Abstract
We have identified a cohort of zebrafish expressed sequence tags encoding eight Na,K-ATPase α subunits and five β subunits. Sequence comparisons and phylogenetic analysis indicate that five of the zebrafish α subunit genes comprise an α1-like gene subfamily and two are orthologs of the mammalian α3 subunit gene. The remaining α subunit clone is most similar to the mammalian α2 subunit. Among the five β subunit genes, two are orthologs of the mammalian β1 isoform, one represents a β2 ortholog, and two are orthologous to the mammalian β3 subunit. Using zebrafish radiation hybrid and meiotic mapping panels, we determined linkage assignments for each α and β subunit gene. Na,K-ATPase genes are dispersed in the zebrafish genome with the exception of four of the α1-like genes, which are tightly clustered on linkage group 1. Comparative mapping studies indicate that most of the zebrafish Na,K-ATPase genes localize to regions of conserved synteny between zebrafish and humans. The expression patterns of Na,K-ATPase α and β subunit genes in zebrafish are quite distinctive. No two α or β subunit genes exhibit the same expression profile. Together, our data imply a very high degree of Na,K-ATPase isoenzyme heterogeneity in zebrafish, with the potential for 40 structurally distinct α/β subunit combinations. Differences in expression patterns of α and β subunits suggest that many of the isoenzymes are also likely to exhibit differences in functional properties within specific cell and tissue types. Our studies form a framework for analyzing structure function relationships for sodium pump isoforms using reverse genetic approaches.
Na,K-ATPase is an integral membrane protein responsible for establishing and maintaining the electrochemical gradients of sodium and potassium ions across the plasma membrane of virtually all animal cells (Cantley 1981). The enzyme is composed of equimolar amounts of α and β subunits (Jørgensen 1974). The α subunit contains the catalytic site for ATP hydrolysis and is the cellular receptor for cardiac glycoside drugs such as ouabain and digitalis (Jørgensen 1974; Cantley 1981). The β subunit is a glycosylated polypeptide that is an essential component of Na,K-ATPase (Cantley 1981). However, the exact role of the β subunit in the activity of the enzyme has not yet been established.
Na,K-ATPase α and β subunits are each encoded by multigene families. Molecular cloning has identified four α and three β subunit genes in humans and rodents (Levenson 1994; Shamraj and Lingrel 1994; Malik et al. 1998; Underhill et al. 1999). Human genome sequencing projects have not revealed any previously unidentified Na,K-ATPase subunits. The α and β subunit genes are dispersed in the mouse genome, with the exception of the α2 and α4 genes which are very tightly linked on chromosome 1 (Levenson 1994; Besirli et al. 1998; Underhill et al. 1999). Analysis of the expression profiles of α and β subunits indicates that each gene is expressed in a distinct tissue- and cell-specific fashion. The α1 subunit is ubiquitously expressed (Shyjan and Levenson 1989). The α2 subunit shows predominant expression in brain, heart, and skeletal muscle, whereas the α3 subunit is restricted to tissues of neural origin (Shyjan and Levenson 1989; Cameron et al. 1994). Expression of the α4 subunit has so far been detected exclusively in the epididymis and testis (Underhill et al. 1999; Woo et al. 1999). Na,K-ATPase β subunit isoforms also exhibit distinct expression patterns. The β1 and β3 subunits are ubiquitously expressed, whereas the β2 isoform is expressed predominantly in brain (Shyjan and Levenson 1989; Malik et al. 1996, 1998). Chromosomal dispersion of the α and β subunit genes, coupled with their distinct expression patterns, suggests specialized functional roles for each of the α and β subunit isoforms.
Analysis of Na,K-ATPase expressed in heterologous systems has suggested that α/β subunit interaction is promiscuous (Jewell and Lingrel 1991; Lemas et al. 1994; Crambert et al. 2000), and that as many as 12 structurally distinct isoenzymes may exist. However, the functional significance for this high degree of isoenzyme diversity remains enigmatic. Comparison of the biochemical properties of alternate α/β subunit combinations has so far revealed only subtle functional differences between isoenzymes (Jewell and Lingrel 1991; Crambert et al. 2000). The analysis of mice carrying targeted mutations in the α1 and α2 subunit genes has suggested important functional differences between these genes in the regulation of cardiac contractility. The basis for these functional differences has not yet been clearly explained.
To better understand the function of distinct Na,K-ATPase isoenzymes, we have initiated studies designed to characterize Na,K-ATPase genes expressed in the zebrafish, Danio rerio. Zebrafish is an excellent model system for studying vertebrate development and gene function. In addition, reverse genetic techniques have been developed that provide powerful approaches to the analysis of the physiological function of expressed genes. We describe the cloning and characterization of a cohort of sodium pump genes from the zebrafish. Of this cohort, eight cDNAs encode α subunit isoforms and five encode distinct β subunit isoforms. We have mapped the positions of the α and β subunit genes in the zebrafish genome, and examined expression profiles of each of the genes in a panel of zebrafish tissues. Identification of the members of the zebrafish Na,K-ATPase multigene family provides an initial framework for experiments designed to investigate the functional properties of these isoforms using this model system.
RESULTS
Identification and Characterization of Zebrafish Na,K-ATPase α Subunits
We used BLAST (Altschul et al. 1990) and keyword searches of the GenBank database to identify zebrafish expressed sequence tags (ESTs) encoding Na,K-ATPase α subunits. We sequenced the corresponding clones and identified overlapping regions. In cases where clones were unavailable or did not contain a complete open reading frame (ORF), full-length sequences were generated by a combination of reverse transcriptase–polymerase chain reaction (RT–PCR) and 5′-RACE (rapid amplification of cDNA ends). This approach resulted in the identification of full-length cDNAs encoding eight distinct α subunits. BLAST analysis of the deduced amino acid sequences indicates that each zebrafish clone exhibits greatest similarity with vertebrate Na,K-ATPase α subunits. As shown in Table 1, the zebrafish α subunits share 77%–92% amino acid identity with rat Na,K-ATPase α1, α2, and α3 subunits, and 72%–78% identity with the rat α4 subunit. The sequences are considerably less similar to α subunits of other P-type ATPases including the gastric H,K-ATPase (61%–64% amino acid identity). Pairwise comparisons of zebrafish to rat α subunits indicate that zebrafish α subunit clones 1, 3, 4, 7, and 8 are most similar to the rat α1 subunit, clone 2 is most similar to the rat α2 subunit, whereas clones 5 and 6 are most similar to the rat α3 subunit. However, for six of the eight zebrafish clones, similarities to the rat α1, α2, and α3 subunits differ by no more than 3%. Except for the α3-like clones 5 and 6, simple sequence comparisons fail to provide a clear picture of which rat and fish genes are likely to be orthologous.
Table 1.
Pairwise Comparisons between Zebrafish and Rat Na,K-ATPase α Subunits
| Clone 1 | Clone 2 | Clone 3 | Clone 4 | Clone 5 | Clone 6 | Clone 7 | Clone 8 | |
|---|---|---|---|---|---|---|---|---|
| Rat α 1 | 89 | 85 | 79 | 82 | 85 | 85 | 88 | 87 |
| Rat α 2 | 86 | 86 | 78 | 81 | 85 | 85 | 86 | 84 |
| Rat α 3 | 86 | 84 | 77 | 80 | 90 | 92 | 85 | 84 |
| Rat α 4 | 78 | 77 | 72 | 74 | 76 | 76 | 77 | 77 |
Numbers represent percent amino acid identity.
To better understand the relationship among zebrafish α subunits, we conducted sequence comparisons between each pair of zebrafish clones (Table 2). This analysis indicates that the α1-like clones 1, 7, and 8 share 90%–93% identity, whereas clones 5 and 6 share 94% identity. Clones 2, 3, and 4 show no more than 86% identity to any other α subunit clone. Clones 3 and 4 are more similar to clones 1, 7, and 8 than to any of the other clones, whereas clone 2 shares equal similarity to all α subunit clones with the exception of clones 3 and 4. These comparisons suggest that clones 1, 7, and 8, and clones 5 and 6 comprise two distinct subfamilies of zebrafish α subunit genes. Based on comparison with rat α subunits, clones 1, 7, and 8 are likely to represent a subfamily of α1-like genes, whereas clones 5 and 6 appear to represent a subfamily of α3-like genes. Clone 2 cannot be grouped with either subfamily.
Table 2.
Pariwise Comparisons among Zebrafish Na,K-ATPase α Subunit Isoforms
| Clone 2 | Clone 3 | Clone 4 | Clone 5 | Clone 6 | Clone 7 | Clone 8 | |
|---|---|---|---|---|---|---|---|
| Clone 1 | 83 | 81 | 85 | 85 | 85 | 93 | 91 |
| Clone 2 | 77 | 78 | 83 | 83 | 84 | 83 | |
| Clone 3 | 83 | 77 | 76 | 81 | 84 | ||
| Clone 4 | 79 | 79 | 84 | 86 | |||
| Clone 5 | 94 | 85 | 84 | ||||
| Clone 6 | 85 | 84 | |||||
| Clone 7 | 90 |
Numbers represent percent amino acid identity.
A highly variable 10 amino acid-long segment located within the third cytoplasmic domain of the Na,K-ATPase α subunit has been proposed to represent an α subunit isoform-specific signature sequence (Pressley 1992). We therefore compared this region of zebrafish α subunits with the corresponding segment of Na,K-ATPase α subunits from a variety of vertebrate species. As shown in Figure 1, zebrafish clones 3, 4, 7, and 8 possess an α subunit signature sequence highly similar to that of α1 subunits of mammals, birds, amphibians, and teleosts. Although this region of clone 1 is more divergent, clone 1 still clusters with other members of the α1-like subfamily. The α subunit signature sequence in clone 6 is most similar to the corresponding segment found in the rat, chicken, and Tilapia α3 subunit. Clone 5 contains a signature sequence that shares similarities with mammalian α2 and α3 subunits. Clone 2 contains a signature sequence most similar to that of the α subunit from the Electrophorus electric organ and the mammalian and avian α2 subunits. This analysis supports the view that zebrafish clones 1, 3, 4, 7, and 8 are likely to represent orthologs of the mammalian α1 subunit, whereas clones 5 and 6 appear to represent orthologs of the mammalian α3 isoform. Additionally, these results raise the possibility that clone 2 is an ortholog of the mammalian α2 isoform.
Figure 1.
Comparison of zebrafish and other vertebrate Na,K-ATPase α subunits. A segment of the third intracellular domain corresponding to residues 484–514 of the rat α1 subunit is shown. The highly variable signature sequence is shown in bold. Sequences were aligned using CLUSTALW. Ellipses in sequences allow optimal alignment for amino acid insertions/deletions.
Phylogenetic Analysis of α Subunits
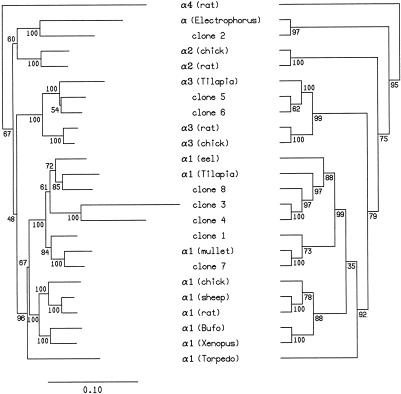
To examine the evolutionary relationships between zebrafish and mammalian Na,K-ATPase α subunit genes, we conducted a phylogenetic analysis of α subunits using maximum parsimony (MP) (Felsenstein 1981) and distance matrix (DM) methods (Fitch and Margoliash 1967). This analysis included the zebrafish, representative mammalian, and all available nonmammalian vertebrate Na,K-ATPase α subunits. The consensus trees generated by this analysis are presented in Figure 2. The DM tree is shown on the left and the MP tree on the right. Within the α1 gene subfamily, the trees generated by both methods exhibit the expected phylogenetic relationships between species, although the basal position of the Torpedo α subunit is supported by only 67% (DM) or 35% (MP) of the trees generated by bootstrap analysis. Zebrafish clones 1, 3, 4, 7, and 8 cluster unambiguously with α1 sequences of other teleosts (DM: 100% of trees, MP: 99% of trees), as well as α1 sequences from all other species (DM: 96% of trees, PM: 92% of trees). Clustering of zebrafish clones 5 and 6 with α3 sequences is also strongly supported (DM: 100% of trees, MP: 99% of trees). However, the two methods produced discordant results for zebrafish clone 2. The DM method clusters clone 2 and the Electrophorus α subunit with the mammalian and avian α2 subunits (60% of trees), whereas this clustering was supported by only 14% of the MP trees. The consensus MP tree suggests an early divergence of clone 2 from the α1, α2, and α3 gene lineages (supported by 75% of MP trees). The results of this phylogenetic analysis indicate that zebrafish Na,K-ATPase α subunits can be grouped into three subfamilies; five can be assigned to the α1-like subfamily (clones 1, 3, 4, 7, and 8) while two can be assigned to the α3-like subfamily (clones 5 and 6). Clone 2 appears to represent a distinct α subunit subfamily, but the phylogenetic analysis does not establish conclusively whether this clone is a true ortholog of the mammalian α2 subunit.
Figure 2.
Phylogenetic analysis of vertebrate Na,K-ATPase α subunits. Consensus tree generated using the distance matrix method is shown at left with the scale of evolutionary distances below. Consensus tree generated using maximum parsimony method is at right. Numbers at each branch point indicate percent support from bootstrap analysis. Na,K-ATPase α subunit sequences were obtained from SwissProt or GenBank: rat α1 (atn_rat), α2 (atn2_rat), α3 (atn3_rat), and α4 (AAB81285); chicken α1 (atn1_chick), α2 (atn2_chick), and α3 (atn3_chick); sheep α1 (atn1_sheep); Bufo marinus α1 (atn1_bufma); Xenopus laevis α1 (atn1_xenla); European eel α1 (atna_angan); Tilipia mossambica α1(AAD11455) and α3 (AAF75108); mullet α1 (atna_catco); Torpedo californica α1 (atna_torca). Electrophorus electricus α subunit sequence is from Kaya et al. (1997).
Identification and Characterization of Zebrafish Na,K-ATPase β Subunits
Zebrafish ESTs encoding multiple Na,K-ATPase β subunits were identified by searches of the GenBank database. By sequencing the corresponding clones, we identified and characterized full-length ORFs encoding five distinct β subunit isoforms. Sequence comparisons indicate that β subunit clones 1a and 1b are orthologs of the vertebrate β1 subunit, sharing 66% identity with each other (Table 3) and 57% identity with the rat β1 subunit (Table 4). Zebrafish β subunit clone 2 shows only 37%–46% identity with the other zebrafish β subunit clones (Table 3). However, this clone shares 74% identity with the rat β2 subunit (Table 4), and therefore appears to represent an ortholog of the mammalian β2 subunit. Zebrafish β subunit clone 3a was identified previously as an ortholog of the mammalian β3 subunit (Appel et al. 1996). Sequence analysis indicates that zebrafish clone 3b shares 59% identity with clone 3a and only 36%–46% identity with any of the other zebrafish β subunit clones (Table 3). These results suggest that zebrafish clone 3b is a second member of the β3 gene subfamily.
Table 3.
Pairwise Comparisons among Zebrafish Na,K-ATPase β Subunit Isoforms
| Clone 1b | Clone 2 | Clone 3a | Clone 3b | |
|---|---|---|---|---|
| Clone 1a | 66 | 41 | 35 | 36 |
| Clone 1b | 37 | 33 | 36 | |
| Clone 2 | 45 | 46 | ||
| Clone 3a | 59 |
Numbers represent percent amino acid identity.
Table 4.
Pairwise Comparisons between Zebrafish and Rat Na,K-ATPase β Subunits
| Clone 1a | Clone 1b | Clone 2 | Clone 3a | Clone 3b | |
|---|---|---|---|---|---|
| Rat β1 | 57 | 57 | 44 | 39 | 37 |
| Rat β2 | 38 | 40 | 74 | 47 | 46 |
| Rat β3 | 37 | 36 | 64 | 50 | 45 |
| Xenopus β3 | 40 | 38 | 53 | 59 | 57 |
Numbers represent percent amino acid identity.
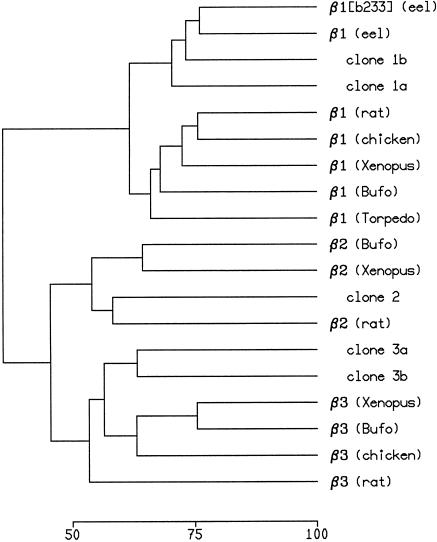
An alignment of the deduced amino acid sequences of the five zebrafish Na,K-ATPase β subunit isoforms is shown in Figure 3. Each of the β subunits contains an amino-terminal cytoplasmic domain, a single predicted transmembrane segment, and an extracellular carboxy-terminal domain containing six conserved cysteine residues and several potential sites for amino-linked glycosylation. A dendrogram showing sequence relationships among the zebrafish and other vertebrate β subunits is presented in Figure 4. This analysis clearly supports the idea that zebrafish clones 1a and 1b are β1 gene subfamily members, clone 2 is a β2 subunit gene, and clones 3a and 3b are members of the β3 gene subfamily.
Figure 3.
Comparison of zebrafish β subunits were aligned using CLUSTALW. Ellipses in sequences allow optimal alignment for amino acid insertions/deletions. Conserved cysteines are shaded; potential amino-linked glycosylation sites are underlined. The single transmembrane (TM) domain is indicated by the solid line above the sequence. Amino acids are numbered to the left of each line.
Figure 4.
Sequence relatedness of Na,K-ATPase β subunits. Dendrogram of amino acid sequence similarity was constructed using the PILEUP program. The scale at the bottom indicated percent identity between β subunits. Sequences were obtained from SwissProt or GenBank: European eel β1 (atnb_angan) and β1 isoform b233 (CAB85586); rat β1 (atnb_rat), β2 (atnc_rat), and β3 (atnd_rat); chicken β1 (atnb_chick) and β3 (atnd_chick); Xenopus laevis β1 (AAA82967), β2 (CAC08235), and β3 (atnd_xenla), Bufo marinus β1 (atnb_bufma), β2 (atnc_bufma), and β3 (atnd_bufma); Torpedo californica β1 (atnb_torca).
Chromosomal Mapping of Zebrafish Na,K-ATPase α and β Subunit Genes
We determined the chromosomal positions of zebrafish Na,K-ATPase genes using two radiation hybrid panels (Geisler et al. 1999; Hukriede et al. 1999) and a meiotic mapping panel (Woods et al. 2000). Twelve genes were mapped on the T51 panel, nine genes on the LN54 panel, and five genes on the HS panel. ESTs corresponding to α subunit clones 1 (AA495128) and 2 (AA494679), and β subunit clone 3a had been mapped previously on the T51 panel (Geisler et al. 1999), and α subunit clones 1 and 8 had been mapped on the HS panel (Woods et al. 2000). A summary of the map positions of individual α and β subunit genes is presented in Table 5. Clones 1, 3, 4, and 8 (which are members of the α1 gene subfamily) all mapped to linkage group (LG) 1 between microsatellite markers z9382 and z9394. These four genes are thus very tightly clustered in the genome. Clone 7, the fifth α1-like gene, mapped to the middle of LG 9 and is closely linked to gabpa and actbb. We confirmed the location of zebrafish α subunit clone 2, which had been mapped previously to LG 2 between microsatellite markers z17291 and z1406 (Geisler et al. 1999). Clone 5, an α3-like gene, mapped to LG 19 between microsatellite markers z7450 and z3782. EST AW281746, corresponding to clone 5, had been mapped independently to LG 19 (http://zfrhmaps.tch.harvard.edu/ZonRHmapper/). Clone 6, the second member of the α3 gene subfamily, mapped to LG 16 between microsatellite markers z3072 and z6921. Clone β1a mapped to LG 6 in close proximity to ndufb5, whereas clone β1b localized to LG 1 between microsatellite markers z5508 and z5058. β subunit clone 2, an ortholog of the mammalian β2 subunit, mapped to LG 23 between microsatellite markers z9351 and z7243. We confirmed the location of clone β3a on LG 2 between microsatellite markers z4733 and z4300 (Geisler et al. 1999), whereas β subunit clone 3b mapped to LG 15 between microsatellite markers z6312 and z9214.
Table 5.
Map Positions of Zebrafish and Orthologous Human and Mouse Na,K-ATPase α and β Subunit Genes
| Clone | Accession no. | Forward primer | Reverse primer | Zebrafish | Human ortholog | Mouse ortholog | |||
|---|---|---|---|---|---|---|---|---|---|
| Locus | LG | Name | Locatione | Name | Chromosome (cM)f | ||||
| α Clone 1 | AF286373 | gtaggtctccctctccaccc | gcgccttcccctactcact | atp1a1a.1 | 1bc | ATP1A1 | 1p13.1 | Atp1a1 | 3 (48.4) |
| α Clone 3 | AF286374 | atacatcctgcgacggaatc | caccagattctttatcgcttttc | atp1a1a.2 | 1a | ||||
| α Clone 4 | AF308598 | gcatggaagttgctgtcaga | ctgcacatgttcccaagaga | atp1a1a.3 | 1a | ||||
| α Clone 7 | AY008375 | gagaaagctcatccttagac | acagcaaaactactggaac | atp1a1b | 9a | ||||
| α Clone 8 | AY008376 | gaggttgggtggaacaagaa | attgactgaaaatgggcaga | atp1a1a.4 | 1a | ||||
| α Clone 2 | AF286373 | ggtgaacgactctccagctc | gaagggtgtgactctggga | atp1a2 | 2bc | ATP1A2 | 1q23-24 | Atp1a2 | 1 (94.2) |
| α Clone 5 | AF308599 | ggatagaagagggaggagg | gtattgcgatgtgttgtggac | atp1a3a | 19ad | ATP1A3 | 19q13.2 | Atp1a3 | 7 (5.5) |
| α Clone 6 | AY008374 | agctcttccgtgctcaagtc | agccgtgtgaaaatacatgc | atp1a3b | 16a | ||||
| β Clone 1a | AF286375 | agccatgggacgttcattta | ccacacgaggcagaaatac | atp1b1a | 6a | ATP1B1 | 1q24.2 | Atp1bl | 1 (86.8) |
| β Clone 1b | AF308597 | cgtgaaattccaatggatgc | aatgcagatgttggcattga | atp1b1b | 1a | ||||
| β Clone 2 | AF286376 | ccgagttcagcgagagagac | cacctcccacaatctcctgt | atp1b2 | 23a | ATP1B2 | 17p13.1 | Atp1b2 | 11 (40) |
| β Clone 3a | X89722 | ttccgggtcctggtgaccgaat | gggttaaaattcaattaaatgttcg | atp1b3a | 2b | ATP1B3 | 3q23 | Atp1b3 | 9 (51) |
| β Clone 3b | AF293369 | tgtccatcacgtccattttg | accttcacgagcacaggttt | atp1b3b | 15ad | ||||
This paper.
Human map positions were obtained from OMIM (http://www3.ncbi.n1m.nih.gov/OMIM).
Mouse map positions were obtained from MGD (http://www.informatics.jax.org).
Based on the above analyses, we propose that the α1-like genes clustered on LG 1 and represented by clones 1, 3, 4, and 8 be designated atp1a1a.1, atp1a1a.2, atp1a1a.3, and atp1a1a.4, respectively. The α1-like gene represented by clone 7 we have named atp1a1b, whereas the gene encoded by clone 2 was named atp1a2. We have named the two α3-like genes represented by clones 5 and 6 atp1a3a and atp1a3b, respectively. Our data indicates that zebrafish possess two Na,K-ATPase β1-like genes (1a and 1b), which we have designated atp1b1a and atp1b1b, respectively. The zebrafish ortholog of the mammalian β2 subunit (clone 2) was named atp1b2, whereas the two zebrafish β3-like clones (3a and 3b) were named atp1b3a and atp1b3b, respectively.
Expression of Zebrafish α and β Subunit Genes
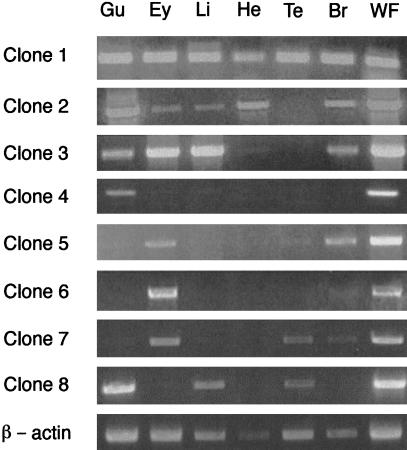
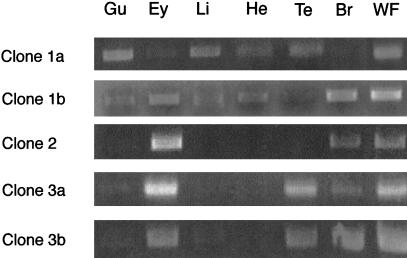
Expression of Na,K-ATPase α and β subunit mRNAs was analyzed by RT–PCR. Total RNA was prepared from a panel of adult zebrafish tissues including gut, eye, liver, heart, testis, and brain and used as template in RT–PCR reactions. The expression profile of zebrafish α subunits is shown in Figure 5. As a control, β-actin transcripts were amplified from individual RNA samples. Each RNA produced an RT–PCR product of the appropriate size, demonstrating the integrity of the samples. Analysis of the PCR products indicates that atp1a1a.1 (clone 1) is ubiquitously expressed, while atp1a2 (clone 2) is expressed in all tissues examined with the exception of testis. Clone 3 (atp1a1a.2), in contrast, is expressed in all tissues with the exception of brain and testis. Expression of atp1a1a.3 (clone 4) is expressed predominantly in gut, atp1a3a (clone 5) mRNA is present in gut, eye, testis, and brain, while atp1a3b (clone 6) mRNA is detected only in eye and brain. Expression of atp1a1b (clone 7) is restricted to eye, brain, and testis, while atp1a1a.4 (clone 8) mRNA is detectable in the gut, liver, and testis. The expression profile of zebrafish β subunit clones is presented in Figure 6. Among the β1-like genes, clone 1b (atp1b1b) is ubiquitously expressed while clone 1a (atp1b1a) is expressed in all tissues except brain. Expression of β subunit clone 2 (atp1b2) is restricted to eye and brain. Clone 3a (atp1b3a) is expressed predominantly in gut, eye, testis, and brain. Clone 3b (atp1b3b) is expressed in all tissues with the exception of heart. These results strongly indicate differential expression patterns for each of the α and β subunit genes, and suggest that each gene is likely to subserve distinct functions.
Figure 5.
Expression of Na,K-ATPase α subunit mRNAs in zebrafish tissues. Total RNA was prepared from adult zebrafish and used as template for RT–PCR. Amplification products were analyzed by electrophoresis on a 1% agarose gel. (Gu) gut; (Ey) eye; (Li) liver; (He) heart; (Te) testis; (Br) brain; (WF) shole fish. β-actin controls are shown at the bottom.
Figure 6.
Expression of Na,K-ATPase β subunit mRNAs in zebrafish tissues. Total RNA was prepared from adult zebrafish and used as template for RT–PCR. Amplification products were analyzed by electrophoresis on a 1% agarose gel. (Gu) gut; (Ey) eye; (Li) liver; (He) heart; (Te) testis; (Br) brain; (WF) whole fish.
DISCUSSION
We have identified and cloned eight Na,K-ATPase α subunit and five β subunit genes from zebrafish. Sequence comparisons and phylogenetic analysis indicate that five of the zebrafish α subunit genes form an α1-like gene subfamily, two correspond to the α3 subfamily, and one appears to encode an α2-like subunit. Among the β subunit genes, two are orthologs of the mammalian β1 isoform, one represents a β2 ortholog, and two are orthologous to the mammalian β3 subunit. The presence of orthologous genes corresponding to at least two α and three β subunit isoforms in both zebrafish and mammals clearly implies that gene duplications producing multiple α and β gene subfamilies predate the last common ancestor of teleosts and mammals.
We have utilized gene mapping to further study the evolution of zebrafish Na,K-ATPase α and β subunit genes. Map positions of Na,K-ATPase genes and syntenic markers in zebrafish and humans are shown in Table 6. (Detailed genetic maps [M1–M5] are available as on-line supplementary material at http://www.genome.org.) In general, the mapping results tend to support the phylogenetic analysis. For example, the two zebrafish α3-like genes map to LG 16 and LG 19. Each of these linkage groups contains orthologs of additional human genes that map to Hsa 19q13. The zebrafish β1-like genes map to LG 1 and LG 6, and appear to share conserved syntenies with a set of human genes located on Hsa 1q. The β3-like genes map to LG 2 and LG 15 and are syntenic with orthologs of a group of human genes that map to Hsa 3q. The zebrafish α1-like genes consist of one gene on LG 9 and a cluster of four genes tightly linked on LG 1. In humans, ATP1A1 and CASQ2 are tightly linked near the centromere on Hsa 1p, and the orthologous zebrafish genes on LG 9 are also tightly linked. These results suggest the possibility that a segment of Hsa 1p and LG 9 may contain additional genes that are syntenic in both species. The cluster of α1-like genes on LG 1 represents a more complicated scenario. Conserved synteny between LG 1 and Hsa 1 involves several genes located on Hsa 1q, whereas ATP1A1 maps to Hsa 1p13. However, the presence of zebrafish duplicates of EN1 and DLX2 on LG 1 and LG 9 supports the view that these are duplicate chromosome segments (Postlethwait et al. 2000). In the case of the zebrafish β2 gene, which maps to LG 23 and Hsa 17p13, no additional genes have been identified that are syntenic with atp1b2 in both species. Instead, genes that are linked to ATP1B2, such as TP53, are located on LG 5 and LG 15 in the zebrafish genome (Postlethwait et al. 2000).
Table 6.
Markers Syntenic with Na,K-ATPase Genes in Zebrafish and Human
| Name | Zebrafish gene | Human ortholog | ||
|---|---|---|---|---|
| Accession No. | Map positiona | Name | Map position | |
| atp1a1a.1-.4 | 1-0.0 | ATP1A1 | 1p13.1 | |
| atp1b1b | 1-21.8 | ATP1B1 | 1q24.2 | |
| oct1 | AF030519 | 1-0.0 | POU2F1 | 1q22-q23 |
| (b1zf1) | AW174816 | 1-0.0 | BLZF1 | 1q24 |
| (capn9) | AI544664 | 1-91.5 | CAPN9 | 1q42 |
| atp1a2 | 2-108.6 | ATP1A2 | 1q21-q23 | |
| (sec2211) | AI497465 | 2-40.4 | SEC22L1 | 1q21.2- |
| (mpz) | AW174967 | 2-108.6 | MPZ | 1q22-q23 |
| cp1a2 | U10330 | 2-55.8 | PLA2G4A | 1q25 |
| (arf1) | AI558333 | 2-12.3 | ARF1 | 1q42 |
| (fh) | AA555477 | 2-155.3 | FH | 1q42.1 |
| atp1b3a | 2-50.0 | ATP1B3 | 3q23 | |
| tfa | AI497391 | 2-50.0 | TF | 3q21 |
| (copb2) | AI415987 | 2-95.6 | COPB2 | 3q23 |
| (d1g1) | AI723230 | 2-12.3 | DLG1 | 3q29 |
| (loc57018) | AW165033 | 2-87.8 | LOC57018 | 3q |
| (arpc4) | AA542497 | 2-50.0 | ARPC4 | 3p |
| (dkfzp564k247) | AI477599 | 2-7.3 | DKFZP564K247 | 3p |
| (epha3) | AW175051 | 2-95.6 | EPHA3 | 3p11.2 |
| atp1b1a | 6-65.3 | ATP1B1 | 1q24.2 | |
| (mypt2) | AW173939 | 6-59.6 | MYPT2 | 1q32.1 |
| atp1a1b | 9-66.5 | ATP1A1 | 1p13.1 | |
| (casq2) | AA497210 | 9-60.5 | CASQ2 | 1p13.3-p11 |
| (ddah1) | AI722688 | 9-110.0 | DDAH1 | 1p22 |
| atp1b3b | 15-32.7 | ATP1B3 | 3q23 | |
| gap43 | L27645 | 15-132.5 | GAP43 | 3q13 |
| chd | AF034606 | 15-127.8 | CHRD | 3q27 |
| atp1a3b | 16-22.1 | ATP1A3 | 19q13.2 | |
| gsk3a | AJ223501 | 16-30.0 | GSK3A | 19q13 |
| (kiaa0306) | AI584937 | 16-30.0 | KIAA0306 | 19q13 |
| apoeb | Y13652 | 16-40.9 | APOE | 19q13 |
| (ceacam1) | AI353126 | 16-32.8 | CEACAM1 | 19q13.2 |
| (rps9) | AI353657 | 16-72.5 | RPS9 | 19q13.4 |
| (dnase2) | AI330733 | 16-99.2 | DNASE2 | 19p13.2 |
| atp1a3a | 19-33.8 | ATP1A3 | 19q13.2 | |
| (erf) | (AI658144) | 19-36.4 | ERF | 19q13 |
| (apoea) | (AI544900) | 19-51.6 | APOE | 19q13 |
| atp1b2 | 23-124.4 | ATP1B2 | 17p13.1 | |
Linkage group and position (in cM) on HS map.
The only zebrafish Na,K-ATPase subunit for which the mammalian ortholog has not been clearly resolved, either by sequence comparisons or phylogenetic analysis, is α subunit clone 2 (atp1a2). Comparative mapping, however, provides compelling evidence that the mammalian α2 subunit gene and zebrafish clone 2 are in fact orthologous. Our data indicate that clone 2 maps to zebrafish LG 2. In zebrafish, atp1a2 is tightly linked to copa and mpz and the orthologs of these three genes in mouse are within an interval of less than 2 cM. In addition, several other loci show conserved syntenies with these chromosomes (Table 6). These results strongly indicate that zebrafish clone 2 and ATP1A2 are orthologous genes, but have diverged to such an extent that their sequence relationships are no longer clearly apparent. Such a relationship has been described for the zebrafish hlx3 and murine Dbx2 homeodomain genes (Seo et al. 1999). It will be of considerable interest to determine whether sequence divergence between the zebrafish and mammalian α2 genes has led to functional differences between the mammalian and zebrafish α2 subunit polypeptides.
Analysis of the zebrafish genome indicates that zebrafish have undergone an additional round of genome duplication compared to mammals (Amores et al. 1998; Postlethwait et al. 1998). In the absence of gene loss, we would predict that each mammalian Na,K-ATPase α and β subunit gene should have two zebrafish orthologs. Consistent with this view, we have identified two zebrafish orthologs of the rat α3, β1 and β3 subunit genes. In contrast, we have found only single zebrafish orthologs of the rat α2 and β2 subunits, suggesting that the second α2 and β2 orthologs were lost during zebrafish evolution. It has been established that the degradation of one of two copies of a duplicate gene is a common phenomenon in gene evolution (Haldane 1933; Li 1980; Watterson 1983), whereas recent work suggests that many duplicate genes have been lost during zebrafish evolution (Postlethwait et al. 2000). Alternatively, it is possible that these genes are still present in the zebrafish genome, but are not represented in the cDNA libraries used to generate the zebrafish ESTs. Interestingly, five of the zebrafish α subunit genes appear to be orthologous to the mammalian α1 gene. Thus for the α1 gene subfamily, there must have been additional gene duplication events in addition to the genome-wide duplication. Because our mapping data places four of the α1-like genes within a tight cluster on LG 1, we surmise that these loci have arisen by local, tandem duplications that have arisen after the hypothesized genome duplication event. Retention of a high proportion of Na,K-ATPase α and β subunit gene duplicates suggests that these genes are more likely to possess specialized, rather than redundant, functional properties. In this context, it should be noted that a zebrafish ortholog of the mammalian α4 subunit has not as yet been identified. The mammalian α4 subunit is expressed exclusively in testis, a tissue from which no zebrafish ESTs have been reported. It will be interesting to determine whether this α subunit isoform is present in zebrafish spermatozoa.
The expression patterns of Na,K-ATPase α and β subunit genes in zebrafish are quite distinctive, with no two α or β subunit genes exhibiting the same expression profile. The expression of α subunit genes is particularly striking in liver, where mRNAs corresponding to four of the eight zebrafish α subunit isoforms are detected. This contrasts with the situation in rodents, where the α1 subunit is the only isoform that appears to be expressed (Shyjan and Levenson 1989). In mammals, the α3 gene is expressed predominantly within neurons (Cameron et al. 1994) and in retinal photoreceptors (Schneider et al. 1991). Both of the zebrafish α3 orthologs show expression in brain and eye. However, clone 5 (atp1a3a) is also expressed in gut, suggesting an additional role for this isoform in the digestive tract. More precise localization of the two α3 genes in zebrafish will be required to determine whether the α3 genes are likely to perform similar functions in mammals and fish. The expression pattern for the five zebrafish α1-like genes is also somewhat surprising. Each α1 gene shows an overlapping yet distinctive expression pattern. Expression of clone 1 is ubiquitous. Clone 4 (atp1a1a.3) is expressed in all tissues except brain, whereas clone 3 (atp1a1a.2) is expressed in all tissues except brain and testis. Expression of clone 7 (atp1a1b) is restricted to brain, eye, and testis, whereas expression of clone 8 (atp1a1a.4) is only detected in gut, liver, and testis among organs tested. These expression profiles are consistent with the view that the α1-like genes most likely subserve distinct functions. In situ hybridization analysis of α1-like mRNA expression should help to discriminate whether these genes are differentially expressed within a tissue. Differential cellular localization would provide further support for the view that these genes are not merely redundant but play specialized physiological roles in zebrafish.
A fundamental unresolved issue concerning Na,K-ATPase is why there are so many different α and β subunit isoforms. In mammals, each of the four α subunits appears capable of associating with any of the three β subunits, potentially giving rise to ⋝12 structurally distinct isoenzymes. The potential for isoenzyme heterogeneity is even greater in zebrafish. If each zebrafish α subunit can associate with each of the five β subunit isoforms, 40 different zebrafish Na,K-ATPase isoenzymes could potentially be formed. It seems unlikely that all 40 α/β subunit combinations are actually formed in vivo. Analysis of α and β subunit expression within specific cell types should provide a more precise picture of which α/β subunit combinations may actually be produced within a given tissue or cell type. It also seems highly unlikely that all α/β subunit combinations form holoenzymes that are functionally equivalent. The view that at least some of the isoenzymes express distinct functional properties is a more appealing hypothesis. Functional characterization of mammalian Na,K-ATPase has failed to delineate clear biochemical differences between isoenzymes. Furthermore, no naturally occurring mutations in mouse or human sodium pump genes have yet been described. It has thus been difficult to associate a physiological or behavioral deficit with the function of a particular isoenzyme. The identification of the cohort of Na,K-ATPase genes expressed in zebrafish facilitates the application of conditional knockdown approaches to elucidate the roles these genes play in zebrafish development and physiology.
METHODS
Cloning of Zebrafish α and β Subunits
cDNAs encoding zebrafish Na,K-ATPase α and β subunits were identified by searches of the GenBank EST database. Clones were obtained from Genome Systems, Research Genetics, or RZPD (Resource Center for the German Genome Project, Berlin). ESTs that were unavailable from these sources were generated by RT–PCR using protocols described in the SuperScript RT–PCR kit (Life Technologies). Clones that did not encode a complete ORF were extended using a 5′-RACE kit (Life Technologies) using whole adult zebrafish mRNA as template. cDNAs were sequenced using an ABI 377 automated DNA sequencer.
α subunit clone 1 (atp1a1a.1; GenBank accession no. AF286372) was obtained from Genome Systems and corresponds to ESTs AA495128 and AA495435 (GenBank). The cDNA is 3474 bp in length, whereas the complete ORF spans nucleotides 174–3260. α subunit clone 2 (atp1a2; GenBank accession no. AF286373) is a 3889-bp long cDNA containing a complete ORF that spans nucleotides 101–3154. Nucleotides 1461–3889 were determined by sequencing clone ICRFp524B1612 (RZPD) which corresponds to EST AA494679 (GenBank). Nucleotides 568–1460 were generated by RT–PCR and the 5′ end generated by 5′-RACE using nested primers complementary to nucleotides 622–642 and 582–601. α subunit clone 3 (atp1a1a.2; GenBank accession no. AF286374) is a 3235-bp long cDNA containing a complete ORF that spans nucleotides 62–3133. Nucleotides 296–3255 were obtained by sequencing clone MPMGp609H2236Q.1 (RZPD) which corresponds to EST AI878291 (GenBank). The 5′ end of the cDNA was generated by 5′-RACE using nested primers complementary to nucleotides 311–332 and 344–363. α subunit clone 4 (atp1a1a.3; GenBank accession no. AF308598) is a 3275-bp long cDNA containing a complete ORF that spans nucleotides 71–3145. Nucleotides 623–3275 were determined by sequencing clone UCDMp574K2119Q2 (RZPD) which corresponds to EST AW184276 (GenBank). The 5′ end of the cDNA was obtained by 5′-RACE using nested primers complementary to nucleotides 706–723 and 753–772. α subunit clone 5 (atp1a3a; GenBank accession no. AF308599) was generated by RT–PCR using primers based on the sequence of ESTs AW281430 and AW281746 (GenBank). The cDNA is 3664 bp in length and the complete ORF spans nucleotides 185–3256. α subunit clone 6 (atp1a3b; GenBank accession no. AY008374) was generated by RT–PCR. The cDNA is 3309-bp long and the complete ORF spans nucleotides 18–3089. Two overlapping cDNA fragments spanning nucleotides 284–3309 were generated by RT–PCR using primers based on the sequences of ESTs AW280277, AW077565, and AW279681 (GenBank). The 5′ end of the cDNA was generated by 5′-RACE using nested primers complementary to nucleotides 234–303 and nucleotides 317–338. α subunit clone 7 (atp1a1b; GenBank accession no. AY008375) was generated by RT–PCR. The cDNA is 3284 bp in length and the complete ORF spans nucleotides 15–3092. Two overlapping cDNA fragments spanning nucleotides 237–3284 were generated by RT–PCR based on the sequences of ESTs AW281423, AW077399, and AW077182 (GenBank). The 5′ end of the cDNA was generated by 5′-RACE using nested primers complementary to nucleotides 584–564 and nucleotides 358–341. α subunit clone 8 (atp1a1a.4; GenBank accession no. AY008376) is a 3262-bp long cDNA containing a complete ORF that spans nucleotides 76–3150. Nucleotides 112–3262 were determined by sequencing clone CHBOp575E1024Q3 (RZPD) which corresponds to EST AW422519 (GenBank). The 5′ end of the cDNA was generated by 5′-RACE using nested primers complementary to nucleotides 351–371 and 149–168.
β subunit clone 1a (atp1b1a; GenBank accession no. AF286375) was obtained from Research Genetics and corresponds to EST AI384184 (GenBank). The cDNA is 1477-bp long and the complete ORF spans nucleotides 71–991. β subunit clone 1b (atp1b1b; GenBank accession no. AF308597) was obtained from RZPD (Clone number UCDMp611D1320) and corresponds to EST AW280780 (GenBank). The cDNA is 2384 nucleotides long and the complete ORF spans nucleotides 120–1028. β subunit clone 2 (atp1b2; GenBank accession no. AF286376) was obtained from Research Genetics and corresponds to EST AI629140 (GenBank). The cDNA is 1076 nucleotides in length and the complete ORF spans nucleotides 127–984. β subunit clone 3b (atp1b3b; GenBank accession no. AF293369) was obtained from Research Genetics and corresponds to EST AI722718 (GenBank). The cDNA is 1755 nucleotides long and the complete ORF spans nucleotides 80–907. β subunit clone 3a (atp1b3a; GenBank accession no. X89722) has been described previously by Appel et al. (1996).
Phylogenetic Analysis
Full-length Na,K-ATPase α and β subunit amino acid sequences were aligned using either the PILEUP (Devereux et al. 1984), or CLUSTALW (Thompson et al. 1994) programs. Phylogenetic analysis of Na,K-ATPase α subunits by maximum parsimony and distance matrix methods was performed using the Phylip suite of programs (version 3.573c) described by Felsenstein (1981). Regions in which alignments were ambiguous due to amino acid insertions or deletions were omitted, leaving 993 amino acids available for phylogenetic analysis (aligned with amino acids 30–497 and 501–1023 of rat α1). Maximum parsimony trees were calculated using PROTPARS. Evolutionary distance trees were constructed by using the algorithm of Fitch and Margoliash (1967). For each method, tree reliability was estimated by analysis of 100 half jackknife subreplicates. Trees were rooted using the Na,K-ATPase α subunit from sea urchin, Hemicentrotus pulcherrimus (Mitsunaga-Nakatsubo et al. 1996).
Chromosomal Localization of α and β Subunit Genes
Zebrafish α and β subunit genes were mapped using the Goodfellow T51 radiation hybrid (RH) panel (Kwok et al. 1998), the Ekker LN54 RH panel (Hukriede et al. 1999), and the heat shock (HS) double haploid meiotic mapping panel (Kelly et al. 2000; Woods et al. 2000). The T51 panel was purchased from Research Genetics. PCR products specific for each zebrafish α and β subunit gene were amplified using primers corresponding to unique 3′ UTR sequences. Primers were designed using the program Primer3 (S. Rozen and H.J. Skaletski, unpubl., data available at http://www-genome.wi.mit.edu/genome_software/other/primer3.html). A complete list of primers used to determine linkage assignments is detailed in Table 5. PCR reactions were performed in duplicate on the RH panels using conditions described previously (Geisler et al. 1999; Hukriede et al. 1999). PCR reaction products were fractionated on 2% agarose gels, and each sample scored for presence or absence of the zebrafish-specific amplicon. Linkage assignments were computed using the resources of the Tübingen Zebrafish Mapping Facility (http://www.map.tuebingen.mpg.de) and the Igor Dawid Web site (http://mgchd1.nichd.nih.gov:8000/zfrh/beta.cgi). Sodium pump genes were mapped on the HS panel using single strand conformation polymorphism analysis as described by Woods et al. (2000). One of the primers in each PCR reaction was end-labeled using γ-32P ATP and products were detected by autoradiography after fractionation on acrylamide gels. The strain distribution patterns on the HS panel were analyzed using MapManager (http://mcbio.med.buffalo.edu/mapmgr.html). All loci (with the exception of atp1b1a) were mapped on at least two of the three mapping panels. Loci mapped only on the RH panels were placed on the HS map by interpolation of markers mapped on both panels. Comparative mapping of sodium pump genes between Danio rerio and Homo sapiens was performed as described previously (Postlethwait et al. 2000).
Expression of Zebrafish α and β Subunit mRNAs
Several tissues including gut, eye, liver, heart, testis, and brain were dissected manually from adult zebrafish. Tissues were homogenized in TRIzol Reagent (Life Technologies) and total RNA extracted according to the method of Chomczynski and Sacchi (1987). Organ-specific RNA (0.3–0.5 μg) was used as template to generate single stranded cDNA using the SuperScript First Strand Synthesis kit (Life Technologies). One twentieth of the reaction product was used in each PCR reaction. PCR was carried out with Platinum Pfx DNA Polymerase (Life Technologies) using a RoboCycler Gradient Temperature Cycler (Stratagene). An initial 3 min denaturation step at 94°C was followed by 34 cycles at 94°C for 30 s, 55–60°C for 30 s, and 72°C for 1 min. A final elongation step was carried out at 72°C for 10 min. PCR products were analyzed by electrophoresis on a 1% agarose gel. (Sequences of gene-specific primers used for RT–PCR are available as supplementary material [Table A] at http://www.genome.org.)
Acknowledgments
This work was supported by NIH grants HL 39263 (to R.L.), CA 73935 (to K.C.C.), and 1-G20-RR11724 (to J.H.P.). We are especially grateful to Dr. Jessica Moore for help with zebrafish dissections, and Dr. Robert Geisler for computing linkage assignments.
NOTE ADDED IN PROOF
After submission of this manuscript, we sequenced a second Na,K-ATPase β2 subunit (atp1b2b, accession no. AF373976) with 74% amino acid identity to atp1b2 (now renamed atp1b2a).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL rlevenson@hmc.psu.edu; FAX (717) 531-5013.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.186001.
REFERENCES
- Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amores A, Force A, Yan Y-L, Joly L, Amemiya C, Ho RK, Langeland J, Prince V, Wang Y-L, Westerfield M, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Appel C, Gloor S, Schmalzing G, Schachner M, Bernhardt R. Expression of a Na,K-ATPase beta3 subunit during development of the zebrafish central nervous system. J Neurosci Res. 1996;46:551–564. doi: 10.1002/(SICI)1097-4547(19961201)46:5<551::AID-JNR4>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Besirli CG, Gong T-WL, Lomax MI. The Atp1b3 gene for the Na,K-ATPase β3 subunit maps to mouse chromosome 9, and a related gene, Atp1b3-rs, maps to mouse chromosome 3. Mamm Genome. 1998;9:171–172. doi: 10.1007/s003359900713. [DOI] [PubMed] [Google Scholar]
- Cameron R, Klein L, Shyjan AW, Rakic P, Levenson R. Neurons and astroglia express distinct subsets of Na,K-ATPase α and β subunits. Mol Brain Res. 1994;21:333–343. doi: 10.1016/0169-328x(94)90264-x. [DOI] [PubMed] [Google Scholar]
- Cantley LC. Structure and mechanism of the Na,K-ATPase. Curr Top Bioenerg. 1981;11:201–237. [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-choloform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Crambert G, Hasler U, Beggah AT, Yu C, Modyanov NN, Horisberger JD, Lelievre L, Geering K. Transport and pharmacological properties of nine different human Na, K-ATPase isozymes. J Biol Chem. 2000;275:1976–1986. doi: 10.1074/jbc.275.3.1976. [DOI] [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Fitch WM, Margoliash E. Construction of phylogenetic trees. Science. 1967;155:279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- Geisler R, Rauch GJ, Baier H, van Bebber F, Brobeta L, Dekens MP, Finger K, Fricke C, Gates MA, Geiger H, et al. A radiation hybrid map of the zebrafish genome. Nat Genet. 1999;23:86–89. doi: 10.1038/12692. [DOI] [PubMed] [Google Scholar]
- Haldane J. The part played by the recurrent mutation in evolution. Am Nat. 1933;67:5–9. [Google Scholar]
- Hukriede N, Joly L, Tsang M, Miles J, Tellis P, Epstein J, Barbazuk W, Li F, Paw B, Postlethwait J, et al. Radiation hybrid mapping of the zebrafish genome. Proc Natl Acad Sci. 1999;96:9745–9750. doi: 10.1073/pnas.96.17.9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell EA, Lingrel JB. Comparison of the substrate dependence properties of the rat Na,K-ATPase α1, α2, and α3 isoforms expressed in HeLa cells. J Biol Chem. 1991;266:16925–16930. [PubMed] [Google Scholar]
- Jørgensen PL. Purification and characterization of (Na+ + K+)-ATPase. IV. Estimation of the purity and of the molecular weight and polypeptide content per enzyme unit in preparations from the outer medulla of rabbit kidney. Biochim Biophys Acta. 1974;356:53–67. doi: 10.1016/0005-2736(74)90293-4. [DOI] [PubMed] [Google Scholar]
- Kaya S, Yokoyama A, Imagawa T, Taniguchi K, Froehlich JP, Albers RW. Cloning of the eel electroplax Na,K-ATPase α subunit. Ann NY Acad Sci. 1997;834:129–131. doi: 10.1111/j.1749-6632.1997.tb52238.x. [DOI] [PubMed] [Google Scholar]
- Kelly P, Chu F, Woods I, Ngo-Hazelett P, Cardozo T, Huang H, Kimm F, Liao L, Yan Y-L, Zhou Y, et al. Genetic linkage mapping of zebrafish genes and ESTs. Genome Res. 2000;10:558–567. doi: 10.1101/gr.10.4.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok C, Korn RM, Davis ME, Burt DW, Critcher R, McCarthy L, Paw BH, Zon LI, Goodfellow PN, Schmitt K. Characterization of whole genome radiation hybrid mapping resources for non-mammalian vertebrates. Nucleic Acids Res. 1998;26:3562–3566. doi: 10.1093/nar/26.15.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemas MV, Yu HV, Takeyasu K, Kone B, Fambrough DM. Assembly of the Na,K-ATPase α-subunit isoforms with Na,K-ATPase β-subunit isoforms and H,K-ATPase β-subunit. J Biol Chem. 1994;269:18651–189655. [PubMed] [Google Scholar]
- Levenson R. Isoforms of the Na,K-ATPase: Family members in search of function. Rev Physiol Biochem Pharmacol. 1994;123:1–45. doi: 10.1007/BFb0030902. [DOI] [PubMed] [Google Scholar]
- Li W-H. Rate of gene silencing at duplicate loci: A theoretical study and interpretation of data from tetraploid fishes. Genetics. 1980;95:237–258. doi: 10.1093/genetics/95.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik N, Canfield VA, Beckers M, Gros P, Levenson R. Identification of the mammalian Na,K-ATPase β3 subunit. J Biol Chem. 1996;271:22754–22758. doi: 10.1074/jbc.271.37.22754. [DOI] [PubMed] [Google Scholar]
- Malik N, Canfield V, Sanchez-Watts G, Watts AG, Scherer S, Beatty BG, Gros P, Levenson R. Structural organization and chromosomal localization of the human Na,K-ATPase β3 subunit gene and pseudogene. Mamm Genome. 1998;9:136–143. doi: 10.1007/s003359900704. [DOI] [PubMed] [Google Scholar]
- Mitsunaga-Nakatsubo K, Yamazaki K, Hatoh-Okazaki M, Kawashita H, Okamura C, Akasaka K, Shimada H, Yasumasu I. cDNA cloning of Na,K-ATPase α-subunit from embryos of the sea urchin, Hemicentrotus pulcherrimus. Zoolog Sci. 1996;13:833–841. doi: 10.2108/zsj.13.833. [DOI] [PubMed] [Google Scholar]
- Postlethwait J, Yan Y-L, Gates M, Horne S, Amores A, Brownlie A, Donovan A, Egan E, Force A, Gong Z, et al. Vertebrate genome evolution and the zebrafish gene map. Nat Genet. 1998;18:345–349. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- Postlethwait J, Woods I, Ngo-Hazelett P, Yan Y, Kelly P, Chu F, Huang H, Hill-Force A, Talbot W. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 2000;10:1890–902. doi: 10.1101/gr.164800. [DOI] [PubMed] [Google Scholar]
- Pressley TA. Phylogenetic conservation of isoform-specific regions within a subunit of Na,K-ATPase. Am J Physiol. 1992;262:C743–C751. doi: 10.1152/ajpcell.1992.262.3.C743. [DOI] [PubMed] [Google Scholar]
- Schneider BG, Shyjan AW, Levenson R. Co-localization and polarized distribution of Na,K-ATPase α3 and β2 subunits in photoreceptor cells. J Histochem Cytochem. 1991;39:507–517. doi: 10.1177/39.4.1848572. [DOI] [PubMed] [Google Scholar]
- Seo H-C, Nilsen F, Fjose A. Three structurally and functionally conserved Hlxgenes in zebrafish. Biochim Biophys Acta. 1999;1489:323–335. doi: 10.1016/s0167-4781(99)00187-6. [DOI] [PubMed] [Google Scholar]
- Shamraj OI, Lingrel JB. A putative fourth Na,K-ATPase α-subunit gene is expressed in testis. Proc Natl Acad Sci. 1994;91:12952–12956. doi: 10.1073/pnas.91.26.12952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyjan AW, Levenson R. Antisera specific for the α1, α2, α3 and β subunits of the Na,K-ATPase: Differential expression of α and β subunit in rat tissue membranes. Biochemistry. 1989;28:4531–4535. doi: 10.1021/bi00437a002. [DOI] [PubMed] [Google Scholar]
- Thompson, J., Higgins, D., and Gibson, T. 1994. http://www.ebi.ac.uk/clustalw
- Underhill AD, Canfield VA, Dahl JP, Gros P, Levenson R. The Na,K-ATPase α4 gene (Atp1a4) encodes a ouabain-resistant α subunit and is tightly linked to the α2 gene (Atp1a2) on mouse chromosome 1. Biochemistry. 1999;38:14746–14751. doi: 10.1021/bi9916168. [DOI] [PubMed] [Google Scholar]
- Watterson G. On the time for gene silencing at duplicate loci. Genetics. 1983;105:745–766. doi: 10.1093/genetics/105.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo AL, James PF, Lingrel JB. Characterization of the fourth α isoform of the Na,K-ATPase. J Membrane Biol. 1999;169:39–44. doi: 10.1007/pl00005899. [DOI] [PubMed] [Google Scholar]
- Woods IG, Kelly PD, Chu F, Ngo-Hazelett P, Yan Y-L, Huang H, Postlethwait JH, Talbot WS. A comparative map of the zebrafish genome. Genome Res. 2000;10:1903–1914. doi: 10.1101/gr.10.12.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]