Abstract
We use quantum mechanics/molecular mechanics (QM/MM) simulations to study the cleavage of the ribonucleic acid (RNA) backbone catalyzed by ribonuclease H. This protein is a prototypical member of a large family of enzymes that use two-metal catalysis to process nucleic acids. By combining Hamiltonian replica exchange with a finite-temperature string method, we calculate the free energy surface underlying the RNA cleavage reaction and characterize its mechanism. We find that the reaction proceeds in two steps. In a first step, catalyzed primarily by magnesium ion A and its ligands, a water molecule attacks the scissile phosphate. Consistent with thiol-substitution experiments, a water proton is transferred to the downstream phosphate group. The transient phosphorane formed as a result of this nucleophilic attack decays by breaking the bond between the phosphate and the ribose oxygen. In the resulting intermediate, the dissociated but unprotonated leaving group forms an alkoxide coordinated to magnesium ion B. In a second step, the reaction is completed by protonation of the leaving group, with a neutral Asp132 as a likely proton donor. The overall reaction barrier of ~15 kcal mol−1, encountered in the first step, together with the cost of protonating Asp132, is consistent with the slow measured rate of ~1–100/min. The two-step mechanism is also consistent with the bell-shaped pH dependence of the reaction rate. The non-monotonic relative motion of the magnesium ions along the reaction pathway agrees with X-ray crystal structures. Proton transfer reactions and changes in the metal ion coordination emerge as central factors in the RNA cleavage reaction.
Keywords: QM/MM, free energy, enzyme, catalysis, umbrella sampling, RNase H, proton transfer, reaction coordinate, string method, Hamiltonian replica exchange, two-metal ion catalysis, phosphate hydrolysis, RNA cleavage
Introduction
Phosphate ester hydrolysis is an essential reaction in biology, playing a central role in nucleic acid processing, energy transduction, and signaling. Despite the importance of this ubiquitous reaction, the mechanistic details of phosphate ester cleavage remain relatively poorly understood. In particular, the mechanisms underlying the catalytic action of the proteins and the role of ion coordination in metal-catalyzed phosphate cleavage are not fully characterized. In addition, there is an active debate concerning possible intermediates. Based on experimental findings, a one-step mechanism has been proposed,1 whereas theoretical calculations indicated the possibility of a two-step reaction with a transient phosphorane intermediate.2,3 Other major unsolved problems concern the proton transfer (PT) processes that take place during the phosphate hydrolysis reaction.4,5 In many systems, the search is ongoing for the residues that participate in the PT steps involved in activating the water nucleophile or hydroxide ion, and in protonating the leaving group. Only limited experimental information is available concerning the PT steps in the enzymatic reaction. Much of this information is indirect, with mutations of the protein sequence and chemical modifications of the nucleic acid shedding some light on the possible PT events occurring as part of the overall reaction. To uncover the mechanistic details of the phosphate cleavage reaction, the structures of the transition states and the corresponding protonation states of the participating species have to be identified.
We focus on the catalytic reaction of the RNase H enzyme, a member of the superfamily of polynucleotide transferases. RNase H catalyzes the nonspecific hydrolysis of the RNA strand of an RNA.DNA hybrid via two-metal ion catalysis. This catalytic mechanism is ubiquitous amongst nucleic acid processing enzymes6,7 and ribozymes.8 The RNase H active site is homologous to those of retroviral integrases, transposases, argonautes, Holliday-junction resolvases, and 3′ exonucleases. RNase H thus provides an excellent model system to study the enzymatic hydrolysis aided by two divalent metal ions for which experimental9–15 and theoretical2,3,16–18 investigations have provided high-resolution structures, detailed mutational data, and modeled possible reaction mechanisms.
The intense studies of RNase H are also motivated by the close similarity of its active site to that of human immunodeficiency virus (HIV) reverse transcriptase (HIV-RT).11 HIV-RT is a primary target for antiretroviral drugs because both its polymerase and RNase H domains are essential for viral replication. A better understanding of the two-metal catalyzed hydrolysis reaction in RNase H thus promises guidance in the design of novel anti-HIV drugs that target HIV-RT beyond its polymerase function.14
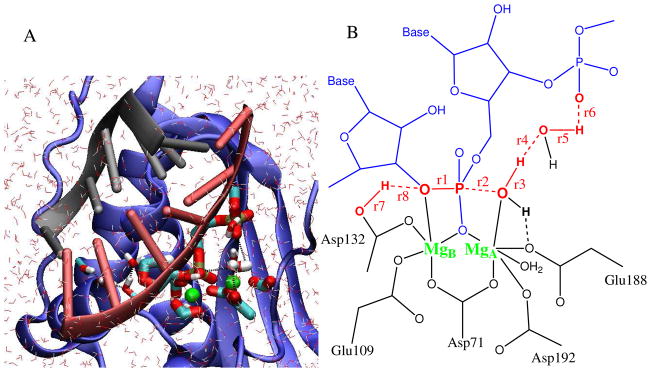
Both structural and functional similarities between HIV-RT, human RNase H and Bc. halodurans (Bh) RNase H have been demonstrated in a recent study.11 Here, we focus on Bh RNase H because of the availability of high-resolution crystallographic structures for this bacterial enzyme.10 In these structures, the RNA.DNA duplex substrate is positioned for the cleavage of the phosphate ester bond, and the active site includes the two catalytic Mg2+ ions (Figure 1A). Crystal structures of mimics of reaction intermediates and of the product complex12 provide additional reference frames of the reaction pathway.
Figure 1.
QM/MM simulation system. (A) Complex between RNase H and an RNA.DNA duplex in solution. The RNA chain is shown in red, the DNA in grey, and the protein in blue. The cleaved phosphate, the nucleophilic water, and side chains of key residues in the active site are shown in atomistic rendering. Mg2+ ions are shown in green. (B) Schematic of the RNase H active site. Distances r1, r2, …, r8 entering the reaction coordinates are shown in red (see section on free energy simulations in Methods).
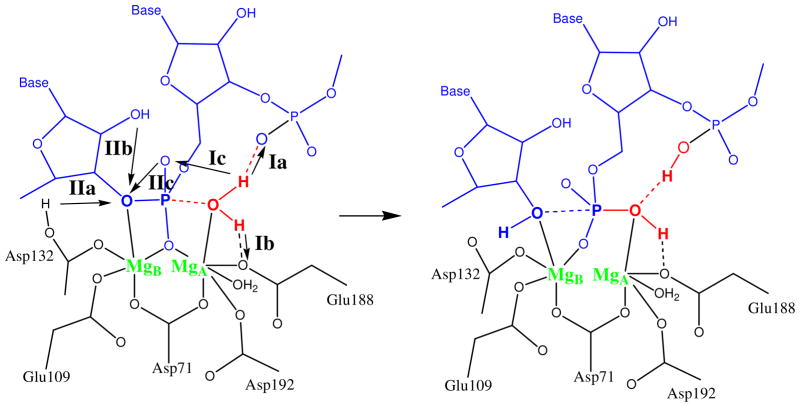
The catalytic reaction of RNase H and possible PT pathways have been studied in quantum mechanical/molecular mechanical (QM/MM) simulations using density functional theory (DFT) calculations with the BLYP functional combined with Car-Parrinello molecular dynamics,2 and using DFT/B3LYP with constrained energy minimizations and approximate free energy estimation.3 De Vivo et al.2 found that in Bh RNase H the reaction followed a two-step AN+DN mechanism. The proton abstracted from the attacking water was transferred to the pro-RP oxygen of the scissile phosphate (pathway Ic in Figure 2) via three water molecules to form a phosphorane intermediate. This proton was subsequently shuttled to the leaving group, again via water molecules (pathway IIb in Figure 2). Elsässer and Fels3 also reported a two-step reaction with a phosphorane intermediate, again with the pro-RP oxygen of the scissile phosphate accepting the proton of the attacking water molecule.
Figure 2.
Schematic representation of the active site coordination. Arrows in the reactant state representation indicate possible PT pathways in processes I and II.
Here we study the mechanism of the RNase H enzymatic reaction on the basis of QM/MM simulations19 with the B3LYP20 density functional. We determine the multidimensional free energy surface underlying the catalytic reaction by combining Hamiltonian replica exchange (RE),21 umbrella sampling (US), and finite-temperature string method calculations.22 Our simulations show a reaction pathway that differs significantly from those of the earlier computational studies, but is consistent with the structural changes seen in experiment, the measured rate of the reaction and its unusual pH dependence, and the effects both of thiol substitutions on the RNA and of protein mutations.
Methods
Structural Model
The QM/MM calculations were initiated from the X-ray crystal structure of RNase H complexed with a hybrid RNA.DNA duplex substrate solved 1.85 Å resolution, with Protein Data Bank (PDB) code 1ZBI.10 This complex structure was solvated by adding water and ions, and prepared for simulations as described earlier.17 For the initial model we chose D132N (PDB code 1ZBI) instead of D192N (PDB code 1ZBL) because of its substantially better crystallographic resolution (1.85 Å versus 2.20 Å), and a mutation distant from metal ion A where the reaction is initiated.
QM/MM Simulations
The equilibrated classical system was trimmed to a sphere of 32 Å radius centered at the Mg2+ ions. The simulation sphere was neutralized by removing excess chloride ions. In contrast to the hammerhead ribozyme,23 the small changes in the crystal structures of RNase H unbound, reactant, intermediate, and product states10,12 suggest that its active site is relatively rigid, without large protein backbone movements during catalysis. We used the Q-Chem program24 to perform the QM calculations at the B3LYP20 6-31+G(d) DFT level of theory. Test calculations with a larger basis set are presented in Figure S1. The QM system was coupled with the CHARMM program using full electrostatic embedding.25 Standard link atom treatment was used to connect the QM and MM regions, adding hydrogen atoms for the missing ligands. The system was further equilibrated with QM/MM simulations, and full QM/MM minimizations were carried out to determine starting structures for subsequent free energy simulations. The QM/MM dynamics was run with a Langevin thermostat at a temperature of 300 K with 1 fs time step. The total times used for analysis (combining US and string simulations) are 128 and 283 ps, respectively, with Asp132 protonated and unprotonated.
The QM region included the RNA backbone from the ribose immediately upstream of the cleaved phosphodiester bond to the first and second phosphate group downstream (i.e., in the 3′ direction). In addition, the two Mg2+ ions were treated quantum mechanically, together with the carboxy-groups of their ligands Asp71, Glu109, Asp132, Glu188, and Asp192, by representing the Asp residues as formic acid and the Glu residues as acetic acid. Finally, nine and eleven water molecules were included in the QM region of the simulations with unprotonated and protonated Asp132, respectively. With 91 and 98 atoms in the QM regions of the systems with unprotonated and protonated Asp132, respectively, including hydrogen atoms, the QM regions are substantially larger than in our earlier study.17 Unless noted otherwise, all results refer to simulations with Asp132 protonated.
Free Energy Simulations
To determine the free energy surface underlying the reaction, we performed US simulations using harmonic bias potentials with spring constants of 100 kcal mol−1 Å2) placed along the reaction coordinates. The reaction coordinates were chosen as linear combinations of the length differences between bonds formed and broken in the electron transfer (ET) and PT steps. In our calculations, we considered the following coordinates: ET=r1–r2, PT1a=r3–r4, PT1b=r5–r6, PT2=r7–r8, where r1 is the distance between the phosphorus and the ribose O3′ oxygen of the cleaved P-O bond, r2 is the distance between the phosphorus and the oxygen of the nucleophilic water oxygen, r3 is the distance between this oxygen and the hydrogen atom of the nucleophilic water donated to the intermediate water, r4 is the distance between this hydrogen and the oxygen of the intermediate water, r5 is the distance between the oxygen of the intermediate water and its hydrogen donated to the downstream phosphate, r6 is the distance between this hydrogen and the pro-Rp oxygen of the downstream phosphate group, r7 is the distance between the unliganded carboxy-oxygen of Asp132 and its proton, and r8 is the distance between this Asp132 proton and the O3′ leaving oxygen (Figure 1B). To accelerate equilibration and sampling, we ran the US simulations for the different windows in parallel and coupled them with Hamiltonian RE,21 attempting exchanges of the structures in neighboring windows at intervals of 20 fs.
The data from the US and string simulations described below were jointly unbiased using a histogram-free version of the “multidimensional weighted histogram analysis method” (WHAM).26 We determined the free energy Fi of simulation i by iterative solution of
| (1) |
The Fi values were calculated iteratively, with a convergence threshold of 0.0001 kcal mol−1. Eq 1 was used to unbias data jointly from US and string simulations, which were both harmonically restrained. xk,m, represents configuration k in simulation m (1≤ m ≤ N ), nm is the number of configurations in simulation m, Um (x) is the harmonic constraint energy corresponding to the bias in simulation m, kB is Boltzmann’s constant, and Fi=1=0 by definition.
With converged Fi, estimates of the unnormalized probability density p(q) along any reaction coordinate q(x) can be obtained from
| (2) |
where δ(q) is Dirac’s delta function. In practice, we binned the data points along the reaction coordinate q, defining δ(q(x)− qi) =1 for x in the bin centered at qi and zero otherwise. The free energy profile (or potential of mean force) along q is then G(q) = − kBT ln p(q), up to an arbitrary constant.
String method calculations
To determine a minimum-free energy path, we adapted the finite-temperature string method.22 The initial string connecting reactant and product states is defined as a curve in the M-dimensional space of collective coordinates, , as in ref. The string is parameterized by its reduced length ξ, 0 ≤ ξ ≤ 1, with distances defined by the arc-length using a Euclidian metric in the space of the collective coordinates. The -s are continuous functions that describe the change of the i-th collective coordinate, with corresponding to the reactant state and to the product state. In our string simulations, N equidistant images (i.e., system configurations with collective coordinates corresponding to a particular string position) were distributed along the string, ξ1 =0 <ξ2 <…< ξN =1. We then ran 100-fs QM/MM simulations for each image i, with harmonic restraining potentials centered at S0(ξi) acting on the collective coordinates. By fitting high order polynomials (up to order 10) to the average collective coordinates of each image, an updated string was constructed that was again divided into N segments of equal arc-length. This procedure of running restrained simulations and updating the string was iterated until the changes of all collective coordinates fell below a given threshold. Note that in this procedure the string endpoints were allowed to move and could thus relax toward stationary points in the space of collective coordinates.
The string calculations were implemented using 3 to 5 coordinates, depending on the protonation processes involved in the reaction. For the first step of the reaction with unprotonated Asp132, the string was constructed in the space of ET, PT1a and PT1b as described above, chosen to capture the water attack on the phosphate group, water deprotonation, and backbone bond breaking. In the simulations with a protonated Asp132, we expanded the space of the string calculations. To complete the bond-breaking step by protonation of the leaving group, we added the PT2 coordinate describing the PT from Asp132 to the O3′ of the leaving group. The dimensionality of the string was further increased to 5 by treating the bond breaking (r7) and bond forming (r8) proton-oxygen distances independently, to correctly sample this PT process.
Results and Discussion
Deprotonation of the attacking water
RNase H cleaves the RNA backbone through a nucleophilic attack on the phosphate group. Here we considered the case of water acting as the nucleophile. To explore the order of the water deprotonation and backbone bond breaking processes, and to identify the energetically feasible pathways, we performed extensive energy minimizations of the QM/MM system. In the minimizations, we drove the nucleophilic attack by restraining the difference in lengths of the formed and broken bonds (between the water oxygen and phosphorus atoms, and between the phosphorus and the O3′ oxygen of the ribose). With this reaction coordinate, we did not a priori specify the group accepting a proton from the attacking water, allowing us to obtain an unbiased representation of this key reaction step.
We consistently found that the initial deprotonation of water proceeded via pathway Ia (Figure 2) with proton transfer to the downstream phosphate group. We performed extensive minimizations, in which the reaction is repeatedly driven back and forth by varying the restraint on the difference in bond lengths. While approaching the phosphorus, the attacking water molecule transferred one of its protons to the downstream phosphate before the P-O3′ bond was broken. This PT took place both when the attacking water molecule formed a direct hydrogen bond to the accepting oxygen of the downstream phosphate, and when their interaction was mediated by an intervening hydrogen-bonded water molecule.
We performed additional energy minimizations to test alternative mechanisms of the nucleophilic attack. To probe pathway Ib (Figure 2), in which the proton is transferred to Glu188, we performed calculations for a reduced QM region without the downstream phosphate group. This phosphate group could thus no longer accept the proton from the water in the QM region. As a result, we found that the preferred proton pathway was towards the Glu188 group. To test whether this pathway is competitive with pathway Ia, we reinitiated calculations from a minimized reactant state structure in which the nucleophilic water was hydrogen bonded to Glu188, but with the downstream phosphate returned to the QM region. However, minimization performed with this initial condition again led to the product of pathway Ia, with the proton being transferred from the water to the downstream phosphate. We obtained the same pathway also when the carboxylic acids in the QM region were modeled with longer carbohydrate chains (using acetic acid and propionic acid for the Asp and Glu residues, respectively). Jointly, these results indicate that pathway Ia is energetically the most favorable. Consequently, we performed further simulations to map the free energy profile of this reaction process.
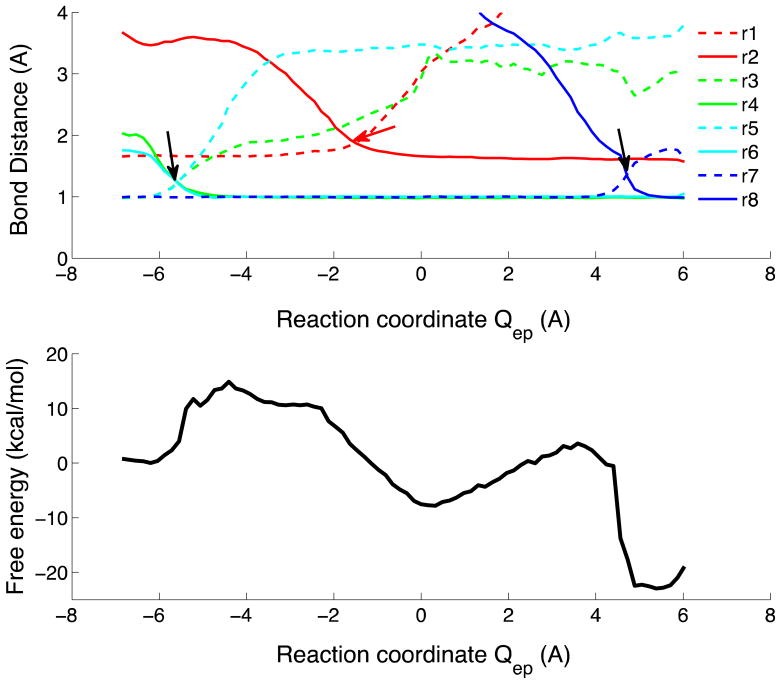
Figures 3 and 4 show the 1D and 2D free energy profiles for pathway Ia, calculated for the system with Asp132 protonated. A histogram-free implementation of the multidimensional WHAM method was used to combine the data from the RE/US and string simulations. The transition state barriers for this reaction step are about ~15 kcal mol−1. A barrier of about the same height (17 kcal mol−1) was obtained with Asp132 unprotonated (Figure S2). Movie 1 in Supporting Information (SI) provides an animation of the reaction.
Figure 3.
Reaction coordinate changes and 1D free energy profile for the full catalytic reaction along the reaction coordinate Qep=ET+(PT1a+PT1b)/2+PT2 of the protonated Asp132 system. (Top) The solid lines show the bond forming distances, the dashed lines show the bond breaking distances, as described in the text. (Bottom) Free energy profile projected onto the Qep reaction coordinate.
Figure 4.
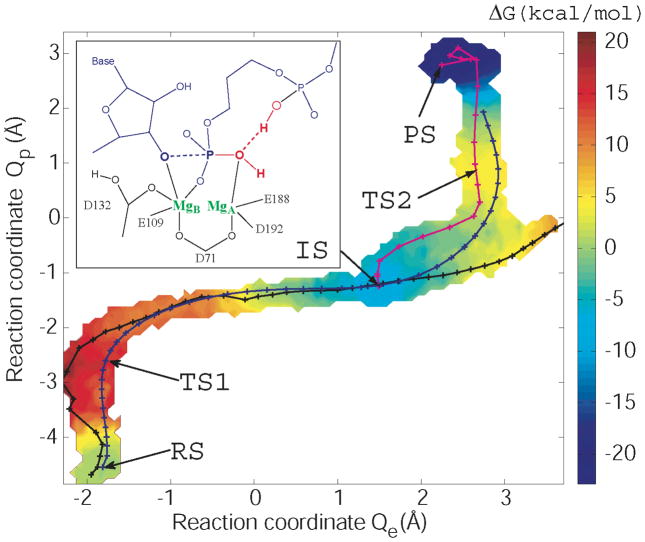
2D free energy surface and reaction strings for the complete catalytic reaction with a protonated Asp132, obtained by projection onto coordinates Qe=ET probing bond formation and breaking, and Qp=(PT1a+PT1b)/2+PT2 probing PT steps. The black curve shows the initial string; the blue and purple curves show the converged strings for the entire reaction and for the final step, respectively. The inset shows a schematic of the active site coordination.
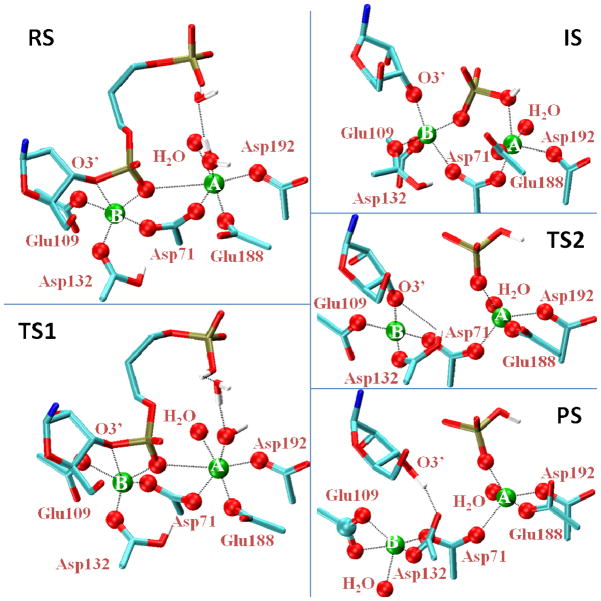
At the transition state TS1 of process I, a hydroxide ion is formed by PT from the nucleophilic water to the downstream phosphate (Figure 5). The shoulder in the free energy profiles immediately to the right of TS1 in Figures 3 and 4 corresponds to a state in which the hydroxide ion is fully formed but not yet bonded to the phosphorus atom. The subsequent formation of this bond, and the simultaneous breakage of the P-O3′ bond (marked ET in Figure 3 top) entail no significant additional barrier. In our simulations with Asp132 protonated, we thus did not observe a phosphorane intermediate. Instead, an intermediate state (IS) was formed with the P-O3′ broken, but the ribose O3′ not yet protonated (Figure 5).
Figure 5.
Representative structures along the catalytic reaction of RNase H using protonated Asp132. The reactant state and first transition state are shown on the left (top to bottom), with the intermediate state, second transition state, and product state on the right. The downstream phosphate is only shown for the initial reaction steps (left). Dashed lines indicate select interactions. Oxygen atoms (red) within 2.6 Å of the Mg2+ ions A or B (green) are shown as spheres.
The formation and stability of this intermediate were found to depend on the protonation state of Asp132. Consistent with our earlier simulations using only a small QM system,17 we found that with Asp132 unprotonated, the OH− attack did not result in breakage of the P-O3′ bond. Instead, the reaction stalled by forming a metastable phosphorane species immediately after TS1 (Figure S2). Without the proton on Asp132, this intermediate was highly unstable, consistent with the calculated free energy surface and also with recent simulations probing this pathway.3 In unrestrained simulations with Asp132 unprotonated, the intermediate decayed on a picosecond timescale and the reactants were fully recovered in their original form with the water molecule as the nucleophile, not an OH− ion. With the phosphorane intermediate being highly unstable, the protonation of the leaving group thus emerges as a key second process to complete the overall reaction.
Pathway Ia for the nucleophile deprotonation is supported by chemical modifications of the RNA backbone. In the homologous RNase HI of E. coli, the substitution of sulfur for the pro-Rp oxygen of the downstream phosphate group, 3′ to the scissile phosphodiester bond, lowered the catalytic rate almost ten-fold.28 This large effect of a modification outside the immediate active site is consistent with pathway Ia, in which the downstream pro-Rp oxygen accepts a proton. In contrast, the alternative pathway Ic observed in recent simulations2,3 requires an indirect explanation of this effect,2 with the pro-Rp oxygen of the downstream phosphate stabilizing the nucleophilic water. Moreover, in pathway Ic the pro-Rp oxygen of the scissile phosphate becomes protonated, in apparent contradiction to experiments that show no rate decrease with sulfur substituted for this oxygen.29 Another alternative, pathway Ib, is also questionable in Bh RNase H, where the mutation E188Q had relatively little effect on the kinetics.10,12 In light of both experiment and our simulations, pathway Ia is thus a likely route for the deprotonation of the nucleophilic water.
Protonation of the leaving group
Completion of the reaction requires the protonation of the leaving group. Stoichiometrically, this step consumes one of the two protons of the attacking water. However, in our intermediate (Figure 5), one of the protons is bound to the downstream phosphate, and the other to the scissile phosphate but pointing away from the O3′ acceptor. In previous simulations using a small QM system,17 we did not observe PT from the protonated pro-Rp oxygen atom of the scissile phosphate to the dissociated ribose O3′. In an earlier study of polymerase III, a similar challenge arose and the leaving group could not be protonated with a surmountable energy barrier.5 While it is possible that PT from the downstream or scissile phosphate groups to the leaving group occurs on a long time scale, it seems likely that a specific donor protonates the ribose.
We found that PT from Asp132 to the O3′ provides a kinetically viable pathway for protonation of the leaving group. To examine the role of Asp132 as a possible proton donor, we extended the free energy and string simulations from the intermediate state to the product state. To resolve this process, we increased the number of coordinates from four (PT1a and PT1b to describe the first PT, ET to describe the phosphate bond formation/breaking, and PT2 for the last PT, see also Figure 3) to five (PT1a, PT1b, ET, PT2 bond forming distance r8, PT2 bond breaking distance r7) by explicitly taking into account both the bond forming and the bond breaking distances of the PT from the Asp132 to the leaving group.
The resulting 1D and 2D free energy surfaces (Figures 3 and 4) show barriers of ~12 kcal mol−1 for the transition from IS to the final product state. Our simulations show that Asp132 passes its proton to the leaving group by a rotation of its sidechain (see Movie 2 in SI). This motion is tightly coupled to the motion of the Mg2+ ion in the B site. As Asp132 rotates, the ion moves together with the unprotonated oxygen of the carboxylic acid group, to which it is strongly bound. This ion motion opens up a pathway for the proton to pass from Asp132 to the oxygen of the ribose coordinating metal ion B. After the proton is transferred, the ribose dissociates from the ion to form the reaction product.
Experiments support our observation that a protonated Asp132 can serve as the direct proton donor to the ribose. The increased binding affinity of RNase H to the RNA.DNA substrate in the D132N mutant,10 with the Asn mutation mimicking a protonated Asp, suggests that the pKa of Asp132 is indeed shifted up in the enzyme-substrate complex. A role of Asp132 as proton donor was also suggested30 on the basis of structures with only a single Ca2+ ion bound. In a recent study of a bacterial polymerase, a protonated aspartic acid in the active site was found to serve as a proton donor.4 Consistent with a proton donor function of Asp132, the conservative D132N mutation abolishes catalysis.10,12
Nevertheless, the protonation of the leaving group could also proceed through alternative pathways. One possibility is protonation directly from the surrounding solution. However, the O3′ oxygen does not form direct hydrogen bonds with water throughout the simulations and also the catalytic rate peaks at pH~8,15,31,32 which would disfavor (but not necessarily exclude) protonation from the surrounding solvent. An indirect PT pathway is via the 2′OH of the sugar in the 5′ direction of the cleavage site (pathway IIb in Figure 2). This pathway is supported by the fact that there is a conserved serine (Ser133) residue coordinating a conserved water molecule (present in, e.g., PDB structures 1ZBL, 1ZBI, 2G8I, 2G8F, 2R7Y) in a hydrogen bonding structure with the 2′OH (Figure S3). While Ser133 was found to be non-essential for catalysis,33 studies suggested that the ribose 2′OH group may participate both as proton donor and as proton acceptor.29,34 This 2′OH group also forms a hydrogen bonding chain with a single file of water molecules that is in contact with bulk water. These water molecules in turn form hydrogen bonds with the next sugar group in the 5′ direction of the RNA substrate chain. This sugar group interacts with the conserved Lys180, which was suggested to be involved in the catalysis.35 In SI, we show that the proton transfer from a hydronium ion via the ribose 2′OH group to the leaving group occurs on a picosecond time scale, suggesting that this pathway could indeed provide a possible alternative to protonation via Asp132.
Reaction pathway and rate
The results of our free energy calculations and of string method simulations consistently show a two-step mechanism for the RNA cleavage reaction (Figure 3 and Figure 4). In the first reaction process, an intermediate is formed in which the sugar-phosphate bond is broken, but the leaving group is unprotonated and coordinated to metal B. As part of this first process, the OH− nucleophile is formed by transferring the proton of the attacking water molecule to the downstream phosphate. Then the bond between the OH− and the phosphorus atom forms while the bond between the phosphorus atom and the ribose breaks. A free energy barrier of ~15 kcal mol−1 (17 kcal mol−1 with unprotonated Asp132) is encountered right after the proton transfer step (Figure 4). Interestingly, this barrier and the order of events in the first process are nearly the same with Asp132 protonated (Figures 3 and 4) or unprotonated (Figure S2). The main difference is that with Asp132 protonated the intermediate structure formed immediately after bond breaking corresponds to a hydrolyzed phosphate and an alkoxide ribose coordinated to a Mg2+ ion, and not the metastable phosphorane formed with the unprotonated Asp132.
In a second process, the reaction is completed by the transfer of a proton to the leaving group. The formation of a distinct intermediate with a dissociated but unprotonated ribose is consistent with the unusual pH dependence of the measured rates. The RNase H enzyme superfamily exhibits a bell-shaped pH dependence with an optimum pH value of about 8.15,31,32 Acidic pH would slow down process I, with protonated acceptor groups reducing the rate of water deprotonation; basic pH would slow down process II, with Asp132 (or other donors involved in protonating the leaving group) being deprotonated. In our calculations, we find that these two processes have barriers of comparable height (~15 and ~12 kcal mol−1), with statistical uncertainties estimated at ~2–3 kcal mol−1 from analyses using subsets of data. Slowing down either process would thus affect the overall rate. The bell-shaped pH profile can therefore be accounted for at least qualitatively by our double-proton-transfer mechanism.36 In contrast, if the metastable intermediate were a phosphorane,2,3 with a rapid internal PT from the scissile phosphate to the leaving group, one would require extraneous effects to account for the slow-down observed at basic pH.
The overall barrier of ~15 kcal mol−1, encountered in process I, is consistent with the measured rate after factoring in the energetic cost of protonating Asp132 at up to 4 kcal mol−1 for a pKa between 4 and 7. (The reason for including at least part of this cost is that with Asp132 unprotonated, process I did not complete in our calculations.) For a kinetic prefactor of ~1/ps, the experimental rates of ~100 min−1 and ~1 min−1 for E. coli37,38 and human37,39 RNase H1 at 37°C correspond to barrier heights of ~16.7 and ~19.5 kcal mol−1, respectively, assuming further that product release is fast. We expect the rate and barrier of the Bh enzyme to be closer to that of the faster E. coli RNase H1.
Magnesium ion coordination and motion
Changes in the coordination of the two Mg2+ ions are thought to be a major factor in the catalytic lowering of the reaction barrier for RNA backbone cleavage by RNase H. A remarkable structural feature emerging from the crystal structures is a strained pentacoordinated Mg2+ ion B in the reactant state.12 Product formation has been suggested12 as a means to release the strain, driving the system toward a more common hexagonal coordination of the Mg2+ ion B.
In our simulations, the pentacoordinated structure of Mg2+ ion B was retained in the reactant state. However, as the reaction progressed towards the intermediate state, the magnesium coordination changed to a more symmetric pentacoordinated structure, in case of both the protonated Asp132 and the deprotonated Asp132 simulations (Figure S4). We monitored this symmetry change by following the axial angle of the trigonal bipyramid between the leaving group, the Mg2+ ion, and the oxygen of the Asp71 as a function of the reaction coordinate (Figure S4B). During the reaction, the angle changed from 150 degrees in the reactant state to 170 and 90 degrees in the intermediate and product states, respectively. We also found that in the intermediate state the distance between the leaving group and the Mg2+ ion B decreased significantly to values approaching those of the other four ligands (Figure S4A). In the final product state, the protonated ribose separated from Mg2+ ion B, substituted as a ligand by a conserved water molecule located in a pocket near Glu109 (Figure 5 panel PS). Interestingly, both metal ions A and B were pentacoordinated in the intermediate state for the protonated Asp132 (Figure 5). A pentacoordinated metal ion A was also seen in polymerase III simulations.5 In the RNase H simulations with unprotonated Asp132, metal ion A remained hexacoordinated.
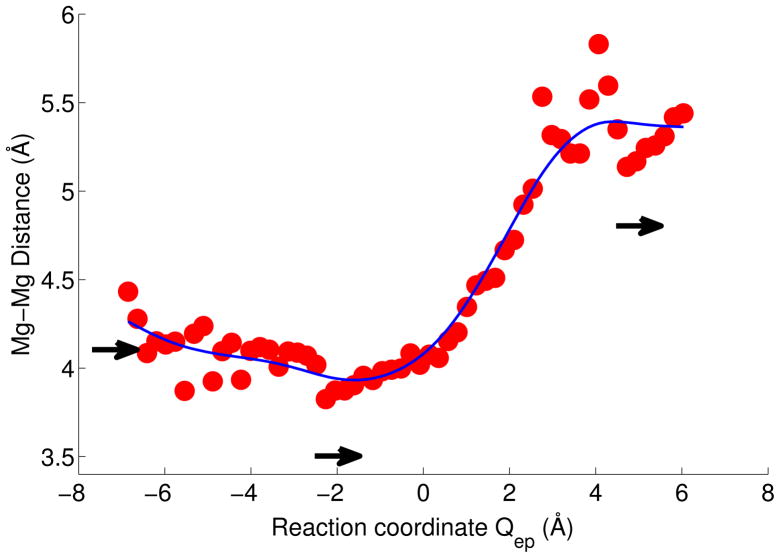
As the reaction progressed, the Mg-Mg distance changed in our simulations from about 4.2 Å in the reactant state to about 3.8 Å in the transition state TS1 and intermediate state IS with Qep between -4 and 0 Å (Figure 6). From the intermediate state towards the product state, the distance increased to about 5.3 Å. These simulation results are in near quantitative agreement with experiment.10,12 The crystal structures also show a contraction of the Mg-Mg distance from reactant-like states (4.1 Å) to the ‘intermediate’, transition-state-like complex (3.5 Å), followed by an increase in the product-state complex (4.8 Å).10,12 A likely cause of this nonmonotic change in the metal ion distance is their tight coordination to the incoming nucleophilic oxygen and to the outgoing ribose oxygen, whose motions the ions follow. Similar metal ion movements were observed in theoretical calculations of other enzymes using two-metal ion catalysis.40,41
Figure 6.
Mg2+-Mg2+ distance along the reaction coordinate Qep=ET+(PT1a+PT1b)/2+PT2 for the catalytic reaction of the protonated Asp132 system. Arrows indicate the Mg2+-Mg2+ distances in reactant, transition-state like, and product structures10,12 (PDB codes 1ZBL, 2G8H and 2G8F, and 2G8V, respectively; left to right). The data points are calculated from reweighted string simulations and the curve is a spline fit to guide the eye.
Differences from Earlier Simulations of RNase H
The reaction pathways and the character of the intermediate in our simulations differ significantly from those in the earlier studies by De Vivo et al.2 and by Elsässer and Fels,3 as discussed above. One possible factor is the use of the BLYP42,43 density functional by De Vivo et al.2, which has been reported to lower the high barriers for PT between donor and acceptor atoms at larger distances.44–46 In contrast, here PT dominates the motion over the top of the barriers. Our 2D free energy surface in Figure 4, with phosphate bond formation/breaking coordinates on the horizontal axis and PT coordinates on the vertical axis, shows that the saddles at the two transition states (for the formation of the intermediate and the final product, respectively) are both oriented parallel to the PT direction, suggesting that the PT reactions commit the system to the formation of intermediate and product states, respectively. The PT reactions were also found to be central elements of the two-metal catalysis in a bacterial polymerase.4 We note, however, that bulk solvation effects may be underestimated in our QM/MM calculations of a non-periodic system, as compared to the fully periodic calculations of De Vivo et al.2
Differences in the hydrogen bonding pattern near metal A provide another possible explanation for the differences in the PT rates here and in Ref. 2, due to the use of different initial structures (D132N and D192N with PDB codes 1ZBI and 1ZBL, respectively). In particular, there are differences in the conformation of Glu188 that could in principle affect the PT pathway in process I. In the D132N structure used here (PDB code 1ZBI), Glu188 is bound to metal ion A, whereas in the D192N structure it is switched out, forming a strong hydrogen bond with the amide group of the mutated D192N residue (PDB code 1ZBL, used in the simulations of Ref. 2). To explore the influence of the Glu188 position, we performed additional calculations of an E188A mutant based on the D132N structure, designed to mimic a conformation in which Glu188 is not interacting directly with metal ion A (as in D192N). As shown in Movie 3 in SI, this mutation did not alter the reaction mechanism in process I. A quantitative characterization of the E188A mutation would require a full free energy calculation. In this regard, it would also be interesting to test whether the side-chain position of Glu188 is coupled to the protonation state of Asp132.
Concluding Remarks
The RNase H enzyme catalyzes the hydrolysis of the phosphate ester bond of the RNA backbone in an RNA.DNA duplex substrate in a nonspecific manner. Formally, the hydrolysis reaction requires (Figure 2, process I) a general base to deprotonate the attacking water molecule, and (Figure 2, process II) a general acid to protonate the leaving group. In our simulations with protonated Asp132, we found that the reaction proceeded in two steps. However, the intermediate found here in the case of protonated Asp132 was not a phosphorane, but a cleaved phosphate with a deprotonated ribose alkoxide coordinated to Mg2+ ion B. A central role of an alkoxide nucleophile intermediate was previously suggested for the two-metal catalyzed RNA backbone cleavage reaction of the hammerhead ribozyme.8
Our results, and those of earlier simulations,2,3 jointly suggest that there are several competing PT transfer pathways that may be comparable in their energetics. Based on our simulations, we propose that the deprotonation of the water nucleophile in process I takes place via the downstream phosphate group. We also found that in process II the leaving group can be protonated via the Asp132 residue. The reasons for the two PT reactions to be slow are different in processes I and II. The initial PT is slow because of the high energetic cost to deprotonate a water molecule. In contrast, the second PT is slow because the acceptor group is bound to magnesium ion B and not involved in hydrogen bonds, except to the O2′ group of the sugar. PT from the protonated Asp132 therefore requires a partial dissociation of the alkoxide from the metal ion, and also a significant change in the Asp132 side chain conformation. We note that for the PT in process II we did find a more facile route from a hydronium ion via the sugar O2′ group. However, this alternative is limited by the formation of the hydronium, again resulting in a slow overall reaction, as seen in experiment.39
Our finding of an intermediate with a protonated downstream pro-Rp oxygen helps explain both the strong effect of a sulfur substitution on the catalytic rate,28 and the absence of an effect of a sulfur substitution at the pro-Rp oxygen of the scissile phosphate.29 Our proposed double proton transfer mechanism and its energetics are also consistent with the bell shaped pH profile.36 Our simulations also account for the overall barrier inferred from experiments, with catalytic rates of ~100 min−1 and ~1 min−1 for E. coli37,38 and human37,39 RNase H1, respectively.
Central to the catalytic function of RNase H are the magnesium ions, and the changes in their coordination structure.10 In our simulations, the coordination of the magnesium ions agreed well with the crystal structures. In particular, we obtained pentacoordinated metal ions at both sites (A and B) at the intermediate structures. We also found that the coordination around the metal ion in the B site becomes more symmetric as the intermediate state is reached. This change in the coordination to a more typical structure for magnesium could possibly be a major factor in the overall reaction.
Our calculations show that in RNase H, the catalytic effect is achieved by exploiting the Mg2+ coordination. The Mg2+ ions not only positioned the attacking water and the scissile phosphate, but also trapped the alkoxide intermediate that formed after bond cleavage. As the reaction progressed, we found that the strained coordination geometry of the Mg2+ ions in the reactant state relaxed from an asymmetric pentacoordinated structure to a symmetric trigonal bipyramid at the intermediate state. Metal ion coordination thus contributes to the catalytic efficiency, helping to lower the reaction barrier in RNase H, and possibly in other enzymes using two-metal ion catalysis.
Supplementary Material
Acknowledgments
We thank Drs. Bernard Brooks, Yihan Shao, and Lee Woodcock for their valuable help. This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, and used the computational resources of the biowulf cluster.
Footnotes
Supporting Information Available: Reactant state structure stability, protonation of leaving group via ribose O2′, effect of E188A mutation, complete author list of reference 24, basis set effects, free energy surface, sequence alignment, magnesium ion coordination, and two movies of the catalytic reaction. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Krakowiak A, Owczarek A, Koziolkiewicz M, Stec WJ. Chem Bio Chem. 2002;3:1242. doi: 10.1002/1439-7633(20021202)3:12<1242::AID-CBIC1242>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 2.De Vivo M, Dal Peraro M, Klein ML. J Am Chem Soc. 2008;130:10955. doi: 10.1021/ja8005786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elsässer B, Fels G. Phys Chem Chem Phys. 2010;12:11081. doi: 10.1039/c001097a. [DOI] [PubMed] [Google Scholar]
- 4.Venkatramani R, Radhakrishnan R. Protein Sci. 2010;19:815. doi: 10.1002/pro.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cisneros GA, Perera L, Schaaper RM, Pedersen LC, London RE, Pedersen LG, Darden TA. J Am Chem Soc. 2009;131:1550. doi: 10.1021/ja8082818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pingoud V, Wende W, Friedhoff P, Reuter M, Alves J, Jeltsch A, Mones L, Fuxreiter M, Pingoud A. J Mol Biol. 2009;393:140. doi: 10.1016/j.jmb.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 7.Yang W. Nat Struct Mol Biol. 2008;15:1228. doi: 10.1038/nsmb.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lott WB, Pontius BW, von Hippel PH. Proc Natl Acad Sci USA. 1998;95:542. doi: 10.1073/pnas.95.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nowotny M, Cerritelli SM, Ghirlando R, Gaidamakov SA, Crouch RJ, Yang W. EMBO J. 2008;27:1172. doi: 10.1038/emboj.2008.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Cell. 2005;121:1005. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 11.Nowotny M, Gaidamakov SA, Ghirlando R, Cerritelli SM, Crouch RJ, Yang W. Mol Cell. 2007;28:264. doi: 10.1016/j.molcel.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 12.Nowotny M, Yang W. EMBO J. 2006;25:1924. doi: 10.1038/sj.emboj.7601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanaya S. FEBS J. 2009;276:1481. doi: 10.1111/j.1742-4658.2009.06906.x. [DOI] [PubMed] [Google Scholar]
- 14.Schultz SJ, Champoux JJ. Virus Res. 2008;134:86. doi: 10.1016/j.virusres.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsunaka Y, Haruki M, Morikawa M, Oobatake M, Kanaya S. Biochemistry. 2003;42:3366. doi: 10.1021/bi0205606. [DOI] [PubMed] [Google Scholar]
- 16.Zhou T, Sun L, Humphreys J, Goldsmith EJ. Structure. 2006;14:1011. doi: 10.1016/j.str.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 17.Rosta E, Woodcock HL, Brooks BR, Hummer G. J Comput Chem. 2009;30:1634. doi: 10.1002/jcc.21312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dal Peraro M, Spiegel K, Lamoureux G, De Vivo M, DeGrado WF, Klein ML. J Struct Biol. 2007;157:444. doi: 10.1016/j.jsb.2006.10.019. [DOI] [PubMed] [Google Scholar]
- 19.Warshel A, Levitt M. J Mol Biol. 1976;103:227. doi: 10.1016/0022-2836(76)90311-9. [DOI] [PubMed] [Google Scholar]
- 20.Becke AD. J Chem Phys. 1993;98:5648. [Google Scholar]
- 21.Fukunishi H, Watanabe O, Takada S. J Chem Phys. 2002;116:9058. [Google Scholar]
- 22.EW, Ren W, Vanden-Eijnden E. J Phys Chem B. 2005;109:6688. doi: 10.1021/jp0455430. [DOI] [PubMed] [Google Scholar]
- 23.Radhakrishnan R. Biophys J. 2007;93:2391. doi: 10.1529/biophysj.107.104661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shao Y, et al. Phys Chem Chem Phys. 2006;8:3172. doi: 10.1039/b517914a. [DOI] [PubMed] [Google Scholar]
- 25.Woodcock HL, Hodoscek M, Gilbert ATB, Gill PMW, Schaefer HF, III, Brooks BR. J Comput Chem. 2007;28:1485. doi: 10.1002/jcc.20587. [DOI] [PubMed] [Google Scholar]
- 26.Kumar S, Rosenberg JM, Bouzida D, Swendsen RH, Kollman PA. J Comput Chem. 1992;13:1011. [Google Scholar]
- 27.Zhu F, Hummer G. Proc Natl Acad Sci USA. 2010;107:19814. doi: 10.1073/pnas.1009313107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haruki M, Tsunaka Y, Morikawa M, Iwai S, Kanaya S. Biochemistry. 2000;39:13939. doi: 10.1021/bi001469+. [DOI] [PubMed] [Google Scholar]
- 29.Uchiyama Y, Miura Y, Inoue H, Ohtsuka E, Ueno Y, Ikehara M, Iwai S. J Mol Biol. 1994;243:782. doi: 10.1016/0022-2836(94)90047-7. [DOI] [PubMed] [Google Scholar]
- 30.Babu CS, Dudev T, Casareno R, Cowan JA, Lim C. J Am Chem Soc. 2003;125:9318. doi: 10.1021/ja034956w. [DOI] [PubMed] [Google Scholar]
- 31.Bastock JA, Webb M, Grasby JA. J Mol Biol. 2007;368:421. doi: 10.1016/j.jmb.2007.01.083. [DOI] [PubMed] [Google Scholar]
- 32.Kashiwagi T, Jeanteur D, Haruki M, Katayanagi K, Kanaya S, Morikawa K. Protein Eng. 1996;9:857. doi: 10.1093/protein/9.10.857. [DOI] [PubMed] [Google Scholar]
- 33.Kanaya S, Kohara A, Miura Y, Sekiguchi A, Iwai S, Inoue H, Ohtsuka E, Ikehara M. J Biol Chem. 1990;265:4615. [PubMed] [Google Scholar]
- 34.Shigenori I, Shin K, Makoto W, Eiko O, Haruki N. FEBS Lett. 1995;368:315. [Google Scholar]
- 35.Yoshikawa M, Iwasaki H, Kinoshita K, Shinagawa H. Genes Cells. 2000;5:803. doi: 10.1046/j.1365-2443.2000.00371.x. [DOI] [PubMed] [Google Scholar]
- 36.Stanford NP, Halford SE, Baldwin GS. J Mol Biol. 1999;288:105. doi: 10.1006/jmbi.1999.2673. [DOI] [PubMed] [Google Scholar]
- 37.Wu HJ, Lima WF, Crooke ST. J Biol Chem. 2001;276:23547. doi: 10.1074/jbc.M009676200. [DOI] [PubMed] [Google Scholar]
- 38.Kanaya E, Kanaya S. Eur J Biochem. 1995;231:557. doi: 10.1111/j.1432-1033.1995.0557d.x. [DOI] [PubMed] [Google Scholar]
- 39.Lima WF, Wu H, Nichols JG, Prakash TP, Ravikumar V, Crooke ST. J Biol Chem. 2003;278:49860. doi: 10.1074/jbc.M306543200. [DOI] [PubMed] [Google Scholar]
- 40.Dupureur CM. Curr Opin Chem Biol. 2008;12:250. doi: 10.1016/j.cbpa.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 41.Wang L, Yu X, Hu P, Broyde S, Zhang Y. J Am Chem Soc. 2007;129:4731. doi: 10.1021/ja068821c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Becke AD. Phys Rev A. 1988;38:3098. doi: 10.1103/physreva.38.3098. [DOI] [PubMed] [Google Scholar]
- 43.Lee C, Yang W, Parr RG. Phys Rev. 1988;B37:785. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 44.Barone V, Orlandini L, Adamo C. Chem Phys Lett. 1994;231:295. [Google Scholar]
- 45.Geissler PL, Van Voorhis T, Dellago C. Chem Phys Lett. 2000;324:149. [Google Scholar]
- 46.Sadhukhan S, Muñoz D, Adamo C, Scuseria GE. Chem Phys Lett. 1999;306:83. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.