Abstract
Multiple signaling pathways ultimately modulate the epigenetic information embedded in the chromatin of gene promoters by recruiting epigenetic enzymes. We found that, in estrogen-regulated gene programming, the acetyltransferase CREB-binding protein (CBP) is specifically and exclusively methylated by the coactivator-associated arginine methyltransferase (CARM1) in vivo. CARM1-dependent CBP methylation and p160 coactivators were required for estrogen-induced recruitment to chromatin targets. Notably, methylation increased the histone acetyltransferase (HAT) activity of CBP and stimulated its autoacetylation. Comparative genome-wide chromatin immunoprecipitation sequencing (ChIP-seq) studies revealed a variety of patterns by which p160, CBP, and methyl-CBP (meCBP) are recruited (or not) by estrogen to chromatin targets. Moreover, significant target gene-specific variation in the recruitment of (1) the p160 RAC3 protein, (2) the fraction of a given meCBP species within the total CBP, and (3) the relative recruitment of different meCBP species suggests the existence of a target gene-specific “fingerprint” for coregulator recruitment. Crossing ChIP-seq and transcriptomics profiles revealed the existence of meCBP “hubs” within the network of estrogen-regulated genes. Together, our data provide evidence for an unprecedented mechanism by which CARM1-dependent CBP methylation results in gene-selective association of estrogen-recruited meCBP species with different HAT activities and specifies distinct target gene hubs, thus diversifying estrogen receptor programming.
Keywords: CREB-binding protein, CARM1/PRMT4, arginine methylation, estrogen signaling, genome-wide binding sites, gene networks
Nuclear hormone receptors (NRs) are ligand-activated transcription factors (TFs) that bind to cognate DNA response elements of target genes. They control gene expression by recruiting a repertoire of multicomponent enzymatic machineries to remodel the chromatin structure and assemble the transcriptional machinery. In the absence of ligands, some NRs are located in the nucleus, bind to the DNA response elements of target genes, and recruit corepressor complexes that contain histone deacetylases, which induce a repressive chromatin state at the target locus. The NR subfamily of steroid hormone receptors, unless exposed to certain antagonists, does not generally recruit corepressor complexes. Instead, these NRs are either located in the cytoplasm and/or kept inactive by chaperones. Yet, for all NRs, agonist binding triggers a common principle; namely, allosteric changes that lead to the exposure of receptor surface(s) for interaction with coactivators and dissociation of corepressors from the nonsteroid NRs. While a plethora of NR coactivators has been described, most of the structure–function studies have been done with the paradigmatic p160/SRC family (comprising SRC1/NCoA1, SRC2/TIF2/GRIP-1/NCoA2, and SRC3/RAC3/pCIP/AIB1/ACTR/TRAM/NCoA3) (Gronemeyer et al. 2004; Rosenfeld et al. 2006; Gronemeyer and Bourguet 2009; York and O'Malley 2010). The SRC coactivators provide, in turn, a scaffold for tethering secondary coactivators such as coactivator-associated arginine methyltransferase 1 (CARM1/PRMT4) and CREB-binding protein (CBP) to target genes. CARM1/PRMT4 (hereafter referred to as CARM1) belongs to the family of protein arginine methyltransferases of type I that catalyzes the formation of asymmetric dimethylarginine (Bedford and Clarke 2009). CARM1 was first identified in a yeast two-hybrid screen for proteins interacting with the p160 steroid coactivator GRIP1/TIF2 and was shown to interact with the activation domain 2 of all three p160 family members. CARM1 cooperates with the acetyltransferase CBP when recruited by p160 proteins via their activation domain 1 (for review, see DY Lee et al. 2005). Thus, p160 proteins constitute a platform for the formation of a multicomponent complex that contains distinct histone-modifying enzymatic activities: CBP is known to activate transcription by acetylating histone tails on specific lysine residues, whereas CARM1 methylates histone H3 on Arg 17 and Arg 26, which correlates with NR-mediated gene activation (Ma et al. 2001; Daujat et al. 2002).
Arginine methylation is not restricted to post-translational modification of histones. CARM1 also methylates a variety of nonhistone proteins (Bedford and Clarke 2009), including coactivators such as CBP/p300 (Xu et al. 2001; Chevillard-Briet et al. 2002; YH Lee et al. 2005) and p160 family members (Feng et al. 2006; Naeem et al. 2007). Different domains of CBP and p300 can be methylated by CARM1 in vitro. Methylation within the KIX domain of p300 abrogates its interaction with CREB (Xu et al. 2001), while methylation of CBP in a region further downstream has no influence on CREB signaling but is important for estrogen receptor (ER)-dependent transcription (Chevillard-Briet et al. 2002). Notably, methylation of p300 in this region is required for response to DNA damage (Lee et al. 2011). Finally, methylation of p300 within the p160 interaction domain (GBD, for GRIP-binding domain) has been proposed to disrupt the p300/p160 complex in vitro (YH Lee et al. 2005).
In the present study, we investigated the role of CBP methylation by CARM1 for ligand-induced CBP recruitment to ER-responsive target genes. Comparative genome-wide chromatin immunoprecipitation sequencing (ChIP-seq) showed the existence of distinct but overlapping binding site repertoires specific for each methyl-CBP (meCBP) species, suggesting that CBP methylation by CARM1 specifies estrogen-induced subprograms.
Results
Site-specific methylation of CBP by CARM1 in vivo
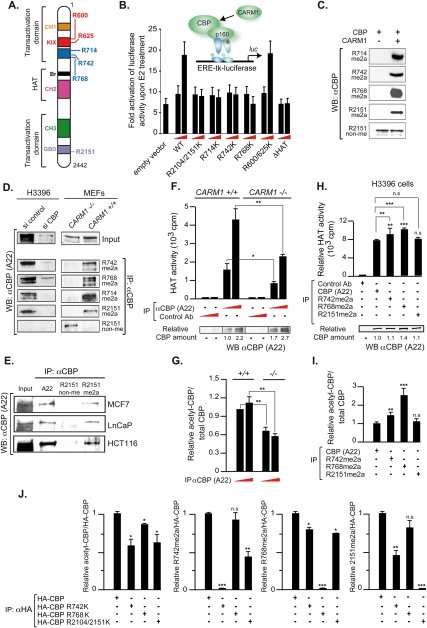
Both CBP and p300 are substrates for CARM1 that methylate six arginines in CBP/p300 in vitro (Fig. 1A; Xu et al. 2001; Chevillard-Briet et al. 2002; YH Lee et al. 2005). Transient transactivation assays were performed in estrogen-dependent H3396 breast cancer cells in the presence of a methylation-deficient point mutation of a single (R714K, R742K, and R768K) or two arginine (R2104/2151K) residues that correspond to the methylation site identified in p300 in the GBD (R2142) and to a site that is important for the activity of the p300–GRIP1 complex (Xu et al. 2001; Chevillard-Briet et al. 2002; YH Lee et al. 2005). These experiments revealed that CBP-mediated superactivation of an estrogen response element (ERE)-driven luciferase reporter required the integrity of the histone acetyltransferase (HAT) domain and all CARM1 target sites, with the exception of those in the KIX domain (Fig. 1B). To demonstrate that CARM1 methylates CBP and p300 not only in vitro but also in vivo, we generated monospecific polyclonal antibodies that recognize selectively the CARM1-methylated epitope encompassing CBP arginines required for estrogen-dependent transcription (CBPR714me2a, CBPR742me2a, CBPR768me2a, and CBPR2151me2a) or the epitope comprising nonmethylated R2151 (CBPR2151non-me) by ELISA (Supplemental Material); the antibodies were validated by Western blotting of recombinant in vitro methylated or nonmethylated CBP (Fig. 1C). Immunoprecipitations of endogenous CBP from H3396 cells with anti-meCBP antibodies, followed by immunoblot analysis with the general A22 anti-CBP antibody, demonstrated that CBP is methylated on these residues in vivo (Fig. 1D, left panel). Knockdown of CBP by RNAi obliterated immunodetection of all methyl-selective antibodies, thus confirming that they recognized CBP epitopes. Analysis of mouse embryo fibroblasts (MEFs) lacking CARM1 (Yadav et al. 2003) demonstrated that CARM1 is the only enzyme that methylates CBP on these residues in vivo and confirmed antibody selectivity. Indeed, no meCBP was immunoprecipitated from CARM1−/− MEFs using antibodies specific for the various asymmetrically dimethylated arginines, while CBP was readily detected with the identical antibodies in immunoprecipitations from CARM1+/+ MEFs (Fig. 1D, right panel). While CBP nonmethylated at R2151 (CBPR2151non-me) was detected in CARM1−/− MEFs, no such species was observed in CARM1+/+ or H3396 cells, which confirms the antibody selectivity for nonmethylated R2151. CBPR2151non-me was also absent in MCF7, LnCaP, HCT116, and several other cells (Fig. 1E; data not shown). Thus, in cells expressing functional CARM1, CBPR2151 is mainly, if not exclusively, methylated.
Figure 1.
CBP is exclusively methylated by CARM1 in vivo and methylation increases its HAT activity. (A) CBP methylation sites identified in vitro. (CH1-3) Zinc fingers 1–3; (Br) bromodomain. (B) Transient transactivation assays of an estrogen-responsive luciferase reporter in H3396 breast cancer cells transfected with 100 ng or 1 μg of CBP or its methylation-deficient mutants. (C) Recombinant CBP fragments nonmethylated or methylated in vitro by CARM1 were detected by immunoblotting with the antibodies recognizing the meCBP/non-meCBP species as indicated. (D) Immunoprecipitations of H3396 nucleofected with siRNAs against CBP or control siRNA (left panel) or of CARM1−/− and CARM1+/+ extracts (right panel) with the indicated antibodies followed by immunoblotting with the general CBP-A22 antibody. (E) MCF7, LnCaP, and HCT116 nuclear extracts were immunoprecipitated with the indicated antibodies, and precipitated proteins were analyzed by immunoblot with CBP-A22 antibody. (F,G) Immunoprecipitations of extracts from CARM1−/− and CARM1+/+ MEFs with CBP (A22) or a control antibody. (Top panel) Different amounts of the identical immunoprecipitation were subjected in parallel to HAT assays using histone H3 peptide as a substrate, and to immunoblotting with CBP A22 or an antibody recognizing autoacetylated CBP. (Bottom panel) CBP amounts were quantified using ImageJ, and graphs were from at least three independent experiments. (H,I) Immunoprecipitations of H3396 cell extracts with CBP (A22) and antibodies recognizing CBPR742me2a, CBPR768me2a, and CBPR2151me2a or a control antibody. Immunoprecipitations were subjected in parallel to HAT assays and to immunoblotting with CBP (A22) or an antibody recognizing CBP autoacetylation, and quantifications were as in F and G. (J) Immunoprecipitations with an anti-HA antibody of H3396 cell extracts transfected with HA-CBP wild type or HA-CBP mutated on R742, R768, or R2104/2151 into lysines were subjected to immunoblotting with antibodies recognizing CBP autoacetylation, CBPR742me2a, CBPR768me2a, and CBPR2151me2a and quantifications relative to CBP wild type were as in F and G. Statistically significant differences are shown: (*) P < 0.01; (**) P < 0.001; (***) P < 0.001.
CBP methylation increases its HAT activity and its autoacetylation
The acetyltransferase (HAT) activity of CBP is important for coactivation and phosphorylation can modulate its enzyme activity and substrate specificity (Ait-Si-Ali et al. 1998; Huang et al. 2007). To test whether methylation regulates its HAT activity, CBP was immunoprecipitated from CARM1−/− and CARM1+/+ MEFs, and HAT assays were performed with a histone H3 peptide as a substrate. Interestingly, the HAT activity of CBP from CARM1−/− MEFs was markedly decreased relative to that from wild-type MEFs (Fig. 1F), indicating that CARM1-dependent CBP methylation increases its HAT activity. As autoacetylation regulates HAT activity of p300/CBP (Thompson et al. 2004; Black et al. 2006; Arif et al. 2007), we tested whether methylation regulates CBP HAT activity by altering autoacetylation. Indeed, Western blots with an antibody recognizing autoacetylated CBP in immunoprecipitations from CARM1+/+ or CARM1−/− MEFs revealed that, relative to total CBP, the fraction of autoacetylated CBP was strongly increased when CARM1 was expressed (Fig. 1G). That the same CARM1+/+ MEFs also display a largely enhanced HAT activity of CBP (Fig. 1F) supports a mechanism by which methylation by CARM1 increases CBP autoacetylation, which in turn enhances its HAT activity.
To analyze whether specific methylation sites were involved in modulating HAT activity, antibodies recognizing CBP methylated selectively at R742, R768, or R2151 were used to immunoprecipitate CBP that was modified on these sites in H3396 cells in vivo. Notably, methylation at R768—and, to a lesser extent, R742—increases HAT activity as compared with global CBP (Fig. 1H). Not surprisingly, methylation at R2151 did not have any impact on HAT activity relative to global CBP, as our study indicates that most, if not all, CBP is methylated at this site. Finally, immunoprecipitation of the various methyl species used for HAT assays that were immunoblotted in parallel for CBP autoacetylation showed a direct correlation between CBP HAT activity and autoacetylation (Fig. 1I). Immunoprecipitations of H3396 cells transfected with either wild-type or mutant CBPs further supported that each CBP methylation site is required for efficient HAT activity. The mutations R742K and R2104K/R2151K—and, to a lesser extent, R768K—impaired CBP autoacetylation, revealing that the integrity of these residues is important for CBP HAT activity (Fig. 1J, left panel, note that similar results were obtained with the single CBPR2151K mutant). Interestingly, immunoblotting the same immunoprecipitations with antibodies recognizing CBP methylated selectively at R742, R768, or R2151 revealed a cross-talk between these sites. Indeed, mutations R742K and R2151K strongly affected reciprocal methylation, while R768K decreased methylation at R742 and R2151 moderately (Fig. 1J). Altogether, our data support a model in which the methylation and integrity of CBP at R742, R768, and R2151 stimulate CBP HAT activity by increasing its autoacetylation.
Methylation and interaction of CBP with p160 proteins are critically required for recruitment to ERα target genes
The synergy between CARM1 and CBP for steroid hormone receptor coactivation depends on the integrity of CARM1-dependent CBP methylation in its central region (R714, R742, and R768) and in the GBD (R2151) (Fig. 1A,B; Chen et al. 2000; Chevillard-Briet et al. 2002; YH Lee et al. 2005). As it has been proposed that methylation of the p300 GBD plays an important role in NR coactivator complex assembly (YH Lee et al. 2005), we investigated whether methylation affects recruitment of CBP to the prototypic estrogen-responsive TFF1 promoter in H3396 cells, which express high levels of ERα and CBP (Supplemental Fig. 1A–C). That estrogen induced the level of “active” chromatin marks at the TFF1 promoter (including H3K9ac, CARM1-dependent H3R17me2a, and the recruitment of ERα, TIF2, RAC3, CARM1, CBP, and p300) (Supplemental Fig. 1D–F) confirmed a functional estrogen response in H3396 cells.
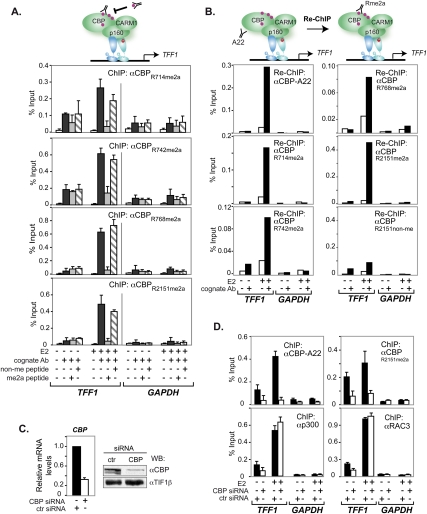
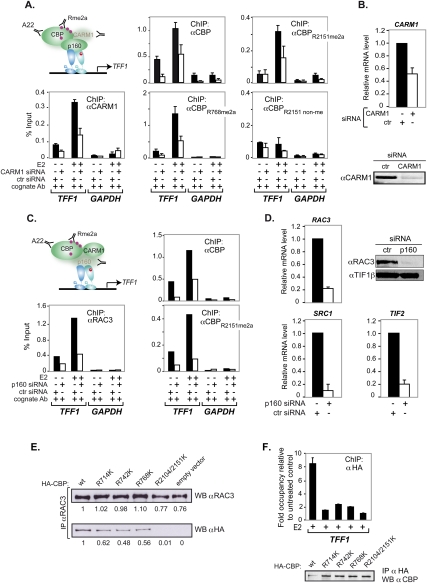
ChIP analyses demonstrated that estrogen recruited all methylated CBP species to TFF1 (Fig. 2A). Similar results were obtained for other ERα target genes (Supplemental Fig. 2). Competition with the cognate R-methylated but not the nonmethylated peptides dramatically reduced the ChIP signals, confirming the selective recognition of meCBP species (Fig. 2A). Re-ChIPs proved that the antibodies indeed recognized CBP rather than protein(s) harboring identical/cross-reacting epitopes (Fig. 2B). Knocking down CBP in H3396 cells demonstrated that CBP deficiency alters CBPR2151me2a recruitment, but not that of p300 or RAC3, and excluded cross-reaction with p300 (Fig. 2C,D). Identical results were obtained with the other methyl-selective CBP antibodies, confirming recruitment of all four methylated CBP species to TFF1 (data not shown). As expected, CARM1 knockdown decreased total CBP and meCBP recruitment by estrogen to TFF1 (Fig. 3A,B). No compensatory recruitment of CBPR2151non-me was seen, thus providing initial evidence indicating that CBP methylation is important for binding to TFF1 (see also below).
Figure 2.
Methylated CBP was recruited to the TFF1 promoter. (A) H3396 cells, treated with E2 or vehicle (EtOH) for 1 h, were subjected to ChIP with the indicated antibodies. Bound DNA was amplified by real-time PCR with specific primers for TFF1 and GAPDH promoters. For peptide competition, 100-fold molar excess of either the corresponding cognate peptide or an irrelevant control peptide was added to the antibody prior to ChIP. The means ± standard deviations are from at least two independent experiments. (B) H3396 cells were subjected to a first ChIP using the general CBP antibody (A22), and complexes were eluted and subjected to a second immunoprecipitation with or without specific antibody as indicated. Bound DNA was amplified as in A. (C) Knockdown of CBP monitored by RT–PCR (left panel) and Western blotting (right panel). CBP mRNA levels are shown relative to those of 36B4. Immunoblots were done with A22, and TIF1β was the loading control. (D) H3396 cells were nucleofected with siRNAs directed against CBP or a control siRNA (ctr). After 48 h, cells were treated with E2 or vehicle (EtOH) for 1 h and subjected to ChIP with or without the indicated antibodies. Bound DNA was amplified as in A.
Figure 3.
CARM1-dependent methylation and interaction with p160/SRC coactivators were required for ligand-induced CBP recruitment to TFF1. (A) H3396 cells were nucleofected with a siRNA against CARM1 or a control siRNA. After 48 h, cells were treated with E2 or vehicle for 1 h and ChIP assays were done with the indicated antibodies. Bound DNA was amplified by real-time PCR with specific primers for TFF1 and GAPDH promoters. The means ± standard deviations are from at least two independent experiments. (B, top) Knockdown of CARM1 monitored by RT–PCR. CARM1 mRNA levels are displayed relative to 36B4 mRNA. (Bottom panel) Expression of CARM1 was assessed by immunoblotting. (C) H3396 cells were nucleofected with a siRNA against p160 or a control siRNA (ctr). After 48 h, cells were treated with estrogen (E2) or vehicle for 1 h and ChIP assays were done with the indicated antibodies. Bound DNA was amplified by real-time PCR as in A. (D) Knockdown of p160s monitored by RT–PCR and immunoblotting. P160 mRNA levels are displayed relative to 36B4 mRNA. (Top right panel) Expression of RAC3 was assessed by immunoblotting, and TIF1β was the loading control. (E) Immunoprecipitations with an anti-RAC3 antibody of H3396 cell extracts transfected with HA-CBP wild type or HA-CBP mutated on R714, R742, R768, or R2104/2151 into lysines were subjected to immunoblotting with antibodies recognizing either RAC3 (top panel) or HA (bottom panel). Quantifications of RAC3 and HA-CBP were done relative to the expression in CBP wild-type condition using ImageJ software. (F) H3396 cells stably infected with retroviral vector expressing HA-CBP wild type or the methylation-deficient CBP mutants HA-CBPR714K, HA-CBPR742K, HA-CBPR768K, and HA-CBPR2104/2151K were treated with E2 or vehicle (EtOH) for 1 h and subjected to ChIP with an anti-HA antibody. Bound DNA was amplified by real-time PCR with specific primers for TFF1 and DPP10 promoters; DPP10 was used as a cold region for ChIP reference. Results are expressed as FO relative to DPP10 relative to ethanol controls. (Bottom panel) Amounts of expressed HA-CBP and methylation-deficient mutants were determined by immunoblotting with anti-HA antibody.
It is commonly believed that p160 proteins act as assembly factors for CBP recruitment, but formal proof was lacking. The collective knockdown of all three p160s significantly reduced estrogen-dependent binding of global CBP and CBPR2151me2a to TFF1, revealing unequivocally their involvement in CBP recruitment (Fig. 3C,D). Coimmunoprecipitations (co-IPs) with antibodies directed against global CBP (A22), CBPR742me2a, CBPR768me2a, or CBPR2151me2a confirmed that CBP methylated at R742, R768, or R2151 is fully capable of interacting with RAC3 in vivo (data not shown). The above results show CBP recruitment to TFF1 in a p160 and CARM1-dependent fashion in vivo. Importantly, co-IPs of transiently expressed HA-tagged CBP mutants R714K, R742K, or R768K revealed significantly impaired in vivo RAC3 binding of the mutants and a complete loss of RAC3 binding of CBPR2104K,2151K as compared with the corresponding HA-CBP wild type (Fig. 3E). Moreover, ChIP experiments performed on TFF1 and several ER target genes in H3396 cells stably expressing CBP or CBP methylation-deficient mutants (R714K, R742K, R768K, and R2104K/2151K) revealed that CBP wild type but none of the mutants was efficiently recruited to ER target promoters, albeit some residual recruitment was still observed (Fig. 3F; data not shown). Together, these results demonstrate that the integrity of these CARM1-methylated arginines is important for RAC3 coactivator interaction, and thus for efficient CBP recruitment to ER target genes.
Genome-wide binding patterns of meCBP species upon ER activation
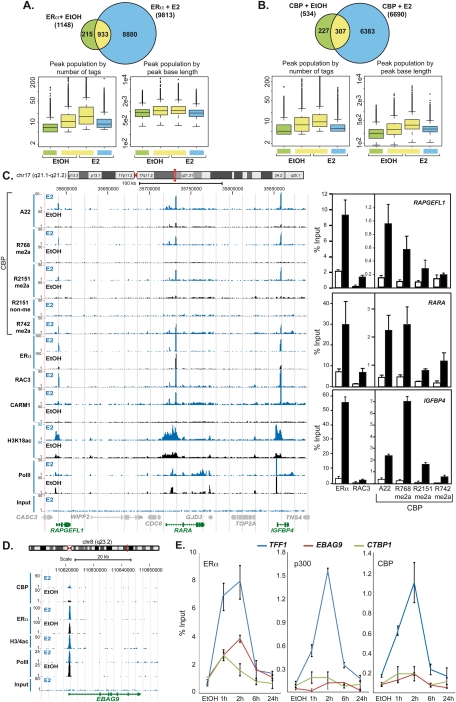
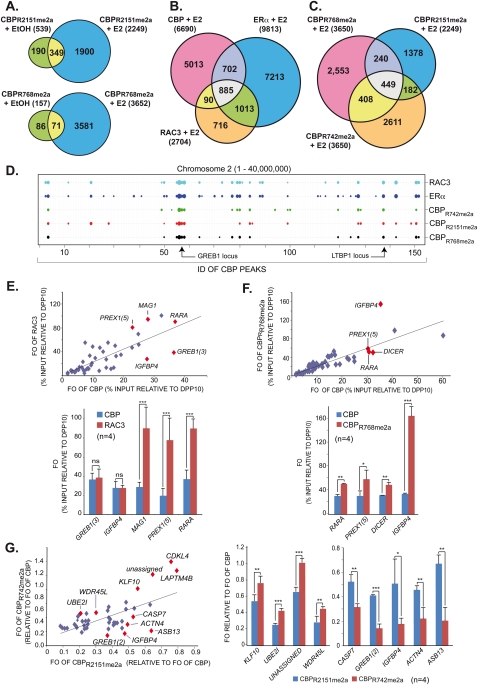
Comparative chromatin-binding profiles for ERα, RAC3, CARM1, CBP and its CARM1-methylated species, global H3 and H3K18 acetylation (a histone mark that is catalyzed by CBP and p300), and RNA polymerase II in the presence and absence of estrogen were established by ChIP-seq. Peak prediction was performed by MACS using P-values optimized by receiver operating characteristic (ROC) curve analyses from the comparison of 68 loci with the experimentally determined results obtained by ChIP coupled to real-time PCR (ChIP-qPCR) (see the Supplemental Material). A 1-h pulse of estrogen led to a dramatic nearly 10-fold increase of ERα-binding sites due to de novo ERα recruitment to nearly 9000 new sites, and resulted concomitantly in a large increase of loci (>6600) occupied by CBP (Fig. 4A,B). ChIP-seq profiles were extensively validated by ChIP-qPCR, as depicted for the loci encompassing RAPGEFL1-RARA-IGFBP4, GREB1, or DICER (Fig. 4C; Supplemental Fig. 3A–C) and a large number of additional binding sites (Supplemental Material). Comparison with the ERα cistrome mapped in MCF7 (Carroll et al. 2006; Lin et al. 2007; Welboren et al. 2009) revealed a core of 284 commonly identified sites and 4915 sites found by at least two groups (Supplemental Fig. 3D). The two groups using ChIP-seq report 2333 common sites even though different cell lines (H3396 and MCF7) were used. Note that, in the present study, the colocalization patterns apparent from the comparative global mapping of CBP, its methylated species, ERα, and RAC3 reveal the high precision of the present binding site analysis and confirm at the same time the absence of nonmethylated CBPR2151 at a global scale.
Figure 4.
Genome-wide binding patterns of estrogen-recruited ERα and CBP species. (A,B) Venn diagrams and box plots revealing the numbers of peaks called by MACS from the ChIP-seq experiments for ERα and CBP. Parameters for optimal sensitivity and specificity of peak detection by MACS version 1.3.7.1 were derived from ROC curves (Supplemental Material). Peak size and overlap analysis was done with ChordChart. The Venn diagrams display peak numbers detected for the indicated conditions; “E2” indicates 1 h of treatment. The box plots illustrate the characteristics of peak populations with respect to number of tags or peak base lengths in base pairs. (C, left panel) Signal peak representation in UCSC (University of California at Santa Cruz) Genome Browser format of ChIP-seq profiles for ERα, CARM1, RAC3, CBP (A22), and the indicated R-methylated or nonmethylated CBP species after 1 h of estrogen or vehicle exposure. Patterns of histone H3K18 acetylation and RNA polymerase II binding are shown for comparison. (Right panels) Binding sites near the RAPGEFL1, RARA, and IGFBP4 genes were validated independently by ChIP-qPCR. (D) Comparative binding site patterns for CBP, ERα, RNA polymerase II, and H3/4 acetylation at the putative EBAG9 promoter. (E) Kinetics of estrogen-induced binding of ERα, CBP, and p300 to TFF1, EBAG9, and CTBP1 in H3396 cells treated with E2 or vehicle for the indicated time. Chromatin-immunoprecipitated DNA was amplified by real-time PCR with binding site-specific primers. The means ± standard deviations are from at least two independent experiments.
That apo-ERα and CBP were detected at 1148 and 534 loci, respectively, implies that, under hormonally naive conditions, CBP was not strongly engaged by other transcription regulators, at least not to an extent comparable with its recruitment by holo-ERα (Fig. 4A,B). Interestingly, estrogen had a rather different effect on the binding characteristics of pre-existing and induced sites. As expected, the mean number of tags increased for pre-existing sites, but de novo binding sites displayed, on average, fewer tag counts, and sites that got lost had the lowest amounts of tags. In the case of CBP, but not ERα, the average size of isolated tags increased upon ligand exposure for preoccupied sites. This suggests that preoccupied loci are poised to respond more rapidly and/or more efficiently to the hormonal stimulus. The increased size of CBP-binding sites is compatible with a model in which holo-ERα initiates local events that lead to recruitment of additional CBP-interacting factors, thereby stabilizing a complex that occupies a larger region.
Estrogen action induces different patterns of cofactor binding to target sites
Comparative analysis of ChIP-seq profiles revealed a locus-specific variability in the recruitment of cofactors by ERα. For the majority of known ER target genes, ERα recruitment occurs in a hormone-dependent manner concomitantly with the binding of coactivator RAC3, CBP, and its methylated species, resulting in promoter-proximal histone H3K18 acetylation, while polymerase II-binding profiles differ only for a few loci (Fig. 4C; Supplemental Fig. 3A–C). However, we noted several alternative binding paradigms. While TFF1 binds efficiently ERα, RAC3, CBP, and p300, the ER-binding fragment-associated antigen 9 (EBAG9), an estrogen-inducible putative regulator of tumor progression, and CTBP1 bind ERα at the transcriptional start sites (TSSs) but display weak RAC3 (data not shown) and little if any binding of CBP or p300 (Fig. 4D,E). Despite the apparent absence of these two HATs, there is a strong peak of locally defined histone H3/4 acetylation and polymerase II binding to the same site, suggesting that ERα recruits a distinct HAT. Another rather unique example of differential binding between meCBP species is CARM1, which binds in the presence of estrogen mainly CBPR742me2a at the TSS and several exonic sites correlating with peaks of histone H3/4 acetylation (Supplemental Fig. 4A). Another unusual binding paradigm was observed at the CBP gene (CREBBP) itself, which bound ERα and meCBP species, particularly CBPR742me2a, in the absence of a ligand. Estrogen destabilized (me)CBP binding but did not affect that of ERα (Supplemental Fig. 4B). Together, these results reveal that, in contrast to the simplified view according to which holo-ERα recruits coactivators, which in turn recruit HATs, there is a plethora of alternative binding modes that impact on the efficiency of recruitment of these factors, most likely reflecting promoter complexity and chromatin constitution and dynamics.
Estrogen induces distinct binding patterns of meCBP species
As for CBP, estrogen exposure of H3396 cells resulted in de novo binding of meCBP species to ERα target sites. A total of 2249 sites, of which 539 pre-existed, became occupied by CBPR2151me2a, and 3652 sites were bound by CBPR768me2a, of which very few pre-existing sites (157) were seen for this species (Fig. 5A). Not all ERα-binding sites recruited RAC3 and/or CBP, as 885 sites were co-occupied by all three factors, while 1013 and 702 sites were shared solely by ERα and either RAC3 or CBP, respectively (Fig. 5B). The 90 sites co-occupied by RAC3 and CBP are candidates for ERβ-dependent recruitment or indirectly induced binding. Among the three meCBP species, all combinations of binding site co-occupancy were observed with 449 sites commonly occupied by CBPR742me2a, CBPR768me2a, and CBPR2151me2a (Fig. 5C). However, between 182 and 408 sites were co-occupied by only two meCBPs and >1000 sites were bound by only one of the species. To illustrate the global binding patterns of the various factors and CBP species, we developed ChordCharts (Supplemental Material), which can be weighted according to the binding site-associated P-values or tag numbers (Fig. 5D). Such ChordCharts visualize site-selective recruitment and possible divergent functionality/signaling of these various meCBP species.
Figure 5.
Differential global estrogen-induced recruitment patterns of methylated CBP species. (A) Venn diagrams displaying pre-existing and de novo generated chromatin-binding sites in H3396 at global scale for CBPR2151me2a and CBPR768me2a after 1 h of estrogen or vehicle treatment. (B) Venn diagrams as in A revealing E2-induced recruitment of CBP, ERα, and RAC3 to common and divergent binding sites. (C) Venn diagram as in B illustrating the recruitment of CBPR768me2a, CBPR2151me2a, and CBPR742me2a to common and divergent binding sites upon E2 treatment. (D) ChordChart representation of binding patterns for the indicated factors at chromosome 2 (1–40,000,000 bp). The X-axis comprises the cumulative CBP IDs of consecutive peaks along the chromosome. Peaks for the different factors are represented by weighted dots according to the number of tags generating a peak. Binding sites at GREB1 and LTBP1 are indicated. (E) Scatter plot (top panel) derived from ChIP-qPCR validations (bottom panel) of E2-induced binding sites identified by ChIP-seq displaying FOs (relative to DPP10 site 1) of CBP relative to those of RAC3 at multiple loci, some of which are indicated; only ERα-binding loci were considered. (F) Scatter plot as in E but revealing divergent FOs of CBP and CBPR768me2a (relative to DPP10 site 1) at E2-induced ERα-binding sites. (G) Differential recruitment of methylated CBP species to individual ERα target genes. Scatter plot (left panel) derived from ChIP-qPCR validations (right panels) for E2-induced ERα-binding loci. The scatter plot displays normalized FOs for CBPR2151me2a relative to CBPR742me2a. FOs for the meCBP species were normalized relative to the FOs of bulk CBP at the same site, thereby taking into account variations of total CBP binding. Statistically significant differences are shown: (*) P < 0.01; (**) P < 0.001; (***) P < 0.0001.
To validate the existence of divergent site-specific binding patterns, we used quadruplicate ChIP-qPCR. Scatter plots of close to 50 binding sites revealed an astounding heterogeneity. Comparing the DPP10-calibrated fold occupancies (FOs) at a given site for CBP and RAC3 revealed a spectrum of relative occupancies in which, for example, the MAG1 site exhibited a threefold higher occupancy for RAC3 than the IGFBP4 site (Fig. 5E). Note the statistically highly significant differences between RAC3 and CBP FOs for MAG1, PREX1(5), and RARA, while these FOs are not significantly different for GREB1(3) and IGFBP4. Comparing the FOs of CBP and its R768me2a species revealed a much higher abundance of CBPR768me2a at IGFBP4 than at RARA, PREX1(5), and DICER sites (Fig. 5F). Similar conclusions about the heterogeneity of binding site occupancies can be drawn for the methylated CBP species. To this end, FOs for CBPR742me2a and CBPR2151me2a were calibrated relative to those of total CBP; the corresponding scatter plot reveals very clearly a large spectrum of relative binding occupancies of the two species for different loci (Fig. 5G). Together, the above data show an enormous variability of the relative bindings of different factors (i.e., TFs and cognate coregulators) and, moreover, of epigenetically modified HAT cointegrators to ERα target genes.
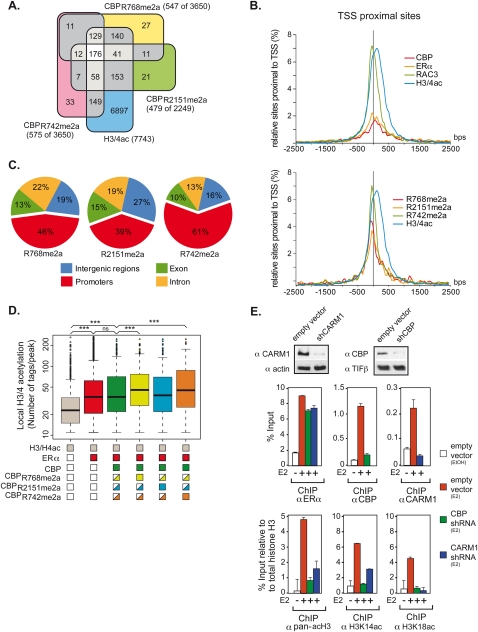
Different global positioning and divergent HAT activity of meCBP species at TSS
HATs are enriched at TSSs (Wang et al. 2009). Plotting binding sites found 10 kb upstream of and downstream from TSSs (Fig. 6A) revealed a dual peak of ERα: one at a position where RAC3 and meCBP species also peak, and a second one ∼100 base pairs (bp) further downstream, while total TSS-proximal CBP hits the highest point between the ERα peaks (Fig. 6B). H3/4 acetylation peaks at the position of the downstream peak of ERα but shows a shoulder at the TSS position. Comparing promoter acetylation with the presence of a particular CBP species revealed major differences (Fig. 6C). While <40% of the sites bound by CBPR2151me2a also revealed acetylated histones, >60% of the loci were H3/4-acetylated when CBPR742me2a was recruited to this site. CBPR768me2a binding correlated with acetylation of 46% of the bound sites. While these results revealed a correlation between the recruitment of a particular meCBP to a given promoter region and the likelihood of that region to be acetylated, it did not allow a direct correlation between the extent of acetylation and the binding of a meCBP species. To obtain this information, the average number of tags showing H3/4 acetylation was compiled for all species at a genome-wide level (Fig. 6D). The recruitment of ERα to a locus even in absence of CBP strongly increased the acetylation level of that site, and the corecruitment of ERα and CBP did not significantly change acetylation levels. This indicates that, in the majority of cases, ERα binding results in recruitment of a HAT other than CBP, and that holo-ERα binding without any HAT corecruitment is the exception. Importantly, however, the average level of histone acetylation was increased significantly over that seen at ERα-occupied sites when either CBPR768me2a or CBPR742me2a was corecruited. These results are fully in line with the decreased HAT activity of CBP observed in in vitro assays with CARM1−/− cells (Fig. 1F) and the increased activity found in immunoprecipitations in H3396 cells with specific antibodies against CBPR768me2a or CBPR742me2a as compared with total CBP (Fig. 1H). The above data confirm at a global level that methylation of CBP at R742 and/or R768 increases its HAT activity in vivo.
Figure 6.
Global analysis reveals meCBP-specific positioning and HAT activity. (A) Venn diagrams showing the number of common peaks of meCBP species ±10 kb around the TSS. (B) Density plots of the sites around the TSS are shown for CBP, ERα, RAC3, H3/4ac, and the meCBP species. (C) Global distribution of meCBP species. Only sites binding a single meCBP species were considered. (D) Correlation between global H3/4 acetylation and recruitments of ERα and (me)CBP. The box plot shows the peak populations for loci that display peaks for H3/4ac only (gray); H3/4ac and ERα (red); H3/4ac, ERα, and CBP (A22) (dark green); H3/4ac, ERα, and CBPR768me2a (light green); H3/4ac, ERα, and CBPR2151me2a (light blue); and H3/4ac, ERα, and CBPR742me2a (orange). Painted boxes below the X-axis indicate the presence of signal and half-painted boxes indicate the presence of other CBP species at some of these loci. H3/4 acetylation is increased statistically significant (P < 0.0001; Mann-Whitney test) at loci containing ERα alone and at those containing ERα together with (me)CBP. Populations of peaks at loci containing CBPR768me2a (yellow) or CBPR742me2a (orange) exhibit a statistically significant increase of H3/4 acetylation if compared with the entire population of CBP-positive sites (revealed by the A22 antibody; green) or loci (blue) occupied by CBPR2151me2a (P < 0.0001; Mann-Whitney test). (E, top panel) Cell extracts from H3396 cells transiently infected with either empty PLKO lentivirus or PLKO containing shRNAs directed against CBP or CARM1 were subjected to immunoblotting with anti-CBP and anti-CARM1 antibody 72 h post-infection. TIF1β and actin were used as loading controls for CBP and CARM1, respectively. (Bottom panels) H3396 infected with shRNAs directed against CBP and CARM1 were subjected to ChIP assays with the indicated antibodies after 1 h of treatment with E2 or vehicle (EtOH). Bound DNA was amplified by qPCR with primers specific to TFF1 promoter. Error bars correspond to standard deviations.
In order to further support that the divergent HAT activities at the most proximal TSSs are due mainly to differential methylation of CBP and are not mediated by another HAT, subclones of H3396 cells expressing lentiviral shRNAs against CARM1 or CBP to decrease endogenous CARM1 and CBP protein levels were generated. These cell lines were used in ChIP experiments on TFF1 and several other ER target genes with antibodies against ERα, CBP, CARM1, pan-acetylated histone H3, and histone H3 acetylated on K14 or K18, two well-documented histone H3 marks that are catalyzed by CBP and p300 in vivo (Ferrari et al. 2008; Horwitz et al. 2008). Our data showed that acetylation of histone H3 at K14 and K18 was strongly decreased in the absence of CBP (shCBP) but also in the absence of CARM1 (shCARM1) (Fig. 6E; data not shown), demonstrating that both CBP and CARM1 are involved in histone H3 acetylation at E2-responsive promoters and that CBP methylation is directly or indirectly responsible for the acetylation status of histones on chromatin.
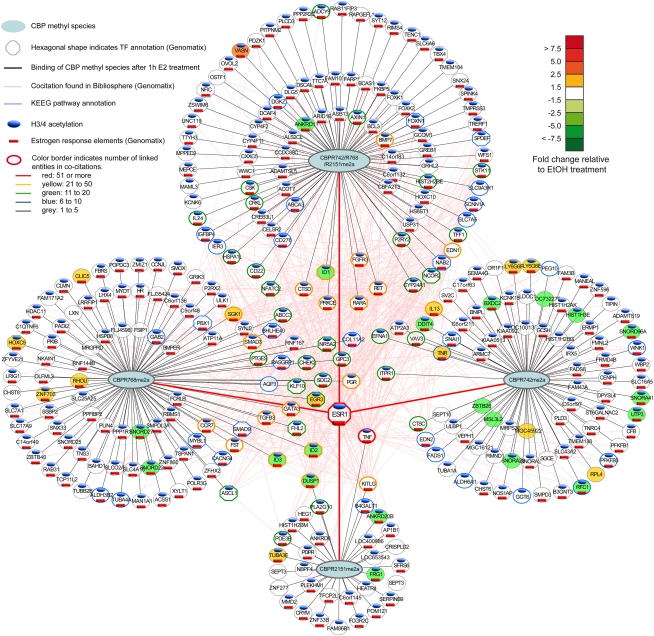
meCBP species specify distinct gene hubs within the estrogen-regulated gene network
Transcription profiling at 1, 2, 6, and 24 h after estrogen exposure identified ∼1100 genes that were regulated by estrogen directly or indirectly by >1.5-fold (Supplemental Fig. 5A). A comparison between this gene set and genes to which ERα and the meCBP species had been recruited at 1 h identified ∼400 genes (Fig. 7; Supplemental PowerPoint file) that are likely to correspond to primary targets. Indeed, the large majority of these genes harbor EREs (red bars below names in Fig. 7; Supplemental PowerPoint file) and display acetylation at their TSS (blue bars above names in Fig. 7; Supplemental PowerPoint file). Note that, in this respect, de novo motif discovery for the chromatin-binding sites revealed, as expected, EREs for all meCBP species (data not shown). The other estrogen-regulated genes are likely to comprise CBP-independent and indirectly regulated ER target genes. Among the CBP-dependent ER target genes (referred to as “ER-CBP regulon”), we separated four classes of regulated gene sets: those recruiting all three species, and three sets that bound CBP methylated at only a single site. Analysis of these sets revealed major functional differences. Most notably, the CBPR768me2a gene set comprised nearly 20% of TFs, while 2% of TFs were found in the CBPR742me2a gene set (Supplemental Fig. 5B). Moreover, the fraction of down-regulated genes was significantly higher (50%) when only CBPR742me2a was recruited upon estrogen exposure than CBPR768me2a (31%). This was particularly obvious at the 6-h time point, when the highest gene regulatory activity was measured. Genes to which all three species were recruited comprised 22% of TFs and were generally up-regulated (87%) by estrogen (Supplemental Fig. 5B–D).
Figure 7.
Illustration of the ER-meCBP regulon and its functional status 1 h after estrogen treatment. A comprehensive illustration of the regulon up to 24 h after treatment and of the pathways in which the various ER targets are involved is shown as a Supplemental PowerPoint file. The regulon was constructed by crossing transcriptomics profiles at 1, 2, 6, and 24 h after estrogen induction (only genes with >1.5-fold changes of expression were considered) with the ChIP-seq data at 1 h, considering genes in 10 kb vicinity of meCBP binding sites. The four hubs comprise regulated genes that bind either all three meCBP species simultaneously or only one of the three species alone. Note that, in the case of triple species binding, it is not possible to distinguish between three separate meCBP molecules and a single CBP bearing all three methyl tags. Within the regulon, TFs are indicated as well as the regulation status (color-coded), the presence of histone acetylation in the vicinity of the TSS, and the presence of EREs (codes are specified in the figure). The connectivity of the various genes in the ER-meCBP regulon was derived from cocitation analysis and is depicted by the color of the gene circle according to the code given in the figure. This connectivity is illustrated by lines connecting the various genes, and places most of the TFs in the center of the regulon.
An investigation of the relationships between the genes of the ER-CBP regulon by cocitation (Genomatix) and KEGG pathway analysis (DAVID) revealed an intricate network of temporally regulated ER target genes to which single or multiple meCBP species are recruited. This network can be deconvoluted into hubs according to the binding pattern of meCBP species (displayed as circles in Fig. 7; Supplemental PowerPoint file) and a central component of highly cocited targets that comprise TFs (e.g., EGR3, KLF10, GATA3, FLH2, ID1, SMAD1, and MYBL1), including nuclear receptors (e.g., ER, PGR, RARA, and NR5A2), cytokines (e.g., TNF and IL13), growth factors or growth factor receptors (e.g., TGFB3, KITLG, and FGFR), kinases (e.g., EFNA1, RET, PRKCE, and SGK1), cell cycle regulators (e.g., CHEK2), or other types of pleiotropic regulators (e.g., ITPR1, DUSP1, RNF167, SDC2, and RASGRP1). It is worth noting that factors belonging to the same gene family were found to involve differential recruitment of meCBP species. For example, the progesterone receptor gene associated with CBPR768me2a only, while all three meCBP species were recruited to the promoter of the retinoic acid receptor gene. Similary, eight snoRNAs genes recruited either CBPR768me2a or CBPR742me2a. The above deconvolution of the ER-CBP regulon reveals apparent subprogramming of the estrogen-induced gene program, which is based on the differential recruitment of methylated CBP species to a particular target site.
Discussion
Numerous studies in model systems have shown that chromatin acetylation generally precedes transcription (Cosma 2002) and that formation of a transcription preinitiation complex involves recruitment of CBP/p300 chromatin remodeling complexes as a critical early step (Black et al. 2006). However, one of the questions that has been very poorly addressed, especially at the genome-wide level, is how chromatin binding of an incoming signal like the holo-ER is diversified such that subsets of genes are coordinately regulated in a gene- and temporally controlled manner, as has been visualized early on in Drosophila (Ashburner et al. 1974). To gain insight into the early events of signal diversification, we set out to study the interplay between the different epigenetic enzymes corecruited by ERα/p160 complexes, CBP, and CARM1, initially on the prototypic TFF1 target gene and at the genome-wide scale to reveal the consequences of CARM1-mediated CBP methylation (Chevillard-Briet et al. 2002).
Our results reveal that CBP methylation is required for estrogen-dependent recruitment of CBP to ERα target genes, as knocking down CARM1 resulted in significantly reduced recruitment of total CBP and also of CBPR2151me2a to TFF1, paralleling the decreased cellular levels of this meCBP species; yet, no compensatory chromatin recruitment of CBPR2151non-me was seen despite its increased cellular levels. Moreover, in contrast to their wild-type parent, the methylation-deficient CBP mutants were not efficiently recruited to estrogen target genes, most likely due to a decreased or impaired binding to the coactivator RAC3 in vivo. These results suggest that methylated arginines of CBP may correspond to a docking surface for factors required to stabilize CBP at target gene chromatin. Indeed, CBP mutated at R714, R742, and R768 was not recruited to class II transactivator (CIITA) target sites in transient ChIP assays (Zika et al. 2005). However, the development of antibodies recognizing selectively the corresponding nonmethylated epitopes is required to prove this hypothesis.
It has been suggested that methylation of p300R2142 destabilizes binding to GRIP1 (YH Lee et al. 2005). No such function could be attributed to the corresponding R2151 of CBP. CBPR2151me2a was rather more efficiently coimmunoprecipitated with endogenous RAC3 than the general A22 anti-CBP antibody, whereas no signal was seen when antibodies directed against nonmethylated CBPR2151 were used (data not shown). The absence of detectable amounts of CBPR2151non-me and the estrogen-induced recruitment of its methylated counterpart to cognate promoters support the notion that methylation of R2151 enhances or is required for RAC3/p160 interaction; this notion is fully supported by a decreased/impaired RAC3 binding of the corresponding R to K CBP mutants. Whether methylation of the p160 interaction domains of CBP has a different function than for p300 remains to be established, as well as the role of CBPR2151non-me.
Comparative global ChIP-seq profiling of CBP species, ERα, RAC3, polymerase II, and global H3K18 acetylation revealed an astounding complexity of estrogen-induced events in this H3396 breast cancer cell line model, as visualized by ChordCharts. While the majority of prototypic estrogen target genes such as TFF1, CTSD, and GREB1 shows the “classical” recruitment mode of ERα, RAC3, CBP, and meCBP species in a similar hormone-dependent manner with varying amounts of gene-specific binding in a hormonally naive medium, the global analysis reveals major differences for each of these factors. Indeed, while the estrogen-inducible genes EBAG9 and CTBP1 bind apo-ERα (and polymerase II) and recruit additional receptors in a ligand-dependent manner, the amounts of recruited CBP, meCBP, CARM1, and p300 are negligible compared with other genes despite a strong peak of H3/4 acetylation; these genes are likely to operate in a CBP/p300-independent manner by engaging a distinct HAT. RAC3-free CBP-recruiting estrogen target genes may use the p160 homologs SRC1 and/or TIF2/GRIP1, supporting gene selectivity of p160s. Sites devoid of ERα but occupied by RAC3 are indicative of ERβ or other RAC3-using TFs targeting this locus.
An important result of this study is that CBP methylation by CARM1 (1) is a gene-specific feature and (2) increases local acetylation in the presence of CBPR742me2a or CBPR768me2a but not of CBPR2151me2a, as most if not all CBP is methylated at this site. Indeed, ChIP-qPCR-based scatter plots of genes selected from the comparative ChIP-seq profiles confirmed that there is a large variation in the amount of a particular meCBP species recruited by estrogen to a given target. Similarly, also the relative abundance of CBP and the p160 RAC3 can vary in a gene-specific manner. In addition to altered HAT activity, it is tempting to speculate that methylation may provide novel surfaces for interaction with other factors or for stabilization of assembled complexes. A functional diversity of meCBP species is supported by the observations that CBP HAT activity is differently affected by methylation at R742, R768, or R2151, and that CBPR742me2a/CBPR768me2a binding to promoters increases its probability of being acetylated. The observed cross-talk for methylation at R742 and R2151 suggests coregulation of CARM1 activity at these sites that may correspond to a composite signal recognized by (a) specific “reader(s).” The variation in both the recruitment of CBP relative to RAC3 and gene-specific recruitment of a particular (set of) meCBP species allows for a large heterogeneity, which could account for certain features in the timing and response intensity of a given target gene, thereby setting up an orchestrated and diversified response initiated by a single small molecular chemical signal.
Crossing of ChIP-seq and transcriptomics profiling, together with the corresponding gene annotation, turned out to be a powerful method to deconvolute the estrogen-induced gene program. Among the target genes that involve meCBP recruitment (the ER-CBP gene regulon), we observed an astounding diversification of an intricate estrogen-regulated gene network in H3396 cells. Functional diversification involved, for example, the fraction of TF genes to which a given meCBP species bound; the majority of TF targets bound CBPR768me2a or all three meCBP species, while CBPR742me2a recruitment involves hardly any TF genes. Similarly, CBPR742me2a is recruited as frequently to estrogen down-regulated genes as to up-regulated genes, while the presence of all three methylations is highly predictive for transcription activation. The observation of a cross-talk between the methylation sites supports the view of a regulated “writing” of these marks. While, at present, we have no technical approach to decipher the complete set of factors bound to a target site at a given point in time, we believe that (some of) the factors—including epigenetic and transcription regulatory marks and machineries—that make up the identity of a given chromatin site will direct the substrate selectivity of CARM1. In turn, the set of meHATs at a given site is likely to contribute to signaling, together with the other factors, to generate the ultimate temporally controlled transcriptional readout. It is tempting to speculate that CARM1-methylated CBP residues correspond to docking sites for “readers,” which altogether generate the observed transcriptional input at a given target site.
Cocitation analysis reveals a tight connection between the meCBP hubs in which TFs play, as expected, a dominant role. However, similarly central are cytokines, growth factors or growth factor receptors, kinases, and cell cycle regulators. Including pathway annotation and kinetics of transcription activation (Supplemental PowerPoint file) provides a comprehensive view of how ER and (methylated) coregulator recruitment ultimately generates a temporally controlled transcription response that can be deconvoluted into multiple hubs of genes. A multidimensional analysis of TF and coregulator binding, histone modification, and polymerase recruitment/activity, together with the corresponding (nascent) transcription profiling, including noncoding RNAs, will ultimately reveal the details of the cognate gene program and its key elements that may be altered in pathology.
CBP displays features of a tumor suppressor (for review, see Iyer et al. 2004), and a high frequency of human follicular lymphoma, diffuse large B-cell lymphoma, and relapsed lymphoblastic leukemias display deletion/mutation of CREBBP (Mullighan et al. 2011; Pasqualucci et al. 2011). As also CARM1 may be deregulated in cancer (Hong et al. 2004), it is possible that aberrant methylation of CBP may be observed in cancer, either as a marker or even linked to the transformation process. Indeed, our preliminary tissue microarray analysis reveals a deregulation of the epi-enzyme cross-talk in several types of cancers, and suggests a link between abnormal CBP methylation and disease progression. Given the impact of CARM1 and of CBP in key growth regulatory circuits, and the role of CBP in cell differentiation (Kawasaki et al. 1998), it will be interesting to assess the relevance of site-selective CBP aberrant expression/mislocalization in cancer.
Materials and methods
Cell lines, reagents, and antibodies
H3396 cells were maintained in RPMI 1640 supplemented with 10 mM HEPES, 10% fetal bovine serum (FBS), and antibiotics. Clone 3 (CARM1−/−) and clone 13 (CARM1+/+) of MEFs (Yadav et al. 2003), a gift from Dr. M. Bedford, were cultured in DMEM supplemented with antibiotics and 10% fetal calf serum. ERα (HC20), CBP (A22), p300 (N15), HA (12CA5), and RAC3 (M397) antibodies were from Santa Cruz Biotechnology; anti-acetylCBP was from Cell Signaling; ChIP-grade antibodies against H3K14ac (07-353) was from Millipore, and H3K18ac (ab1191) was from Abcam. FuGENE 6 transfection reagent was purchased from Roche, and β-estradiol was purchased from Sigma Chemicals.
Generation and purification of antibodies
Peptides used for immunization of rabbits encompass the arginine residue of interest either in the unmodified form or asymmetrically dimethylate (Supplemental Material). All peptides were coupled to ovalbumin, validated by SDS-PAGE, and injected into rabbits. Samples of the rabbit serum drawn before immunization (preimmune control) and at different time points after immunization and boost were tested by Western blotting using nonmethylated or in vitro methylated CBP fragments. Monospecific polyclonal antibodies were obtained by differential dual affinity chromatography using sulfolink resins prepared according to the manufacturer's protocol (Pierce). For example, anti-CBPR768me2a antibodies were obtained by positive selection from serum passed over a first column containing the immobilized ISPS(Rme2a)MPQPC peptide. Serum was applied by gravity flow, washed with Tris-buffered saline, and eluted with 0.1 M glycine-HCl (pH 2.5), and the eluate was neutralized with 1 M Tris-HCl (pH 8.5). Antibodies cross-reacting with the cognate nonmethylated peptide were removed by negative selection on a second column containing the immobilized ISPSRMPQPC peptide. Antibody titer and specificity were determined by ELISA using not only methyl-arginine and non-methyl-arginine peptides but also peptides containing the other potential methylation sites (Supplemental Material). Antibodies showing sufficient selectivity in ELISA were selected and used in this study.
Plasmids and recombinant proteins
pGEX CBP (amino acids 1869–2441) was a kind gift from A. Harel-Bellan and pGEX CBP (amino acids 685–774) was described previously (Chevillard-Briet et al. 2002). GST fusion proteins were expressed in BL21 bacteria as explained before (Vandel et al. 2001). pRSETB CBP (amino acids 685–774) was made by digestion of pGEX CBP (amino acids 685–774) by HindIII/BamH1 and introduction into pRSETB cut by HindIII/BglII. pCMV-HA-CBP and pCMV-HA-CBPΔHAT were provided by A. Harel-Bellan. pCMV-HA-CBPR714K, pCMV-HA-CBPR742K, pCMV-HA-CBPR768K, pCMV-HA-CBPR600,625K, and pCMV-HA-CBPR2104,2151K were obtained by in vitro site-directed mutagenesis with GeneEditor (Promega) and were reintroduced into pCMV-HA-CBP cut by AflII–BglII or Xba1 (R2104/2151K mutant). Fragments containing the single mutants were subcloned from pCMV-HA-CBPR714K, pCMV-HA-CBPR742K, pCMV-HA-CBP768K, and pCMV-HA-CBP2104K/2151K into pBABE-HA-CBP hygro with BstBI to generate the corresponding pBABE-HA-CBP mutant vectors. The pBabe-HA-CBP vectors were used to generate stable H3396 cell lines expressing HA-CBP wild type and the methylation-deficient mutants. shRNA against human CARM1 and CBP were designed from Sigma-Aldrich and were cloned into lentiviral vector pLKO.1-puro (Addgene).
Immunoprecipitation, Western blotting, RT–PCR, siRNA, and shRNA tranfections
Immunoprecipitation and co-IP experiments were performed as described (Fauquier et al. 2008). For RT–PCR, total RNA was extracted from cells using GenElute RNA extraction kit (Sigma). Two micrograms of total RNA were reverse-transcribed using AMV-RTase (Roche) with oligo(dT) (New England Biolabs) as primers for 1 h at 42°C according to standard procedures. cDNAs were then diluted 10-fold and analyzed by real-time PCR using the Roche LC480 LightCycler device with sequence-specific primers and SYBR Green QPCR MasterMix (Qiagen). Primers for amplification of target genes are listed in the Supplemental Material. All qPCR values were normalized relative to 36B4 mRNA, and standard deviations were calculated.
For siRNA transfection, 5 × 106 cells were electroporated with siRNA using the Cell Line Nucleofector kit (Amaxa Biosystem) according to the manufacturer's instructions. siRNA-mediated knockdown efficiency was assessed by qPCR and immunoblotting. All experiments were performed 48 h after nucleofection. siRNA sequences are given in the Supplemental Material. The control siRNA used was the Allstar Negative Control siRNA from Qiagen.
For shRNA transfections and virus production, pLKO.1 lentiviral vector containing or not containing shRNAs directed against CBP and CARM1 was transfected with the packaging plasmids pLP1, pLP2, and pLP/VSVG (Invitrogen) using FuGENE HD Transfection Reagent (Roche) in HEK293T cells according to the manufacturer's instructions. After 48 h, the supernatant of HEK293T was used to infect H3396 cells, and knockdown efficiency was monitored 72 h later by Western blotting.
Generation of stable H3396 cell lines
The retroviral plasmids expressing pBABE-HA-CBP and the mutants were transfected in HEK293T cells along with pCLII Ampho helper plasmid (Imgenex) with FuGENE6 transfection reagent (Roche). After 48 h, the medium was collected and filtered through a 0.4-μm filter before being used to infect H3396 cells. Cells were then treated with hygromycin B and expanded. Stable integration and expression of the plasmids were monitored by qPCR and immunoblotting with an HA antibody.
Methylation and acetylation assays
In vitro methylation assays of CBP fragments (GST or His fusion proteins of CBP) with recombinant CARM1 have been described (Chevillard-Briet et al. 2002). Acetyltransferase activity of CBP was assessed after immunoprecipitation in CARM1+/+ and CARM1−/− MEFs. Immunoprecipitations were performed with either anti-CBP-A22 antibody or an irrelevant antibody. Immunoprecipitations were mixed with 3 μg of histone H3 peptide (residues 1–22) and 0.3 μM [3H]-acetyl-coenzyme A (GE Healthcare). Reactions were performed for 45 min at 30°C in 50 mM Tris (pH 8.0), 150 mM NaCl, 5 mM EDTA, and 0.5% NP40 before being spotted on P81 (Whatman). Filters were rinsed twice for 1 h with 50 mM Na2CO3 (pH 9.0), and histone H3 acetylation was determined by liquid scintillation counting.
ChIP, ChIP-seq, re-ChIP, and peptide competitions
ChIP assays were performed according to standard protocols, and the conditions used for the various antibodies are detailed in the Supplemental Material. Quantitative real-time PCRs of bound promoters were performed with 2 μL of DNA, sequence-specific primers, and the SYBR Green QPCR MasterMix (Qiagen). The primer pairs used are listed in the Supplemental Material. For re-ChIPs, immunocomplexes were eluted in 10 mM DTT for 30 min at 37°C. The eluate was diluted 50 times in ChIP dilution buffer for reimmunoprecipitation with a second antibody. For peptide competition experiments, antibodies were preincubated with a 100-fold molar excess of peptide for 1 h at 4°C. The antibody–peptide mixture was then added to the soluble chromatin followed by ChIP. ChIP-seq and the data analysis pipeline are described in the Supplemental Material.
Acknowledgments
We thank Dr. A Harel-Bellan and Dr. M. Bedford for plasmids and cells, and J.M. Garnier for constructing pBABE-HA-CBP and mutants. We are grateful to Audrey Furst and Céline Bonhomme for excellent technical assistance. We thank Gilles Duval for generating rabbit sera and Pascal Eberling for peptide synthesis. S.S.W., M.W., and L.F are recipients of an ARC fellowship, and D.G.C. and M.W. are fellows of the Fondation pour la Recherche Médicale. This work was supported by grants from the European Union (EPITRON CT2005-518417) (to H.G.), the Institut National du Cancer (to H.G. and L.V.), and the Ligue contre le Cancer (to H.G.; équipe labellisée). It was also supported by an “Action Thématique et Incitative sur Programme” from the Centre National de la Recherche Scientifique and grants from the Fondation pour la Recherche Médicale, the Association pour la Recherche contre le Cancer, and the Université Paul Sabatier in Toulouse to L.V.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.619211.
Supplemental material is available for this article.
References
- Ait-Si-Ali S, Ramirez S, Barre FX, Dkhissi F, Magnaghi-Jaulin L, Girault JA, Robin P, Knibiehler M, Pritchard LL, Ducommun B, et al. 1998. Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature 396: 184–186 [DOI] [PubMed] [Google Scholar]
- Arif M, Kumar GV, Narayana C, Kundu TK 2007. Autoacetylation induced specific structural changes in histone acetyltransferase domain of p300: probed by surface enhanced Raman spectroscopy. J Phys Chem B 111: 11877–11879 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Chihara C, Meltzer P, Richards G 1974. Temporal control of puffing activity in polytene chromosomes. Cold Spring Harb Symp Quant Biol 38: 655–662 [DOI] [PubMed] [Google Scholar]
- Bedford MT, Clarke SG 2009. Protein arginine methylation in mammals: who, what, and why. Mol Cell 33: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JC, Choi JE, Lombardo SR, Carey M 2006. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol Cell 23: 809–818 [DOI] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat Genet 38: 1289–1297 [DOI] [PubMed] [Google Scholar]
- Chen D, Huang SM, Stallcup MR 2000. Synergistic, p160 coactivator-dependent enhancement of estrogen receptor function by CARM1 and p300. J Biol Chem 275: 40810–40816 [DOI] [PubMed] [Google Scholar]
- Chevillard-Briet M, Trouche D, Vandel L 2002. Control of CBP co-activating activity by arginine methylation. EMBO J 21: 5457–5466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma MP 2002. Ordered recruitment: gene-specific mechanism of transcription activation. Mol Cell 10: 227–236 [DOI] [PubMed] [Google Scholar]
- Daujat S, Bauer UM, Shah V, Turner B, Berger S, Kouzarides T 2002. Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr Biol 12: 2090–2097 [DOI] [PubMed] [Google Scholar]
- Fauquier L, Duboe C, Jore C, Trouche D, Vandel L 2008. Dual role of the arginine methyltransferase CARM1 in the regulation of c-Fos target genes. FASEB J 22: 3337–3347 [DOI] [PubMed] [Google Scholar]
- Feng Q, Yi P, Wong J, O'Malley BW 2006. Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol Cell Biol 26: 7846–7857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari R, Pellegrini M, Horwitz GA, Xie W, Berk AJ, Kurdistani SK 2008. Epigenetic reprogramming by adenovirus e1a. Science 321: 1086–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronemeyer H, Bourguet W 2009. Allosteric effects govern nuclear receptor action: DNA appears as a player. Sci Signal 2: pe34 doi: 10.1126/scisignal.273pe34 [DOI] [PubMed] [Google Scholar]
- Gronemeyer H, Gustafsson JA, Laudet V 2004. Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov 3: 950–964 [DOI] [PubMed] [Google Scholar]
- Hong H, Kao C, Jeng MH, Eble JN, Koch MO, Gardner TA, Zhang S, Li L, Pan CX, Hu Z, et al. 2004. Aberrant expression of CARM1, a transcriptional coactivator of androgen receptor, in the development of prostate carcinoma and androgen-independent status. Cancer 101: 83–89 [DOI] [PubMed] [Google Scholar]
- Horwitz GA, Zhang K, McBrian MA, Grunstein M, Kurdistani SK, Berk AJ 2008. Adenovirus small e1a alters global patterns of histone modification. Science 321: 1084–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WC, Ju TK, Hung MC, Chen CC 2007. Phosphorylation of CBP by IKKα promotes cell growth by switching the binding preference of CBP from p53 to NF-κB. Mol Cell 26: 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer NG, Ozdag H, Caldas C 2004. p300/CBP and cancer. Oncogene 23: 4225–4231 [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Eckner R, Yao TP, Taira K, Chiu R, Livingston DM, Yokoyama KK 1998. Distinct roles of the co-activators p300 and CBP in retinoic-acid-induced F9-cell differentiation. Nature 393: 284–289 [DOI] [PubMed] [Google Scholar]
- Lee DY, Teyssier C, Strahl BD, Stallcup MR 2005. Role of protein methylation in regulation of transcription. Endocr Rev 26: 147–170 [DOI] [PubMed] [Google Scholar]
- Lee YH, Coonrod SA, Kraus WL, Jelinek MA, Stallcup MR 2005. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc Natl Acad Sci 102: 3611–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Bedford MT, Stallcup MR 2011. Regulated recruitment of tumor suppressor BRCA1 to the p21 gene by coactivator methylation. Genes Dev 25: 176–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Vega VB, Thomsen JS, Zhang T, Kong SL, Xie M, Chiu KP, Lipovich L, Barnett DH, Stossi F, et al. 2007. Whole-genome cartography of estrogen receptor α binding sites. PLoS Genet 3: e87 doi: 10.1371/journal.pgen.0030087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Baumann CT, Li H, Strahl BD, Rice R, Jelinek MA, Aswad DW, Allis CD, Hager GL, Stallcup MR 2001. Hormone-dependent, CARM1-directed, arginine-specific methylation of histone H3 on a steroid-regulated promoter. Curr Biol 11: 1981–1985 [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA, Heatley SL, Holmfeldt L, Collins-Underwood JR, Ma J, et al. 2011. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature 471: 235–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeem H, Cheng D, Zhao Q, Underhill C, Tini M, Bedford MT, Torchia J 2007. The activity and stability of the transcriptional coactivator p/CIP/SRC-3 are regulated by CARM1-dependent methylation. Mol Cell Biol 27: 120–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, Kasper LH, Lerach S, Tang H, Ma J, et al. 2011. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature 471: 189–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld MG, Lunyak VV, Glass CK 2006. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev 20: 1405–1428 [DOI] [PubMed] [Google Scholar]
- Thompson PR, Wang D, Wang L, Fulco M, Pediconi N, Zhang D, An W, Ge Q, Roeder RG, Wong J, et al. 2004. Regulation of the p300 HAT domain via a novel activation loop. Nat Struct Mol Biol 11: 308–315 [DOI] [PubMed] [Google Scholar]
- Vandel L, Nicolas E, Vaute O, Ferreira R, Ait-Si-Ali S, Trouche D 2001. Transcriptional repression by the retinoblastoma protein through the recruitment of a histone methyltransferase. Mol Cell Biol 21: 6484–6494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K 2009. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138: 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welboren WJ, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FC, Span PN, Stunnenberg HG 2009. ChIP-seq of ERα and RNA polymerase II defines genes differentially responding to ligands. EMBO J 28: 1418–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Chen H, Du K, Asahara H, Tini M, Emerson BM, Montminy M, Evans RM 2001. A transcriptional switch mediated by cofactor methylation. Science 294: 2507–2511 [DOI] [PubMed] [Google Scholar]
- Yadav N, Lee J, Kim J, Shen J, Hu MC, Aldaz CM, Bedford MT 2003. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc Natl Acad Sci 100: 6464–6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- York B, O'Malley BW 2010. Steroid receptor coactivator (SRC) family: masters of systems biology. J Biol Chem 285: 38743–38750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zika E, Fauquier L, Vandel L, Ting JP 2005. Interplay among coactivator-associated arginine methyltransferase 1, CBP, and CIITA in IFN-γ-inducible MHC-II gene expression. Proc Natl Acad Sci 102: 16321–16326 [DOI] [PMC free article] [PubMed] [Google Scholar]