Abstract
Linkage disequilibrium (LD) is a proven tool for evaluating population structure and localizing genes for monogenic disorders. LD-based methods may also help localize genes for complex traits. We evaluated marker–marker LD using 43 microsatellite markers spanning chromosome 20 with an average density of 2.3 cM. We studied 837 individuals affected with type 2 diabetes and 386 mostly unaffected spouse controls. A test of homogeneity between the affected individuals and their spouses showed no difference, allowing the 1223 individuals to be analyzed together. Significant (P < 0.01) LD was observed using a likelihood ratio test in all (11/11) marker pairs within 1 cM, 78% (25/32) of pairs 1–3 cM apart, and 39% (7/18) of pairs 3–4 cM apart, but for only 12 of 842 pairs more than 4 cM apart. We used the human genome project working draft sequence to estimate kilobase (kb) intermarker distances, and observed highly significant LD (P < 10−10) for all six marker pairs up to 350 kb apart, although the correlation of LD with cM is slightly better than the correlation with megabases. These data suggest that microsatellites present at 1-cM density are sufficient to observe marker–marker LD in the Finnish population.
Linkage disequilibrium (LD), the nonrandom association between alleles of linked markers, reflects the size of chromosomal segments remaining intact in a population. LD analysis has proved powerful for high-resolution mapping of disease genes for monogenic disorders, including cystic fibrosis (Rommens et al. 1989) and diastrophic dysplasia (Hastbacka et al. 1994). In the Finnish population, LD has narrowed the candidate gene interval for many recessive disorders that have one or a few predisposing alleles at a single locus (Peltonen et al. 1999).
In principle, LD also may aid in positional cloning of genes for complex traits in founder populations, particularly if one or a small number of founder alleles contributes substantially to disease risk, and if the density of markers is sufficiently high. Marker–marker LD may provide an upper bound for the needed marker density by demonstrating the presence of chromosomal regions inherited together since the population was founded or subjected to a bottleneck.
The limited available experimental data report varying evidence of LD over cM distances. In one study of 50 Finns, up to one half of the marker pairs within 2 cM showed LD at P < 0.05 (Peterson et al. 1995). However, a study of the X chromosome in 80 Finnish males reported that only 2 of 16 marker pairs up to 2 cM apart showed significant LD (P < 0.05) (Laan and Paabo 1997). On chromosomes 5, 6, and 8, in an average of 986 haplotypes from Irish schizophrenia pedigrees, significant LD (P < 0.05) was observed in 81% of marker pairs within 1 cM and 35% of pairs 1 to 2 cM apart (Kendler et al. 1999). A genome-wide survey of LD performed with 5048 microsatellites in 54 independent chromosomes from European, Utah, and Amish CEPH families detected significant (P < 0.01) LD in ∼4% of markers within 4 cM of one another (Huttley et al. 1999). Recently, Eaves et al. assessed LD in a 6.5-cM region of chromosome 18q21 in samples of 800 chromosomes each from Finland, Sardinia, the United States, and the United Kingdom (Eaves et al. 2000). They found LD extending up to 1 cM, with somewhat greater LD in the Finns and Sardinians. Additional data are needed in various populations and genomic regions to estimate the extent and variability of LD and to assess whether the presence of LD will be useful for fine-mapping complex disease genes.
Type 2 diabetes is a common disorder that causes considerable morbidity and mortality throughout the world. Evidence for a genetic component in type 2 diabetes has been obtained using twin and family history studies (Newman et al. 1987; Rich 1990; Kaprio et al. 1992). Several groups have identified linkage signals for type 2 diabetes affection status (Ehm et al. 2000), and one underlying gene has been identified (Horikawa et al. 2000). The Finland–United States Investigation of NIDDM Genetics (FUSION) study aims to identify susceptibility genes for type 2 diabetes and related quantitative traits in the Finnish population. In this study, we ascertained type 2 diabetes-affected sibling pairs and additional relatives (Valle et al. 1998). A 10-cM genome scan on >2000 individuals revealed several regions of suggested affected sib-pair linkage (Ghosh et al. 2000), including portions of both arms of chromosome 20 (Ghosh et al. 1999). To better assess the importance of these regions, we genotyped a total of 43 markers at 2.3 cM average density on this chromosome. These data provide the opportunity to assess the degree of marker–marker LD and the distances over which LD extends. We present here microsatellite marker-to-marker LD across chromosome 20 in 1223 individuals. We show that significant intermarker LD is virtually always observed for marker pairs up to 1 cM apart, and is generally absent for marker pairs at distances greater than 4 cM apart.
RESULTS
Forty-three microsatellite markers spanning chromosome 20 were genotyped on 837 unrelated Finns affected with type 2 diabetes and 386 of their spouses. Estimated genetic positions of the markers are shown in Table 1. The 43 markers have an average estimated heterozygosity of 0.77 and average spacing of 2.3 cM across the 99.2-cM chromosome. Table 1 also shows the number of alleles we observed for each marker and the number of alleles we analyzed when low-frequency alleles were pooled.
Table 1.
Genetic Map and Marker Statistics
| Marker | Map position (cM) | H | Alleles observeda | Alleles analyzedb |
|---|---|---|---|---|
| S103 | 0.0 | 0.70 | 8 | 5 |
| S117 | 0.5 | 0.87 | 15 | 8 |
| S906 | 5.1 | 0.78 | 12 | 6 |
| S193 | 9.1 | 0.72 | 13 | 4 |
| S889 | 10.0 | 0.82 | 19 | 8 |
| S482 | 11.8 | 0.69 | 9 | 4 |
| S849 | 14.8 | 0.69 | 17 | 5 |
| S905 | 18.6 | 0.64 | 11 | 4 |
| S892 | 20.6 | 0.86 | 12 | 8 |
| S846 | 20.6 | 0.73 | 10 | 5 |
| S115 | 22.1 | 0.64 | 7 | 4 |
| S851 | 25.5 | 0.76 | 14 | 6 |
| S917 | 26.2 | 0.87 | 16 | 8 |
| S894 | 30.4 | 0.83 | 12 | 7 |
| S189 | 32.4 | 0.69 | 10 | 6 |
| S898 | 37.3 | 0.74 | 18 | 5 |
| S114 | 42.7 | 0.83 | 11 | 7 |
| S912 | 48.2 | 0.83 | 12 | 7 |
| S477 | 48.9 | 0.74 | 11 | 5 |
| S874 | 52.1 | 0.79 | 16 | 6 |
| S195 | 52.7 | 0.85 | 16 | 7 |
| S909 | 53.4 | 0.70 | 16 | 5 |
| S107 | 58.1 | 0.84 | 15 | 6 |
| S170 | 59.6 | 0.80 | 17 | 8 |
| S96 | 62.4 | 0.82 | 13 | 7 |
| S119 | 65.7 | 0.80 | 10 | 6 |
| S481 | 65.9 | 0.83 | 13 | 8 |
| S836 | 66.9 | 0.83 | 12 | 7 |
| S888 | 67.7 | 0.90 | 20 | 8 |
| S886 | 68.1 | 0.86 | 10 | 7 |
| S197 | 69.1 | 0.75 | 11 | 6 |
| S178 | 70.2 | 0.78 | 11 | 7 |
| S866 | 71.8 | 0.85 | 19 | 8 |
| S196 | 74.5 | 0.79 | 23 | 4 |
| S857 | 76.3 | 0.84 | 13 | 8 |
| S480 | 79.2 | 0.75 | 11 | 6 |
| S840 | 80.2 | 0.80 | 18 | 7 |
| S211 | 80.2 | 0.65 | 13 | 5 |
| S120 | 83.2 | 0.83 | 18 | 7 |
| S100 | 85.8 | 0.74 | 12 | 5 |
| S102 | 87.7 | 0.44 | 5 | 4 |
| S171 | 96.1 | 0.80 | 13 | 6 |
| S173 | 99.2 | 0.73 | 13 | 7 |
H, Heterozygosity.
Number of alleles observed in an average of 5114 individuals per marker, typed as part of the FUSION study.
Number analyzed when alleles with a frequency below 5% are pooled.
To determine if the affected individuals and their spouses showed different haplotype frequencies, a permutation test of heterogeneity was performed for all 903 possible pairs of the 43 markers. Among these pairs, 12 were significant at the 0.01 level, consistent with randomness. Further, the distribution of the 903 P-values appeared to be uniform across the interval zero to 1 (data not shown). These results suggested no important differences in haplotype frequencies exist between the affected individuals and controls, allowing us to pool data across the two samples.
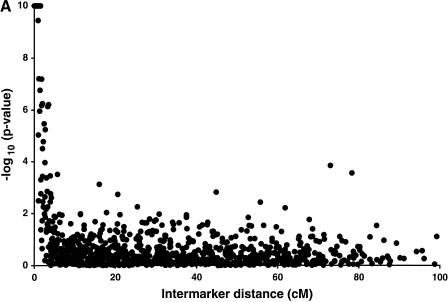
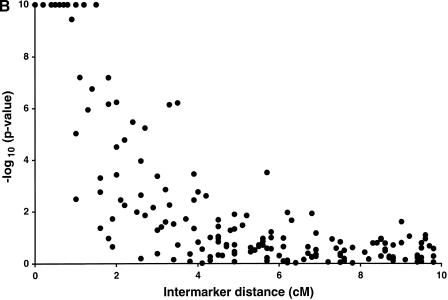
After pooling the diabetic individuals and their spouses to generate a sample of 1223 individuals, we analyzed all 903 possible marker pairs using a likelihood ratio test for LD. We plotted the −log10 P-values of all 903 comparisons in the combined sample as a function of estimated intermarker distance (Fig. 1a). A total of 55 marker pairs are significant at the 0.01 level (−log10 P-value of 2), compared to nine significant results expected by chance in the absence of LD. LD between marker pairs less than 10 cM apart are shown in greater detail (Fig. 1b). We observed significant LD between all marker pairs within 1 cM, 78% between 1 and 2 cM, 79% between 2 and 3 cM, and 39% between 3 and 4 cM (Table 2). Only 1.4% (12/842) of marker pairs greater than 4 cM apart show significant LD at the 0.01 level, slightly greater than that expected by chance alone.
Figure 1.
Presence of LD on chromosome 20 in 1223 Finns. −log10(P-values) for a likelihood ratio statistic, limited at 10−10. Each point represents one pair of markers; (a) all 903 possible pairs of 43 markers, (b) 168 pairs of markers <10 cM apart.
Table 2.
Number of Marker Pairs in LD
| Intermarker distance (cM) | n | P < 0.01 | P < 0.001 | ||||
|---|---|---|---|---|---|---|---|
| spouses | affecteds | combined | spouses | affecteds | combined | ||
| <1 cM | 11 | 11 | 11 | 11 | 10 | 11 | 11 |
| 1–2 cM | 18 | 9 | 13 | 14 | 7 | 10 | 12 |
| 2–3 cM | 14 | 2 | 8 | 11 | 1 | 4 | 7 |
| 3–4 cM | 18 | 4 | 6 | 7 | 1 | 2 | 4 |
| ≥4 cM | 842 | 17 | 16 | 12 | 0 | 3 | 4 |
| Total in 99.2 | 903 | 43 | 54 | 55 | 19 | 30 | 38 |
P-values from a likelihood ratio test for LD. n, number of marker pairs. The spouse, affected, and combined samples consist of 386,837 and 1223 individuals, respectively.
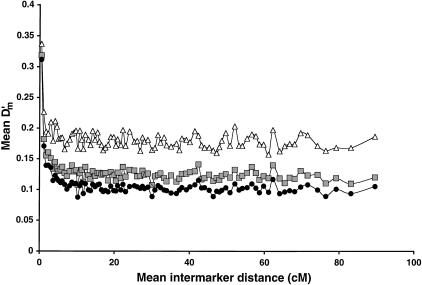
We also measured LD using a D‘ statistic modified for multiallelic markers (Hedrick 1987). The rise in D‘ values for intermarker distances up to 4 cM is similar to the likelihood ratio P-values (Fig. 2). Importantly, this modified D‘ statistic is inversely related to sample size, as can be observed by comparing samples of 386 spouse controls and 837 affecteds to the combined sample. For distant marker pairs not expected to exhibit LD, the multiallelic D‘ statistic approaches asymptotes that are larger for smaller samples (Fig. 2).
Figure 2.
Strength of LD on chromosome 20 in 1223 Finns. Multiallelic D‘ statistic for 386 spouses (open triangles), 837 individuals with type 2 diabetes (shaded squares), and the combined sample of 1223 (black circles). Each point represents 10 marker pairs, and is plotted at the mean intermarker distance for those pairs.
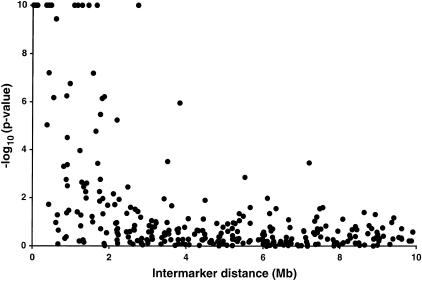
The extent of LD depends on recombination rather than physical intermarker distance, although the presence of LD over kilobase distances can be used during positional cloning studies to determine the marker density needed to detect LD. We used finished and unfinished genomic sequence to estimate the physical distances between our markers. We evaluated the presence of significant LD for all 903 possible pairs of 43 markers, and show marker pairs up to 10 megabases (Mb) apart (Fig. 3). Significant LD at the 0.01 level is observed for 69% (20/29) of markers up to 1 Mb apart, 51% (18/35) between 1 and 2 Mb apart, 12% (4/33) between 2 and 3 Mb apart, and 6% (2/36) between 3 and 4 Mb apart. Only 1.4% (11/770) of markers greater than 4 Mb apart showed significant LD. Strongly significant LD (P < 10−10) was observed for all six marker pairs up to 350 kb apart, and 75% (9/12) for pairs up to 500 kb apart. As expected, the correlation between LD and physical distance (Spearman rank correlation r = −0.56) is less strong than with genetic distance (r = −0.63) for markers up to 10 cM apart.
Figure 3.
LD versus physical distance. −log10(P-values) for a likelihood ratio statistic, limited at 10−10. Each point represents one pair of markers. Two hundren ninety marker pairs up to 10 Mb apart are shown.
DISCUSSION
We analyzed marker–marker LD across all of chromosome 20 using a large sample of 1223 unrelated Finnish subjects. We observed significant (P < 0.01) marker–marker LD for all marker pairs within 1 cM of each other and continued elevated LD for distances up to 4 cM (Fig. 1). We obtained similar conclusions when analyzing the data based on significance level 0.001 (Table 2) or 0.05 (Fig. 1). We observed significant LD (P < 0.01) at estimated physical distances up to 4 Mb, with strongly significant LD (P < 10−10) at distances up to 350 kb. We did not detect regions of chromosome 20 that exhibited obviously higher or lower LD (data not shown), although the variable spacing of our markers did not provide ideal data to address this issue.
Our findings of significant LD cannot be compared directly to results obtained from most other studies using microsatellite markers because the likelihood ratio test is strongly affected by sample size. In particular, our large sample size enables quite sensitive detection of LD. The expected value of the likelihood ratio statistic is proportional to sample size, while P-values are affected even more strongly by sample size. This characteristic is consistent with the smaller number of significant marker pairs observed in the smaller sample of 386 spouses (Table 2).
The likelihood ratio statistic shown here detects the presence but does not evaluate the strength of LD. To measure the strength of LD and to compare our data with a recent report (Eaves et al. 2000), we computed a multiallelic extension of the D‘ statistic (Fig. 2). Although this statistic is valid for the published comparison of samples with the same size that was typed using the same markers, we realized that the statistic is not valid for comparison with our data because it is strongly affected by sample size and allele frequencies. In general, smaller sample sizes show larger multiallelic D‘ values. At intermarker distances well beyond those where we would reasonably observe LD, we would expect the multiallelic D‘ values of different sized samples to approach the same asymptote. However, multiallelic D‘ values in our sample of 386 individuals approached an asymptote almost twice as large as that observed for 1223 individuals. We also investigated a recently reported statistic (Zhao et al. 1999) that has the advantage of providing a quantity that appears near mean zero under the null hypothesis of linkage equilibrium; however, the variance of this statistic still depends on sample size. We are not aware of a good summary measure that enables accurate comparison of LD between studies that use different sample sizes and markers with differing numbers of alleles and differing allele frequencies.
We have previously reported evidence for linkage to type 2 diabetes on chromosome 20 in our Finnish sample (Ghosh et al. 1999). For LD analysis, one might expect heterogeneity when combining case and control samples. This would likely be true for a simple Mendelian disease in the region of the disease locus. However, for a complex disorder such as type 2 diabetes where a particular susceptibility variant alone likely has only limited independent impact on disease risk, we would expect little effect of susceptibility variants on LD along the chromosome. Thus, the similar extent of LD among individuals affected with diabetes compared to spouse controls (Table 2) and the homogeneity of haplotype frequency estimates between these groups is not surprising.
LD may depend on population substructure, which may be advantageous in the search for genes conferring susceptibility to a complex trait such as diabetes. Rare disease alleles in families from the north and east of Finland travel on haplotypes as large as 15 cM (de la Chapelle and Wright 1998), suggesting that it may be possible to identify diabetes-associated haplotypes in similar geographic subsets. Yet a recent study found no difference between LD in 50 males from southwestern and eastern regions of Finland, founded ∼2000 and ∼400 years ago, respectively (Jorde et al. 2000). Two recent reports detected only modestly increased LD for the isolated Finnish and Sardinian populations compared to other European or American samples for microsatellites (Eaves et al. 2000) and single nucleotide polymorphisms (SNPs) (Taillon-Miller et al. 2000), respectively. Additional comparative data will help clarify whether LD is increased in various founder populations, enabling LD to be detected using smaller sample sizes or a lower density of markers.
Although methods to genotype SNPs are becoming easier and more accessible (Chen et al. 1998; Ryan et al. 1999; Griffin and Smith 2000), a high-density map of microsatellite markers remains extremely useful for studies of complex traits. The presence of multiple alleles allows haplotypes to be inferred more easily, generating more power to detect LD, although microsatellite marker mutation breaks down conserved haplotypes, and can also reduce the extent of LD. A significant challenge for assessing LD with microsatellite markers is the difficulty in generalizing LD statistics to multiple alleles, as described above.
We evaluated marker-to-marker LD as part of our investigation of disease variant-to-marker LD. Ideally, a threshold level of LD obtained between neighboring markers would indicate that the interval had been effectively scanned for an associated disease variant, although whether such a threshold can be found remains unclear. It seems highly likely that an interval lacking significant marker-to-marker LD has not yet been scanned thoroughly. Thus, to proceed with fine-mapping of a type 2 diabetes susceptibility gene on chromosome 20, we will add markers to regions where less marker-to-marker LD is observed while searching for alleles or haplotypes associated with disease. Additional empirical data are needed to determine the marker density required for association studies of complex diseases.
METHODS
Sample
The FUSION study design and first phase of sample recruitment have been described previously (Valle et al. 1998). Briefly, we sampled 580 families ascertained through a type 2 diabetes-affected sibling pair, and collected additional affected and unaffected relatives. A second cohort includes additional family members as well as a separate set of 275 families. All individuals studied are believed to be of Finnish heritage based on their birthplaces and their grandparents’ birthplaces within Finland. In this report we used genotype data from 837 unrelated affected individuals and 386 unrelated spouses. Two hundred six of these spouses tested unaffected by oral glucose tolerance test (OGTT) in our study, 160 reported themselves to be unaffected, and 20 had an unknown phenotype.
Markers
We selected 43 microsatellite markers, including 39 dinucleotide and 4 tetranucleotide repeats, from chromosome 20 maps and genotyped them as previously described (Ghosh et al. 1997). We estimated heterozygosities and the number of alleles from an average of 1142 unrelated individuals per marker. These heterozygosities are very similar to those we reported previously (Ghosh et al. 1999), but differ slightly because we analyzed an expanded sample. Data for all markers are consistent with Hardy-Weinberg equilibrium.
Maps
We estimated sex-averaged genetic maps from combined data on 983 individuals in 205 FUSION-extended families and from cleaned genotypes of CEPH pedigrees (Broman et al. 1998). The genetic map used in these analyses differs slightly from a previously reported map (Ghosh et al. 1999) because additional FUSION data have now been incorporated. This marker order is consistent with the available mapping data produced by the Chromosome 20 Mapping Group at the Sanger Centre, and were obtained from the World Wide Web at http://www.sanger.ac.uk/HGP/Chr20/.
We estimated a physical map using chromosome 20 clone sequences available from the Sanger Centre and GenBank as of October 30, 2000. Most sequence data were produced by the human Chromosome 20 Sequencing Group at the Sanger Centre, and can be obtained from ftp://ftp.sanger.ac.uk/pub/human/chr20/. Other data were obtained from GenBank ftp://ncbi.nlm.nih.gov/genbank/genomes/H_sapiens/CHR_20/. We determined the nucleotide positions of our markers on individual clones using electronic PCR and BLAST (http://www.ncbi.nlm.nih.gov/), and used an unpublished clone order (P. Deloukas, pers. comm.) to generate a contiguous map. We assumed the size of six contig gaps to be 100 kb each, and the size of 12 unfinished BACs to be their current sequence length. We also assumed that 100 nucleotides of each clone overlaps the adjacent clone, and that all finished clones are numbered according to their orientation on the chromosome. The resulting physical map, which starts at marker D20S103 and ends at marker D20S173, is 59 Mb long.
Tests of Linkage Equilibrium between Pairs of Markers
We formed 903 = (43*42)/2 marker pairs by considering all possible pairs of the 43 microsatellite markers. For each marker pair, we performed joint analyses for the 837 diabetic individuals and their 386 spouse controls. Maximum likelihood estimates for allele frequencies were obtained for each of the 43 markers by allele counting. Maximum likelihood estimates of haplotype frequencies for the 903 marker pairs were obtained by employing the expectation-maximization (E-M) algorithm. We used a likelihood ratio statistic to test for LD. This statistic compared the maximum probability of the observed sample of genotypes allowing for LD (based on estimates of haplotype frequencies) to the maximum probability of the observed sample assuming linkage equilibrium (based on estimates of haplotype frequencies calculated as products of the relevant allele frequency estimates). The distribution of −2 times the natural logarithm of this likelihood ratio is asymptotically distributed as χ2, with degrees of freedom equal to (n1−1)(n2−1), where n1 was the number of alleles used for the first marker, and n2 was the number of alleles used for the second marker. To reduce the effects of rare alleles and to preserve the power to reject the null hypothesis of linkage equilibrium by limiting the degrees of freedom of the test statistic, we pooled alleles with frequencies less than 0.05.
Prior to pooling the data from the affected individuals and their spouses, we carried out a likelihood ratio test of homogeneity of haplotype frequencies for each pair of markers. For this test we estimated the haplotype frequencies separately for the affected individuals and their spouses using the E-M algorithm. The likelihood ratio statistic compared the product of the maximum likelihoods for the two samples to the maximum likelihood for the two samples analyzed jointly. Again, alleles with frequencies <0.05 in the joint sample were pooled. Because of small estimated haplotype frequencies, we assessed significance levels using a permutation test. For each of the 903 marker pairs, we generated 100 replicate samples by permutation. To construct these permuted samples, we randomly permuted affection status of the 1223 individuals, keeping the marker data the same. For each permuted sample, we calculated the likelihood ratio test for homogeneity of haplotype frequencies. Based on the 100 permuted samples for a marker pair, we estimated the P-value in the test for homogeneity of haplotype frequencies as the proportion of permuted-data statistics greater than the observed-data statistic.
To measure the strength of disequilibrium, we used Lewontin's D‘ (Lewontin 1964) modified for multiple alleles (Hedrick 1987). For two-allele markers, D‘ is the standardized disequilibrium value that takes the usual disequilibrium coefficient P(AiBj) − P(Ai)P(Bj) and divides it by its maximal possible value. Given multiple alleles, we calculate the weighted average of the D‘ values where the weights are the products of the corresponding allele frequencies. That is,
D‘ = Σi Σj pi qj |Dij‘|,
where pi and qj are allele frequencies at the two loci of interest, and Dij‘ is the standardized disequilibrium coefficient based on alleles Ai and Bj.
ACKNOWLEGMENTS
We thank the Finnish citizens who volunteered to participate in the FUSION study. This project was made possible by intramural funds from the National Human Genome Research Institute (project no. OH95-C-N030). The work in Finland was partially supported by the Finnish Academy (38387 and 46558). K.L.M. is a recipient of a Burroughs Wellcome Fund Career Award in the Biomedical Sciences. E.M.L. was supported previously by NIH training grant HG00040. R.M.W. was supported previously by an individual NRSA from the NIH (DK09525), and is now supported by a Career Development Award from the ADA. M.B. is supported by NIH grant HG00376.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL boehnke@umich.edu; FAX (734) 763-2215.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.173201.
REFERENCES
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL. Comprehensive human genetic maps: Individual and sex-specific variation in recombination. Am J Hum Genet. 1998;63:861–869. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Livak KJ, Kwok PY. A homogeneous, ligase-mediated DNA diagnostic test. Genome Res. 1998;8:549–556. doi: 10.1101/gr.8.5.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Chapelle A, Wright FA. Linkage disequilibrium mapping in isolated populations: The example of Finland revisited. Proc Natl Acad Sci. 1998;95:12416–12423. doi: 10.1073/pnas.95.21.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaves IA, Merriman TR, Barber RA, Nutland S, Tuomilehto-Wolf E, Tuomilehto J, Cucca F, Todd JA. The genetically isolated populations of Finland and Sardinia may not be a panacea for linkage disequilibrium mapping of common disease genes. Nat Genet. 2000;25:320–323. doi: 10.1038/77091. [DOI] [PubMed] [Google Scholar]
- Ehm MG, Karnoub MC, Sakul H, Gottschalk K, Holt DC, Weber JL, Vaske D, Briley D, Briley L, Kopf J, et al. Genomewide search for type 2 diabetes susceptibility genes in four American populations. Am J Hum Genet. 2000;66:1871–1881. doi: 10.1086/302950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Karanjawala ZE, Hauser ER, Ally D, Knapp JI, Rayman JB, Musick A, Tannenbaum J, Te C, Shapiro S, et al. Methods for precise sizing, automated binning of alleles, and reduction of error rates in large-scale genotyping using fluorescently labeled dinucleotide markers. Genome Res. 1997;7:165–178. doi: 10.1101/gr.7.2.165. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Watanabe RM, Hauser ER, Valle T, Magnuson VL, Erdos MR, Langefeld CD, Balow J, Jr, Ally DS, Kohtamaki K, et al. Type 2 diabetes: Evidence for linkage on chromosome 20 in 716 Finnish affected sib pairs. Proc Natl Acad Sci. 1999;96:2198–2203. doi: 10.1073/pnas.96.5.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Watanabe RM, Valle TT, Hauser ER, Magnuson VL, Langefeld CD, Ally DS, Mohlke KL, Silander K, Kohtamaki K, et al. The Finland–United States investigation of non-insulin-dependent diabetes mellitus genetics (FUSION) study. I. An autosomal genome scan for genes that predispose to type 2 diabetes. Am J Hum Genet. 2000;67:1174–1185. [PMC free article] [PubMed] [Google Scholar]
- Griffin TJ, Smith LM. Single-nucleotide polymorphism analysis by MALDI-TOF mass spectrometry. Trends Biotechnol. 2000;18:77–84. doi: 10.1016/s0167-7799(99)01401-8. [DOI] [PubMed] [Google Scholar]
- Hastbacka J, de la Chapelle A, Mahtani M, Clines G, Reeve-Daly M, Daly M, Hamilton B, Kusumi K, Trivedi B, Weaver A, et al. The diastrophic dysplasia gene encodes a novel sulfate transporter: positional cloning by fine-structure linkage disequilibrium mapping. Cell. 1994;78:1073–1087. doi: 10.1016/0092-8674(94)90281-x. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. Gametic disequilibrium measures: Proceed with caution. Genetics. 1987;117:331–341. doi: 10.1093/genetics/117.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikawa Y, Oda N, Cox NJ, Li X, Orho-Melander M, Hara M, Hinokio Y, Lindner TH, Mashima H, Schwarz PE, et al. Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet. 2000;26:163–175. doi: 10.1038/79876. [DOI] [PubMed] [Google Scholar]
- Huttley GA, Smith MW, Carrington M, O’Brien SJ. A scan for linkage disequilibrium across the human genome. Genetics. 1999;152:1711–1722. doi: 10.1093/genetics/152.4.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorde LB, Watkins WS, Kere J, Nyman D, Eriksson AW. Gene mapping in isolated populations: New roles for old friends? Hum Hered. 2000;50:57–65. doi: 10.1159/000022891. [DOI] [PubMed] [Google Scholar]
- Kaprio J, Tuomilehto J, Koskenvuo M, Romanov K, Reunanen A, Eriksson J, Stengard J, Kesaniemi YA. Concordance for type 1 (insulin-dependent) and type 2 (non-insulin-dependent) diabetes mellitus in a population-based cohort of twins in Finland. Diabetologia. 1992;35:1060–1067. doi: 10.1007/BF02221682. [DOI] [PubMed] [Google Scholar]
- Kendler KS, MacLean CJ, Ma Y, O'Neill FA, Walsh D, Straub RE. Marker-to-marker linkage disequilibrium on chromosomes 5q, 6p, and 8p in Irish high-density schizophrenia pedigrees. Am J Med Genet. 1999;88:29–33. doi: 10.1002/(sici)1096-8628(19990205)88:1<29::aid-ajmg5>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Laan M, Paabo S. Demographic history and linkage disequilibrium in human populations. Nat Genet. 1997;17:435–438. doi: 10.1038/ng1297-435. [DOI] [PubMed] [Google Scholar]
- Lewontin RC. The interaction of selection and linkage. I. General considerations; Heterotic models. Genetics. 1964;49:49–67. doi: 10.1093/genetics/49.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman B, Selby JV, King MC, Slemenda C, Fabsitz R, Friedman GD. Concordance for type 2 (non-insulin-dependent) diabetes mellitus in male twins. Diabetologia. 1987;30:763–768. doi: 10.1007/BF00275741. [DOI] [PubMed] [Google Scholar]
- Peltonen L, Jalanko A, Varilo T. Molecular genetics of the Finnish disease heritage. Hum Mol Genet. 1999;8:1913–1923. doi: 10.1093/hmg/8.10.1913. [DOI] [PubMed] [Google Scholar]
- Peterson AC, Di Rienzo A, Lehesjoki AE, de la Chapelle A, Slatkin M, Freimer NB. The distribution of linkage disequilibrium over anonymous genome regions. Hum Mol Genet. 1995;4:887–894. doi: 10.1093/hmg/4.5.887. [DOI] [PubMed] [Google Scholar]
- Rich SS. Mapping genes in diabetes. Genetic epidemiological perspective. Diabetes. 1990;39:1315–1319. doi: 10.2337/diab.39.11.1315. [DOI] [PubMed] [Google Scholar]
- Rommens JM, Iannuzzi MC, Kerem B, Drumm ML, Melmer G, Dean M, Rozmahel R, Cole JL, Kennedy D, Hidaka N, et al. Identification of the cystic fibrosis gene: Chromosome walking and jumping. Science. 1989;245:1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- Ryan D, Nuccie B, Arvan D. Non-PCR-dependent detection of the factor V Leiden mutation from genomic DNA using a homogeneous invader microtiter plate assay. Mol Diagn. 1999;4:135–144. doi: 10.1016/s1084-8592(99)80037-x. [DOI] [PubMed] [Google Scholar]
- Taillon-Miller P, Bauer-Sardina I, Saccone NL, Putzel J, Laitinen T, Cao A, Kere J, Pilia G, Rice JP, Kwok PY. Juxtaposed regions of extensive and minimal linkage disequilibrium in human Xq25 and Xq28. Nat Genet. 2000;25:324–328. doi: 10.1038/77100. [DOI] [PubMed] [Google Scholar]
- Valle T, Tuomilehto J, Bergman RN, Ghosh S, Hauser ER, Eriksson J, Nylund SJ, Kohtamaki K, Toivanen L, Vidgren G, et al. Mapping genes for NIDDM. Design of the Finland–United States Investigation of NIDDM Genetics (FUSION) Study. Diabetes Care. 1998;21:949–958. doi: 10.2337/diacare.21.6.949. [DOI] [PubMed] [Google Scholar]