Abstract
The data for the public working draft of the human genome contains roughly 400,000 initial sequence contigs in ∼30,000 large insert clones. Many of these initial sequence contigs overlap. A program, GigAssembler, was built to merge them and to order and orient the resulting larger sequence contigs based on mRNA, paired plasmid ends, EST, BAC end pairs, and other information. This program produced the first publicly available assembly of the human genome, a working draft containing roughly 2.7 billion base pairs and covering an estimated 88% of the genome that has been used for several recent studies of the genome. Here we describe the algorithm used by GigAssembler.
On May 24, 2000, the public Human Genome Project staged the first “freeze” of all currently available sequence data, coordinated by the director, Francis Collins, Greg Schuler at the National Center for Biotechnology Information, Adam Felsenfeld at the National Human Genome Research Institute, and the twenty primary public human sequencing centers (Box 1). Public database accessions for ∼22,000 shotgun-sequenced clones were selected for this freeze, mostly bacterial artificial chromosome (BAC) clones (International Human Genome Sequencing Consortium 2001). The sequence contigs were extracted from these accessions and cleaned up as necessary by Schuler. We will refer to these contigs as “initial sequence contigs”. There were ∼375,000 such initial sequence contigs. The complete public human genome sequence is not projected to be available until 2003. To get a useable working draft in the short term, it was necessary to order and orient these initial sequence contigs along the 20 unfinished autosomes and two sex chromosomes as best as the data permitted, detecting overlaps and building larger sequence contigs where possible. Chromosomes 21 (Hattori et al. 2000) and 22 (Dunham et al. 1999) had already been finished and nearly complete ordered and oriented euchromatic sequence was available for them.
Box 1. The Human Genome Sequencing Consortium
The twenty institutions that form the Human Genome Sequencing Consortium include: Baylor College of Medicine, Houston, Texas, USA (http://www.hgsc.bcm.tmc.edu/); Beijing Human Genome Center, Institute of Genetics, Chinese Academy of Sciences, Beijing, China (http://hgc.igtp.ac.cn/); Cold Spring Harbor Laboratory, Lita Annenberg Hazen Genome Center, Cold Spring Harbor, New York, USA (http://nucleus.cshl.org/genseq/lita_annenberg_hazen_genome_cent.htm); Gesellschaft für Biotechnologische Forschung mbH, Braunschweig, Germany (http://genome.gbf.de/); Genoscope, Evry, France (http://www.genoscope.fr/); Genome Therapeutics Corporation, Waltham, Massachusetts, USA (http://www.genomecorp.com/); Institute for Molecular Biotechnology, Jena, Germany (http://genome.imb-jena.de/); Joint Genome Institute, U.S. Department of Energy, Walnut Creek, California, USA (http://www.jgi.doe.gov/); Keio University, Tokyo, Japan (http://www-alis.tokyo.jst.go.jp/HGS/); Max Planck Institute for Molecular Genetics, Berlin, Germany (http://seq.mpimg-berlin-dahlem.mpg.de/); RIKEN Genomic Sciences Center, Saitama, Japan (http://hgp.gsc.riken.go.jp/); The Sanger Centre, Hinxton, UK (http://www.sanger.ac.uk/HGP/); Stanford Genome Technology Center, Palo Alto, California, USA (http://www-sequence.stanford.edu/); Stanford Human Genome Center, Palo Alto, California, USA (http://shgc.stanford.edu/); University of Oklahoma's Advanced Center for Genome Technology, Oklahoma, USA (http://www.genome.ou.edu/); University of Texas Southwestern Medical Center at Dallas, Texas, USA (http://www3.utsouthwestern.edu/index.htm); University of Washington Genome Center, Seattle, Washington, USA (http://www.genome.washington.edu/UWGC/); Multimegabase Sequencing Center, Institute for Systems Biology, Seattle, Washington, USA (http://www.systemsbiology.org/); Whitehead Institute for Biomedical Research, MIT, Cambridge, Massachusetts, USA (http://www-genome.wi.mit.edu/); and the Washington University Genome Sequencing Center, St. Louis, Missouri, USA (http://genome.wustl.edu/gsc/).
A group led by Robert Waterston at the Washington University Genome Sequencing Center (WUGSC) created a map of the large insert clones from the May 24 freeze, based on the genome-wide physical map they had developed of ∼300,000 clones, primarily using fingerprint overlaps, but also employing information from radiation hybrid, genetic, YAC–STS, and cytogenetic maps, as well as BAC end matches (International Human Genome Mapping Consortium 2001; International Human Genome Sequencing Consortium 2001). A clone fingerprint is defined by the set of sizes of fragments from the clone created by restriction enzyme digest and measured by gel electrophoresis. Sets of clones containing statistically significant overlaps between their fingerprints are grouped into fingerprint clone contigs. Sequenced clones from a fingerprint clone contig are used for the sequence assembly. The May 24 map of sequenced clones consisted of some 1700 fingerprint clone contigs, each with an approximate chromosomal location, plus a few additional contigs that could not be reliably placed on a chromosome. The end points of the individual sequenced clones, as well as their overlaps and relative order along the chromosome, were only very roughly determined in these fingerprint clone contigs. Thus, the problem of clone order needed to be solved along with the problem of initial sequence contig order and orientation. Initial sequence contigs from different clones within a fingerprint clone contig often showed long sequence overlaps, giving strong evidence of clone order, but not giving an entirely unambiguous signal because of the occasional presence of near exact duplicated regions (Ji et al. 2000).
These two problems were somewhat intertwined: Clone order information from the fingerprint map was needed during the assembly of the initial sequence contigs within a fingerprint clone contig, and this assembly led to refined clone order.
Further evidence of initial sequence contig order and orientation was obtained from sequence matches between the initial sequence contigs and mRNA or EST sequences. These matches help to order and orient the initial sequence contigs that contain the exons of a gene, even if these exons are separated by quite long introns, improving the usefulness of the working draft for the study of genes. A greater number of matches could be found between the initial sequence contigs from the shotgun-sequenced clones from the freeze and the paired ends of ∼500,000 BAC clones that were only end-sequenced (Zhao 2000). These matches also provide order and orientation information for the initial sequence contigs, but can be misleading because a significant percentage of the BAC end sequences are mispaired (Zhao 2000).
A greedy (Cormen et al. 1990) algorithm, called GigAssembler, was developed to use the initial sequence contig, map, mRNA, EST, and BAC end data to assemble the genome sequence of the May 24 freeze (Kent and Haussler 2000). The resulting assembly, produced in mid June, consisted of 2,182,660,273 base pairs covering about 70% of the genome. This was quickly followed by an assembly covering significantly more of the genome using the data from the June 15 freeze. Since that time, further new human sequence has been added to the public databases, and new freezes have been declared periodically. The GigAssembler algorithm has developed further as well during that time. Starting with the September freeze, matches from the paired end sequences of plasmid clones were added to improve the ordering and orientation. These were taken from approximately one million genome-wide random reads from the Genome Center at the Whitehead Institute for Biomedical Research (WIBR) in the assembly of the September 5 freeze, and about one million further reads from the Sanger Centre and WUGSC in the October 7 freeze, made available through the SNP Consortium at http://snp.cshl.org. In addition, ordering and orientation information for the initial sequence contigs that is contained in some of the public database accessions was extracted and used by GigAssembler in later assemblies, along with information from assembled contigs of finished sequence (“NT contigs”). A history of the assemblies is given in Table 1.
Table 1.
Working Drafts Produced by GigAssembler
| Freeze | Input (GB) | Output (GB) | % of genome covered |
|---|---|---|---|
| May 24, 2000 | 3.3 | 2.2 | 70 |
| June 15, 2000 | 3.6 | 2.5 | 82 |
| July 17, 2000 | 4 | 2.7 | 87 |
| September 5, 2000 | 4.1 | 2.7 | 87 |
| October 7, 2000 | 4.2 | 2.7 | 88 |
| December 12, 2000 | 4.3 | 2.7 | 90 |
| April 1, 2001 | 4.5 | 2.8 | 92 |
The total size of all initial sequence contigs that are input to the assembly and the total size of the contigs produced by the assembly, both in gigabases, are shown for the assemblies produced by GigAssembler. Percent of the genome covered is estimated as described in the International Human Genome Sequencing Consortium (2001) As the quality of the input data improved, the amount of artifactual duplication was reduced, resulting in a higher increase in the percentage of the genome covered relative to the increase in the total size of the contigs of the assembly.
The first release to the public was July 7, 2000, on http://genome.ucsc.edu, and included both the assembly of the May 24 freeze and the substantially larger assembly of the June 15 freeze. All subsequent assemblies have been available at that Web site as well.
The purpose of this paper is to describe the algorithm used by GigAssembler. A description of the assembly itself, and the discoveries that have been made using the assembled working draft sequence, is given in the paper on the working draft genome by the International Sequencing Consortium and related papers (Bentley et al. 2001; Bock et al. 2001; Cheung et al. 2001; Clayton et al. 2001; Fahrer et al. 2001; Futreal et al. 2001; International Human Genome Sequencing Consortium 2001; International SNP Map Working Group 2001; Li et al. 2001; Murray and Marks 2001; Nestler and Landsman 2001; Pollard 2001; Riethman et al. 2001; Tupler et al. 2001; Wolfsberg et al. 2001; Yu 2001).
There are many algorithms that assemble reads from subclones of a single BAC, PAC, cosmid, or other clone, or from a whole-genome shotgun of a relatively small and not strongly repetitive genome (Bonfield et al. 1995; Sutton et al. 1995; Huang 1996; Huang and Madan 1999; P. Green, http://www.genome.washington.edu/UWGC/analysistools/phrap.htm). The sequencing centers primarily used PHRAP (Green, unpubl. software) to assemble shotgun reads of subclones of a large insert clone into initial sequence contigs. Myers has pioneered an alternative approach to the assembly of larger genomes, working directly from paired whole-genome shotgun reads (Anson and Myers 1999). This method, embodied in the Celera assembler, was successful for the Drosophila melanogaster genome (Myers et al. 2000) and has been applied to the human genome as well. There has not yet been an opportunity for us to compare the results of the Celera assembly to GigAssembler's assembly.
Most successful assembly algorithms, whether for individual large insert clones or for whole genomes, have been based on greedy methods, and all have certain features in common. They begin by looking for sequence overlaps among the reads to be assembled and build up sequence contigs by making the best overlaps first. All must have some heuristics to avoid being confused by repetitive sequence. Some, like CAP (Huang 1996; Huang and Madan 1999), have heuristics to detect chimeric reads in their input, that is, reads composed of sequence from two or more nonadjacent places in the genome. Some, like the Celera assembler, CAP3 (Huang 1996), and GigAssembler, build scaffolds that consist of several ordered and oriented sequence contigs separated by sequence gaps. One distinguishing feature of GigAssembler is the variety of information it uses in building scaffolds. Each sequence gap in a GigAssembler scaffold can be bridged by a plasmid end pair, BAC end pair, mRNA, or EST, or can be derived directly from ordering information present in a public database accession. The Bellman-Ford algorithm is used to check distance constraints on each scaffold (Cormen et al. 1990; Bonfield et al. 1995). Unlike most previous assemblers, GigAssembler was designed to work as a second-stage assembler, using the above information along with sequence overlaps to assemble the entire human genome, after PHRAP or another assembly method has been used to separately assemble the reads of each draft-sequenced large insert clone into a set of initial sequence contigs. GigAssembler operates at a very large scale, assembling 2.7 billion bases of the human genome from 4.3 billion bases of input in about 400,000 initial sequence contigs, 1.1 billion bases of EST sequence, 1.8 billion bases of paired plasmid reads, and 0.4 billion bases of BAC end sequences.
RESULTS AND DISCUSSION
Assembly Process Overview
The assembly proceeds according to the following major steps:
Decontaminating and repeat masking the sequence.
Aligning mRNA, EST, BAC end, and paired plasmid reads against initial sequence contigs. On a cluster of 100 Pentium III (866 MhZ) CPUs running Linux, this takes about three days.
Creating an input directory structure using Washington University map and other data. This step takes about an hour on a single computer.
For each fingerprint clone contig, aligning the initial sequence contigs within that contig against each other. This takes about three hours on the cluster.
Using the GigAssembler program within each fingerprint clone contig to merge overlapping initial sequence contigs and to order and orient the resulting sequence contigs into scaffolds. This takes about two hours on the cluster.
Combining the contig assemblies into full chromosome assemblies. This takes about twenty minutes on one computer.
The steps will be described in more detail below.
Decontaminating and Repeat Masking the Sequence
For the most part, the sequencing centers themselves remove bacterial, vector, and other contaminants from the large insert clones before they are deposited in the international public databases. Greg Schuler at NCBI does an additional decontamination step that helps assure that even the older sequenced clones are screened for newly identified contaminants such as bacterial transposons and phages. Checks for rodent contamination and sequences that are a mixture of multiple clones are also made. At the end of this we receive three files containing the sequence for roughly 30,000 clones in roughly 400,000 initial sequence contigs with known contaminants removed. We also receive a list of dubious clones that are excluded from the assembly. To ease further processing we break up the three large files into a separate file for each clone. We run RepeatMasker (http://ftp.genome.Washington.edu/RM/RepeatMasker.html) using the -q (quick) setting on each clone. The repeat masking is done on our computer cluster and takes about 12 hours.
Alignment of mRNA, ESTs, BAC Ends, and Paired Reads
Alignments of mRNA, ESTs, BAC ends, and paired plasmid (queries) versus the clone initial sequence contigs (target) are the raw material for constructing bridges to order and orient the sequence contigs. We developed a program, psLayout, for this purpose. In brief, psLayout follows the following steps:
Build up an index of all of the 10-mers in a 30-Mb subset of the target sequence, excluding simple repeating elements from this index;
Break up the query sequence into overlapping 500-base regions;
Look up the 10-mers that occur in the 500-base region in the index and identify clusters of matching 10-mers in the target sequence that represent likely areas of homology;
Do a detailed alignment between the 500-base region and the sections of the target sequence identified in step 3. In the case of mRNAs and ESTs this alignment is done in a fashion that tolerates introns;
Stitch together the alignments from overlapping 500-base regions using a dynamic programming algorithm.
PsLayout reports all alignments above a certain minimal quality between a collection of query sequences and a collection of database sequences. PsLayout distinguishes bases contained in repetitive DNA from other bases. All bases in the genomic sequences that RepeatMasker classifies as having 85% or more base identity with a repeating element are considered repetitive, and the remaining bases are considered unique. Unlike many other matching protocols, the repetitive bases are not masked out, but are kept and matches involving them are tallied separately in the alignment scores. A run of matching repetitive bases provides little or no evidence of a genuine overlap between a query and a database sequence, but a run of mismatching repetitive bases can in some circumstances provide strong evidence against a genuine overlap.
As an additional measure to help minimize the confounding effect of repeats, especially low-copy-number repeats, which may not be masked by RepeatMasker, the alignments are also passed through a near best in genome filter. In the human genome there exist many regions that have been duplicated over the course of evolutionary time. Most of these duplications are old enough that they have diverged from each other in >1% of their bases. In the draft sequence there are overlapping initial sequence contigs from different BAC clones that cover the same region of the genome. These overlapping initial sequence contigs only differ from each other where sequencing errors have occurred, typically in substantially <1% of the bases as the initial sequence contigs are assembled from 4× or higher shotgun coverage, and the reads they are based on tend to have been created on modern and well-maintained equipment. On the other hand, the EST and BAC end sequences are single reads from a large variety of sources, and many were done using older more error prone sequencing procedures. A 5% sequencing error rate is not uncommon in these sequences. To help distinguish between true matches and spurious matches caused by pseudogenes and other near identically repeated regions we only retain those matches that are “near best in genome”. Alignments are merged and sorted so that all alignments sharing a common query sequence appear together. A score based largely on percentage base identity is given to each alignment. For each region of the query the best scoring alignment is recorded. Alignments are discarded if they have 1% or greater divergence than the best scoring alignment. For example, if an EST aligned to clone A with 96.5% base identity, to clone B with 96% base identity, and to clone C with 95% base identity, the alignment to clones A and B would be kept and the alignment to clone C would be discarded. As genomic coverage approaches 100%, the probability of the true match being among those matches that are near best in the genome gets closer to one. In cases of highly similar paralogous genes, some cross-mapping of ESTs may occur, but it is usually locally consistent and thus does not adversely affect the assembly.
Creating the Directory Structure
The GigAssembler program itself operates on a single fingerprint clone contig at a time. Currently, the contigs vary from <100,000 bases to >60,000,000 bases. GigAssembler reads 13 input files in addition to however many clone sequence files are in the contig and produces 11 output files for each contig. The input and output files are described in Kent and Haussler (2000). A collection of a half dozen programs prepare the input from various sources including the Washington University map, the alignments described above, and information provided by NCBI. The result of this is a directory structure with one directory per chromosome and one subdirectory per fingerprint clone contig. The GigAssembler program can then be run in parallel, each contig being assembled by a separate CPU.
The GigAssembler Program
Here we briefly outline the key steps in GigAssembler program itself.
-
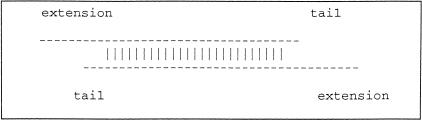
Build merged sequence contigs (called “rafts”) from overlapping initial sequence contigs. An initial sequence contig is either a sequence contig from the accession of a draft clone or a constructed contig of finished sequence (an “NT contig”) prepared at NCBI by Greg Schuler. Ideally, the alignment joining two initial sequence contigs would look like that displayed in Figure 1, where vertical bars indicate matching bases from the region that is considered to align. This region could include some mismatches and indels, but for simplicity, these are not shown. Here the alignment goes to one end of each sequence. However, in actual initial sequence contig data, because of the presence of low-quality data at the ends of many initial sequence contigs, this is often not the case, even for initial sequence contigs that should be joined. To classify the unmatched end regions, it is useful to introduce a little terminology, as illustrated in Figure 2.
The nonaligning parts of these two sequences are at the ends and can be divided into ‘tails’ and 6‘extensions’ as labeled in Figure 2. The longer unaligned end on one side or another of an alignment is the extension; the shorter is the tail.
To build a raft, GigAssembler assigns a score to each aligning pair. The alignments are then processed using the following procedure with the best scoring alignments processed first.
- If neither initial sequence contig in the alignment is in a raft, make a raft out of the two.
-
If one initial sequence contig is in a raft but not the other, the other initial sequence contig is added to the raft if it does not conflict with what is already in the raft. Consider the raft of the initial sequence contigs A, B, C, and an alignment involving C and D in Figure 3.The parts of D marked with +s are known to be consistent with the raft because of the CD alignment. The parts marked with dashes are acceptable because they extend the raft. However, the parts marked with question marks must also agree with the raft so far. This is checked by looking for alignments between D and the other members of the raft (alignments between D and B, and between D and A). If the alignments check out, D would be added to the raft.
-
If one initial sequence contig is in a raft and the other in another raft, the two rafts are merged, using a procedure like that described for case b.The scoring function is crucial here. It is not unusual for the data to conflict. It is important that especially the first joins be based on the strongest matches. The current scoring function strongly favors overlaps that are unique, weakly favors overlaps that are repeat masked, strongly discourages sequence mismatches and inserts within the aligning blocks, and moderately discourages tails. Alignments below a certain threshold of the scoring function are not used to build rafts. The actual C code for the scoring function is given in Kent and Haussler (2000).
-
Build sequenced clone contigs (called “barges”) from overlapping clones. This procedure is somewhat similar to raft building. Barges are called barges because they are similar to rafts, but are built from larger pieces.
To build a barge, the overlap between each sequenced clone is calculated by looking at the alignments that were used (but not the ones that were rejected) by the raft-building stage. The clone overlap is the sum of all of the initial sequence contig overlaps. Note that the clone overlap gives us relative position but not relative orientation of the two clones as the orientation of initial sequence contigs within a clone is not necessarily consistent.
The overlaps are used to order the clones in the following manner.- Clones that are completely enclosed by another clone are put aside.
- A clone is selected and the most overlapping other clone is joined with it to initialize an ordered list of clones.
- Given an ordered list of clones ABCD, though there are still clones in the fingerprint clone contig that significantly overlap clones in this list, the clone X that overlaps as much as possible with another element on the list is selected and inserted into the list as follows:
- If X overlaps A but not B, the order becomes XABCD.
- If X overlaps A and B, and overlap(A,B) < overlap(A,X) and overlap(A,B) < overlap(B,X) then the order becomes AXBCD. Here, overlap(A,B) is the total number of bases in the overlaps between the initial sequence contigs of A and the initial sequence contigs of B.
- Otherwise, steps i and ii are repeated, shifting the list so that C is considered in place of B and B in place of A.
- If there are still clones left in the fingerprint clone contig that have not been added to the ordered list after the iterations of the above steps cease, then a new barge is started to accommodate the remaining clones.
- The order of the clones in each barge is compared to the fingerprint map, and if the barge looks backwards the order of its clones is reversed.
- Barge coordinates are given in the following manner:
- The first clone is given an offset of zero.
- For the nth clone, Offset(n + 1) = Offset(n) + Size(n) − Overlap(n,n + 1), where the size of a clone is defined as the total length of all its initial sequence contigs.
-
(f) Clones that are completely enclosed are given the coordinate:Offset(inner) = Offset(outer) + (Size(outer) − Size(inner))/2
Assign default coordinates to initial sequence contigs. The default coordinate of an initial sequence contig is just the barge offset of the clone it is in plus its start position in the accession for the clone. Default coordinates are then assigned to a raft based on the average of the coordinates of the initial sequence contigs making up the raft.
-
Build a “raft-ordering” graph. This is a directed graph with two types of nodes: rafts and sequenced clone end points. An edge from one node to another implies the first node comes before the second. Associated with each edge is also a range of distances allowed between the centers of the objects represented by the two nodes.
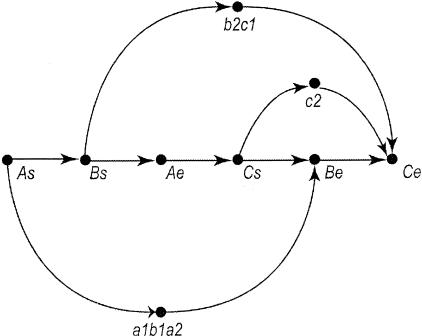
As an example, consider the sequenced clones A, B, C containing the initial sequence contigs a1, a2, b1, b2, c1, and c2 laid out in Figure 4.
The rafts in this figure are a1b1a2, b2c1, and c2. The initial graph would just contain the ends of the clones. Representing the start and end of clone A as As and Ae this is represented in Figure 5. Adding the rafts gives what is depicted in Figure 6.
Add information from mRNAs, ESTs, paired plasmid reads, BAC end pairs, and ordering information from the sequencing centers. This information is used to connect rafts in the ordering graph in a three-step process—building a ‘bridge’ out of alignments of other data with initial sequence contigs, scoring the bridge, and then adding bridges one at a time, best scoring first, to the ordering graph. A bridge defines order and orientation of the initial sequence contigs, as well as an allowable range of distances between them. The score function for bridges is the sum of two factors. The first factor is based on the type of the information. mRNA information is given the highest weight, then paired plasmid reads, information provided by the sequencing centers, ESTs, and BAC end matches, in that order. The second factor is based on the strength of the underlying alignment and is very similar to the score used for building rafts. Bridges that would conflict with the graph as constructed so far are rejected. Conflicts are detected using Bellman-Ford algorithm as described in the note below.
Walk the bridge graph to get an ordering of rafts. Each bridge is walked in the order of the default coordinates assigned in step 3 subject to the constraint that if a raft has predecessors, all the predecessors must be walked before the raft is walked.
- A sequence path through each raft is built as follows:
- Find the longest, most finished initial sequence contig that passes though each section of the raft.
- Put the best initial sequence contig for the first part of the raft into the sequence path.
- Find an alignment between the best initial sequence contig for the first part of the raft and the best initial sequence contig for the second part of the raft.
- Search for a ‘crossover point’ in the alignment where it would be reasonable to switch the sequence path to the next initial sequence contig. This crossover point is ideally 250 bases from the end of the larger, more finished initial sequence contig, but may be adjusted depending on the exact alignment.
- Repeat steps c and d to extend the sequence path until the end of the raft is reached.
Build the final sequence for the fingerprint clone contig by inserting the appropriate number of Ns between raft sequence paths. Currently 100 Ns are inserted between rafts that are part of the same barge, 50,000 Ns between barges that are bridged, and 100,000 Ns between unbridged barges.
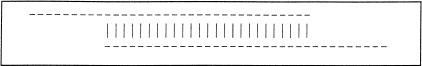
Figure 1.
Two sequences overlapping end to end. The sequences are represented as dashes. The aligning regions are joined by vertical bars. End-to-end overlap is an extremely strong indication that two sequences should be joined into a contig.
Figure 2.
Two sequences with tails. The nonaligning regions on either side can be classified into ‘extensions’ and ‘tails.’ Short tails are fairly common even when two sequences should be joined into a contig because of poor quality sequence near the ends and occasional chimeric reads. Long tails, however, are generally a sign that the alignment is merely due to the sequences sharing a repeating element.
Figure 3.
Merging into a raft. A contig (‘raft’) of three sequences: A, B, and C has already been constructed by GigAssembler. The program now examines an alignment between sequence C and a new sequence, D, to see whether D should also be added to the raft. The parts of D marked with +s are compatible with the raft because of the C/D alignment. The program must also check that the parts of D marked with ?s are compatable with the raft by examining other alignments.
Figure 4.
Three overlapping draft clones: A, B, and C. Each clone has two initial sequence contigs. Note that initial sequence contigs a1, b1, and a2 overlap as do b2 and c1.
Figure 5.
Ordering graph of clone starts and ends. This represents the same clones as in Fig. 4. (As) The start of clone A; (Ae) the end of clone A. Similarly Bs, Be, Cs, and Ce represent the starts and ends of clones B and C.
Figure 6.
Ordering graph after adding in rafts. The initial sequence contigs shown in Fig. 4 are merged into rafts where they overlap. This forms three rafts: a1b1a2, b2c1, and c2. These rafts are constrained to lie between the relevant clone ends by the addition of additional ordering edges to the graph shown in Fig. 5.
Assessment of the Accuracy of the Orientation and Order
We created two test sets to assess the accuracy of the October 7 freeze working draft as assembled by GigAssembler. The first, called FinishedContigs, is a collection of 24 clone contigs with a total of 145 clones taken from chromosomes 7, 12, 14, 17, 20, 21, and 22 for which we have finished sequence spanning the entire clone contig. The number of clones per clone contig varies from 4 to 13. We obtained a draft version for 88 of these clones by looking for a previous version of the finished clone in GenBank. The second test set, called Scrambled22, was generated by Ray Wheeler at Neomorphic, Inc. by taking the 12 finished sequence contigs from chromosome 22 and randomly choosing a tiling path of 233 ‘synthetic’ BACs covering them. The sequence of each synthetic BAC was then ‘draftified‘ by the introduction of gaps, indels, and substitutions in a way that the statistics on the resulting initial sequence contigs reasonably matched the statistics from initial sequence contigs of real draft BACs. Finally, the initial sequence contigs were given random order and orientation.
We ran the GigAssembler algorithm on all of the clone contigs from both of these test data sets and compared its predicted order and orientation for the initial sequence contigs to the true order and orientation of the initial sequence contigs, as can be derived from the finished sequence. We measured the orientation agreement as the fraction of initial sequence contigs that were oriented correctly. The average orientation agreement for the FinishedContigs test set was 0.90, and varied from 0.50 (near random guessing) to 1.0 (perfect) among the 23 clone contigs. Performance degraded as the number of initial sequence contigs per draft clone increased and the size of the initial sequence contigs decreased. On the 12 contigs of the Scrambled22 test set, the average orientation agreement was about 0.87 and varied from 0.71 to 1.0.
To measure the accuracy of the predicted order of the initial sequence contigs, we counted the number of violations of monotonicity in the order of the starting positions of the initial sequence contigs. A violation of monotonicity occurs at initial sequence contig A when the initial sequence contig following A in the predicted order in fact should come before A. For example, if the correct order of the initial sequence contigs is 1,2,3,4,5,6,7 and the predicted order is 1,5,2,4,7,3,6 then there are two violations, at initial sequence contigs A = 5 and A = 7. We measure order agreement as the fraction of initial sequence contigs in the predicted order where violations do not occur (excluding the last initial sequence contig in the predicted order, which cannot have a violation). In the above example, the order agreement is 4/6. The average order agreement for the Finished Contigs test set was 0.85, and varied from 0.50 to 1.0 among the 23 clone contigs. On the 12 contigs of the Scrambled22 test set, the average order agreement was 0.83 and varied from 0.74 to 0.93.
Conclusions
GigAssembler could be improved in several ways. A mechanism to detect misassemblies or chimerism in the initial sequence contigs could be quite helpful, perhaps along the lines of the mechanism developed for CAP2 (Huang 1996). Improved use of clone end information might lead to better barge construction in Step 2 of the algorithm as well (very recent modifications to GigAssembler to include this information have borne this out). Both of these improvements would reduce the rate of artifactual duplication in the draft assembly, which occurs when valid overlaps are not detected and thus the same region of the genome is assembled in two different parts of the assembly. It was estimated that 3% of the October 7 working draft genome sequence represented artifactual duplication, some caused by lack of complete assembly by GigAssembler and some by missed overlaps between fingerprint clone contigs (International Human Genome Sequencing Consortium 2001). More use could be made of PHRAP quality scores for individual bases during the assembly as well. Finally, it would be helpful to have some methods to impose an upper bound on the total assembled size of a large insert clone, and to eliminate initial sequence contigs from the assembly entirely when it appears that they don’t belong with the clone, or have other serious problems. Cross contamination from other clones is one source of initial sequence contigs that don't belong. It is estimated to be rare (International Human Genome Sequencing Consortium 2001) but is a significant problem for draft-sequenced clones because it is difficult to detect and eliminate until the clones are “topped up” to higher levels of coverage.
The assembly of the working draft of the human genome, although still imperfect, has permitted significant research to go forward, rather than wait years for the finishing of the sequence. In particular, having an assembly has allowed the construction of genome-wide gene prediction sets, and the side-by-side comparison of different kinds of genome annotation, including chromosomal band locations, STS positions for genetic, radiation hybrid, YAC–STS and cytogenetic maps, GC content, density of various repeat families, CpG islands, ESTs, mRNAs, SNPs, and both known and predicted genes (International Human Genome Sequencing Consortium 2001). These comparisons are possible through the on-line genome browsers at UCSC (http://genome.ucsc.edu/goldenPath/hgTracks.html) and ENSEMBL (http://www.ensembl.org), both of which use the GigAssembler assembly. Some of the discoveries that have been made using these and other tools are described elsewhere (Bentley et al. 2001; Bock et al. 2001; Cheung et al. 2001; Clayton et al. 2001; Fahrer et al. 2001; Futreal et al. 2001; International Human Genome Sequencing Consortium 2001; International SNP Map Working Group 2001; Li et al. 2001; Murray and Marks 2001; Nestler and Landsman 2001; Pollard 2001; Riethman et al. 2001; Tupler et al. 2001; Wolfsberg et al. 2001; Yu 2001). However, the Web pages at http://genome.ucsc.edu are currently averaging >40,000 page requests per day, hence we suspect that the bulk of the new discoveries that this work has enabled have yet to be reported.
Acknowledgments
D.H. acknowledges support from DOE Grant F603–99ERG2849, NSF Grant DBI-9808007, Dean Patrick Mantey, and Chancellor M.R.C. Greenwood for the 100-node computational cluster. Thanks to Alan Zahler for his advice and encouragement. We thank Scot Kennedy, Terry Furey, Ray Wheeler, Aaron Tomb, Patrick Gavin, Nick Littlestone, and Paul Tatarsky for help testing the sequence and for technical support, and Ann Pace and Maria Guarino for administrative support. We thank Bob Waterston, Eric Lander, Francis Collins, Ewan Birney, Greg Schuler, John Sulston, and the rest of the genome analysis group for their advice and additional support.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Notes
The raft-ordering graph built in Steps 4 and 5 specifies a partial order on the midpoints of the rafts from a fingerprint clone contig. Additional constraints on this ordering are given by the distance ranges allowed for each bridge between rafts. The entire collection of partial order and distance constraints is represented by a conjunction of difference constraints, as defined, for example, in Cormen et al. (1990), pages 540–543. Each distance constraint has the form x − y ≤ B, where x and y are variables representing the midpoints of two rafts, and B is a constant, representing a bound on the number of base pairs that separate x and y. If it can be inferred from the partial order that x comes before (5′ of) y, then we can specify difference constraints that represent both an upper bound B and a lower bound b on the distance between x and y. The upper bound is expressed as y − x ≤ B. The lower bound is expressed with the distance constraint x − y ≤ −b. However, if the order of x and y are unknown, then only an upper bound B on the distance that separates them can be specified as part of a conjunction of distance constraints. This is specified by the two constraints x − y ≤ B and y − x ≤ B. A lower bound would require a disjunction of two distance constraints. Finally, if it is known that x comes before y but no distance bounds are known, then this fact can be represented by the distance constraint x − y ≤ 0.
The conjunction of distance constraints associated with a raft ordering graph has a feasible solution if and only if there is a placement of the rafts such that their midpoints are linearly ordered in a manner consistent with the partial order in the raft-ordering graph and the distances between the midpoints of bridged pairs of rafts are in the allowed distance ranges. The Bellman-Ford algorithm for single source shortest paths can be used to determine if a conjunction of distance constraints has a feasible solution in time proportional to the number of constraints (bridges) times the number of variables (rafts), as shown in Cormen et al. (1990). A similar approach was used for a physical mapping problem by Thayer et al. (1999). The feasibility problem for conjunctions of difference constraints is a special case of the linear programming problem and can also be solved by linear programming algorithms, but these can be slower.
In the GigAssembler algorithm, bridges between rafts are added incrementally in a greedy fashion, and the Bellman-Ford procedure is only used to test feasibility of a new bridge. Because a lower bound cannot be enforced on the distance between the midpoints of two unordered rafts in a conjunction of difference constraints, the solution of the conjunction of difference constraints may give a positioning of the midpoints of the rafts that puts some pairs of rafts too close, possibly overlapping them in a way that contradicts the sequence data. That is why Step 6 is necessary. It would be more elegant to combine disjunctions and conjunctions of distance constraints in specifying the raft layout problem, but unfortunately such a combination leads to an NP-hard feasibility problem (Papadimitriou and Steiglitz 1982). Thus, our final layout step employs a simple heuristic approach, rather than an integrated optimization of all constraints.
Footnotes
E-MAIL kent@biology.ucsc.edu; FAX (831) 459-4829.
Article published on-line before print: Genome Res., 10.1101/gr.183201.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.183201.
REFERENCES
- Anson E, Myers G. Proc. RECOMB '99, Lyon, France. 1999. Algorithms for whole genome shotgun sequencing; pp. 1–9. [Google Scholar]
- BAC Resource Consortium. Integration of cytogenetic landmarks into the draft sequence of the human genome. Nature. 2001;409:953–958. doi: 10.1038/35057192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Deloukas P, Dunham A, French L, Gregory SG, Humphrey SJ, Mungall AJ, Ross MT, Carter NP, Dunham I, et al. The physical maps for sequencing human chromosomes 1, 6, 9, 10, 13, 20 and X. Nature. 2001;409:942–943. doi: 10.1038/35057165. [DOI] [PubMed] [Google Scholar]
- Bock JB, Matern HT, Peden AA, Scheller RH. A genomic perspective on membrane compartment organization. Nature. 2001;409:839–841. doi: 10.1038/35057024. [DOI] [PubMed] [Google Scholar]
- Bonfield JK, Smith KF, Staden R. A new DNA sequence assembly program. Nucleic Acids Res. 1995;23:4992–4999. doi: 10.1093/nar/23.24.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung VG, et al. Integration of cytogenetic landmarks into the draft sequence of the human genome. Nature. 2001;409:953–958. doi: 10.1038/35057192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton JD, Kyriacou CP, Reppert SM. Keeping time with the human genome. Nature. 2001;409:829–831. doi: 10.1038/35057006. [DOI] [PubMed] [Google Scholar]
- Cormen TH, Leiserson CE, Rivest RL. Introduction to algorithms. Cambridge, MA: MIT Press; 1990. [Google Scholar]
- Dunham I, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, Smink LJ, Ainscough R, Almeida JP, Babbage A, et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- Fahrer AM, Bazan JF, Papathanasiou P, Goodnow CC. A genomic view of immunology. Nature. 2001;409:836–838. doi: 10.1038/35057020. [DOI] [PubMed] [Google Scholar]
- Futreal PA, Kasprzyk A, Briney E, Mullikin JC, Wooster R, Stratton MR. Cancer and genomics. Nature. 2001;409:850–852. doi: 10.1038/35057046. [DOI] [PubMed] [Google Scholar]
- Hattori M, Fujiyama A, Taylor TD, Watanabe H, Yada T, Park HS, Toyoda A, Ishii K, Totoki Y, Choi DK, et al. The DNA sequence of human chromosome 21. Nature. 2000;405:311–319. doi: 10.1038/35012518. [DOI] [PubMed] [Google Scholar]
- Huang W, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. An improved sequence assembly program. Genomics. 1996;33:21–31. doi: 10.1006/geno.1996.0155. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- International Human Genome Mapping Consortium. A physical map of the human genome. Nature. 2001;409:934–941. doi: 10.1038/35057157. [DOI] [PubMed] [Google Scholar]
- International SNP Map Working Group. A map of human genome sequence variation containing 1.4 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- Ji Y, Eichler EE, Schwartz S, Nicholls RD. Structure of chromosomal duplicons and their role in mediating human genomic disorders. Genome Res. 2000;10:597–610. doi: 10.1101/gr.10.5.597. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Haussler DH. Technical Report: UCSC-CDRL-00–17, Dec. 2000. 2000. GigAssembler: An algorithm for the initial assembly of the human genome working draft. [Google Scholar]
- Li WH, Gu Z, Wang H, Nekrutnko A. Evolutionary analyses of the human genome. Nature. 2001;409:847–849. doi: 10.1038/35057039. [DOI] [PubMed] [Google Scholar]
- Murray AW, Marks D. Can sequencing shed light on cycling? Nature. 2001;409:844–846. doi: 10.1038/35057033. [DOI] [PubMed] [Google Scholar]
- Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, Flanigan MJ, Kravitz SA, Mobarry CM, Reinert KHJ, Remington KA, et al. A whole-genome assembly of Drosophila. Science. 2000;287:868–877. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- Nestler EJ, Landsman E. Learning about addiction from the human draft genome. Nature. 2001;409:834–835. doi: 10.1038/35057015. [DOI] [PubMed] [Google Scholar]
- Papadimitriou CH, Steiglitz K. Combinatorial optimization. Englewood Cliffs, NJ: Prentice-Hall, Inc.; 1982. [Google Scholar]
- Pollard TD. Genomics, the cytoskeleton and motility. Nature. 2001;409:842–843. doi: 10.1038/35057029. [DOI] [PubMed] [Google Scholar]
- Riethman HC, Xiang Z, Paul S, Morse E, Hu XL, Flint J, Chi HC, Grady DL, Moyzis RK. Integration of telomeric sequences with the draft human genome sequence. Nature. 2001;409:948–951. doi: 10.1038/35057180. [DOI] [PubMed] [Google Scholar]
- Sutton GG, White O, Adams MD, Kerlavage AR. TIGR assembler: A new tool for assembling large shotgun sequencing projects. Genome Sci Tech. 1995;1:9–19. [Google Scholar]
- Thayer EC, Olson MV, Karp RM. Error checking and graphical representation of multiple-complete-digest (MCD) restriction-fragment maps. Genome Res. 1999;9:79–90. [PMC free article] [PubMed] [Google Scholar]
- Tupler R, Perini G, Green MR. Expressing the human genome. Nature. 2001;409:832–833. doi: 10.1038/35057011. [DOI] [PubMed] [Google Scholar]
- Wolfsberg TG, Schuler GD, McEntyre J. Guide to the draft genome. Nature. 2001;409:824–826. doi: 10.1038/35057000. [DOI] [PubMed] [Google Scholar]
- Yu A, Zhau C, Fan Y, Jang W, Mungall AJ, Deloukas P, Olsen A, Doggett NA, Ghebranious N, Broman KW. Comparison of human genetic and sequence-based physical maps. Nature. 2001;409:951–953. doi: 10.1038/35057185. [DOI] [PubMed] [Google Scholar]
- Zhao S, Malek J, Mahairas G, Fu L, Nierman W, Venter JC, Adams MD. Human BAC ends quality assessment and sequence analyses. Genomics. 2000;63:321–332. doi: 10.1006/geno.1999.6082. [DOI] [PubMed] [Google Scholar]