction defect. J Med Genet 43: 817–821, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Plant LD, Bowers PN, Liu Q, Morgan T, Zhang T, State MW, Chen W, Kittles RA, Goldstein SN.A common cardiac sodium channel variant associated with sudden infant death in African Americans, SCN5A S1103Y. J Clin Invest 116: 430–435, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Poelzing S, Forleo C, Samodell M, Dudash L, Sorrentino S, Anaclerio M, Troccoli R, Iacoviello M, Romito R, Guida P, Chahine M, Pitzalis M, Deschênes I.SCN5A polymorphism restores trafficking of a Brugada syndrome mutation on a separate gene. Circulation 114: 368–376, 2006 [DOI] [PubMed] [Google Scholar]
- 18. Splawski I, Timothy KW, Tateyama M, Clancy CE, Malhotra A, Beggs AH, Sagnella GA, Kass RS, Keating MT.Variant of SCN5A sodium channel implicated in risk of cardiac arrhythmia. Science 297: 1333–1336, 2002 [DOI] [PubMed] [Google Scholar]
- 19. Tan BH, Valdivia CR, Rok BA, Ye B, Ruwaldt KM, Tester DJ, Ackerman MJ, Makielski JC.Common human SCN5A polymorphism have altered electrophysiology when expressed in Q1077 splice variants. Heart Rhythm 2: 741–747, 2005 [DOI] [PubMed] [Google Scholar]
- 20. Tan BH, Valdivia CR, Song C, Makielski JC.Partial expression defect for the SCN5A missense mutation G1406R depends on splice variant background Q1077 and rescue by mexiletine. Am J Physiol Heart Circ Physiol 291: H1822–H1828, 2006 [DOI] [PubMed] [Google Scholar]
- 21. Van Norstrand DW, Tester DJ, Ackerman MJ.Over-representation of the pro-arrhythmic, sudden death predisposing sodium channel polymorphism, S1103Y, in a population-based cohort of African American sudden infant death syndrome. Heart Rhythm 5: 712–715, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang DW, Desai RR, Crotti L, Arnestad M, Insolia R, Pedrazzini M, Ferrandi C, Vege A, Rognum T, Schwartz PJ, George AL., JrCardiac sodium channel dysfunction in sudden infant death syndrome. Circulation 115: 368–376, 2007 [DOI] [PubMed] [Google Scholar]
- 23. Wang X, Xu R, Abernathey G, Taylor J, Alzghoul MB, Hannon K, Hockerman GH, Pond AL.Kv11.1 channel subunit composition includes Mink and varies developmentally in mouse cardiac muscle. Dev Dyn 237: 2430–2437, 2008 [DOI] [PubMed] [Google Scholar]
- 24. Ye B, Valdivia CR, Ackerman MJ, Makielski JC.A common human SCN5A polymorphism modifies expression of an arrhythmia causing mutation. Physiol Genomics 12: 187–193, 2003 [DOI] [PubMed] [Google Scholar]