Abstract
Recently we have explored the use of knockout/congenic mouse strains for isolating and mapping quantitative trait loci (QTLs). Because most knockout strains have been bred to be B6.129 congenic strains, they can be used to test for QTLs in the targeted chromosomal area as long as there is a genetic difference between B6 and 129. Thus, we have tested a number of knockout/congenic strains in a series of behavioral tests in which mouse performance has a significant genetic component. We have also developed a breeding scheme for distinguishing the effects of background flanking genes from the targeted ablation. In screening several knockout/congenics, we have found at least one that harbors a behavioral QTL in the 129 chromosomal segment. The position of this QTL was confirmed subsequently by several F1 crosses.
Mice with ablations of single genes have been extremely useful in analyzing gene function. They have revolutionized our ability to test new hypotheses about the nature of human diseases, especially ones that are caused by genetic mutations. However, because of the way that these strains are generally derived, they also can be used to map complex traits. In essence, many established knockout mice are also congenic strains that have retained a short piece of 129 genetic material surrounding the target ablated locus. Commonly, when knockout mice are made, embryonic stem cells derived from the 129 strain of mice are used. Once these stem cells become germline, the mice are routinely backcrossed to the B6 strain for a number of generations (usually 6–10). This backcrossing scheme yields a congenic strain in which most background genes are derived from B6 but the chromosomal segment flanking the target gene remains of 129 origin. There are >50 of these knockout/congenic strains commercially available. Thus, these knockout/congenic strains represent a potential resource for mapping genes surrounding the target (or knocked out) locus. To investigate this resource, we have used these strains to map genes affecting certain behaviors, including open field activity. We selected open field activity as it has successfully been used as a measure of exploratory behavior and emotionality in mice (Flint et al. 1995; Gershenfeld et al. 1997; Koyer et al. 2000). The positions of any potential QTLs were then confirmed by additional crosses.
RESULTS
We have tested a number of knockout/congenic mouse strains in hopes of detecting background flanking 129 genes that affect behavioral characteristics. In past studies, we have found that the 129S1/SvImJ (129) strain performs differently from C57BL/6J (B6) on a variety of behavioral tests (Bolivar et al. 2000a; Cook et al. 2001; Bolivar et al., in press). Others have found differences between 129 and B6 strains as well (Logue et al. 1997; Owen et al. 1997b; Balogh et al. 1999; Paulus et al. 1999; Tarantino et al. 2000). This evidence supports our search for 129-derived chromosomal regions that may influence behavioral phenotypes. Although the 129 source of embryonic stem cells in many of the knockouts may be from a different 129 substrain than the one used in our initial experiments, this problem can be resolved by follow-up breeding studies.
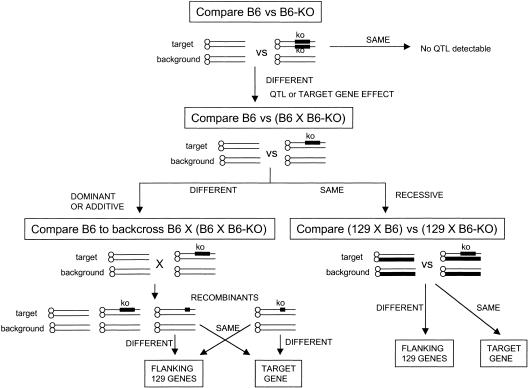
We developed a testing scheme to enable the separation of flanking and target gene effects on behavioral performance (Fig. 1). The first step in this scheme is determining if there is a difference between B6 and B6-KO (B6 with homozygous knockout region). If there is a difference, then B6 is compared to (B6 X B6-KO) F1. A difference at this stage would be indicative of dominance (or additive) inheritance of the trait. In that case, backcrossing should be used to separate the flanking region from the target gene. If there is no difference between B6 and (B6 X B6-KO) F1, then inheritance is recessive and a comparison between (129 X B6) versus (129 X B6-KO) will distinguish between target locus and flanking region effects.
Figure 1.
Strategy for determining whether a specific phenotype is the result of the ablated target gene or the flanking 129 genes. Comparisons are made to determine the cause of the genetic effect (target or flanking) as well as its mode of inheritance — dominant, additive or recessive. Lines represent chromosomes; target indicates the chromosome with the mapped target locus; background indicates the other 19 chromosomes. KO indicates that the chromosome contains the targeted ablation. Heavy lines indicate that the origin of the chromosomal segment is 129. Thin lines indicate B6 chromosomal segment origin. B6-KO indicates that a strain is homozygous for the targeted ablated or knocked out region. In a B6 congenic/knockout strain, after N9, >99.8% of the unlinked genes are of B6 origin. Arrows indicate the next step to be taken in scheme.
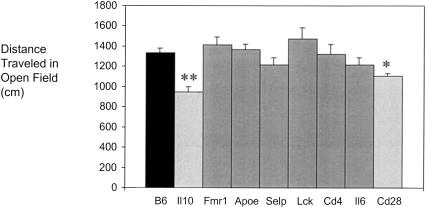
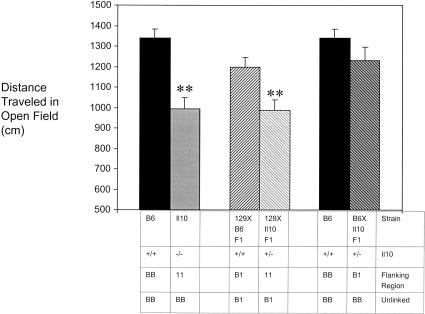
To test this scheme, we screened several knockout/congenic mouse strains in a battery of behavioral tests including open field, zero maze, and fear conditioning. Only open field activity is reported here. Several of these strains have targeted mutations in genes known to influence behavior (e.g., Apoe, Fmr1 and Il6) (for review, see Bolivar et al. 2000b); however, these latter strains did not behave differently from B6 in our open field test. We found that at least two knockout/congenic strains, B6-Il10−/− (N10F14) and B6-Cd28−/− (N12F8), behaved differently from B6 in the open field (Fig. 2). As B6-Il10−/− performance differed the most from B6, it was selected to test our scheme. Under this strategy, having established that B6 differs from B6-Il10−/−(Fig. 3), B6 mice were then compared to (B6 X B6-Il10−/−) mice. As these two groups did not differ from each other (Fig. 3), we hypothesized a recessive genetic effect (Fig. 1). To determine whether this recessive trait is due to flanking 129 genes or the ablated target gene, (129 X B6-Il10−/−) F1 mice were compared to (129 X B6) F1 mice. (129 X B6-Il10−/−) F1 and (129 X B6) F1 mice performed differently in the open field (Fig. 3), thus indicating flanking 129 genes and not Il10 are responsible. This conclusion is not surprising based on the known function of Il10 (Kuhn et al. 1993).
Figure 2.
Open field performance of inbred and knockout/congenic mouse strains. The knockout mouse strains tested included B6-Il10tm1Cgn (N10F14), B6-Fmr1tm1Cgr (N9), B6-Apoetm1Unc (N10F19), B6-Selptm1Bay (N10F14), B6-Lcktm1Mak (N12), B6-Cd4tm1Knw (N10F8), B6-Il6tm1Kopf (N11F7), and B6-Cd28tm1Mak (N12F8). Knockout mice are all on a B6 background and in at least the N9 generation of backcrossing. They are indicated by the name of the ablated target gene. Twelve male mice of each strain were tested. Results are shown + s.e.m. There was a significant effect of strain (F8,99 = 4.236, P<.001). Fisher's posthoc tests were then used to make pairwise comparisons. Single asterisks indicate P<0.05; double asterisks indicate P<0.01 in ANOVA analysis using STATVIEW.
Figure 3.
Open field performance of F1 and knockout/congenic mice. Twenty-four male mice of each strain were tested. Results are shown + s.e.m. There was a significant effect of strain (F4,115 = 7.733, P<.001). Fisher's posthoc tests were then used to make pairwise comparisons. Double asterisks indicate P<0.01 in ANOVA analysis using STATVIEW.
To explore the origin of the deficient behavior in the B6-Il10−/− mice, we performed further genotyping of this strain in the vicinity of the target gene, Il10, on Chromosome 1 (Table 1). As predicted, a considerable segment of the chromosome remains of 129 origin. This is expected based on the usual methods used to produce congenic strains (for review, see Flaherty 1981; Silver 1995). Furthermore, our results strongly indicate that the gene or genes controlling this open field activity must reside in this region and be polymorphic between 129 and B6. In a backcross of the B6-Il10−/− knockout/congenic to B6, open field activity segregated with markers adjacent to Il10 on Chromosome 1 (P < 0.01). This observation confirms that this trait is governed by a gene or genes linked to Il10 and not by an unlinked contaminant. These findings agree with several previous studies that map QTLs for emotionality to this region (Flint et al. 1995; Caldarone et al. 1997; Gershenfeld et al. 1997; Gershenfeld and Paul 1997; Owen et al. 1997a; Wehner et al. 1997; Talbot et al. 1999; Turri et al. 1999; Koyer et al. 2000). In a recent paper, Talbot et al. (1999) have narrowed the region for the open field behavior QTL on chromosome 1 obtained by Flint et al. (1995) to a 0.8 cM interval. This region overlaps with the 129 flanking region in the B6-Il10−/−knockout/congenic strain.
Table 1.
Genotypings of 129 and II10 Knockout on Chromosome 1
| Strain | cM position of Chromosome 1 markers | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 32.8 | 34.8 | 36.9 | 42.6 | 47.0 | 52.5 | 59.0 | 64.5 | 69.9 | 75.4 | 80.9 | |
| B6 | b | b | b | b | b | b | b | b | b | b | b |
| B6-II10− | b | b | 1 | 1 | 1 | 1 | 1 | 1 | 1 | b | b |
| 129 (six strains)a | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
All 129 substrains typed the same at all Chromosome 1 markers listed above indicating that this region is highly preserved among these substrains. The 129 strains tested were 129P1, 129P3, 129P4, 129S1, 129T2, and 129X1. Markers were (listed from left to right) D1Mit5, D1Mit303, D1Mit19, D1Mit181, D1Mit215, D1Mit440, D1Mit60, D1Mit308, D1Mit495, D1Mit102, and D1Mit501.
b, B5; 1, 129.
DISCUSSION
This scheme of testing knockout/congenic mice for behavioral traits has several distinct advantages over more commonly used breeding protocols. First, it yields a ready-made congenic strain for detecting a behavioral phenotype. Thus, mapping is precise and fine mapping can be performed readily. Second, the use of identical members of the knockout/congenic strain will allow repeat observations that will increase the power of the investigation. Third, problems related to the mixing of genetic backgrounds, normally occurring in a backcross or intercross QTL study, will be reduced since the strains will all be on the B6 background.
By testing a number of knockout/congenic strains of mice, we were able to locate a gene or genes that influence performance in an open field activity test. We confirmed the location of these genes in further breeding studies. This scheme of testing knockout/congenic strains for a number of quantitative traits could have much value in identifying weak genetic influences such as those seen in complex diseases. As long as there is a difference between 129 and B6, this technique can be used for mapping and obtaining a readymade congenic. The new Mouse Phenome Project (designed to characterize phenotypically eight strains of mice including B6 and 129 and sponsored by the National Institutes of Health and The Jackson Laboratory) should provide a larger number of complex trait differences between 129 and B6. In addition, an important side benefit of this effort would be to determine whether certain ablated genes have unknown pleiotropic effects that are not obvious from visual inspection. Moreover, with the rapid addition of new knockout mice to the already available repertoire, it may soon be possible to use these strains for fine mapping as well. If there are several strains with target genes in close vicinity to one another, overlapping 129 chromosomal segments may allow finer map discrimination.
Finally, the mapping of genes influencing complex traits to the 129 passenger chromosomal segment points to the necessity of confirming that the ablated target gene is actually responsible for the phenotype to be studied. Even with knockout strains that have been made congenic, there is still a large flanking region that is derived from the donor strain. The concept that flanking regions in congenic strains could be influencing a particular phenotype is not new and has been pointed out by some of the early workers in mouse genetics (Snell 1948; Green 1966; Boyse 1977). These passenger genes may be particularly problematic when considering complex traits such as behavior, as pointed out by several more recent reviews (Crusio 1996; Gerlai 1996; Wolfer and Lipp 2000). Thus, some results involving knockout strains may be misleading. Polymorphic differences between strains that are governed by genes in the vicinity of the targeted locus may often be responsible.
Because these knockout/congenics provide the researcher with a set of strains, which predominantly differ at a very short stretch of chromosome, they may become a very useful tool in identifying the causative gene(s) as well. With the anticipated completion of the sequencing of the mouse genome, it will be possible to enumerate all of the genes within this chromosomal segment. For a 5-cM stretch, the average number of genes is predicted to be 100 (Claverie 2001). Moreover, because both B6 and 129 genomes are being sequenced (B6 in public domain and 129 in Celera), we will also know all of the polymorphic differences between these two strains in this segment. This should aid greatly our ability to identify candidate genes and to make good hypotheses for future experiments.
METHODS
Animals
We either purchased mice from The Jackson Laboratory or bred them at the Wadsworth Center. We tested all mice between 8 and 16 weeks of age. The Institutional Care and Use Committee approved all procedures. The 129S1/SvImJ substrain (former name 129S3/SvImJ; renamed Feb. 2001) was used for all of our breeding studies.
Open Field
We tested all mice in a Digiscan 16-beam automated activity monitor (42cm X 42cm X 30cm, Accuscan) as previously described (Bolivar et al. 2000a). We used total distance traveled by the mouse during five minutes in a darkened and sound-attenuated chamber as our measure of activity.
Genotyping
We used MIT DNA markers to determine the extent of the 129 flanking region. We performed genotypings by gel electrophoresis and staining of the gels with ethidium bromide as previously described (Caldarone et al. 1997).
Acknowledgments
We thank Barbara Ashe and Kathleen Tartaglia for their expert technical assistance and Kevin Manley for his helpful discussions of the manuscript. This work was supported by a grant from the National Institute of Mental Health, NIH.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL flaherty@wadsworth.org; FAX (518) 474-3181.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.194001.
REFERENCES
- Balogh SA, McDowell CS, Stavnezer AJ, Denenberg VH. A behavioral and neuroanatomical assessment of an inbred substrain of 129 mice with behavioral comparisons to C57BL/6J mice. Brain Res. 1999;836:38–48. doi: 10.1016/s0006-8993(99)01586-3. [DOI] [PubMed] [Google Scholar]
- Bolivar VJ, Caldarone BJ, Reilly AA, Flaherty L. Habituation of activity in an open field: A survey of inbred strains and F1 hybrids. Behav Genet. 2000a;30:285–293. doi: 10.1023/a:1026545316455. [DOI] [PubMed] [Google Scholar]
- Bolivar V, Cook M, Flaherty L. List of transgenic and knockout mice: Behavioral profiles. Mamm Genome. 2000b;11:260–274. doi: 10.1007/s003350010051. [DOI] [PubMed] [Google Scholar]
- Bolivar, V.J., Pooler, O., and Flaherty, L. 2001. Inbred strain variation in contextual and cued fear conditioning behavior. Mamm. Genome (in press). [DOI] [PubMed]
- Boyse EA. The increasing value of congenic mice in biomedical research. Lab Anim Sci. 1977;27:771–781. [PubMed] [Google Scholar]
- Caldarone B, Saavedra C, Tartaglia K, Wehner JM, Dudek BC, Flaherty L. Quantitative trait loci analysis affecting contextual conditioning in mice. Nat Genet. 1997;17:335–337. doi: 10.1038/ng1197-335. [DOI] [PubMed] [Google Scholar]
- Claverie J-M. What if there are only 30,000 human genes? Science. 2001;291:1255–1257. doi: 10.1126/science.1058969. [DOI] [PubMed] [Google Scholar]
- Cook MN, Williams RW, Flaherty L. Anxiety-related behaviors in the elevated zero maze are affected by genetic factors and retinal degeneration. Behav Neurosci. 2001;115:468–476. [PubMed] [Google Scholar]
- Crusio WE. Gene-targeting studies: New methods, old problems. Trends Neurosci. 1996;19:186–187. doi: 10.1016/s0166-2236(96)20023-2. [DOI] [PubMed] [Google Scholar]
- Flaherty L. Congenic strains. In: Foster H, Small J, Fox J, editors. The mouse in biomedical research. New York: Academic Press; 1981. pp. 215–222. [Google Scholar]
- Flint J, Corley R, DeFries JC, Fulker DW, Gray JA, Miller S, Collins AC. A simple genetic basis for a complex psychological trait in laboratory mice. Science. 1995;269:1432–1435. doi: 10.1126/science.7660127. [DOI] [PubMed] [Google Scholar]
- Gerlai R. Gene-targeting studies of mammalian behavior: Is it the mutation or the background genotype? Trends Neurosci. 1996;19:177–181. doi: 10.1016/s0166-2236(96)20020-7. [DOI] [PubMed] [Google Scholar]
- Gershenfeld HK, Neumann PE, Mathis C, Crawley JN, Li X, Paul SM. Mapping quantitative trait loci for open-field behavior in mice. Behav Genet. 1997;27:201–210. doi: 10.1023/a:1025653812535. [DOI] [PubMed] [Google Scholar]
- Gershenfeld HK, Paul SM. Mapping quantitative trait loci for fear-like behaviors in mice. Genomics. 1997;46:1–8. doi: 10.1006/geno.1997.5002. [DOI] [PubMed] [Google Scholar]
- Green EL. Breeding systems. In: Green EL, editor. Biology of the laboratory mouse. New York: McGraw-Hill; 1966. pp. 11–22. [Google Scholar]
- Koyer J, Demarest K, McCaughran J, Jr, Cipp L, Hitzemann R. Identification and time dependent of quantitative trait loci for basal locomotor activity in the BXD recombinant inbred series and a B6D2F2 intercross. Behav Genet. 2000;30:159–170. doi: 10.1023/a:1001963906258. [DOI] [PubMed] [Google Scholar]
- Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- Logue SF, Owen EH, Rasmussen DL, Wehner JM. Assessment of locomotor activity, acoustic and tactile startle, and prepulse inhibition of startle in inbred mouse strains and F1 hybrids: Implications of genetic background for single gene and quantitative trait loci analyses. Neuroscience. 1997;80:1075–1086. doi: 10.1016/s0306-4522(97)00164-4. [DOI] [PubMed] [Google Scholar]
- Owen EH, Christensen SC, Paylor R, Wehner JM. Identification of quantitative trait loci involved in contextual and auditory-cued fear conditioning in BXD recombinant inbred strains. Behav Neurosci. 1997a;111:292–300. doi: 10.1037//0735-7044.111.2.292. [DOI] [PubMed] [Google Scholar]
- Owen EH, Logue SF, Rasmussen DL, Wehner JM. Assessment of learning by the Morris water task and fear conditioning in inbred mouse strains and F1 hybrids: Implications of genetic background for single gene mutations and quantitative trait loci analyses. Neuroscience. 1997b;80:1087–1099. doi: 10.1016/s0306-4522(97)00165-6. [DOI] [PubMed] [Google Scholar]
- Paulus MP, Dulawa SC, Ralph RJ, Geyer MA. Behavioral organization is independent of locomotor activity in 129 and C57 mouse strains. Brain Res. 1999;835:27–36. doi: 10.1016/s0006-8993(99)01137-3. [DOI] [PubMed] [Google Scholar]
- Silver LM. Mouse Genetics. New York: Oxford University Press; 1995. [Google Scholar]
- Snell GD. Methods for the study of histocompatibility genes. J Genet. 1948;49:87–108. doi: 10.1007/BF02986826. [DOI] [PubMed] [Google Scholar]
- Talbot CJ, Nicod A, Cherny SS, Fulker DW, Collins AC, Flint J. High-resolution mapping of quantitative trait loci in outbred mice. Nat Genet. 1999;21:305–308. doi: 10.1038/6825. [DOI] [PubMed] [Google Scholar]
- Tarantino LM, Gould TJ, Druhan JP, Bucan M. Behavior and mutagenesis screens: The importance of baseline analysis of inbred strains. Mamm Genome. 2000;11:555–564. doi: 10.1007/s003350010107. [DOI] [PubMed] [Google Scholar]
- Turri MG, Talbot CJ, Radcliffe RA, Wehner JM, Flint J. High-resolution mapping of quantitative trait loci for emotionality in selected strains of mice. Mamm Genome. 1999;10:1098–1101. doi: 10.1007/s003359901169. [DOI] [PubMed] [Google Scholar]
- Wehner JM, Radcliffe RA, Rosmann ST, Christensen SC, Rasmussen DL, Fulker DW, Wiles M. Quantitative trait locus analysis of contextual fear conditioning in mice. Nat Genet. 1997;17:331–334. doi: 10.1038/ng1197-331. [DOI] [PubMed] [Google Scholar]
- Wolfer DP, Lipp HP. Dissecting the behaviour of transgenic mice: Is it the mutation, the genetic background, or the environment? Exp Physiol. 2000;85:627–634. [PubMed] [Google Scholar]