SUMMARY
MicroRNAs are emerging as important regulators of diverse biological processes and pathologies in animals and plants. While hundreds of human miRNAs are known, only a few have known functions. Here we predict human microRNA functions by using a new method that systematically assesses the statistical enrichment of several miRNA targeting signatures in annotated gene sets such as signaling networks and protein complexes. Some of our top predictions are supported by published experiments, yet many are entirely new or provide mechanistic insights to known phenotypes. Our results indicate that coordinate miRNA targeting of closely connected genes is prevalent across pathways. We use the same method to infer which microRNAs regulate similar targets and provide the first genome-wide evidence of pervasive co-targeting, where a handful of “hub” microRNAs are involved in a majority of co-targeting relationships. Our method and analyses pave the way to systematic discovery of microRNA functions.
INTRODUCTION
MicroRNAs (miRNAs) regulate diverse biological processes in animals and plants (Bushati and Cohen, 2007) and are among the most abundant regulatory factors in the human genome, comprising 3–5% of known human genes (Griffiths-Jones et al., 2008). miRNAs recognize target mRNAs by imperfect base pairing to sites in the 3′ untranslated region (3′ UTR), usually with perfect pairing of the miRNA seed region (nucleotides 2–8), ultimately leading to translational repression and/or mRNA degradation (Bushati and Cohen, 2007). Thousands of human genes are predicted to be targeted by miRNAs (Rajewsky, 2006), suggesting that miRNAs play a pervasive role in the regulation of gene expression.
Although hundreds of human miRNAs have been identified and new ones are continually being discovered (Griffiths-Jones et al., 2008), the function of most miRNAs remains unknown. Increasingly, miRNA expression changes are being linked to phenotypes, but the mechanistic role of the miRNA in the underlying biological network is often unclear. Given that many human miRNAs can target up to thousands of genes, how often do miRNAs target a set of related genes to regulate a specific pathway or process? While recent studies show that a few miRNAs have pathway-specific functions (Xiao and Rajewsky, 2009), earlier work suggests that miRNAs primarily serve to fine-tune and confer robustness upon the expression of many genes (Bartel and Chen, 2004; Farh et al., 2005; Stark et al., 2005).
The prevalence of multiple miRNAs targeting the same gene (“co-targeting”) is also unclear. While many genes contain putative binding sites for multiple miRNAs (Krek et al., 2005; Stark et al., 2005), many putative sites may not be functional in vivo. More specifically, the combinations of miRNAs that function together by regulating common targets are unknown. Knowledge of such co-targeting relationships would also enable one to infer a miRNA’s function from the function of its co-targeting miRNAs.
Typically miRNA function is predicted by assessing whether the predicted targets of a given miRNA are enriched for particular functional annotations. Such an approach has several limitations: (1) target prediction is imperfect and can lead to spurious targets (Rajewsky, 2006); (2) having a subset of one’s favorite pathway genes in the putative target set does not necessarily mean that the miRNA functions in the pathway; (3) predicted target sets are often so large (hundreds to thousands of genes) and have such heterogeneous functional annotations that standard algorithms are not sufficiently sensitive to make high-confidence predictions. Rather than progressing from a miRNA to a potentially spurious target set that may or may not have enriched function, here we introduce a computational method called mirBridge, which starts with a gene set of known function, then assesses whether functional sites for a given miRNA are enriched in the gene set compared to random gene sets with similar properties.
We apply mirBridge to a variety of annotated gene sets for signaling pathways, diseases, drug treatments, and protein complexes. We also use mirBridge to infer miRNA pairs that tend to function together by regulating common targets and use the results to assemble a miRNA-miRNA co-targeting network. Together, our analyses provide: (1) hundreds of miRNA function predictions, many of which are supported by published experiments; (2) genome-wide evidence that many miRNAs coordinately regulate multiple components of pathways or protein complexes; and (3) evidence that miRNA co-targeting is prevalent, with a small number of “hub” miRNA families involved in a large fraction of the co-targeting interactions. Both the mirBridge method and the predictions it has generated can serve as important resources for the future experimental dissection of miRNA functions.
RESULTS
mirBridge: linking miRNAs to gene sets
Many gene sets contain tens to hundreds of putative targets for any particular miRNA. However, for a variety of reasons (e.g. mRNA secondary structure occludes binding, or the miRNA and the target are not expressed together) many target sites are not functional in vivo. The goal of mirBridge is to infer whether an unusually large proportion and number of putative target sites for a miRNA (m) in a given gene set (G) are likely to be functional in vivo. Towards this end, mirBridge computes a score by combining the results of three statistical tests that evaluate different aspects of likely-functional target-site enrichment in G. It is essential that the enrichment of sites in G be compared to enrichment in appropriate control gene sets. Below we describe the individual tests and the method for constructing the control gene sets (see Supplemental Experimental Procedures for details).
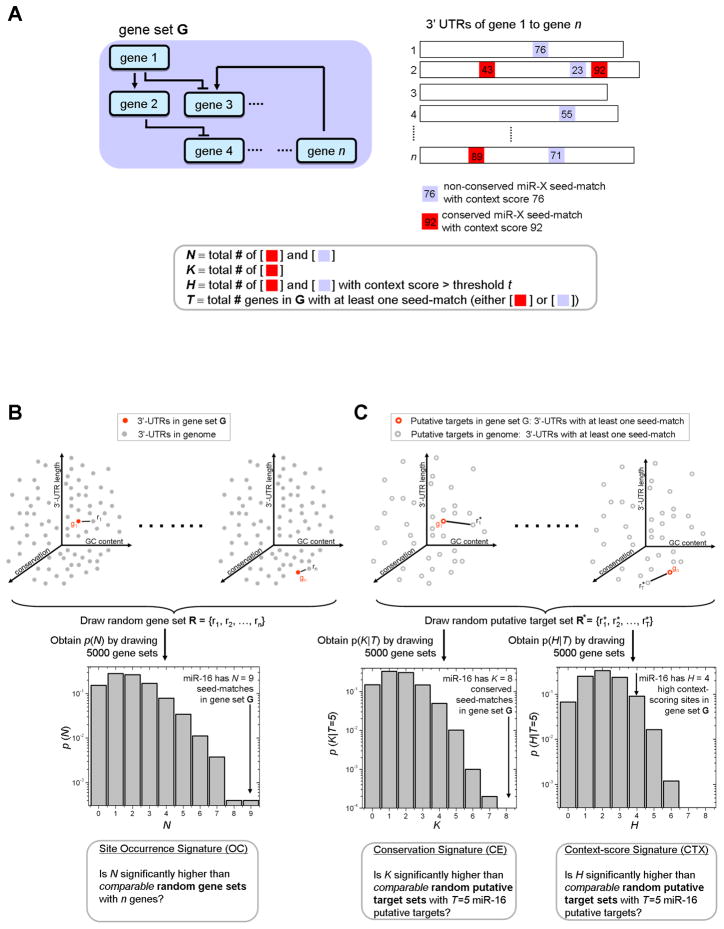
The following definitions are essential to the methodology of mirBridge. First, any gene with one or more seed-matched site for m in its 3′ UTR is deemed a “putative target.” Second, seed-matched sites can be classified into two categories (Figure 1A): “conserved sites” (CS) are sites that are conserved across mammalian genomes; “high-context scoring sites” (HCS) are sites with a context score above a predefined threshold. The context score reflects the likelihood of a seed-matched site to confer repression based on several features, including the distance of the site from the stop codon, accessibility of the site based on secondary structure, and the extent of base pairing beyond the seed (Grimson et al., 2007).
Figure 1. mirBridge overview.
(A) The input to mirBridge is a set of genes. Red and blue squares denote conserved and non-conserved seed-matched sites in the 3′ UTR respectively. The number inside the squares denotes the context score. For each miRNA target sequence of interest, mirBridge computes the N, K, H, and T as illustrated. (B) The procedure for evaluating whether N is significantly higher than that of comparable random gene sets (the OC test). To obtain the null distribution for N, random gene sets with similar 3′ UTR properties were constructed by replacing each gene in the original set (g1, … gn; solid red dots) by a randomly drawn gene (r1, r2, … rn). The probability that ri is drawn to replace gi is inversely proportional to its distance to gi in the 3-D space defined by 3′ UTR length, GC content and general conservation level. The histogram depicts the null distribution of N for miR-16 in the cell-cycle gene set. (C) The procedure for evaluating whether K and H are significantly higher than those of random gene sets containing T putative targets with similar 3′ UTR properties as the putative targets in G (the CE and CTX tests, respectively). The same gene sampling procedure from (B) is used except that only the putative targets in G (empty red dots) are replaced by random putative targets (empty gray dots) so that T is identical across G and the random gene sets. The histograms depict the null distributions of K and H, respectively, for random gene sets with T=5 putative targets for the miR-16 and the cell-cycle gene set.
The first test used by mirBridge, called “conservation enrichment signature” (CE), infers whether the number of CS in G is significantly higher than that of random gene sets containing the same number of putative targets as G. This test is similar to evaluating whether the sites have evolved at a slower rate compared to random putative target sets, but is fundamentally different from prior tests that utilize sequence conservation (Lewis et al., 2005; Stark et al., 2005) (see Supplemental Experimental Procedures). The second test, called “context-score signature” (CTX), evaluates whether the number of HCS is significantly higher than that of random gene sets containing the same number of putative targets as G. The CTX test is designed to detect enrichment of sites in G that are likely functional but not necessarily conserved. The third test, called “site occurrence signature” (OC), evaluates whether the number of putative target sites in G is unusually high compared to random gene sets containing the same number of genes. While target site abundance alone is not necessarily indicative of functional targeting by m, functional targeting enrichment becomes a likely scenario even when G tests as moderately significant for the CE and/or CTX tests. Note that both CE and CTX are based on comparison with random gene sets with the same number of putative targets to detect enrichment in the proportion rather than the number of CS or HCS. This ensures that the comparisons are valid, as gene sets with more putative targets tend to have more CS or HCS. Since true positives are more likely than false positives to test as simultaneously significant across the tests, we combine the three tests and form a composite score (“OC-CE-CTX”) to increase sensitivity without sacrificing specificity.
We developed a nearest-neighbor gene sampling algorithm, motivated by the principle of kernel-based density estimators (Wegman, 1972), to generate random gene sets that are similar to the input gene set with respect to general conservation level, 3′ UTR length, and GC content, which primarily bias the CE, OC, and CTX tests, respectively. Simultaneous adjustment is particularly important because these factors are correlated with each other across genes. Specifically, for the OC test, comparable random gene sets are generated by replacing each member of G with a randomly drawn gene that has similar GC content, 3′ UTR length, and general conservation level (Figure 1B). To ensure that the number of putative targets in the random gene sets is the same as that in G for the CE and CTX tests, the same nearest-neighbor procedure is used but only putative targets in G are replaced by random putative targets (Figure 1C).
Finally, to obtain the OC-CE-CTX p value, the p values of the individual tests are combined using a customized version of the inverse-normal method that corrects for dependencies among tests (Joachim, 1999). When multiple gene sets and/or miRNAs are tested simultaneously, multiple hypothesis testing is corrected by computing the false discovery rate (FDR) using the q-value method (Storey and Tibshirani, 2003). “FDR” and “q value” are used interchangeably below.
Besides 3′ UTR length, GC content, and general conservation, other less apparent factors could bias mirBridge results, but their effects are likely small (see Supplemental Experimental Procedures). The statistical model in mirBridge was also designed to incorporate additional factors if needed – in principle any number of factors can be accounted for by our nearest-neighbor sampling procedure.
mirBridge is fundamentally different from testing whether the number of predicted miRNA targets in a gene set is significantly higher than expected using the Fisher’s Exact Test (FET), a standard way to assess the significance of gene set overlaps. First, mirBridge takes gene set properties into account; second, it combines different and important biological characteristics of target sites; and finally, it uses metrics (CE and CTX) that focus on the proportion of likely-functional target sites instead of the number of predicted target overlaps. In fact, mirBridge has superior sensitivity and specificity compared to FET as shown in the applications below.
Inferring human miRNA functions
To link human miRNA families (miRNAs with a shared seed sequence) to functions, we applied mirBridge to gene sets from (1) canonical signaling pathways from MSigDB (Subramanian et al., 2005); (2) KEGG (Kanehisa and Goto, 2000); (3) human protein complexes from the CORUM database (Ruepp et al., 2008); (4) gene co-expression modules (Segal et al., 2004); (5) Gene Ontology (GO) Biological Process; (6) GO Component; and (7) GO Function (Ashburner et al., 2000). At a FDR cutoff of 0.2, mirBridge predicts 185, 128, 1198, 456, 432, 71, and 175 distinct miRNA-function associations, respectively (Tables S1a–g). Most predictions implicate pathways or protein complexes with multiple putative targets for the miRNA, whereas some have only one (or very few) putative targets containing multiple high-quality sites (e.g. miR-33 and statin pathway). The latter fits the paradigm implied in some recent papers where a miRNA phenotype seems to be accounted for by one (or just a few) targets: “miR-X regulates process Y by targeting gene Z.” However, the prevalence of coordinate targeting of multiple related genes suggests that most miRNAs exert their phenotypic effects by targeting multiple network components.
To facilitate a succinct discussion of such a large set of predictions, Table 1 shows a selection of predictions that either already have support from the literature or wherein the predicted pathway (1) has known activity in the tissue where the miRNA is known to be expressed; or (2) represents core cellular processes (e.g. “apoptosis”) and has a large number of putative targets for the miRNA. We also favor predictions that reoccur in closely related or synonymous gene sets, e.g. “cell cycle” and “G1 to S transition.”
Table 1A.
Selected mirBridge predictions with published evidence (see Table S1 for more details). “High-quality putative targets” are ones with either a conserved or high context-scoring site (see Experimental Procedures).
| miRNA | Function | q value | High-quality putative targets | Evidence |
|---|---|---|---|---|
| 146 | IL1 receptor, NFKB, Toll Like Receptor signaling | 0 | TRAF6, IRAK1, TLR4 | (Jones et al., 2009; Taganov et al., 2006) |
| 15/16/195/424/497 | Cell cycle; G1 to S | 0 | CCNE1, CCND1, CCND3, CCND2, CDC25A | (Linsley et al., 2007; Liu et al., 2008) |
| CCNE1, WEE1, E2F3, CCND1, CCND3, CCND2, CDC25A | ||||
| 29 | Collagen | 0 | COL4A1, COL4A5, COL4A4, COL4A6, COL4A2, COL4A3, FGA | (Li et al., 2009; van Rooij et al., 2008) |
| 7 | ErbB signaling; glioma | 0 | RAF1, EGFR, FRAP1, MAPK1, PIK3CD, PAK1, PIK3R3, RPS6KB1, CAMK2D, PAK2, TGFA, PTK2, CBL, ERBB4, CRKL, MAPK3 | (Kefas et al., 2008; Webster et al., 2009) |
| RAF1, RB1, CALM3, EGFR, FRAP1, MAPK1, PIK3CD, PIK3R3, CAMK2D, TGFA, IGF1R, MAPK3 | ||||
| 7 | insulin signaling | 0.000208 | IRS1, IRS2, RAF1, CALM3, FRAP1, MAPK1, PHKA2, PIK3CD, PIK3R3, RPS6KB1, MKNK1, CBL, FLOT2, PRKAG2, CRKL, SOCS2, PPARGC1A, MAPK3 | (Bravo-Egana et al., 2008; Correa-Medina et al., 2009; Joglekar et al., 2009) |
| 15/16/195/424/497 | Wnt pathway | 0.0356 | FZD10, DVL1, CCND1, PAFAH1B1, PPP2R5C, FZD6, CCND3, DVL3, MAPK9, PRKCI, CCND2, WNT7A, FOSL1, WNT2B | (Bonci et al., 2008) |
| 103/107 | TNF pathway | 0.0522 | HRB, CASP3, TNF, MAP3K7, TNFAIP3, NR2C2 | (Xie et al., 2009) |
| 122 | NO1 pathway | 0.0546 | SLC7A1, RYR2, CALM3, TNNI1 | (Yang and Kaye, 2009) |
| 15/16/195/424/497 | prostate cancer | 0.07345 | CCNE1, AKT3, PIK3R1, MAP2K1, IKBKB, E2F3, RAF1, CCND1, PIK3R3, CHUK, CCNE1, FGFR1, FGFR2, GRB2, FOXO1, IGF1R, BCL2, CREB5, MAPK3 | (Bonci et al., 2008) |
| 135 | TGF beta signaling | 0.07389 | SMAD5, ROCK2, SMURF2, THBS2, ROCK1, SMAD2, FKBP1A, NODAL, PPP2R1B, INHBA, TGFBR1, ACVR1B, BMPR1A, SP1, RPS6KB1, BMPR2, RUNX2, RBX1, SKI | (Li et al., 2008) |
| 34a/449 | Notch signaling | 0.07389 | NOTCH1, DLL1, NUMBL, HDAC1, JAG1, NOTCH2, NOTCH3 | (Ji et al., 2008; Ji et al., 2009) |
| 21 | cytokine-cytokine receptor interaction | 0.0865 | IL12A, CCL20, CCL1, FASLG, TNFRSF11B, TNFRSF10B, IL1B, CCR7, LEPR, BMPR2, XCL1, LIFR, CNTFR, TGFBR2, CXCL5, ACVR2A | (Lu et al., 2009) |
| 1/206 | PIP3 signaling in cardiac myocytes | 0.0977 | IGF1, CREB5, YWHAZ, MET, CDC42, YWHAQ, PTPN1, PREX1 | (Care et al., 2007; Sayed et al., 2007) |
| 17-5p/20/93.mr/106/519.d | cell cycle; G1 to S | 0.122 | CCNE1, CCND1, CDC25A, SMAD3 | (Cloonan et al., 2008; Pickering et al., 2009) |
| CCNG2, RBL1, RPA2, WEE1, E2F1, CCND2, CDKN1A, MCM3, CDC25A, RB1, E2F3, CCND1, CCNE2 | ||||
| 221/222 | breast cancer estrogen signaling | 0.1432 | KIT, CDKN1B, NFYB, SERPINB5, ESR1, THBS1, THBS2 | (Miller et al., 2008; Zhao et al., 2008) |
| 34/449 | BAD pathway (apoptosis) | 0.1499 | KIT, KITLG, BCL2, IGF1, PRKACB | (Chang et al., 2007; Cloonan et al., 2008; He et al., 2007) |
| let-7/98 | breast cancer estrogen signaling | 0.1595 | CYP19A1, NGFB, CDH1, TP53, CDKN1A, FASLG, PPIA, THBS1, DLC1, PAPPA, IL6, DLC1, DST, PAPPA, CND1 | (Schultz et al., 2007) |
| let-7/98 | G1 to S | 0.1871 | E2F6, TP53, PRIM2, CDKN1A, CDC25A, RB1, CCND2, CCND1, CCND2, CCND2 | (Schultz et al., 2008; Yu et al., 2007) |
mirBridge is sensitive to biological signals and can independently uncover known miRNA functions
Although mirBridge is not trained on any dataset of known miRNA functions, several of the top hits already have experimental support in the literature (Table 1A), such as the association of miR-16 with the cell cycle, Wnt signaling and prostate cancer (Calin et al., 2005; Linsley et al., 2007) (Figure S1A). This is also an example where mirBridge links a disease and the pathways underlying its pathology: miR-16 has been shown to work through the Wnt pathway to function as a tumor suppressor in prostate cancer (Bonci et al., 2008). Analogously, miR-7 hits the ErbB pathway in glioblastoma (Kefas et al., 2008; Webster et al., 2009); miR-221/222 hits the estrogen signaling pathway in breast cancer (Miller et al., 2008; Zhao et al., 2008); and let-7 hits the G1-S cell-cycle pathway in breast cancer (Schultz et al., 2008; Yu et al., 2007). mirBridge can also implicate a pathway of interest given the tissue specificity of a miRNA: miR-7 is predicted to regulate the insulin receptor pathway and is known to be highly expressed in insulin-producing cells of pancreatic islets (Bravo-Egana et al., 2008; Correa-Medina et al., 2009; Joglekar et al., 2009). mirBridge also independently uncovered feedback loops: miR-146 is predicted to target several upstream signaling genes in the NF-kB pathway while its transcription is known to be activated by NF-kB (Taganov et al., 2006) (Figure S1B). Another notable prediction supported by the literature is miR-34 targeting BCL2 and several additional anti-apoptotic genes in the BAD pathway (Chang et al., 2007; Cloonan et al., 2008; He et al., 2007). This prediction provides an attractive hypothesis for how miR-34 upregulation could lead to apoptosis. In sum, these results are reassuring and indicate that mirBridge can capture biologically relevant signals.
mirBridge is significantly more sensitive than the standard approach of evaluating gene set overlaps using FET. For instance, when FET is applied to the canonical pathway gene sets, only five predictions can be made at the 0.2 FDR cutoff (Table S1h); all five have FDRs above 0.18 and only one has support from the literature (miR-16 and the Gleevec pathway given that miR-16 is associated with leukemia). Furthermore, none of the top mirBridge predictions supported by published experiments were uncovered. For example, for miR-16, none of the cell-cycle related pathways are ranked near the top even if we ignore the statistical significance and order the pathways within each miRNA family by their q values (the top cell-cycle related entry has rank 54, q=0.55). These results suggest that mirBridge can better uncover biologically relevant signals than FET.
It is important to note that the comprehensiveness of our predictions is dependent on the gene sets used. Some known miRNA functions are not in our predicted list because the appropriate gene set(s) were not included in the analysis. For example, miR-200 is known to function in the epithelial-mesenchymal transition (Burk et al., 2008; Gregory et al., 2008a; Korpal et al., 2008a; Park et al., 2008), but none of the gene sets used in our analysis captures this process. However, when mirBridge is applied to genes whose function annotation in the GeneCards database includes “epithelial-mesenchymal transition,” miR-141/200a has the lowest q value among all miRNAs (q=0.08).
To further assess the ability of mirBridge to predict known miRNA functions independently, we compiled eight additional miRNA phenotypes from the literature and applied mirBridge to seemingly relevant gene sets from KEGG or GeneCards (Table S2). Of nine phenotypes, four miRNA-gene set p values are significant and two are marginally significant (Table 2). In a multiple hypothesis testing context where all miRNAs are tested simultaneously for the phenotype gene set, however, only two would have been predicted at a FDR cutoff of 0.2 even though the desired miRNA ranks at or near the top for all four of the significant cases. This suggests that for these specific gene sets, mirBridge is sensitive to the relevant biological signals but lacks sufficient statistical power after multiple-testing correction. It follows that the hundreds of low-FDR predictions that are made by mirBridge are compelling candidates for experimental follow-up given that these emerged in the simultaneous testing of thousands of miRNA-gene set combinations. We expect the statistical power of mirBridge to continue to improve as additional genomes and knowledge of miRNA-target interactions become available.
Table 2.
Testing mirBridge on several known phenotypes compiled from the literature. The q values were computed based on simultaneous testing across miRNA seeds for the gene set. Black: significant; blue: marginally significant; gray: not significant. See also Table S2.
| miRNA | Known function | p | q | rank (out of 143 seed-matched motifs) | References |
|---|---|---|---|---|---|
| 141/200a | epithelial-mesenchymal transition | 0.0018 | 0.08 | 1 | (Burk et al., 2008; Gregory et al., 2008b; Korpal et al., 2008b; Park et al., 2008) |
| 21 | apoptosis | 0.006 | 0.39 | 1 | (Chan et al., 2005) |
| 155 | B cell receptor signaling | 0.007 | 0.29 | 5 | (Thai et al., 2007) |
| 181 | T cell receptor signaling | 0.008 | 0.07 | 5 | (Li et al., 2007) |
| 34 | P53 pathway | 0.04 | 0.32 | 14 | (Chang et al., 2007; He et al., 2007; Raver-Shapira et al., 2007; Tarasov et al., 2007) |
| 223 | granulocyte differentiation | 0.07 | 0.62 | 15 | (Johnnidis et al., 2008) |
| let-7 | non-small cell lung cancer | 0.55 | 0.63 | 68 | (Esquela-Kerscher et al., 2008; Johnson et al., 2007; Kumar et al., 2008) |
| 122 | cholesterol biosynthesis | 0.83 | 0.99 | 103 | (Krutzfeldt et al., 2005) |
| 133 | cardiac hypertrophy | 0.94 | 0.86 | 134 | (Care et al., 2007) |
We also sought to understand cases where mirBridge failed to predict the correct functions. Closer examination of the three failed cases in Table 2 suggests that for let-7 and miR-133 the gene sets used do not capture the biology relevant to the miRNA targeting. The cell cycle may be a key pathway through which let-7 exerts its effect on lung cancer (Esquela-Kerscher et al., 2008; Kumar et al., 2008; Schultz et al., 2008), but the non-small cell lung cancer gene set lacks most cell cycle genes and other postulated targets such as HMGA2 and MYC (let-7 does hit the G1-S cell-cycle transition pathway; Table 1A). Similarly for miR-133 and cardiac hypertrophy: two out of the three known targets relevant to the phenotype are not in the GeneCards set (CDC42 and WHSC2; (Care et al., 2007)). Finally, for miR-122, it turns out that inhibition of miR-122 by antagomir treatment tends to downregulate, rather than upregulate, cholesterol biosynthetic genes (Krutzfeldt et al., 2005), suggesting that the effect of miR-122 on cholesterol biosynthetic genes is indirect. Thus the insignificant mirBridge p value for miR-122 and cholesterol biosynthesis genes is not surprising.
mirBridge provides many new miRNA function predictions
The majority of mirBridge predictions are not yet directly supported by existing experiments (Tables 1B and S1–4). Some pathways predicted in common for multiple miRNAs seem particularly compelling because the miRNAs are known to be co-regulated. For example, the apoptosis pathway is predicted for miR-23 and -24, which are different in sequence but are co-expressed from the same cluster (Chhabra et al., 2009). Some predictions seem reasonable based on the function of the miRNA host gene. For example, the statin/cholesterol homeostasis pathway is linked to miR-33, which is embedded in an intron of a transcription factor that regulates cholesterol synthesis and uptake (Figure S1C). Other predictions seem plausible based on known miRNA functions with similar developmental placement and timing. For example, axon guidance pathways are predicted for miR-124, which has already been shown to positively regulate neurogenesis (Cheng et al., 2009; Visvanathan et al., 2007). Consistently, miR-124 was linked to the SNARE protein complex as it putatively targets VAMP3, a component of SNARE, via three conserved and high context-scoring sites; VAMP3 is known to function in the docking and fusion of synaptic vesicles with the presynaptic membrane (Sudhof, 2004).
Table 1B.
Selected new mirBridge miRNA function predictions (see Table S1 for details). Same format as Table 1A
| miRNA | Function | q value | High-quality putative targets |
|---|---|---|---|
| 33 | statin pathway | 0.00155 | ABCA1, HMGCR |
| 203 | G alpha i pathway | 0.00532 | MAPK1, JUN, PCLD, SRC, MYEF2, F2RL2, PLD2, EPHB2 |
| 23 | apoptosis | 0.00801 | CHUK, APAF1, CASP7, CASP3, BCL2, BIRC4, IRF1, BNIP3L, LMNB1 |
| 205 | tight junction | 0.01195 | PRKCE, CLDN11, EPB41, CNKSR3, INADL, YES1, VAPA, MAGI2, PARD6G, CLDN8, PTEN, PRKCH, MLLT4, ACTB, PRKCA, PARD6B |
| 187 | antigen processing and presentation | 0.02192 | KIR2DL2, KIR2DL1, KIR2DS2, KIR2DS4, KIR2DS5, KIR2DL4, KIR2DL3, IFNA2, KIR2DL5A |
| 219 | nuclear receptors | 0.02806 | THRB, NR2C2, NR1I2, NR5A2, NR3C1, NR2C2, ESR1 |
| 17-5p/20/93.mr/106/519.d | JNK MAPK pathway | 0.0377 | MAP3K2, MAP3K5, MAP3K9, GAB1, MAP3K12, NR2C2, ZAK, DUSP8, MAPK9, DUSP10, MAP3K3, MAP3K11 |
| 124.2/506 | axon guidance | 0.04983 | CHP, ROCK1, NFATC1, SEMA6D, ITGB1, GNAI1, NRAS, GNAI3, SEMA6A, NRP1, NFAT5, EPHB4, PLXNA3, EPHA3, EPHA2, SEMA5A, ROCK2, SRGAP3, EFNB3, EFNB1, NCK2, GNAI2, SEMA6C, EFNB2 |
| 34a/449 | glycosphingolipid biosynthesis | 0.05144 | FUT1, FUT5, FUT9, GCNT2, B4GALT2 |
| 128 | GnRH signaling | 0.05144 | PRKY, PRKX, MAPK14, MAP2K7, PLCB1, MAP2K4, ADCY8, ADCY2, GRB2, HBEGF, EGFR, CDC42, ADCY6 |
| 24 | cytokine-cytokine receptor interaction | 0.05203 | IFNG, EDA2R, TNFRSF19, CCR4, FASLG, IL10RB, IL1A, CCR1, PDGFRA, PDGFRB, EDA, PDGFC, TNFSF9, IL2RA, IL21R, CX3CR1, IL8RB, EDAR, CCL18, TNFRSF1A, IL1R1, IL8RA, IL29, IL2RB, ACVR1B, FLT1, IL22RA2, IL19, TNF, CSF1R, CNTFR, CLCF1, |
| 33 | PGC-1α pathway | 0.0541 | YWHAH, CAMK4, PPARA, MEF2C, CAMK2G, PPP3CB, CAMK2G, PPARGC1A |
| 375 | purine metabolism | 0.0544 | PDE4A, PDE8A, PDE5A, PDE7B, ADCY9, PDE4D, PDE10A, POLR3G, PDE4D, POLR2, AADCY6, PDE11A |
| 141/200a | EGF/PDGF pathway | 0.0637 | GRB2, MAP2K4, STAT5A, EGFR, PRKCB1, CSNK2A1, JUN, TAL1 |
| 142-5p | ubiquitin mediated proteolysis | 0.07389 | VHL, UBE2D1, SMURF1, CUL2, UBE2A, WWP1, CUL3, UBE2B, CDC23, UBE2E3 |
| 101 | ubiquitin mediated proteolysis | 0.07681 | UBE2D1, UBE2D2, UBE2A, VHL, UBE2D3, UBE2G1, FBXW11, FBXW7, CUL3 |
| 142-3p | regulation of actin cytoskeleton | 0.07816 | ITGAV, APC, MYLK, RAC1, WASL, MYH10, ROCK2, ITGB8, CRK, CFL2, FGF23, MYH9 |
| 19 | Ca signaling | 0.07827 | EDNRB, ADRB1, GRM1, CALM1, CACNA1C, GRIN2A, CHP, SLC25A6, SLC8A1, ADCY7, ITPR1, PDE1C, ADCY1, ATP2B4, ADCY9, PRKACB, PLCB1, SPHK2, ERBB4, ITPKB, PTK2B |
| 24 | apoptosis | 0.1148 | BNIP3L, BCL2L11, BIRC4, FASLG, NFKBIE, HELLS, RIPK1, TRAF1, CASP10, TNFRSF1A, TNF, IRF1, IRF5 |
| 135 | integrin pathway | 0.122 | ROCK2, ITGA1, ITGA2, ARHGEF7, PTK2, ARHGEF6, ROCK1, AKT3, PLCG1, PAK7, ANGPTL2, RHO |
| 93.HD/291-3P/294/295/302/372/373/520 | nuclear receptors | 0.1342 | VDR, NR2C2, PPARA, ESR1, NR4A2, NR2E1, NPM1, NR2F2 |
| 27 | statin pathway | 0.1396 | ABCA1, LDLR, LPL, HMGCR |
| 33 | cell cycle | 0.1555 | CDK6, CCND2, CDC25A, RB1 |
| 383 | O-glycan biosynthesis | 0.1658 | GALNT13, GALNT11, GCNT4, GALNT1, GALNT7 |
| 148/152 | inositol phosphate metabolism | 0.1677 | SYNJ1, PTEN, PI4KA, PIK3CA, ITPKB, PLCB1, ITPK1 |
| 25/32/92/363/367 | phosphatidylinositol signaling | 0.1916 | SYNJ1, PTEN, ITPR1, BMPR2, PIP5K3, PIP5K1C, PIK3R1, PIK3R3, RPS6KA4, PRKAR2B, PCTK1, PRKCE, PIP4K2C, RPS6KB1, CALM3 |
| 30-3p | ubiquitin mediated proteolysis | 0.1928 | UBE2J1, UBE2K, UBE2G1, UBE2D1, UBE2D3 |
| 153 | insulin receptor signaling | 0.1934 | GRB2, PIK3R1, RPS6KA3, RPS6KB1, SORBS1, CAP1, IRS2, FOXO1, AKT3 |
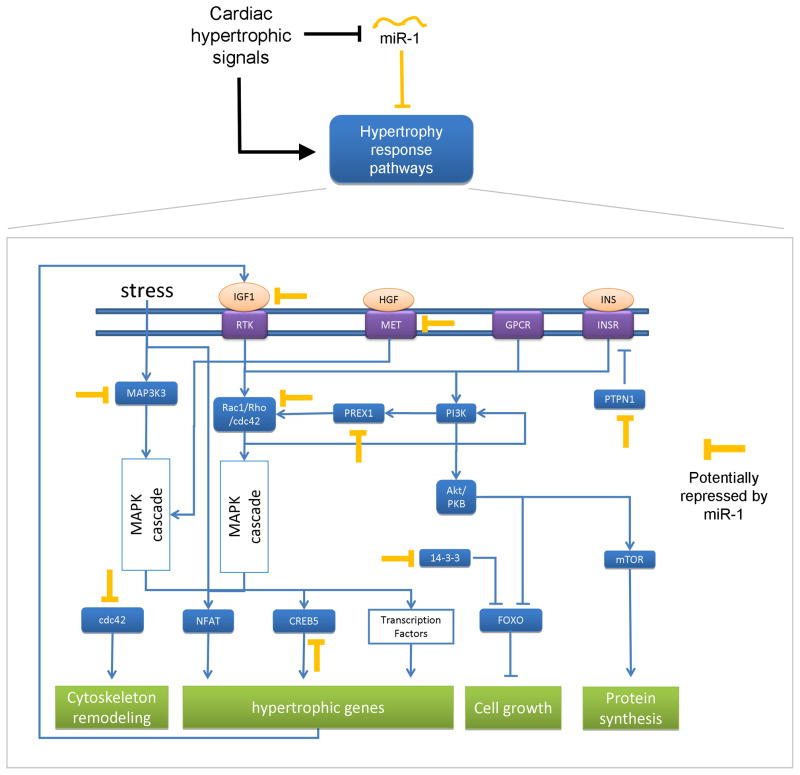
mirBridge predictions can also provide mechanistic interpretations of published experiments. For example, it is known that activation of PIP3 signaling leads to the hypertrophic response in cardiac myocytes and that miR-1 expression is down-regulated upon hypertrophic stress (Care et al., 2007; Heineke and Molkentin, 2006; Sayed et al., 2007). mirBridge links miR-1 to the PIP3 pathway, and the putative miR-1 targets in the pathway are all pro-hypertrophic except PTPN1 (Table 1A), suggesting that the down-regulation of miR-1 helps to drive pathway activation (Figure 2). Post-transcriptional repression by miR-1 could allow these genes to be transcribed at higher (or leaky) levels without triggering a hypertrophic response, such that a reduction in miR-1 expression would suffice to rapidly activate signaling at multiple levels. For example, de-repression of the most downstream factors (e.g. CDC42) could quickly lead to sarcomere remodeling, a first step in the hypertrophic response (Nagai et al., 2003). Increasing levels of upstream factors coupled with positive feedback loops would intensify the response.
Figure 2. miR-1 and PIP3 signaling in cardiac hypertrophy.
The orange repressive arrows depict high-quality putative targets of miR-1 in the PIP3 pathway in cardiac myocytes (see Experimental Procedures). The rest of the network is based on known interactions compiled from the literature (Heineke and Molkentin, 2006). See Figure S1 for network diagrams of other selected predictions discussed in the text.
We envision a useful application of mirBridge would be to probe function of interest guided by the expression profile of likely relevant miRNAs. Since we are interested in neurotransmitter and axon guidance pathways, we applied mirBridge to manually curated gene sets for these pathways and examined miRNAs known to be expressed in the brain (see Supplemental Experimental Procedures). miR-218 is a top hit for both GABA and glutamate gene sets (q=0.025 and 0.033, respectively). That these two neurotransmitter activities may be regulated by the same miRNA is intriguing given that glutamate and GABA are, respectively, the major excitatory and inhibitory neurotransmitters, and that the latter can be enzymatically converted from the former. In addition, encouraged by the recent finding that miR-218 is enriched at synapses of hippocampal neurons, we tested a gene set for the presynaptic cytomatrix protein complex (Siegel et al., 2009). miR-218 is the top hit with a q value of 0.0008. In sum, the mirBridge hits for these gene sets extend early experimental findings to implicate miR-218 as potential regulators of neuronal activity at hippocampal synapses.
miRNA co-targeting is prevalent
Our miRNA-pathway map indicates that some miRNAs function in the same pathway(s) by targeting a similar set of genes. Indeed, many miRNAs may function together (via “co-targeting”) to regulate target-gene expression. To assess the prevalence of co-targeting and infer which miRNAs are co-targeting partners, we next used sets of genes likely regulated by particular miRNAs to create a miRNA-to-miRNA mapping. Specifically, our inputs to mirBridge were the predicted target sets (PTS) of 73 deeply conserved human miRNA families. We call a miRNA family Y a “co-targeting partner” of a miRNA family X if at least one of Y’s seed-matched sequences has a significant mirBridge q value in the PTS of X and denote the relationship as “X→Y.” We predicted co-targeting relationships for all ordered pairs of the 73 families (73 × 72 = 5256 distinct pairs).
Our results indicate that miRNA co-targeting is prevalent: 221 distinct X→Y co-targeting relationships are inferred at a FDR cutoff of 0.2 (Table S3a). A subset of these predictions correspond to miRNA genomic clusters (Yu et al., 2006), such as the miR-19b-2/106a cluster on Xq26.2 and the miR-17-18-19a-20-92 cluster on 13q31.3 (Table S3b). Co-targeting pairs in close genomic proximity are not surprising: these miRNAs are polycistronic and co-expressed, and are thus likely to function together to regulate common targets. In fact, clustered miRNAs are enriched for co-targeting relationships: when X and Y are members of a genomic cluster, they are predicted as co-targeting partners 25% of the time compared to 3% when X and Y are not clustered. Consequently, the median q-value of clustered pairs is significantly lower than that of unclustered ones (p < 2.1 × 10−7, Mann-Whitney Test; see Table S3c for the clusters used in this analysis), indicating that our method for detecting co-targeting is sensitive, specific, and capable of uncovering biologically relevant signals.
If our predictions reflect bona fide biological signals, we also expect a significant percentage of the X→Y pairs to possess mutual co-targeting relationships, i.e. each miRNA’s putative binding sites would have a score below the FDR cutoff in the other miRNA’s PTS. Indeed, 96/221 (43%) of the X→Y predicted pairs do. While the remaining 57% of the X→Y pairs do not have the corresponding Y→X pairs falling below the FDR cutoff, there is nonetheless a significant correlation between their q values (Spearman correlation = 0.42 (p=0); Figure S2). Also, the reciprocal (Y→X) q values of significant X→Y pairs are lower than those of pairs with q values greater than 0.2 (p < 5 × 10−140 Mann-Whitney Test). The general reciprocation of co-targeting scores indicates that a significant percentage of our predictions are specific and that the signals we are detecting are likely biologically relevant.
We also tested whether co-targeting relationships could be inferred from gene set overlaps, where the X→Y q value was computed using FET on the number of genes shared between the PTSs of the miRNA family pair. This analysis failed to provide informative results because almost all tested pairs have a significant q value: 2264 (86%) and 2628 (100%) of the pairs have a q value of less than 0.05 by using the Bonferroni and FDR correction, respectively. This suggests that a core set of genes are frequently predicted as targets for many miRNA family pairs; these likely correspond to genes with highly conserved 3′ UTRs and/or low GC content, properties that favor a gene being predicted as a target using Targetscan. This result strongly suggests that the degree of PTS overlap is not sufficiently specific to detect authentic co-targeting relationships, whereas mirBridge has superior specificity and thus able to provide biologically relevant signals as shown above.
Network analysis of co-targeting interactions
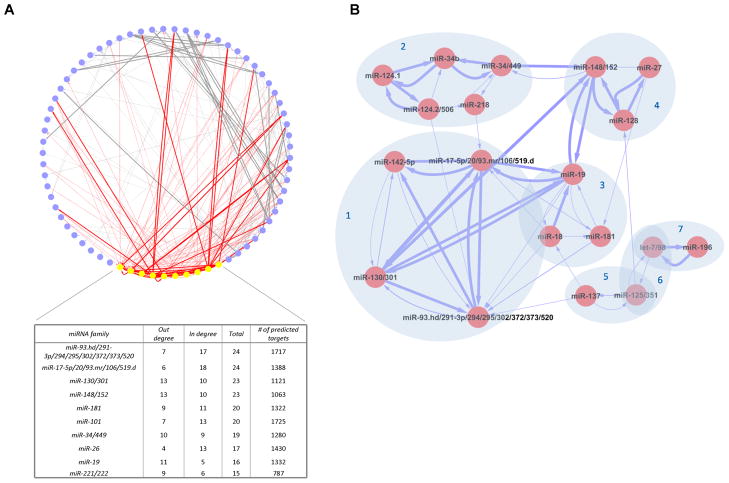
Our co-targeting predictions can naturally be organized as a network where the nodes are miRNA families and the directed edges between nodes denote the X→Y predictions. A network representation enables examination of connectivity patterns beyond pairwise interactions. We first checked whether the edges in the network are evenly distributed across nodes or concentrated around a few nodes (“hubs”). Strikingly, the edges connecting the 10 most connected nodes (out of 69 nodes with at least one adjacent edge) account for more than 55% (123/221) of the edges in the network (Figure 3A and Table S3d). While overall the size of a miRNA family’s PTS is correlated to its connectivity ranking (p = 10−6 Spearman correlation), this correlation becomes insignificant when restricted to families with at least 900 predicted targets (p > 0.1). Since only six of the top 40 most-connected families have fewer than 900 predicted targets, the size of a miRNA family’s PTS alone cannot explain the connectivity pattern among the top 40 families. The hub miRNA families probably have functions in diverse contexts. For example, some hubs have a large number of members and therefore are likely to have more diverse functions depending on the spatial-temporal expression of individual miRNAs (e.g. miR-93.hd/291-3p/294/295/302/372/373/520).
Figure 3. The miRNA-cotargeting network inferred by mirBridge.
The thickness of the edges is proportional to −log (q). (A) The ten most connected nodes and the adjacent edges are highlighted in yellow and red, respectively. (B) Examples of highly interconnected subnetworks. See also Figure S3.
We reasoned that groups of tightly inter-connected nodes might represent miRNAs that perform similar functions. To identify such groups we used a graph clustering tool that ignores edge weights to identify tightly interconnected nodes (Bader and Hogue, 2003) (Figure 3B). We find that subnetwork 1 has four families and is the largest and most highly interconnected; three of the families (miR-17-5p, -130, -93.hd) are among the most connected families (Figure 3A). This subnetwork is also well-connected to subnetwork 3 (miR-18, -19, -181), probably because miR-17-18-19-20 are co-expressed from a polycistronic transcript. The miR-17 cluster is known to be overexpressed in a number of human cancers, including B-cell tumors, while miR-142 is also highly expressed in B cells (Chen and Lodish, 2005; Mendell, 2008). Their shared PTS is enriched for genes in developmental processes (p < 3.8 × 10−5), consistent with the miR-17 cluster’s function in the development of B cells, the heart and lungs (Mendell, 2008; Ventura et al., 2008). Our linking of the miR-142 and miR-130/301 families—whose functions are largely unknown—to the miR-17 cluster suggests that these miRNA families also participate in similar developmental and oncogenic processes.
DISCUSSION
We have introduced a systematic method for inferring miRNA functions by assessing the enrichment of likely functional target sites in gene sets. Key features of mirBridge include combining test metrics that detect different aspects of functional targeting, and a sampling algorithm for removing gene-set biases to improve estimation of statistical significance. Hundreds of human miRNA-function associations were inferred by mirBridge; some are reassuringly supported by published experiments, but many are not previously known and/or provide mechanistic insights beyond published data.
Our results provide hints about the general principles of miRNA-mediated regulation in networks. While some miRNAs could act as global regulators by repressing up to thousands of targets genome-wide (Lewis et al., 2005), many appear to have pathway-specific functions, and these miRNAs tend to target multiple genes in the same pathway. Typically, the predicted targets of the miRNA are genes that drive pathway activity in a coherent direction (e.g. miR-16 targeting of G1-to-S-promoting genes). Such coordinate targeting could partially explain how individual miRNAs can be potent effectors of pathway activity even though the amount of repression conferred by miRNAs tends to be modest for any single target (Baek et al., 2008; Selbach et al., 2008; Xiao and Rajewsky, 2009). As was observed earlier (Martinez et al., 2008; Tsang et al., 2007), some of our predictions (e.g. miR-1) involve miRNAs mediating feedback and feedforward loops, whose functions include protein homeostasis and signal amplification, respectively. For example, miRNAs could be “master” regulators of pathways and thus serve as effective therapeutic targets because positive feedbacks could amplify small changes in protein concentration conferred by miRNA targeting of multiple genes. Our analysis also indicates that miRNAs can function in, and mediate cross-talk among, multiple canonical pathways, such as miR-16’s potential roles across the cell cycle and Wnt pathways to coordinately regulate cellular growth and proliferation.
mirBridge also facilitates context-specific target prediction: one can first predict which pathways a miRNA regulates and then compile high-quality putative targets within a pathway. This strategy may be especially effective for miRNAs that function in only a few pathways, as targets predicted genome-wide may have low specificity (Lewis et al., 2005). Additional filtering can be used to strengthen the target predictions, for example, by requiring that the putative target and the miRNA be significantly correlated in their expression using miRNA-mRNA expression data sets (Lu et al., 2005) (Table S1i).
In addition to providing functional links across miRNAs, our human miRNA-miRNA map provides, to the best of our knowledge, the first genome-wide evidence that miRNA co-targeting is prevalent, and that a handful of hub miRNA families are involved in a large fraction of the co-targeting connections. The abundance of co-targeting further suggests that while individual miRNAs may provide only modest levels of repression, combinatorial targeting by multiple miRNAs (Krek et al., 2005) can potentially achieve a wide range of target-level modulations. Given that multiple miRNAs are expressed at different levels in any given cell type, individual genes can evolve combinations of miRNA binding sites to optimize expression levels across cell types (Bartel and Chen, 2004). miRNA target sites are short and could thus be acquired or lost relatively quickly over evolution to fine-tune gene expression levels.
Designating a group of miRNAs as “co-targeting” does not necessarily imply that these miRNAs are co-expressed so as to regulate their common targets at the same time and place. In fact, the exact opposite is also likely: different miRNAs are responsible for controlling a given set of targets in different contexts. In general, a combination of the above scenarios is likely for individual cases, and additional data (e.g. miRNA and target expression profiles) are needed to further dissect the mechanistic basis of individual co-targeting predictions.
mirBridge is currently limited to assessing enrichment at the level of miRNA families using seed-matched motifs. But this is largely due to our lack of general understanding of miRNA-target interaction beyond seed pairing and features captured by the context score. In principle, the mirBridge methodology is general and can be applied to any combinations of gene sets, sequence motifs and site scoring metrics, including non-miRNA motifs, such as those involved in regulating mRNA stability. Given mirBridge’s ability to simultaneously correct for multiple gene-set biases, and the increasing availability of genomes and annotated gene sets, mirBridge is poised to serve as a key resource for the comprehensive functional dissection of miRNAs and other regulatory sequence motifs in genomes.
EXPERIMENTAL PROCEDURES
Seed-matched site compilation
miRNA family memberships, 3′ UTR sequences, seed-matched sites and their context scores and conservation status were downloaded from TargetScan (http://www.targetscan.org/vert_40/). For each known human gene, the number of seed-matched sites for each miRNA family, the number of those that are conserved, and the context score were computed. Since the context score depends on the full miRNA sequence, the context score for a miRNA family is defined as the average of all human members of that family.
mirBridge
The method as described in the text was implemented in Matlab. More details and related discussions can be found in Supplementary Experimental Procedures.
miRNA function analysis
Canonical signaling pathway and KEGG gene sets were downloaded from http://www.broad.mit.edu/gsea/msigdb/index.jsp. The cancer, CORUM, and GO sets were downloaded from http://robotics.stanford.edu/~erans/cancer/, http://mips.helmholtz-muenchen.de/genre/proj/corum, and NCBI Gene, respectively. To reduce noise and avoid spurious annotations, we only used GO annotations with experimental and peer-reviewed evidence. A miRNA-gene set prediction requires at least one of the miRNA seed motifs (m2-8 and/or m7-A) to test as significant in the gene set. The q value reported for individual miRNAs corresponds to the q value of the seed motif with the smaller p value.
miRNA family selection
The deeply conserved miRNAs are ones that are conserved across human, mouse, rat, dog and chicken. We focused on these miRNAs because they probably have (1) more conserved functions, (2) a larger number of targets compared to less-conserved miRNAs, and (3) stronger conservation enrichment signals.
Target prediction
Targets were compiled for each miRNA by including genes with at least one conserved seed-match (across human, mouse, rat and dog) or a seed-match with a context score of greater than 68 in the 3′ UTR (see Supplemental Experimental Procedures). Predictions based on context score alone were included because functional target sites can be imperfectly conserved. High-quality putative targets in gene sets (Table 1 and S1) were compiled using the same definition.
X→Y predictions and analysis
mirBridge was applied to the predicted target set of each miRNA family. Only the seed-matched motifs of the 73 families were scored. When both seed-matched motifs of a miRNA family are tested significant, the smaller q value is used as the X→Y q value. Human miRNA clusters were obtained from (Yu et al., 2006).
Predicted target set overlap analysis
The number of overlaps between the predicted target set of each miRNA-family pair was computed. The statistical significance was computed using Fisher’s Exact Test (see Supplemental Experimental Procedures).
Predicted target set and pathway overlap analysis
Similar to above except that (1) all genes that are not predicted as a target for any miRNA were removed from the pathway gene sets; and (2) the population size is taken as the number of genes that are predicted as a target for at least one miRNA family and belong to at least one pathway.
Supplementary Material
Acknowledgments
We thank H. Fraser, D. Muzzey, M. Narayanan and M. Umbarger for comments on the manuscript; J. Zhu for discussions; D. Bartel for the suggestion to examine co-targeting by polycistronic miRNAs; M. Fang for help on importing gene sets. This work was supported by grants from the NSF (PHY-0548484), NIH (R01-GM068957), and by a NIH Director’s Pioneer Award to A.v.O. (1DP1OD003936); J.S.T. was partially supported by a doctoral scholarship from the NSERC of Canada; M.S.E. was supported by a HHMI Predoctoral Scholarship, a Paul and Cleo Schimmel Scholarship, and a grant from the NIH (RO1-CA133404) to Phillip Sharp.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D’Urso L, Pagliuca A, Biffoni M, Labbaye C, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- Bravo-Egana V, Rosero S, Molano RD, Pileggi A, Ricordi C, Dominguez-Bendala J, Pastori RL. Quantitative differential expression analysis reveals miR-7 as major islet microRNA. Biochem Biophys Res Commun. 2008;366:922–926. doi: 10.1016/j.bbrc.2007.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CZ, Lodish HF. MicroRNAs as regulators of mammalian hematopoiesis. Semin Immunol. 2005;17:155–165. doi: 10.1016/j.smim.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra R, Adlakha YK, Hariharan M, Scaria V, Saini N. Upregulation of miR-23a approximately 27a approximately 24-2 cluster induces caspase-dependent and -independent apoptosis in human embryonic kidney cells. PLoS One. 2009;4:e5848. doi: 10.1371/journal.pone.0005848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloonan N, Brown MK, Steptoe AL, Wani S, Chan WL, Forrest AR, Kolle G, Gabrielli B, Grimmond SM. The miR-17-5p microRNA is a key regulator of the G1/S phase cell cycle transition. Genome Biol. 2008;9:R127. doi: 10.1186/gb-2008-9-8-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa-Medina M, Bravo-Egana V, Rosero S, Ricordi C, Edlund H, Diez J, Pastori RL. MicroRNA miR-7 is preferentially expressed in endocrine cells of the developing and adult human pancreas. Gene Expr Patterns. 2009;9:193–199. doi: 10.1016/j.gep.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, Weidhaas JB, Brown D, Bader AG, Slack FJ. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008a;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008b doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol. 2006;7:589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- Ji Q, Hao X, Meng Y, Zhang M, Desano J, Fan D, Xu L. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer. 2008;8:266. doi: 10.1186/1471-2407-8-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L, Xiang D, Desano JT, Bommer GT, Fan D, et al. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One. 2009;4:e6816. doi: 10.1371/journal.pone.0006816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joachim H. A Note on Combining Dependent Tests of Significance. Biometrical Journal. 1999;41:849–855. [Google Scholar]
- Joglekar MV, Joglekar VM, Hardikar AA. Expression of islet-specific microRNAs during human pancreatic development. Gene Expr Patterns. 2009;9:109–113. doi: 10.1016/j.gep.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- Jones SW, Watkins G, Le Good N, Roberts S, Murphy CL, Brockbank SM, Needham MR, Read SJ, Newham P. The identification of differentially expressed microRNA in osteoarthritic tissue that modulate the production of TNF-alpha and MMP13. Osteoarthritis Cartilage. 2009;17:464–472. doi: 10.1016/j.joca.2008.09.012. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kefas B, Godlewski J, Comeau L, Li Y, Abounader R, Hawkinson M, Lee J, Fine H, Chiocca EA, Lawler S, Purow B. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res. 2008;68:3566–3572. doi: 10.1158/0008-5472.CAN-07-6639. [DOI] [PubMed] [Google Scholar]
- Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008a;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008b doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Li Z, Hassan MQ, Jafferji M, Aqeilan RI, Garzon R, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem. 2009;284:15676–15684. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Hassan MQ, Volinia S, van Wijnen AJ, Stein JL, Croce CM, Lian JB, Stein GS. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci U S A. 2008;105:13906–13911. doi: 10.1073/pnas.0804438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond CK, Dai H, et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Fu H, Sun F, Zhang H, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008;36:5391–5404. doi: 10.1093/nar/gkn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol. 2009;182:4994–5002. doi: 10.4049/jimmunol.0803560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NJ, Ow MC, Barrasa MI, Hammell M, Sequerra R, Doucette-Stamm L, Roth FP, Ambros VR, Walhout AJ. A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes Dev. 2008;22:2535–2549. doi: 10.1101/gad.1678608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, Jacob S, Majumder S. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283:29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T, Tanaka-Ishikawa M, Aikawa R, Ishihara H, Zhu W, Yazaki Y, Nagai R, Komuro I. Cdc42 plays a critical role in assembly of sarcomere units in series of cardiac myocytes. Biochem Biophys Res Commun. 2003;305:806–810. doi: 10.1016/s0006-291x(03)00838-6. [DOI] [PubMed] [Google Scholar]
- Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering MT, Stadler BM, Kowalik TF. miR-17 and miR-20a temper an E2F1-induced G1 checkpoint to regulate cell cycle progression. Oncogene. 2009;28:140–145. doi: 10.1038/onc.2008.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38(Suppl):S8–13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Ruepp A, Brauner B, Dunger-Kaltenbach I, Frishman G, Montrone C, Stransky M, Waegele B, Schmidt T, Doudieu ON, Stumpflen V, Mewes HW. CORUM: the comprehensive resource of mammalian protein complexes. Nucleic Acids Res. 2008;36:D646–650. doi: 10.1093/nar/gkm936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res. 2007;100:416–424. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008;18:549–557. doi: 10.1038/cr.2008.45. [DOI] [PubMed] [Google Scholar]
- Segal E, Friedman N, Koller D, Regev A. A module map showing conditional activity of expression modules in cancer. Nat Genet. 2004;36:1090–1098. doi: 10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJ, Kane C, et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11:705–716. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC. The synaptic vesicle cycle. Annu Rev Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RJ, Giles KM, Price KJ, Zhang PM, Mattick JS, Leedman PJ. Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem. 2009;284:5731–5741. doi: 10.1074/jbc.M804280200. [DOI] [PubMed] [Google Scholar]
- Wegman EJ. Nonparametric Probability Density Estimation: I. A Summary of Available Methods. Technometrics. 1972;14:533. [Google Scholar]
- Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- Xie H, Lim B, Lodish HF. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes. 2009;58:1050–1057. doi: 10.2337/db08-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Kaye DM. Mechanistic insights into the link between a polymorphism of the 3′UTR of the SLC7A1 gene and hypertension. Hum Mutat. 2009;30:328–333. doi: 10.1002/humu.20891. [DOI] [PubMed] [Google Scholar]
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Yu J, Wang F, Yang GH, Wang FL, Ma YN, Du ZW, Zhang JW. Human microRNA clusters: genomic organization and expression profile in leukemia cell lines. Biochem Biophys Res Commun. 2006;349:59–68. doi: 10.1016/j.bbrc.2006.07.207. [DOI] [PubMed] [Google Scholar]
- Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, Coppola D, Cheng JQ. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem. 2008;283:31079–31086. doi: 10.1074/jbc.M806041200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.