Abstract
Variants in the IMP2 (insulin-like growth factor 2 [IGF2] mRNA-binding protein 2) gene are implicated in susceptibility to type 2 diabetes. We describe the ability of mammalian target of rapamycin (mTOR) to regulate the cap-independent translation of IGF2 mRNA through phosphorylation of IMP2, an oncofetal RNA-binding protein. IMP2 is doubly phosphorylated in a rapamycin-inhibitable, amino acid-dependent manner in cells and by mTOR in vitro. Double phosphorylation promotes IMP2 binding to the IGF2 leader 3 mRNA 5′ untranslated region, and the translational initiation of this mRNA through eIF-4E- and 5′ cap-independent internal ribosomal entry. Unexpectedly, the interaction of IMP2 with mTOR complex 1 occurs through mTOR itself rather than through raptor. Whereas depletion of mTOR strongly inhibits IMP2 phosphorylation in cells, comparable depletion of raptor has no effect; moreover, the ability of mTOR to phosphorylate IMP2 in vitro is unaffected by the elimination of raptor. Dual phosphorylation of IMP2 at the mTOR sites is evident in the mouse embryo, likely coupling nutrient sufficiency to IGF2 expression and fetal growth. Doubly phosphorylated IMP2 is also widely expressed in adult tissues, including islets of Langerhans.
Keywords: mTOR, IMP2, amino acid-dependent phosphorylation, IGF2, mRNA translation, internal ribosome entry, type 2 diabetes
The insulin-like growth factor 2 (IGF2) is the major determinant of fetal growth in the mouse (Efstratiadis 1998), a function assumed after birth by the growth hormone/IGF1 system. Human and murine IGF2 are encoded by a set of mRNAs that differ only in their 5′ untranslated regions (UTRs) (Supplemental Fig. S1). In rapidly growing rhabdomyosarcoma (RD) cells, a human rhabdomyosarcoma cell line, the IGF2 mRNA designated leader 4 (L4) is constitutively translated, whereas the IGF2 mRNA designated leader 3 (L3) is translated in a rapamycin-inhibitable manner (Nielsen et al. 1995). The IGF2 mRNA-binding proteins (IMPs) were subsequently retrieved by their ability to bind differentially to the 1.2-kb L3 but not the 0.1-kb L4 5′ UTR (Nielsen et al. 1999). The IMPs comprise a family of three closely related 60- to 70-kDa RNA-binding proteins (IMP1–3), each containing two RRM domains followed by four KH domains (Nielsen et al. 2001; Yisraeli 2005). IMPs are oncofetal proteins expressed primarily during development but frequently re-expressed in and contributory to malignancy. IMPs have been independently identified repeatedly, usually as mediators of mRNA translational repression, localization, and/or stabilization. IMP1, for example, is orthologous to the chicken zipcode-binding protein (ZBP1), which binds to a 54-nucleotide (nt) “zipcode” sequence in the β-actin 3′ UTR, suppressing β-actin mRNA translation during its transport to the leading edge of the cell; there, Src tyrosine kinase phosphorylates ZBP1/IMP1 at a site between the second and third KH domains, promoting release of β-actin mRNA and disinhibition of its translation (Hüttelmaier et al. 2005). IMP3 is orthologous to Xenopus Vg1RBP/Vera, which mediates the polarized localization of Vg1 mRNA in oocytes; despite its similarity to IMP1, IMP3 does not bind the β-actin zipcode motif (Mori et al. 2001). IMP2 is most distant and lacks nonmammalian orthologs and little is known of its functions. Recently, SNPs in the human IMP2/IGF2BP2 gene have been associated with an excess risk for type 2 diabetes (Saxena et al. 2007; Scott et al. 2007; Zeggini et al. 2007).
IGF2 L3 mRNA expression in the mouse is reported to occur only during fetal life (Newell et al. 1994; Engström et al. 1998); its translation correlates temporally with the onset of expression of the IMP1,2,3 mRNAs, whose expression turns on abruptly at embryonic day 11.5 (E11.5) and is largely extinguished at birth (Nielsen et al. 1999). This timing coincides closely with the onset of the biologic action of IGF2, inasmuch as IGF2-deficient embryos show no size deficit before E11 (Baker et al. 1993) and overexpression of IGF2 has no impact on embryonic size before E13 (Sun et al. 1997). IMP1-null mice are ∼40% smaller than wild type at birth (Hansen et al. 2004), as compared with the 60% smaller size of IGF2-null mice (Efstratiadis 1998), and extracts from 13.5-d-old IMP1-null embryos exhibit diminished polysomal association of IGF2 L3 and L4 mRNAs (Hansen et al. 2004), suggesting that IMP1 is required for proper expression of IGF2 mRNA, at least during development. Moreover, IMP3 overexpression in K562 leukemia cells has been reported to enhance IGF2 L3 mRNA translation (Liao et al. 2005). Nevertheless, overexpression of IMP1 in NIH3T3 cells represses IGF2 mRNA L3 polysomal association and translation (Nielsen et al. 1999), and IMP1 depletion from K562 cells up-regulates IGF2 mRNA and polypeptide abundance without altering IGF2 mRNA L3 translation (Liao et al. 2004). Thus, the role of various IMPs in the regulation of IGF2 L3 mRNA is unclear, and whether the IMPs participate in the rapamycin-sensitive regulation of IGF2 L3 mRNA translation observed in RD cells is unknown (Nielsen et al. 1999). We therefore sought to identify the molecular basis for the rapamycin sensitivity of IGF2-L3 mRNA translational initiation, and the role of the IMPs.
The target of rapamycin (TOR) is a giant protein kinase that functions in two independently regulated hetero-oligomeric complexes; the rapamycin-sensitive complex is mTOR (mammalian TOR) complex 1 (mTORC1), which contains the polypeptides raptor and mLst8. mTORC1 activity is regulated by nutrients, mitogens, and stress, and its major function is in the regulation of cell and organismal size (Wullschleger et al. 2006). The ability of mTORC1 to regulate cell size—i.e., the accumulation of cell mass—is achieved primarily by enhancing mRNA translation through an increase in the capacity and an up-regulation of the activity of the translational apparatus (Ma and Blenis 2009), and secondarily by restraining macroautophagy (Mehrpour et al. 2010). More specifically, mTORC1 increases protein synthesis by stimulating transcription of ribosomal RNAs and tRNAs (Mayer and Grummt 2006; Shor et al. 2010), and by enhancing translation of several subsets of mRNAs. One mTORC1-regulated mRNA subset is a cohort whose 5′ end begins with a run of five to 14 pyrimidines (5′TOP mRNAs) and that encodes many polypeptides of the translational apparatus itself; e.g., ribosomal proteins and elongation factors (Hornstein et al. 2001). The biochemical mechanism by which mTORC1 up-regulates the translation of 5′TOP mRNAs is not known (Patursky-Polischuk et al. 2009). Another cohort of mTORC1-regulated mRNAs have 5′ UTRs (such as the IGF2 L3 5′ UTR) that contain secondary structures that impede ribosomal scanning and therefore are thought to require the action of the eIF-4F complex for translation to be efficiently initiated; many of these mRNAs encode polypeptides that carry out proproliferative/oncogenic functions (Richter and Sonenberg 2005). The roles of the mTORC1 substrates S6K1 and 4E-BP in the regulation of eIF-4F and in the translation of such mRNAs is well documented. The eIF-4F complex consists of the mRNA cap-binding protein eIF-4E, an RNA helicase (eIF-4A), the 4A stimulatory partner eIF-4B, and the scaffold protein eIF-4G. The binding of eIF-4E to the 4G scaffold brings mRNAs into proximity with the 4A/4B helicase. The latter catalyzes unwinding of complex secondary structures in the 5′ UTRs, which facilitates scanning by the 43S preinitiation complex. S6K1 promotes the activity of eIF-4A through the phosphorylation of 4B and by phosphorylating and promoting the degradation of PDCD4, an inhibitor of 4A. The relatively abundant 4E-BPs bind eIF-4E in a manner that prevents 4E binding to 4G; active mTORC1 catalyzes a processive, multiple phosphorylation of 4E-BP at sites located on both sides of its 4E-binding site, resulting in its dissociation from eIF-4E, freeing the latter to associate with eIF-4G (Gingras et al. 2004). The ability of S6K1 and 4E-BP to be phosphorylated by mTOR depends on their ability to bind to raptor (Hara et al. 2002). Removal of raptor from mTOR greatly reduces mTOR-catalyzed phosphorylation of these substrates in vitro, and mutation of the TOS motif (Schalm and Blenis 2002) on S6K1 and 4E-BP, which mediates their binding to raptor (Nojima et al. 2003), eliminates the ability of these proteins to be phosphorylated by mTORC1 in vivo. Unexpectedly, we found that mTORC1 regulates the translational initiation of the IGF2 L3 mRNA through a mechanism entirely distinct from those outlined above; mTORC1 catalyzes a raptor-independent phosphorylation of the RNA-binding protein IMP2, which promotes the ability of IMP2 to bind to the L3 5′ UTR, enabling the initiation of L3 translation by cap- and eIF-4E-independent internal ribosomal entry.
Results
Translational initiation of mRNAs containing the IGF2 L3 5′ UTR sequence occurs through rapamycin-sensitive internal ribosomal entry
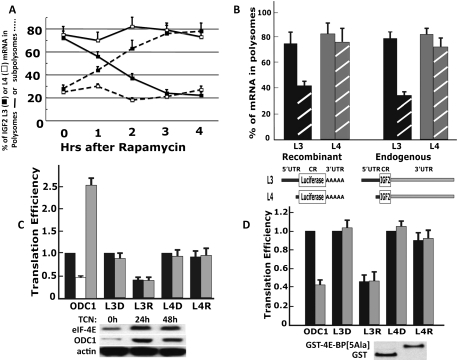
Rapamycin treatment of rapidly growing RD cells does not significantly alter overall polysome abundance (Supplemental Fig. S2), indicating that 3 h of treatment with rapamycin does not substantially inhibit overall translational initiation. We do confirm, however, that rapamycin induces a progressive decrease in the predominantly polysomal localization of the IGF2 L3 mRNA, so that, by 3 h, this mRNA is found predominantly in the subpolysomal region of the sucrose density gradient, consistent with an inhibition of its translational initiation; in contrast, the predominantly polysomal association of the IGF2 L4 mRNA is unaffected by rapamycin (Fig. 1A; Supplemental Fig. S2).
Figure 1.
The rapamycin-sensitive translational initiation of IGF2 L3 mRNA is mediated by its 5′ UTR and is independent of eIF-4E. (A) The effect of rapamycin on the polysomal association of IGF2 L3 and L4 mRNAs in rapidly growing RD cells. Rapamycin (20 nM) was added to RD cells at ∼30% confluency; the cells were harvested at intervals thereafter and extracts were subjected to sucrose density gradient centrifugation. The subpolysomal ([- - -] fractions 1–5, Supplemental Fig. S2) and polysomal ([—] fractions 6–13) regions of the gradient were pooled separately, and each was assayed by qPCR for the content of IGF2 L3 (▪) and L4 (□) mRNAs. The combined results of three experiments are shown ±SEM. (B) The rapamycin sensitivity of IGF2 L3 mRNA translational initiation is conferred by the L3 5′ UTR. The IGF2 L3 and L4 5′ UTR segments were fused to the coding sequences of firefly luciferase (see cartoon) and were transiently transfected into rapidly growing RD cells. After 24 h, cells were treated either with DMSO (filled bars) or rapamycin (20 nM, 3 h.; cross-hatched bars); extracts were separated by sucrose density gradient centrifugation, and the content of endogenous IGF2 L3 (black-filled and hatched bars, right) and L4 (gray-filled and hatched bars, right) mRNAs and of the L3-luciferase (black-filled and hatched bars, left) and L4-luciferase (gray-filled and hatched bars, left) mRNAs in the pooled polysomal fractions was determined by qPCR and is expressed as a percentage of the corresponding total L3 or L4 mRNA in the sample loaded. The combined results of three experiments are shown ±SEM. (C) Overexpression of eIF-4E up-regulates ODC1-luciferase expression but does not alter the expression of L3-luciferase. Plasmids encoding the 5′ UTR of human ODC1 fused to firefly luciferase or L3-firefly luciferase or L4-firefly luciferase, each together with a plasmid encoding Renilla luciferase, were transfected into RD cells that stably express recombinant eIF-4E in a tetracycline-inducible manner. The cells were treated with tetracycline (gray bars) or carrier (black bars) for 24 h. Three hours prior to harvest, cells were treated with DMSO (D) or rapamycin (20 nM, 3 h; R). Thereafter, extracts were assayed for firefly and Renilla luciferase activity and by qPCR for the content of firefly luciferase mRNA. The activity of firefly luciferase was divided by the activity of Renilla luciferase to give a normalized firefly luciferase activity; “translational efficiency” was calculated by dividing the normalized firefly luciferase activity by the measured content of firefly luciferase mRNA, setting to 1 the value of this dividend for the ODC1-luciferase, L3-luciferase, and L4-luciferase conditions in the absence of tetracycline induction or rapamycin treatment (the black bars in ODC1, L3D, and L4D). The unfilled/white bar shows the effect of rapamycin on the expression of ODC1-luciferase in the absence of tetracycline. Each experiment was performed in triplicate, and the combined results of three experiments are shown ±SEM. (D) Overexpression of a nonphosphorylatable mutant of 4E-BP1 suppresses expression of ODC1-luciferase but does not alter expression of L3-luciferase. RD cells were engineered to stably overexpress GST or a GST fused to a nonphosphorylatable 4E-BP1 polypeptide (GST-4E-BP[5Ala]) (Hara et al. 2002). Plasmids encoding ODC1-luciferase, L3-luciferase, or L4-luciferase, each together with a plasmid encoding Renilla luciferase, were transfected into the RD GST (black bars) or GST-4E-BP[5Ala] (gray bars) stable transformants. After 24 h, the cells were treated with DMSO (D) or rapamycin (20 nM, 3 h; R). Translational efficiency was calculated as in C, and set to 1 for the ODC1-luciferase, L3-luciferase, and L4-luciferase conditions in the extracts from cells expressing GST in the absence of rapamycin treatment (the black bars in ODC1, L3D, and L4D). Each experiment was performed in triplicate, and the combined results of three experiments are shown ±SEM.
As noted, the IGF2 L3 and L4 mRNAs differ only in their 5′ UTRs (Supplemental Fig. S1); to determine whether the regulation of the L3 mRNA translational initiation is mediated entirely by this segment, we used a reporter containing the L3 5′ UTR fused upstream of firefly luciferase coding sequences. After transient expression in RD cells, extracts prepared from cells treated with or without rapamycin for 3 h were separated on sucrose density gradients and the relative polysomal abundance of endogenous L3 IGF2 mRNA and L3 luciferase mRNA was measured by quantitative PCR (qPCR). As observed for the endogenous L3 IGF2 mRNA, the recombinant L3 luciferase mRNA is predominantly polysomal in the control cells and shifted to a predominantly subpolysomal localization in the rapamycin-treated cells (Fig. 1B). This behavior indicates that the L3 5′ UTR is sufficient to enable effective, rapamycin-sensitive translational initiation in the absence of the endogenous 3′ UTR, and L3 luciferase can therefore serve as a valid reporter to explore the mechanism of L3 translational initiation and its regulation.
The L3 5′ UTR is ∼1.2 kb and highly structured, a situation thought to indicate a requirement for unwinding through eIF-4E-mediated recruitment of the mRNA to the eIF-4F complex (Richter and Sonenberg 2005). The abundance and availability of eIF-4E is thought to be rate-limiting for the translation of such mRNAs; to examine the role of eIF-4E in the translation of L3-containing mRNAs, we created a pool of stably transduced RD cells that up-regulate the expression of eIF-4E in a tetracycline-inducible manner (Fig. 1C). A threefold increase in eIF-4E polypeptide is evident at 24 h after tetracycline addition; the impact of this modest 4E overexpression is reflected by the marked increase in the abundance of the endogenous ornithine decarboxylase 1 (ODC1) polypeptide, which is encoded by another mRNA with a highly structured 5′ UTR. A recombinant reporter containing the ODC1 5′ UTR fused upstream of firefly luciferase coding sequences cotransfected with a control Renilla luciferase into the RD cells exhibits rapamycin-sensitive expression (Fig. 1C, white bar); 24 h after tetracycline addition, the expression of ODC1-luciferase is increased 2.5-fold (Fig. 1C), demonstrating the enhanced operation of the eIF-4F complex. In contrast, induction of eIF-4E has no effect on the expression of an L3-luciferase (or an L4-luciferase) reporter in the presence or absence of rapamycin (Fig. 1C). It is possible that endogenous levels of eIF-4E already enable maximal translation of L3-luciferase, even in the presence of rapamycin. To explore this possibility, we overexpressed a mutant of 4E-BP1 wherein all five phosphorylation sites are changed to alanine (GST-4E-BP[5Ala]), which enables constitutive binding to and sequestration of endogenous eIF-4E (Hara et al. 2002). Cotransfection with GST-4E-BP[5Ala] suppresses the expression ODC1-luciferase by 60% (Fig. 1D), but has no effect on the expression of L3- or L4-luciferase in the presence or absence of rapamycin (Fig. 1D). These results indicate that, despite its complex secondary structure, the L3-luciferase mRNA is translated in an eIF-4E-independent manner. Notably, ODC1 mRNA, in addition to its cap-dependent translation, also encodes an internal ribosome entry site (IRES) in its 5′ UTR (Pyronnet et al. 2000), perhaps accounting for the incomplete inhibition of its translation by GST-4E-BP[5Ala].
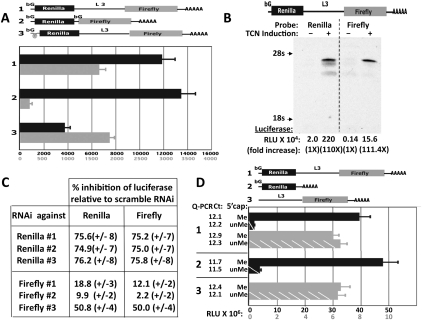
The ability of mRNAs containing an L3 5′ UTR to be translated independently of eIF-4E, despite the complex secondary structure of the L3 segment, raised the possibility that initiation of L3-mRNA translation may proceed by internal ribosomal entry (Gilbert 2010; Komar and Hatzoglou 2011). To evaluate this, we constructed several plasmids encoding dicistronic mRNAs, with the proximal mRNA encoding the human β-globin 5′ UTR situated upstream of Renilla luciferase followed by either the L3 5′ UTR or the β-globin 5′ UTR upstream of firefly luciferase (Fig. 2A). Whereas Renilla luciferase was comparably expressed from both plasmids after transient transfection in RD cells, the expression of firefly luciferase polypeptide was ∼15-fold greater from the internally situated L3 5′ UTR than from the internally placed β-globin 5′ UTR, expression from the latter being negligible. In addition, we inserted a strong hairpin segment immediately beyond the transcriptional start site of the proximal β-globin 5′ UTR; this resulted in a >70% decrease in the expression of Renilla luciferase, but did not diminish expression of firefly luciferase. The expression of dicistronic vectors can be complicated by the presence of cryptic promoters that can drive expression from the distal coding sequence (here, firefly) independently of the proximal (Renilla) sequence, and/or cryptic splice sites that lead to the generation of monocistronic mRNAs. The dicistronic construct shown in Figure 2B was inserted into the TetR-responsive pcDNA5/TO vector and was stably expressed in RD cells expressing TetR; 24 h after addition of tetracycline, there was a 110-fold increase in the activity of both the Renilla and the firefly luciferase (Fig. 2B). Moreover, Northern blot demonstrated the tetracycline induction of a single mRNA, visualized with either a Renilla or a firefly 32P-cDNA probe (Fig. 2B). Thus, the tetracycline-sensitive promoter drives the expression of both luciferases in a quantitatively identical manner from a single mRNA. Further evidence that the same single mRNA mediates the synthesis of both luciferase polypeptides is the ability of several different single RNAis, each directed at one of the luciferase coding sequences, to inhibit the activity of both luciferases to a very similar extent (Fig. 2C). Finally, we used RNA transfection to bypass the possibility of splicing and examine directly the need for a 5′ cap structure capable of binding eIF-4E (Fig. 2D). RNAs encoding the dicistronic construct (β-globin–Renilla luciferase–L3-firefly luciferase) and two monocistronic constructs (β-globin–Renilla luciferase; L3-firefly luciferase) were synthesized in vitro and a 5′ guanosine cap was added either with (N7mG[5′]ppp[2′Om]G) or without (G[5′]pppG) cap methylation (Supplemental Fig. S3). Twenty-four hours after replicate transfections of 100 ng of each RNA into 293T cells, extracts were assayed for luciferase activity and luciferase RNA content. Cap methylation did not differentially affect the abundance of the luciferase mRNA. The ratio of Renilla and firefly luciferase activities measured in extracts of cells transfected with the dicistronic RNA with the methylated cap (Fig. 2D, left) was similar to the ratio seen with DNA transfection (Fig. 2A). Notably, however, the activity generated by the unmethylated (β-globin 5′ UTR)–Renilla luciferase RNA was <5% that observed for the methylated RNA, whereas the activity generated from the internal (L3 5′ UTR)-firefly luciferase sequences was nearly identical whether the dicistronic RNA had a methylated or unmethylated cap. With the monocistronic RNAs, omission of cap methylation resulted in a similar profound inhibition of the activity generated from (β-globin 5′ UTR)–Renilla luciferase RNA, but no change in the activity generated from the (L3 5′ UTR)-firefly luciferase RNA. These results, together with the insensitivity of L3-directed mRNA translation to eIF-4E, provide strong evidence that initiation from the L3 5′ UTR occurs by 5′ cap- and eIF-4E-independent internal ribosomal entry.
Figure 2.
The initiation of IGF2 L3 mRNA translation occurs through internal ribosomal entry. (A) Expression patterns of dicistronic plasmids indicate that translational initiation of mRNAs containing the IGF2 L3 sequences occurs by internal ribosomal entry. Three dicistronic plasmids encoding Renilla luciferase upstream of firefly luciferase, each preceded by the 5′ UTRs shown in the cartoon, were constructed; plasmid 3 is identical to plasmid 1 except for the insertion of a 60-nt hairpin structure (Mauro et al. 2007) as indicated. Twenty-four hours after transfection into RD cells, the activity of Renilla (black bars, black numbers) and firefly (gray bars, gray numbers) luciferase was measured. The results of triplicate transfections are shown. (B) Tetracycline induces a single mRNA and a quantitatively identical fold increase in the Renilla and firefly luciferase activities from a stably expressed dicistronic plasmid containing an internal L3 5′ UTR. RD cells were engineered to stably express the dicistronic plasmid (shown in the cartoon) in a tetracycline-inducible fashion. Extracts were prepared 24 h after the addition of carrier (−) or tetracycline (+). RNA was extracted and luciferase activities were measured. The RNA blot was cut longitudinally in two and hybridized with 32P-labeled cDNA probes against either the Renilla (left) or firefly (right) luciferase coding sequences; an autoradiograph of the reassembled blot and the absolute and relative activities generated from the β-globin–Renilla- and internal L3-firefly-luciferase cDNA are shown. (C) An RNAi against one of the two luciferases encoded by a dicistronic cDNA inhibits the expression of both luciferases to a similar extent. RD cells stably expressing the dicistronic plasmid shown in B were treated with a scramble RNAi or a single RNAi against one luciferase and harvested 48 h later; three different RNAis were employed against each luciferase. The Renilla (or firefly) luciferase activity measured in the extract from the scramble RNAi-treated cells was divided into the Renilla (or firefly) luciferase activity measured from the Renilla (or firefly) RNAi-treated cells. This fraction, subtracted from 1 and multiplied by 100, is shown as the percent inhibition due to the Renilla (or firefly) RNAi. (D) Synthetic monocistronic and dicistronic RNAs containing the IGF2 L3 5′ UTR undergo translational initiation after transient transfection independently of the state of 5′ cap methylation. Capped poly(A) RNA was synthesized in vitro using the mScript mRNA Production system (Epicentre) from dsDNA templates encoding the three constructs indicated. A methylated (N7mG[5′]ppp[2′Om]G; “Me”; solid bars) or unmethylated (G[5′]pppG; “unMe”; hatched bars) 5′ cap was added.One-hundred nanograms of each of the six in vitro transcribed RNA species was transfected in triplicate into 293 cells; 24 h later, extracts were analyzed for the luciferase activities (black bars; black numbers, Renilla; gray bars, gray numbers, firefly) and the abundance of the luciferase RNAs by qPCR (the Ct value is shown). The cap structures, a gel of these six RNAs, and a table containing the actual luciferase measurements are shown in Supplemental Figure S3A–C, respectively.
Rapamycin inhibits translational initiation of L3-mRNAs by inhibiting the binding of IMP2 to L3
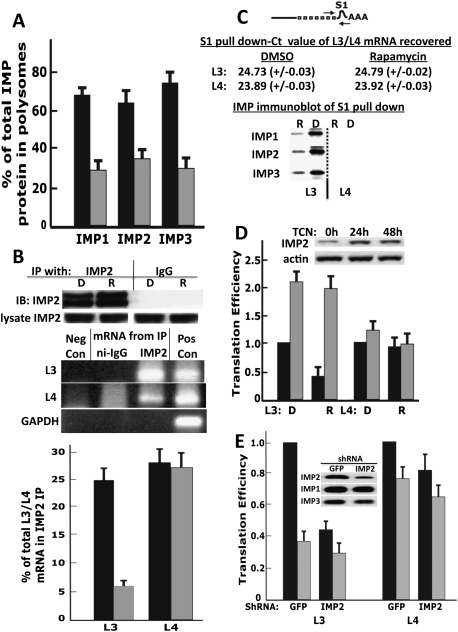
Inasmuch as the translation of L3-containing mRNAs occurs by internal ribosomal entry, the ability of rapamycin to inhibit this process suggests that the initiation of L3-mRNAs may be regulated through a novel and previously undescribed mechanism. Because the IMPs bind selectively to the L3 5′ UTR, we next explored their role in the rapamycin-sensitive translation of L3 IGF2 mRNA. In parallel to the rapamycin-induced shift of the L3 IGF2 mRNA from polysomes to subpolysomal fractions, rapamycin reduces the fraction of endogenous polysome-associated IMP1–3 polypeptides from 60%–75% down to 30%–35% (Fig. 3A). The extent of the rapamycin-induced decrease in polysome-associated IMPs is especially impressive in that the IMPs are known to bind to the shared IGF2 mRNA 3′ UTR (Fig. 3B; Nielsen et al. 1999) and the polysomal association of the IGF2 L4 mRNA is unaffected by rapamycin (Fig. 1A; Supplemental Fig. S2). We therefore sought to determine the effect of rapamycin on the binding of IMP2 specifically to the L3 and L4 mRNAs. Quantitative analysis of the IGF2 mRNA content of IMP2 immunoprecipitates by qPCR shows that rapamycin causes a marked decrease in the amount of IMP2-associated L3 IGF2 mRNA without affecting the abundance of L4 IGF2 mRNA (Fig. 3B, bar graph), indicating that rapamycin selectively disrupts the IMP2-L3 association. To examine this in a reciprocal manner, we appended to the L3-luciferase and L4-luciferase reporters immediately beyond the termination triplet an aptamer (S1) conferring the ability to bind streptavidin (Walker et al. 2008) and expressed each stably in RD cells (Fig. 3C). The polysomal association of these tagged L3- and L4-luciferase mRNAs exhibits a sensitivity to rapamycin comparable with the endogenous L3/L4 mRNAs (Supplemental Fig. S4). The RD transformants stably expressing the S1-tagged mRNAs were treated with DMSO or rapamycin followed by treatment with formaldehyde, extraction, streptavidin affinity purification, and biotin elution of the S1-tagged L3- and L4-containing mRNAs and their associated proteins. After heat inactivation of the formaldehyde cross-links, the samples were subjected to SDS-PAGE. Immunoblot of aliquots normalized for mRNA recovery shows strong bands of IMP1, IMP2, and IMP3 in the L3-luciferase-S1 pull down from the DMSO-treated cells, whereas the abundance of these polypeptides is each greatly reduced in the L3-luciferase-S1 mRNA recovered from the rapamycin-treated cells (Fig. 3C, left). No IMP polypeptide is retrieved with L4-luciferase-S1 (Fig. 3C, right); recall that the IMPs bind to the IGF2 3′ UTR; so, whereas endogenous IMP2 coprecipitates endogenous L4 mRNA (Fig. 3B), the L4-luciferase, lacking the IGF2 3′ UTR, shows no association with the IMPs (Fig. 3C, right). Thus, rapamycin-induced displacement of the IMPs from polysomes is not a passive consequence of the inhibition of translational initiation of the cohort of mRNAs with which the IMPs are associated; rather, rapamycin causes the dissociation of the IMPs from the L3 5′ UTR.
Figure 3.
Rapamycin inhibits translational initiation of L3-mRNAs by inhibiting the binding of IMP2 to L3. (A) Rapamycin displaces the IMP polypeptides from polysomes. Extracts of RD cells treated with DMSO (black bars) or rapamycin (20 nM, 3 h; gray bars) were subjected to sucrose gradient centrifugation. The subpolysomal (fractions 1–5) (Supplemental Fig. S2) and polysomal (fractions 6–13) regions of the gradient were pooled separately, and matched aliquots of each were subjected to SDS-PAGE and immunoblot for IMP1, IMP2, and IMP3. For each IMP blot, the intensity of the subpolysomal and polysomal images were added and divided into the value of the polysomal image and multiplied by 100 to give the percent of that IMP polypeptide in the polysomal fractions. The combined results of five experiments are shown. (B) Rapamycin reduces the amount of IGF2 L3 mRNA bound to IMP2 without affecting IMP2 association with IGF2 L4 mRNA. Extracts of RD cells treated with DMSO (D) or rapamycin (20 nM, 3 h; R) were subjected to immunoprecipitation with anti-IMP2 or nonimmune IgG. (Middle panel) RNA was extracted from the washed immunoprecipitates, and coprecipitating IGF2 L3 and IGF2 L4 mRNAs were quantified by qPCR and are shown in the bar graph (combined results of three experiments). (C) The endogenous IMP polypeptides bind selectively and in a rapamycin-sensitive manner to an L3-luciferase mRNA. An aptamer encoding a streptavidin-binding motif (called S1) was inserted into the L3-luciferase (cartoon) and L4-luciferase plasmids immediately after the stop triplet of the coding region. RD cells stably expressing similar amounts of the recombinant L3-S1mRNAs or L4-S1mRNAs were selected (see Supplemental Fig. S4). Aliquots of extracts prepared from rapidly growing S1 transformants, treated with either DMSO or rapamycin (20 nM, 3 h), were subjected to formaldehyde cross-linking and streptavidin affinity purification as described in the Materials and Methods; the amount of S1-containing mRNAs was quantified by qPCR, and comparable recoveries from cells treated with DMSO or rapamycin was observed. After reversal of the cross-links, the released polypeptides were separated by SDS-PAGE and were subjected to immunoblot for IMP1, IMP2, and IMP3 as shown in the lowest panel. (D) Overexpression of IMP2 in RD cells up-regulates L3-luciferase selectively and abolishes its inhibition by rapamycin. Plasmids encoding L3-luciferase or L4-luciferase, each together with a plasmid encoding Renilla luciferase, were transfected into RD cells that stably express a tetracycline-inducible IMP2, and the cells were treated with tetracycline (gray bars) or carrier (black bars) for 24 h. Three hours prior to harvest, cells were treated with DMSO (D) or rapamycin (20 nM; R). Translational efficiency was calculated as in Figure 1C and set to 1 for the L3-luciferase and L4-luciferase conditions in the extracts from cells not exposed to tetracycline or rapamycin (the black bars in L3D and L4D). Each experiment was performed in triplicate, and the combined results of three experiments ±SEM are shown. (E) Depletion of IMP2 by shRNA strongly suppresses the translation of IGF2-L3 mRNA. Plasmids encoding IGF2-L3-firefly luciferase or IGF2-L4-firefly luciferase, each together with a plasmid encoding Renilla luciferase, were transfected into RD cells stably expressing shRNAs directed against green fluorescent protein (GFP) or IMP2, and the cells were harvested 48 h later. Three hours prior to harvest, cells were treated with DMSO (black bars) or rapamycin (20 nM, gray bars). Extracts were assayed for firefly and Renilla luciferase activity and by qPCR for the content of firefly luciferase mRNA. Translational efficiency was calculated as in Figure 1C and set to 1 for the L3-luciferase and L4-luciferase conditions in the extracts from cells treated with DMSO. Each experiment was performed in triplicate, and the combined results of three experiments are shown ±SEM. The decreases in L3 luciferase and L4 luciferase due to IMP2 shRNA were both significant (P < 0.01 and <0.05, respectively).
To determine whether overexpression of an IMP can overcome the inhibitory action of rapamycin, we created a pool of RD cells that up-regulate the expression of IMP2 in a tetracycline-inducible manner; tetracycline increases IMP2 abundance approximately threefold over endogenous levels by 24 h (Fig. 3D). Up-regulation of IMP2 per se increases the expression of transiently transfected L3-luciferase approximately twofold, and eliminates completely the ability of rapamycin to inhibit L3-luciferase expression (Fig. 3D). Notably, up-regulation of IMP2 has no effect on the expression of L4-luciferase. Depletion of IMP2 using shRNA resulted in a severe inhibition of L3-luciferase activity that was little further suppressed by rapamycin (Fig. 3E). IMP2 depletion also resulted in a 20% inhibition of L4-luciferase activity (P < 0.05) (Fig. 3E). Inasmuch as IMP2 does not bind to L4-luciferase, this modest inhibition may reflect the loss of other IMP2 functions relevant to RNA life cycle or an off-target effect. The strong loss of L3-mRNA translation consequent to IMP2 depletion and the ability of IMP2 overexpression to up-regulate L3-mRNA translation and eliminate its susceptibility to inhibition by rapamycin indicate strongly that IMP2 participates in the rapamycin-sensitive regulation of L3 translation.
IMP2 binds to mTOR but not raptor, and IMP2 binding to mTOR is inhibited by mTORC1 signaling and by IMP2 binding to RNA
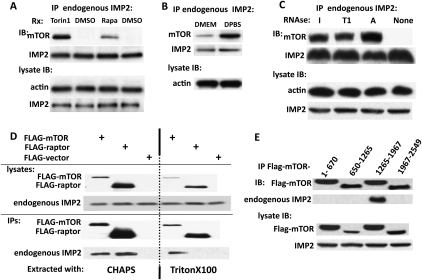
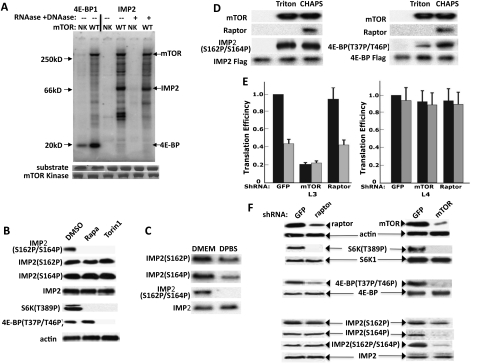
Concomitant with promoting the release of IMP2 from L3, rapamycin promotes the association of IMP2 with the mTOR polypeptide, an association also elicited by treatment of RD cells with the catalytic mTOR inhibitor Torin1 (Fig. 4A). Similarly, withdrawal of amino acids from the medium, a maneuver that selectively inactivates mTORC1 (Avruch et al. 2009), also promotes mTOR binding to IMP2 (Fig. 4B). The ability of IMP2 to bind mTOR appears to be a consequence of the loss IMP2 binding to mRNA, inasmuch as treatment of the RD extracts with a variety of RNases each results in an increased recovery of mTOR in the subsequent IMP2 immunoprecipitate (Fig. 4C). When IMP2 is coexpressed with mTOR and raptor, IMP2 can be retrieved with the mTOR–raptor complex. Surprisingly, when cells are extracted with Triton X-100 to dissociate mTOR from raptor (Hara et al. 2002), IMP2 is recovered exclusively with mTOR (Fig. 4D); IMP2 binds specifically to the mTOR(1265–1967) segment, which is comprised predominantly of the mTOR FAT domain (Fig. 4E; Wullschleger et al. 2006).
Figure 4.
IMP2 binds to the mTOR FAT domain but not raptor, and IMP2 binding to mTOR is inhibited by mTORC1 signaling and IMP2 binding to RNA. (A) The mTOR inhibitors rapamycin and Torin1 promote the binding of IMP2 to mTOR. RD cells were exposed to rapamycin (20 nM, 3 h) or Torin1 (100 nM, 1 h) or for the corresponding times to DMSO. After harvest, immunoprecipitates of endogenous IMP2 were probed for the presence of mTOR. (B) Withdrawal of medium amino acids promotes the binding of IMP2 to mTOR. Rapidly growing RD cells were washed and transferred to fresh DMEM or DPBS; 2 h later, cells were extracted and immunoprecipitates of IMP2 were probed for mTOR. (C) Degradation of RNA promotes the binding of IMP2 to mTOR. Extracts prepared from RD cells were incubated for 10 min at 37°C with the addition of ribonucleases I (1000 U/mL), T1 (200 U/mL), or A (2 μg/mL) or with DMSO. Thereafter, immunoprecipitates of endogenous IMP2 were probed for mTOR. (D) IMP2 binds mTOR but not raptor. RD cells were transfected with Flag-mTOR, Flag-raptor, or Flag vector. After 24 h, cells were extracted with either CHAPS (0.3%; left) or Triton X-100 (1%; right). Extraction with CHAPS preserves, whereas extraction in Triton X-100 abolishes, the mTOR–raptor association. (E) IMP2 binds to an mTOR noncatalytic region comprised predominantly of the FAT domain. Plasmids encoding the Flag-tagged fragments of mTOR shown were transiently expressed in 293T cells, and Flag immunoprecipitates were probed for the presence of endogenous IMP2.
mTORC1 phosphorylates IMP2 at Ser162 and Ser164 in an amino acid-dependent manner in vivo and in a raptor-independent manner in vitro
Inasmuch as raptor is the substrate-binding subunit of mTORC1, the binding of IMP2 selectively to mTOR rather than to raptor is unprecedented for an mTORC1 substrate. Nevertheless, IMP2 immunoprecipitated from rapamycin-treated cells is phosphorylated by mTORC1 in vitro to an extent comparable with that of transiently expressed Flag-4E-BP1 assayed in parallel (Fig. 5A). Notably, several other IMP2-associated proteins are also phosphorylated by mTOR; however, these other in vitro mTOR substrates are greatly reduced or lost entirely if the extract is treated with DNase plus RNase prior to IMP2 immunoprecipitation, indicating that they are associated with IMP2 indirectly through the IMP2-associated mRNAs.
Figure 5.
mTORC1 phosphorylates IMP2 at Ser162 and Ser164 in an amino acid-dependent manner in vivo and in a raptor-independent manner in vitro. (A) mTOR phosphorylates IMP2 and 4E-BP1 to a similar extent in vitro. Flag-mTOR wild type or N2343K (NK) were transiently expressed in 293T cells, extracted with 0.3% CHAPS, immunoprecipitated, washed, and eluted with Flag peptide. Recombinant Flag-4E-BP1 and Flag-IMP2 were extracted with CHAPS from rapamycin-treated 293 cells, immunoprecipitated with anti-Flag with (lanes 5,6 on right) or without prior treatment of the extract with DNase and RNase A, and eluted with Flag peptide. The mTOR kinase wild type was added to lanes 2, 4, and 6, and the inactive mTOR (NK) was added to lanes 1, 3, and 5; the reaction was started by addition of γ32P-ATP and was stopped after 30 min. An autoradiograph is shown. (B) Rapamycin and Torin1 abolish the concurrent phosphorylation of IMP2 Ser162 and Ser164. Phosphopeptide-specific antibodies were generated against IMP2[Ser162P], IMP2[Ser164P], and the doubly phosphorylated IMP2[Ser162P/Ser164P]; the specificity of these antibodies is shown in Supplemental Figure S5. RD cells were treated with rapamycin (200 nM, 1 h), Torin1 (100 nM, 1 h), or DMSO, and extracts were immunoblotted for the phosphorylated forms of IMP2 as well as for S6K1[Thr 389P] and 4E-BP[Thr 37P/46P]. (C) Amino acid withdrawal inhibits the concurrent phosphorylation of IMP2[Ser162/Ser164]. Rapidly growing RD cells were washed and transferred to fresh DMEM or DPBS; extracts were prepared 2 h later and immunoblotted for IMP2 polypeptide and IMP2 phosphorylation sites. (D) mTOR catalyzes the concurrent phosphorylation of IMP2[Ser162/Ser164] in vitro in a raptor-independent manner. Flag-mTOR was expressed in 293 cells and was extracted with either CHAPS (0.3%) or Triton X-100(1%). Flag-4E-BP and Flag-IMP2 were extracted from rapamycin-treated (200 nM, 1 h) 293T. The λ phosphatase was added to the extracts (10 U/mL) with incubation for 10 min at 30°C. The extracts were returned to 4°C and immobilized anti-Flag antibody was added. After 1.5 h, the beads were washed and subjected to Flag peptide elution. Kinase assay was carried out as in A, except nonradioactive ATP was employed and the reaction was analyzed by immunoblot for IMP2[Ser162P/Ser164P] and 4E-BP[Thr 37P/46P]. (E) Depletion of mTOR inhibits expression of IGF2-L3-luciferase, whereas depletion of raptor has no effect. Plasmids encoding IGF2-L3-firefly luciferase or IGF2-L4-firefly luciferase, each together with a plasmid encoding Renilla luciferase, were transfected into RD cells stably expressing shRNAs directed against GFP, mTOR, or raptor and were harvested 48 h later. Three hours prior to harvest, cells were treated with DMSO (black bars) or rapamycin (20 nM, gray bars). Translational efficiency was calculated as in Figure 1C and set to 1 for the L3-luciferase and L4-luciferase conditions in the extracts from cells treated with DMSO. Each experiment was performed in triplicate, and the combined results of three experiments are shown ±SEM. (F) Depletion of mTOR inhibits IMP2 phosphorylation, whereas comparable depletion of raptor does not. Extracts prepared from RD cells stably expressing shRNAs directed against GFP, mTOR, or raptor were immunoblotted for the polypeptides and phosphorylation sites indicated.
To determine whether and where IMP2 is phosphorylated in vivo, IMP2 immunoprecipitated from RD cells (with and without rapamycin treatment) was subjected to tryptic digestion and LC-MS/MS. IMP2 from control cells yielded peptides phosphorylated on Ser162, Ser164, or both Ser162 and Ser164; IMP2 from the rapamycin-treated cells yielded peptides phosphorylated on Ser162 or Ser164 but not the doubly phosphorylated peptide (Supplemental Fig. S5A). Based on these results, we generated phospho-specific antibodies directed against Ser162, Ser164, or the doubly phosphorylated species Ser162P/Ser164P (Supplemental Fig. S5B). Treatment of RD cells with rapamycin (or Torin1) for 1 h appears to have little effect on the abundance of the singly phosphorylated IMP2 species, but eliminates completely the doubly phosphorylated form of IMP2 (Fig. 5B); the responses of S6K1(T389) and 4E-BP (T37/46) phosphorylation to rapamycin and Torin1 are as expected based on previous reports (e.g., Thoreen et al. 2009). Moreover, consistent with its rapamycin sensitivity, the dual phosphorylation of IMP2(Ser162/Ser164) is also strongly inhibited by withdrawal of amino acids from the medium for 2 h and diminished phosphorylation of the individual IMP2 sites (Ser162) and (Ser164) is also evident (Fig. 5C), properties characteristic of mTORC1 substrates. However, in view of the finding that IMP2 binds exclusively to mTOR rather than to raptor, we examined whether, as with 4E-BP, IMP2 phosphorylation by mTOR requires or is facilitated by raptor. Flag-mTOR was extracted with CHAPS so as to retain the mTOR–raptor association or with Triton X-100, which dissociates raptor from mTOR. The raptor-free Flag-mTOR exhibited substantially less ability to phosphorylate 4E-BP1 in vitro as compared with the same amount of Flag-mTOR in complex 1, whereas the lack of raptor had no evident impact on the ability of Flag-mTOR to phosphorylate IMP2 in vitro (Fig. 5D). Consistent with the dispensability of raptor for the mTOR-catalyzed phosphorylation of IMP2, the shRNA-induced depletion of mTOR to ∼20% of control levels severely inhibits L3-luciferase (but not L4-luciferase) activity, whereas depletion of raptor to a similar extent affects neither L3- nor L4 luciferase activity (Fig. 5E). Moreover, shRNA-induced depletion of mTOR strongly reduces IMP2(Ser162/Ser164) dual phosphorylation as well as the individual phosphorylation of IMP2(S164), whereas comparable depletion of raptor has no effect on IMP2 phosphorylation at any site (Fig. 5F). Based on the results in Figures 4 and 5, we conclude that IMP2 is a physiologic mTORC1 substrate whose phosphorylation is largely or entirely independent of raptor.
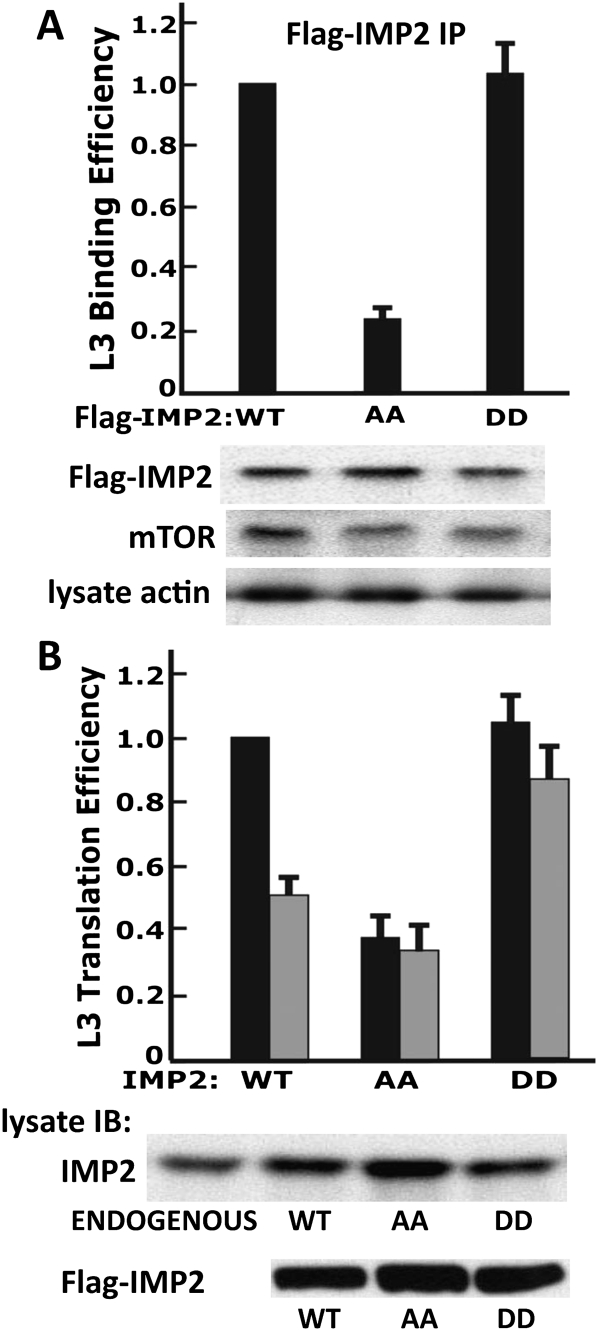
Mutation of IMP2 Ser162 and Ser164 to Ala inhibits IMP2 binding to IGF2 L3 and stimulation of L3 mRNA translation
To evaluate the functional significance of IMP2 phosphorylation for L3 mRNA translation, we transfected RD cells with cDNAs encoding IMP2 wild type [WT], [S162A/S164A], or [S162D/S164D] and examined the ability of the transiently expressed IMP2 variants to bind endogenous IGF2-L3 mRNA and endogenous mTOR (Fig. 6A). Notably, only the IMP2[S162D/S164D] exhibited binding of IGF2 L3 mRNA to an extent comparable with wild-type IMP2[WT], whereas mutation of both Ser162 and Ser164 to Ala greatly reduced IGF2 L3 binding (Fig. 6A). The IMP2[S162A/S164A] or IMP2[S162D/S164D] mutants each bound similar amounts of endogenous mTOR, but somewhat less than that recovered with IMP2[WT] (Fig. 6A). To examine the ability of the IMP variants to enable L3-mediated translation, we selected RD cells stably expressing these Flag-IMP2 variants at comparable levels (Fig. 6B, bottom blot). Initial efforts using the RD cells depleted of endogenous IMP2 by stable expression of 3′ UTR-directed shRNA were unsuccessful because the already slowed growth of these cells was strongly inhibited further by IMP2[S162A/S164A] (data not shown). Parental RD cells were therefore used to select stable transformants expressing IMP2[WT], IMP2[S162A/S164A], or IMP2[S162D/S164D] at levels comparable with (Fig. 6B, top blot, AA) or somewhat less than (Fig. 6B, top blot, WT, DD) those of endogenous IMP2 (Fig. 6B, top blot). L3-firefly luciferase cDNA was cotransfected into these cells with Renilla luciferase, and the expression of firefly luciferase relative to Renilla luciferase, normalized to L3-luciferase mRNA, was measured with and without rapamycin treatment. RD cells expressing Flag-IMP2[WT] exhibit robust expression of L3 luciferase, which, at these modest levels of IMP2[WT] overexpression, continues to be strongly inhibited by rapamycin. In contrast, IMP2[S162A/S164A] overexpressed in amounts similar to that of endogenous IMP2 results in ∼25% the level of L3-luciferase expression as wild-type IMP2[WT] (Fig. 6B). This inhibition may reflect competition for mTOR, inasmuch as IMP2[S162A/S164A] expression inhibited the phosphorylation of endogenous IMP2 (Supplemental Fig. S6). The RD cells expressing IMP2[S162D/S164D] exhibit expression of L3 luciferase comparable with that seen with IMP2[WT]; notably, however, the expression of L3 luciferase in the RD transformants expressing IMP2[S162D/S164D] is completely resistant to inhibition by rapamycin (Fig. 6B). These results indicate that the concurrent phosphorylation of Ser162 and Ser164 is required for the ability of IMP2 to promote L3-directed mRNA translation; moreover, they suggest that Ala substitutions at these IMP2 residues, although diminishing the ability of IMP2 to bind L3 mRNA, convert IMP2 to a potent dominant interfering mutant.
Figure 6.
Mutation of IMP2 Ser162 and Ser164 to Ala inhibits IMP2 binding to IGF2 L3 and stimulation of L3 mRNA translation. (A) Mutation of IMP2[Ser162/164] to Ala but not Asp reduces IMP2 binding to IGF2 L3 mRNA, whereas both IMP2 mutants bind mTOR similarly. Plasmids encoding Flag-tagged IMP2 wild type (WT), IMP2[Ser162Ala/Ser164Ala] (AA), or IMP2[Ser162Asp/Ser164 Asp] (DD) were transiently expressed in RD cells. The cells were extracted 24 h later, Flag-IMP2 variants were immunoprecipitated, and matched aliquots were subjected to phenol/chloroform extraction and measurement of bound IGF2 L3 mRNA by qPCR and immunoblot for the presence of endogenous mTOR. (B) Mutation of IMP2[Ser162/164] to Ala but not Asp reduces the initiation of L3-mRNA translation. RD cells stably expressing Flag-tagged IMP2 wild type (WT), IMP2[Ser162Ala/Ser164Ala] (AA), or IMP2[Ser162Asp/Ser164Asp] (DD) at levels similar to (AA) or less than (WT, DD) endogenous IMP2 (top blot) were transiently transfected with L3-firefly luciferase together with a plasmid encoding Renilla luciferase, and the translational efficiency of L3-luciferase was measured as described in the legend for Figure 2A. The effect of IMP2[Ser162Ala/Ser164Ala] on the phosphorylation of endogenous IMP2 is shown in Supplemental Figure S6.
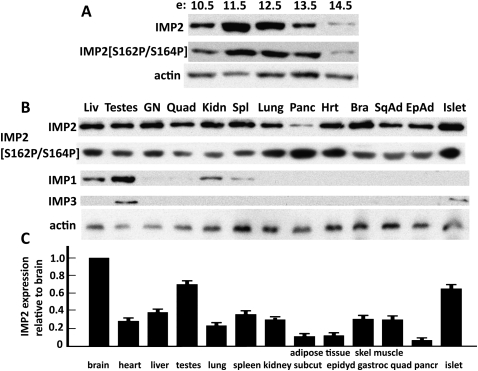
IMP2 and IMP2[Ser162P/Ser164P] are strongly expressed in mouse embryos and adult mouse tissues, and in human islets of Langerhans
The regulation of IMP2 by mTORC1-catalyzed phosphorylation has been elucidated in a human RD cell line. We sought to determine whether, when, and where this modification of IMP2 occurs in a living organism. The IMPs are expressed coordinately during embryonic development in the mouse, appearing around E11.5 (Nielsen et al. 1999). We therefore extracted mouse embryos at several intervals spanning this period and blotted for IMP polypeptides and the presence of phosphorylated forms of IMP2. IMP2 polypeptide is evident at E10.5, peaking at E11.5, and waning progressively thereafter. The relative intensity of IMP2 dual [S162/S164] phosphorylation parallels the IMP2 polypeptide abundance (Fig. 7A). We also surveyed various tissues of the adult mouse for IMP expression (Fig. 7B). As reported previously, all three IMPs are expressed in testes (Hammer et al. 2005); however, only IMP2 exhibits broad general expression at a relatively uniform level. The lowest IMP2 expression is seen in the mouse pancreas; however, a preparation of isolated human islets of Langerhans exhibits substantial levels of IMP2 polypeptide, suggesting that IMP expression in the pancreas may be predominantly in islets. Moreover, IMP2 polypeptides showing concurrent phosphorylation at Ser162 and Ser164 are evident in all of these tissues.
Figure 7.
The expression of IMP2 and IMP2[Ser162P/Ser164P] in mouse embryos, adult mouse tissues, and human islets of Langerhans. (A) IMP2 exhibits concurrent phosphorylation of Ser162 and Ser164 during mouse embryonic development. Whole mouse embryos were harvested at the developmental times indicated and extracted, and aliquots were subjected immunoblot for IMP2, IMP2[Ser162P/Ser164P], and actin. (B) IMP2 uniquely is widely expressed in tissues of the adult mouse and is abundant in human islets of Langerhans. (C) Expression of IMP2 polypeptide relative to mouse brain. As in B, except normalized for total protein content and the combined results of three experiments are shown.
Discussion
Here we show that the initiation of IGF2 L3 mRNA translation in rapidly growing RD cells occurs by eIF-4E- and cap-independent internal ribosomal entry. The RNA-binding protein IMP2, shown previously to bind selectively to the L3 5′ UTR, acts in this process as an essential IRES trans-acting factor (ITAF). The ability of IMP2 to bind the L3 5′ UTR and thus promote L3 translation is strongly dependent on the simultaneous phosphorylation of IMP2 Ser162 and Ser164, residues located between the second RRM domain and first KH domain. The mTOR kinase can catalyze this dual phosphorylation of IMP2 directly in vitro, and, in RD cells and IMP2[Ser162/164], phosphorylation is inhibited by rapamycin and by amino acid withdrawal, establishing mTORC1 as the responsible kinase. Unexpectedly, the interaction of IMP2 with mTORC1 occurs through an association with mTOR itself rather than with raptor, and the ability of mTOR to phosphorylate IMP2 in vitro is unaffected by the elimination of raptor. Several aspects of these findings merit further discussion.
IMP2 Ser162/164 are conserved throughout the mammalian IMP2s, but are not conserved with IMP1 or IMP3 (Supplemental Fig. S5A). Nevertheless, IMP1 and IMP3 are also released from L3 by rapamycin (Fig. 3C); we confirmed that IMP1 is phosphorylated at Ser181 (Brill et al. 2004), a site that, like IMP2 (Ser162/164), is located between the second RRM and first KH domain. Consequently, our ongoing studies seek to determine whether IMP1/3 also serve as mTORC1 substrates and regulators of L3 translation. How phosphorylation at Ser162 and/or Ser164 alters the ability of IMP2 to bind the L3 5′ UTR, and whether it affects IMP2 binding to the IGF2 mRNA 3′ UTR will also require further study. IMP1 deleted of its RRM domains binds target RNAs effectively and colocalizes with wild-type IMP1 in NIH3T3 cells (Nielsen et al. 2002); however, the relevance of IMP1 KH(1–4) binding to the shared IGF2 mRNA 3′ UTR (Nielsen et al. 2004) as compared with the regulatory interaction of IMP2[Ser162P/Ser164P] with the IGF2 mRNA L3 5′ UTR is unclear. Similarly, the identity of the L3 IRES, and the site(s) on the L3 5′ UTR to which IMP2[Ser162P/Ser164P] binds in vivo, are not known. Based on cross-linking of IMP1, IMP2, and IMP3 to RNAs endogenous to 293 cells, a consensus recognition sequence of CAUH (H = A, C, or U) was proposed (Hafner et al. 2010). In vitro, prokaryotic recombinant IMP1 did not bind the L4 5′ UTR, but bound to each of four contiguous fragments derived from the 1164-nt L3 RNA in a Mg++-stimulated manner and with nanomolar affinity, with highest affinity to the pyrimidine-rich segment 729–890; IMP2 and IMP3 bound this segment similarly to IMP1 (Nielsen et al. 1999). Although specific for L3, the relevance of the binding of these L3 fragments in vitro to unphosphorylated IMP1/2/3 with regard to the binding of IMP2[Ser162P/Ser164P] to the intact L3 structure in vivo is not known. Fragmentation of L3 may have altered its structure so as to expose or create IMP-binding sites, while perhaps disallowing an IMP2[Ser162P/Ser164P]-induced structure. The mechanism by which the binding of IMP2[Ser162P/Ser164P] to L3 promotes internal ribosomal entry requires further study. IMP2[Ser162P/Ser164P]-associated polypeptides might facilitate the recruitment of ribosomes to L3; the extent to which eIFs other than 4E are required for L3 translation and the contributions of other RNA BPs remain to be determined. Alternatively or additionally, IMP2[Ser162P/Ser164P] binding may reconfigure L3 so as to facilitate ribosomal binding; ZBP1, for example, has been observed to induce a looping of the β-actin mRNA 3′ UTR (Chao et al. 2010), and thus the IRES on L3 may be generated by an IMP2[Ser162P/Ser164P]-induced reorganization of the L3 structure.
This work also demonstrates the ability of mTORC1, in addition to its well-known control of cap-dependent translation (Richter and Sonenberg 2005; Ma and Blenis 2009), to regulate the cap-independent translational initiation of a cellular mRNA. Expression of IGF2 in the mouse is largely extinguished at birth, whereas the persistent and ubiquitous expression of doubly phosphorylated IMP2 in the adult mouse strongly suggests a continuing functional role. Although IGF2 L3 mRNA is the only target of such regulation identified thus far, it will be of interest to identify the other mRNAs that bind selectively to IMP2[Ser162P/Ser164P] and are coregulated with IGF2 L3 in RD cells, as well as those whose translation is regulated by IMP2[Ser162P/Ser164P] in embryonic development, in β cells and the insulin-responsive tissues of the adult, and in cancers that overexpress IMP2. This mode of translational control may also be relevant in mitosis. There, cap-dependent translation is known to be markedly reduced, yet mTORC1 activity, as reflected by 4E-BP phosphorylation and S6K1 activity, is apparently high (Ramírez-Valle et al. 2010). Cap-independent translation is relatively enhanced during M, and the translation—e.g., of p58 PITSLRE kinase (Cornelis et al. 2000) and ODC (Pyronnet et al. 2000)—by internal ribosomal entry is important to mitotic progression; it will be of interest to determine whether mTORC1-regulated, IMP-dependent translation contributes to mitotic progression.
The finding that IMP2, a direct physiologic substrate of mTORC1, binds to mTOR rather than to raptor is unprecedented. We infer that IMP2(Ser162/164) phosphorylation in the RD cell is catalyzed by intact mTORC1 in vivo because this phosphorylation is exquisitely sensitive to rapamycin and to amino acid withdrawal, behaviors that are characteristic of intact mTORC1. Alternatively, IMP2 may be phosphorylated by a raptor/rictor-free mTOR that remains rapamycin-sensitive and regulated by amino acids. IMP2 lacks a canonical TOS motif or one corresponding to the variant in PRAS40 (FV/MMDE) (Oshiro et al. 2007), binds exclusively to mTOR, and is comparably phosphorylated by mTOR in vitro with or without associated raptor. It is difficult, however, to establish the raptor independence of IMP2 phosphorylation in vivo, inasmuch as raptor is required for cell survival. Nevertheless, it will be of interest to determine whether other physiologic mTORC1 substrates bind primarily to mTOR rather than raptor, and to determine what, if any, differences in regulation are associated with this mode of substrate interaction. One possibility, for example, is that IMP2(Ser162/Ser164) phosphorylation will be insensitive to regulators that operate in part or in whole through interference with the ability of raptor to bind the mTOR substrates.
IMP2 has been implicated recently through genome-wide association studies as a candidate susceptibility gene for type 2 diabetes (Saxena et al. 2007; Scott et al. 2007; Zeggini et al. 2007). The mechanism by which this susceptibility is engendered is not known. Strong epidemiologic evidence links low birth weight to adult susceptibility to type 2 diabetes and cardiovascular disease (Hales and Barker 2001; Barker 2006). In rodent models, maternal and or fetal nutrient insufficiency results in low-birth-weight offspring; postnatal exposure of such offspring to nutrient excess results in a high frequency of diabetes (Simmons 2009). IGF2 (Efstratiadis 1998) and nutrient sufficiency during development (Zeisel 2009) are each critical determinants of fetal growth. Nutrient insufficiency may restrict fetal size in part through IGF2, inasmuch as the expression of the imprinted IGF2 gene in fetal life has been shown in rodent models to be sensitive to maternal and fetal nutrition (Kwong et al. 2006; Sharif et al. 2007). We speculate that variations in IMP2 expression and/or a deficiency in nutrient-driven mTORC1 phosphorylation of IMP2 during development may also result in diminished IGF2 expression in fetal life, low birth weight, and adult susceptibility to type 2 diabetes. Alternatively, or in addition, as IMP2 is the only IMP expressed in β cells and insulin-sensitive tissues in postnatal life, altered expression or regulation of IMP2 by mTORC1 in the adult may be contributory to the pathogenesis of type 2 diabetes. Understanding the regulation of IMP2 and its roles in development and organ function will clarify these issues.
Materials and methods
Cell culture
Human RD cells (American Type Culture Collection CCL-136) were cultured in DMEM medium supplemented with 10% fetal bovine serum at 37°C in 5% CO2.
Dual luciferase reporter assay
Luciferase activity was examined by a dual luciferase reporter assay using the dual luciferase reporter assay kit (Promega) on a TD-20/20 luminometer (Turner Designs). The RD cells were cotransfected with pRL-SV40 Renilla luciferase control vector (Promega). Luciferase activity was measured at 36 h post-transfection.
The plasmids pcDNA-IGF-II L3/luciferase (pcDNA-IGF-II-L3-luciferase) and pcDNA-IGF-II L4/luciferase (pcDNA-IGF-II-L4-luciferase) were kindly provided by Dr. Jan Christiansen. These plasmids contain the firefly luciferase coding region and the complete L3 (1164-base-pair [bp]) or L4 (94-bp) exon, respectively, in the pcDNA3.1 basic vector (Invitrogen). The pcDNA-ODC1-luciferase was generated by insertion of the 5′ UTR of the human ODC1 gene (NM_002539, nt1-334) upstream of firefly luciferase. To make the bicistronic expression vector, human β-globin 5′ UTR (NM_000518, nt1-50), IGF2 L3, and firefly and renilla luciferase coding sequences were ligated into the pcDNA5/TO vector (Invitrogen). The DNA oligonucleotides comprising the 60-nt hairpin structure (Mauro et al. 2007) were synthesized by IDT and inserted into the pcDNA5/TO as a control.
Messenger ribonucleoprotein immunoprecipitation (RIP) assay
Exponentially growing human RD cells were harvested and lysed for 10 min on ice in cold lysis buffer (140 mM KCl, 1.5 mM MgCl2, 20 mM Tris-HCl at pH 7.4, 0.5% Nonidet P-40, 0.5 mM dithiothreitol, 1 U/μL RNase inhibitor, one complete EDTA-free protease inhibitor cocktail tablet). Lysate was centrifuged for 10 min at 12,000 rpm, and supernatant was transferred to a fresh 1.5-mL tube. Total protein concentration in the lysates was measured by Bradford assay. For immunoprecipitation, 2 mg of cytoplasmic lysate proteins were incubated with 500 μL of protein A Dyna magnetic beads precoated with rabbit IgG or anti-human IMP2 antibody for 2 h at 4°C with rotation. Beads were extensively washed with lysis buffer and digested with DNase I and protease K. RNA was extracted with phenol/chloroform and precipitated with ethanol. Real-time PCR was performed to examine RNAs associated with cytoplasmic IMP2 as described above.
S1 tag pull down
The IGFII L3 and L4 were inserted into the Hind III site upstream of the firefly luciferase coding region, which was followed by the S1 streptavidin-binding RNA motif (5′-ACCGACCAGAAUCAUGCAAGUGCGUAAGAUAGUCGCGGGCCGGG-3′) (Walker et al. 2008) in pcDNA5/TO vector. The human RD cell lines stably expressing S1-L3 and S1-L4 growing up to 25%–30% confluence were cross-linked in 0.3% formaldehyde in PBS for 10 min at 37°C (Niranjanakumari et al. 2002). Cells were lysed (150 mM KCl, 10 mM HEPES at pH 7.4, 3 mM MgCl2, 2 mM DTT, 10% glycerol, 0.5% NP-40, protease, RNase inhibitors) for 20 min on ice, sonicated and clarified by centrifugation, precleared with avidin beads, and then incubated with streptavidin beads (Pierce) for 4 h at 4°C; washed extensively with a salt gradient up to 400 mM KCl in the binding buffer with tRNA, glycogen, and 2% NP-40; and eluted for 1 h with 5 mM biotin (Walker et al. 2008). After heat inactivation for 30 min at 60°C (Niranjanakumari et al. 2002), the RNA and proteins were assayed.
In vitro kinase assay
HEK293T cells were transfected with mTOR-Flag using Lipofectamine according to the manufacturer's protocol (Invitrogen). After 36 h, cells were rinsed twice with PBS and lysed in 1 mL of ice-cold lysis buffer (40 mM HEPES at pH 7.4, 2 mM EDTA, 10 mM glycerophosphate, 0.3% CHAPS or 1% Triton X-100, EDTA-free protease inhibitor). After clearing, the supernatant was used to prepare Flag or mTOR endogenous immunoprecipitates. Immunoprecipitates were washed five times with lysis buffer. Kinase assays were performed for 30 min at 30°C. Reactions were stopped by the addition of sample buffer. After boiling the samples for 5 min, proteins were resolved by SDS-PAGE; phosphorylated proteins were then visualized with autoradiography or immunoblotting.
In vitro transcription and RNA transfection
The in vitro transcription and RNA transfections were performed as described (Angel and Yanik 2010). Briefly, T7 promoter-containing dsDNA templates were linearized and capped, and poly(A) RNA was synthesized using the mScript mRNA Production system (Epicentre). To stabilize the mRNA for transient transfection, an N7 methylated cap or an unmethylated cap analog (Epicentre) was added; with the former, 2′O methylation of the first base was also performed (Supplemental Fig. S3). Transcripts were analyzed after poly(A) tailing by denaturing formaldehyde-agarose gel electrophoresis (Supplemental Fig. S3). The in vitro transcribed, capped, and poly(A)-tailed RNAs were transfected with Lipofectamine RNAiMAX (Invitrogen) and, 24 h thereafter, extracts were analyzed for the luciferase activities and the abundance of the luciferase RNAs by qPCR.
Additional descriptions of materials and methods are in the Supplemental Material.
Acknowledgments
We thank T. Ding, Dr. S. Jacobs, Dr. O. Peroni, and Dr. D.Wrighting for assistance with mice; M. Lynch and Dr. E. Schmidt for assistance in sucrose density gradient methods; Dr. S. Vasudevan for suggestions regarding streptavidin-tagged mRNA isolation; Dr. N. Oshiro for discussions; Dr. F.C. Nielsen for IMP1, IMP2, and IMP3 antibodies; and Dr. J. Christiansen for the plasmids pcDNA-IGF-II-L3-luciferase and pcDNA-IGF-II-L4-luciferase. Torin1 was kindly provided by Drs. N.S. Gray and D.M. Sabatini. Human islets were a gift from Dr. J. Markmann and Dr. J. Lei. This work was supported by grants from the NIH (CA73818, DK17776, and DK057521) and ARRA funds.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.2042311.
Supplemental material is available for this article.
References
- Angel M, Yanik MF 2010. Innate immune suppression enables frequent transfection with RNA encoding reprogramming proteins. PLoS ONE 5: e11756 doi: 10.1371/journal.pone.0011756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avruch J, Long X, Ortz-Vega S, Rapley J, Papageorgiou A, Dai N 2009. Amino acid regulation of TOR complex 1. Am J Physiol Endocrinol Metab 296: E592–E602 doi: 10.1152/ajpendo.90645.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker J, Liu JP, Robertson EJ, Efstratiadis A 1993. Role of insulin-like growth factors in embryonic and postnatal growth. Cell 8: 73–82 [PubMed] [Google Scholar]
- Barker DJ 2006. Adult consequences of fetal growth restriction. Clin Obstet Gynecol 49: 270–283 [DOI] [PubMed] [Google Scholar]
- Brill LM, Salomon AR, Ficarro SB, Mukherji M, Stettler-Gill M, Peters EC 2004. Robust phosphoproteomic profiling of tyrosine phosphorylation sites from human T cells using immobilized metal affinity chromatography and tandem mass spectrometry. Anal Chem 76: 2763–2772 [DOI] [PubMed] [Google Scholar]
- Chao JA, Patskovsky Y, Patel V, Levy M, Almo SC, Singer RH 2010. ZBP1 recognition of β-actin zipcode induces RNA looping. Genes & Dev 24: 148–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis S, Bruynooghe Y, Denecker G, Van Huffel S, Tinton S, Beyaert R 2000. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol Cell 5: 597–605 [DOI] [PubMed] [Google Scholar]
- Efstratiadis A 1998. Genetics of mouse growth. Int J Dev Biol 42: 955–976 [PubMed] [Google Scholar]
- Engström W, Shokrai A, Otte K, Granérus M, Gessbo A, Bierke P, Madej A, Sjölund M, Ward A 1998. Transcriptional regulation and biological significance of the insulin like growth factor II gene. Cell Prolif 31: 173–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert WV 2010. Alternative ways to think about cellular internal ribosome entry. J Biol Chem 285: 29033–29038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A-C, Raught B, Sonenberg N 2004. mTOR signaling to translation. Curr Top Microbiol Immunol 279: 169–197 [DOI] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp A-C, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales CN, Barker DJ 2001. The thrifty phenotype hypothesis. BMJ 60: 5–20 [DOI] [PubMed] [Google Scholar]
- Hammer NA, Hansen TO, Byskov AG, Rajpert-De Meyts E, Grøndahl ML, Bredkjaer HE, Wewer UM, Christiansen J, Nielsen FC 2005. Expression of IGF-II mRNA-binding proteins (IMPs) in gonads and testicular cancer. Reproduction 130: 203–212 [DOI] [PubMed] [Google Scholar]
- Hansen TV, Hammer NA, Nielsen J, Madsen M, Dalbaeck C, Wewer UM, Christiansen J, Nielsen FC 2004. Dwarfism and impaired gut development in insulin-like growth factor II mRNA-binding protein 1-deficient mice. Mol Cell Biol 24: 4448–4468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K 2002. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110: 177–189 [DOI] [PubMed] [Google Scholar]
- Hornstein E, Tang H, Meyuhas O 2001. Mitogenic and nutritional signals are transduced into translational efficiency of TOP mRNAs. Cold Spring Harb Symp Quant Biol 66: 477–484 [DOI] [PubMed] [Google Scholar]
- Hüttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH 2005. Spatial regulation of β-actin translation by Src-dependent phosphorylation of ZBP1. Nature 438: 512–515 [DOI] [PubMed] [Google Scholar]
- Komar AA, Hatzoglou M 2011. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle 10: 229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong WY, Miller DJ, Ursell E, Wild AE, Wilkins AP, Osmond C, Anthony FW, Fleming TP 2006. Imprinted gene expression in the rat embryo-fetal axis is altered in response to periconeptional maternal low protein diet. Reproduction 132: 265–277 [DOI] [PubMed] [Google Scholar]
- Liao B, Patel M, Hu Y, Charles S, Herrick DJ, Brewer G 2004. Targeted knockdown of the RNA-binding protein CRD-BP promotes cell proliferation via an insulin-like growth factor II-dependent pathway in human K562 leukemia cells. J Biol Chem 279: 48716–48724 [DOI] [PubMed] [Google Scholar]
- Liao B, Hu Y, Herrick DJ, Brewer G 2005. The RNA-binding protein IMP-3 is a translational activator of insulin-like growth factor II leader-3 mRNA during proliferation of human K562 leukemia cells. J Biol Chem 280: 18517–18524 [DOI] [PubMed] [Google Scholar]
- Ma XM, Blenis J 2009. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 10: 307–318 [DOI] [PubMed] [Google Scholar]
- Mauro VP, Chappell SA, Dresios J 2007. Analysis of ribosomal shunting during translation initiation in eukaryotic mRNAs. Methods Enzymol 429: 323–354 [DOI] [PubMed] [Google Scholar]
- Mayer C, Grummt I 2006. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 25: 6384–6391 [DOI] [PubMed] [Google Scholar]
- Mehrpour M, Esclatine A, Beau I, Codogno P 2010. Overview of macroautophagy regulation in mammalian cells. Cell Res 20: 748–762 [DOI] [PubMed] [Google Scholar]
- Mori H, Sakakibara S, Imai T, Nakamura Y, Iijima T, Suzuki A, Yuasa Y, Takeda M, Okano H 2001. Expression of mouse igf2 mRNA-binding protein 3 and its implications for the developing central nervous system. J Neurosci Res 64: 132–143 [DOI] [PubMed] [Google Scholar]
- Newell S, Ward A, Graham C 1994. Discriminating translation of insulin-like growth factor-II (IGF-II) during mouse embryogenesis. Mol Reprod Dev 39: 249–258 [DOI] [PubMed] [Google Scholar]
- Nielsen FC, Ostergaard L, Nielsen J, Christiansen J 1995. Growth-dependent translation of IGF-II mRNA by a rapamycin sensitive pathway. Nature 377: 358–362 [DOI] [PubMed] [Google Scholar]
- Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC 1999. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol 19: 1262–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen FC, Nielsen J, Christiansen J 2001. A family of IGF-II mRNA binding proteins (IMP) involved in RNA trafficking. Scand J Clin Lab Invest Suppl 234: 93–99 [PubMed] [Google Scholar]
- Nielsen FC, Nielsen J, Kristensen MA, Koch G, Christiansen J 2002. Cytoplasmic trafficking of IGF-II mRNA-binding protein by conserved KH domains. J Cell Sci 115: 2087–2097 [DOI] [PubMed] [Google Scholar]
- Nielsen J, Kristensen MA, Willemoës M, Nielsen FC, Christiansen J 2004. Sequential dimerization of human zipcode-binding protein IMP1 on RNA: a cooperative mechanism providing RNP stability. Nucleic Acids Res 32: 4368–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niranjanakumari S, Lasda E, Brasas R, Carcia-Blanco MA 2002. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods 26: 182–190 [DOI] [PubMed] [Google Scholar]
- Nojima H, Tokunaga C, Eguchi S, Oshiro N, Hidayat S, Yoshino K, Hara K, Tanaka N, Avruch J, Yonezawa K 2003. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J Biol Chem 278: 15461–15464 [DOI] [PubMed] [Google Scholar]
- Oshiro N, Takahashi R, Yoshino K, Tanimura K, Nakashima A, Eguchi S, Miyamoto T, Hara K, Takehana K, Avruch J, et al. 2007. The proline-rich Akt substrate of 40 kDa (PRAS40) is a physiological substrate of mammalian target of rapamycin complex 1. J Biol Chem 282: 20329–20339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patursky-Polischuk I, Stolovich-Rain M, Hausner-Hanochi M, Kasir J, Cybulski N, Avruch J, Rüegg MA, Hall MN, Meyuhas O 2009. The TSC–mTOR pathway mediates translational activation of TOP mRNAs by insulin largely in a raptor- or rictor-independent manner. Mol Cell Biol 29: 640–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyronnet S, Pradayrol L, Sonenberg N 2000. A cell cycle-dependent internal ribosome entry site. Mol Cell 5: 607–616 [DOI] [PubMed] [Google Scholar]
- Ramírez-Valle F, Badura ML, Braunstein S, Narasimhan M, Schneider RJ 2010. Mitotic raptor promotes mTORC1 activity, G(2)/M cell cycle progression, and internal ribosome entry site-mediated mRNA translation. Mol Cell Biol 30: 3151–3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, Sonenberg N 2005. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433: 477–480 [DOI] [PubMed] [Google Scholar]
- Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, et al. 2007. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316: 1331–1336 [DOI] [PubMed] [Google Scholar]
- Schalm SS, Blenis J 2002. Identification of a conserved motif required for mTOR signaling. Curr Biol 12: 632–639 [DOI] [PubMed] [Google Scholar]
- Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, et al. 2007. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316: 1341–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif J, Nakamura M, Ito T, Kimura Y, Nagamune T, Mitsuya K, Okamura K 2007. Food restrictions in pregnant mice can induce changes in histone modifications and suppress gene expression in fetus. Nucleic Acids Symp Ser (Oxf) 51: 125–126 [DOI] [PubMed] [Google Scholar]
- Shor B, Wu J, Shakey Q, Toral-Barza L, Shi C, Follettie M, Yu K 2010. Requirement of the mTOR kinase for the regulation of Maf1 phosphorylation and control of RNA polymerase III-dependent transcription in cancer cells. J Biol Chem 285: 15380–15392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons RA 2009. Developmental origins of adult disease. Pediatr Clin North Am 56: 449–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun FL, Dean WL, Kelsey G, Allen ND, Reik W 1997. Transactivation of Igf2 in a mouse model of Beckwith-Wiedemann syndrome. Nature 387: 809–815 [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS 2009. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem 284: 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SC, Scott FH, Srisawat C, Engelke DR 2008. RNA affinity tags for the rapid purification and investigation of RNAs and RNA–protein complexes. Methods Mol Biol 488: 23–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN 2006. TOR signaling in growth and metabolism. Cell 124: 371–384 [DOI] [PubMed] [Google Scholar]
- Yisraeli JK 2005. VICKZ proteins: a multi-talented family of regulatory RNA-binding proteins. Biol Cell 97: 87–96 [DOI] [PubMed] [Google Scholar]
- Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW, Freathy RM, et al. 2007. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 316: 1336–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel SH 2009. Epigenetic mechanisms for nutrition determinants of later health outcomes. Am J Clin Nutr 89: 1488S–1493S [DOI] [PMC free article] [PubMed] [Google Scholar]