Abstract
We have analyzed the known metabolic enzymes of Escherichia coli in relation to their biochemical reaction properties and their involvement in biochemical pathways. All enzymes involved in small-molecule metabolism and their corresponding protein sequences have been extracted from the EcoCyc database. These 548 metabolic enzymes are clustered into 405 protein families according to sequence similarity. In this study, we examine the functional versatility within enzyme families in terms of their reaction capabilities and pathway participation. In addition, we examine the molecular diversity of reactions and pathways according to their presence across enzyme families. These complex, many-to-many relationships between protein sequence and biochemical function reveal a significant degree of correlation between enzyme families and reactions. Pathways, however, appear to require more than one enzyme type to perform their complex biochemical transformations. Finally, the distribution of enzyme family members across different pathways provides support for the “recruitment” hypothesis of biochemical pathway evolution.
Metabolic enzymes represent one of the most important and extensively studied class of proteins. Consequently, enzymes have been used extensively to address various issues of protein sequence/function relationships. Known metabolic enzyme families exhibit complex patterns of divergent and convergent evolution — many enzyme families usually catalyze a range of biochemical reactions (Jensen and Gu 1996), whereas some of these reactions may also be catalyzed by members of apparently unrelated protein families. A deeper understanding of such subtleties of the sequence-to-function relationship may shed light into the processes of molecular evolution of proteins (Petsko et al. 1993). These studies enable us to address fundamental questions, such as the origins and evolution of biochemical networks, and practical issues, such as function assignment by homology.
For instance, a case study of the distribution of the (βα)8 barrel fold into different pathways appears to suggest a “patchy” mode of evolution for metabolic pathways (Copley and Bork 2000). Another study covering the Enzyme Commission (EC) hierarchy and the distribution of EC numbers across protein fold types addresses the problem of annotation transfer (Hegyi and Gerstein 1999; Wilson et al. 2000). These studies have used protein structure similarities, as three-dimensional structure is generally more conserved than the primary sequence. Although structural similarities permit the detection of very distant homologies, these analyses are confined to homologs of proteins of known structure.
In the absence of protein structure information, sequence comparisons provide a less sensitive but much more comprehensive way of detecting protein function at a genome-wide scale. Function prediction by sequence similarity provides useful hints for the potential cellular roles of proteins in entire genomes. Currently, an average of 60% of the encoded proteins for any genome can be functionally characterized by homology to proteins of known function (Iliopoulos et al. 2000). This assignment procedure, however, often overlooks issues of evolutionary divergence, whereby homologous sequences may have different functions. Such effects may lead to error propagation in sequence databases (Karp 1998). One of the most suitable sets of proteins to address some of these issues is the set of known metabolic enzymes, because of available classification schemes for their functional properties, in terms of the EC reaction hierarchy and pathway participation.
We have performed an extensive correlation of enzyme sequence and function, using the full known metabolic complement of Escherichia coli (Karp et al. 2000). Sequence relationships are represented by the membership of enzymes into protein families on the basis of sequence similarity. Function properties are represented by reaction capabilities and pathway involvement of the corresponding enzymes. We examined (1) functional versatility within enzyme families, that is, the association of enzymes with distinct reactions and pathways and (2) molecular diversity of protein function, that is, the distribution of reactions and pathways across enzyme families. We call these aspects of our analysis the sequence-to-function and the function-to-sequence problems, respectively. The detected patterns of functional versatility and molecular diversity across enzyme families, reactions, and pathways allow the first genome-wide overview of these complex, many-to-many relationships.
RESULTS
We have examined the sequence-to-function problem through the mapping of functional versatility of enzyme families to reaction types (EC numbers) and pathway involvement. The degree to which enzymes span different reactions (section 1) and pathways (section 2) corresponds directly to the structural plasticity and divergence within enzyme families. Furthermore, we have examined the function-to-sequence problem, namely the molecular diversity of protein function in terms of reactions (section 3) and pathways (section 4), using criteria for family membership. The extent to which biochemical reactions and pathways are associated with distinct protein families possibly indicates functional properties that have been invented more than once during evolution.
The 548 small-molecule metabolic enzymes were clustered into 405 enzyme families, with 316 single-member families and the remaining 232 enzymes classified into 89 families. Only 30 families containing 47 enzymes did not match any EC number, therefore mapping these families to reaction types was not possible. Overall, the 548 enzymes were found in 132 pathways and encoded for 422 unique EC numbers, representing 90 oxidoreductases, 133 transferases, 49 hydrolases, 82 lyases, 35 isomerases, and 33 ligases. These six enzyme classes correspond to the first level of the EC hierarchy, respectively.
Functional Versatility of Enzyme Families in Reaction Space
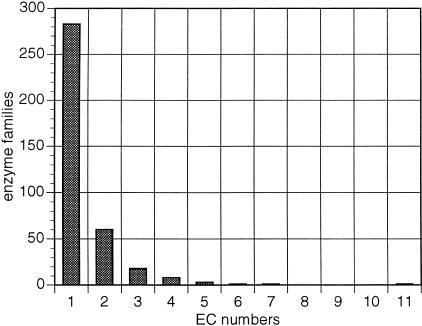
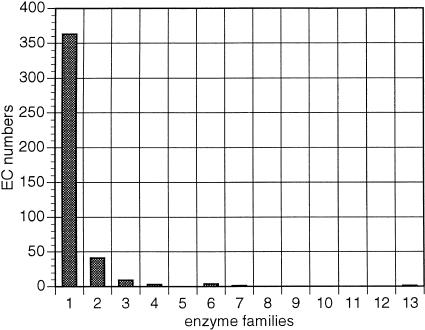
Interestingly, 75% of the total number of enzyme families appear to contain monofunctional enzymes, that is, enzymes known to catalyze a single enzymatic reaction (Fig. 1). This is a very important observation with direct applications to function prediction by sequence similarity, because it suggests that within these families, direct transfer of annotation by similarity can be reliable (desJardins et al. 1997). These monofunctional families include well-known, homologous isozymes such as the Fe/Mn superoxide dismutases (McCord 1976), gluconokinases, and L-asparaginases (Table 1). There is also a number of heteropolymeric enzyme subunits sharing sequence similarity, such as glutamate decarboxylase, formate dehydrogenase, nitrate reductase, and others (Table 1).
Figure 1.
Functional versatility of enzyme families as assessed by reaction types. Frequency distribution of enzyme families (Y-axis) in relation to the number of unique EC numbers (X-axis) these families span.
Table 1.
Enzyme Families with More Than One Member and a Single EC Number
| Reaction type | Enzyme description |
|---|---|
| 1.1.99.5 | P13035 AEROBIC GLYCEROL-3-PHOSPHATE DEHYDROGENASE |
| P13032 ANAEROBIC GLYCEROL-3-PHOSPHATE DEHYDROGENASE SUBUNIT A | |
| 1.2.1.2 | P32174 FORMATE DEHYDROGENASE, CYTOCHROME B556 (FDO) SUBUNIT |
| P24185 FORMATE DEHYDROGENASE, NITRATE-INDUCIBLE, CYTOCHROME | |
| B556 (FDN) SUBUNIT | |
| 1.2.1.12 | P11603 D-ERYTHROSE 4-PHOSPHATE DEHYDROGENASE |
| P06977 GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE A | |
| 1.3.99.1 | P07014 SUCCINATE DEHYDROGENASE IRON-SULFUR PROTEIN |
| P00364 FUMARATE REDUCTASE IRON-SULFUR PROTEIN | |
| 1.6.5.3 | NADH DEHYDROGENASE I SUBUNITS (P33608 CHAIN N NuoN, P33607 CHAIN L NuoL, P31978 CHAIN M NuoM) |
| 1.7.99.4 | P19316 RESPIRATORY NITRATE REDUCTASE 2 GAMMA CHAIN |
| P11350 RESPIRATORY NITRATE REDUCTASE 1 GAMMA CHAIN | |
| 1.15.1.1 | P09157 SUPEROXIDE DISMUTASE [FE] |
| P00448 SUPEROXIDE DISMUTASE [MN] | |
| 2.2.1.2 | P80218 TRANSALDOLASE A |
| P30148 TRANSALDOLASE B | |
| 2.5.1.- | P26601 4-HYDROXYBENZOATE OCTAPRENYLTRANSFERASE |
| P18404 PROTOHEME IX FARNESYLTRANSFERASE | |
| 2.7.1.12 | P46859 THERMORESISTANT GLUCONOKINASE (GLUCONATE KINASE 2) |
| P39208 THERMOSENSITIVE GLUCONOKINASE (GLUCONATE KINASE 1) | |
| 2.7.1.40 | P21599 PYRUVATE KINASE II |
| P14178 PYRUVATE KINASE I | |
| 2.7.1.71 | P24167 SHIKIMATE KINASE I |
| P08329 SHIKIMATE KINASE II | |
| 3.2.1.28 | P37196 PROBABLE CYTOPLASMIC TREHALASE |
| P13482 PERIPLASMIC TREHALASE PRECURSOR | |
| 3.5.1.1 | P18840 L-ASPARAGINASE I |
| P00805 L-ASPARAGINASE II PRECURSOR | |
| 4.1.1.15 | P80063 GLUTAMATE DECARBOXYLASE ALPHA |
| P28302 GLUTAMATE DECARBOXYLASE BETA | |
| 4.1.2.15 | PHOSPHO-2-DEHYDRO-3-DEOXYHEPTONATE ALDOLASE SUBUNITS (P00888 AroF, P00887 AroH, P00886 AroG) |
| 4.1.3.18 | P08143 ACETOLACTATE SYNTHASE ISOZYME I SMALL SUBUNIT |
| P00894 ACETOLACTATE SYNTHASE ISOZYME III SMALL SUBUNIT | |
| 4.2.1.2 | P14407 FUMARATE HYDRATASE CLASS I, ANAEROBIC |
| P00923 FUMARATE HYDRATASE CLASS I, AEROBIC | |
| 6.3.2.4 | P23844 D-ALANINE-D-ALANINE LIGASE A |
| P07862 D-ALANINE-D-ALANINE LIGASE B |
The table is sorted on the basis of the EC number. Reaction type: EC number of the corresponding enzyme family. Enzyme description: SWISS-PROT accession number (Bairoch and Apweiler 2000) and enzyme name description. There are two cases in which the subunit names are not well-specified and gene product names are used instead (reaction type 1.6.5.3 and 4.1.2.1.5).
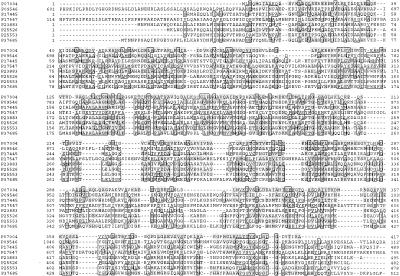
An additional 16% of enzyme families contain two unique EC numbers, as judged by the number of reactions that the family members are known to catalyze. The remaining 9% of enzyme families contain three or more unique EC numbers. There is one enzyme family that contains eight enzymes with 11 unique EC numbers, representing different oxidoreductase reaction types (Fig. 2; Table 2). The common domain of these enzymes corresponds to the pyrroline-5-carboxylate (P5C) dehydrogenase family (Ling et al. 1994).
Figure 2.
Multiple sequence alignment of the common domain of the enzyme family members listed in Table 2. SWISS-PROT accession numbers are given at left. Residue numbers corresponding to the entire protein sequence length are also shown. Identical residues (>50%) are boxed; similar residues (PAM250, >25%) are shaded.
Table 2.
The Enzyme Family with the Highest Known Functional Versatility in the Metabolic Complement of E. coli
| Reaction type | Enzyme description |
|---|---|
| 1.1.1.- | P17547 ALDEHYDE-ALCOHOL DEHYDROGENASE |
| 1.1.1.1 | [ALCOHOL DEHYDROGENASE, ACETALDEHYDE DEHYDROGENASE, PYRUVATE-FORMATE-LYASE DEACTIVASE] |
| 1.2.1.10 | |
| 1.2.1.3 | P23883 PUTATIVE ALDEHYDE DEHYDROGENASE |
| 1.2.1.8 | P17445 BETAINE ALDEHYDE DEHYDROGENASE |
| 1.2.1.16 | P25526 SUCCINATE-SEMIALDEHYDE DEHYDROGENASE [NADP+] |
| 1.2.1.21 | P25553 ALDEHYDE DEHYDROGENASE A |
| 1.2.1.22 | |
| 1.2.1.22 | P37685 ALDEHYDE DEHYDROGENASE B |
| 1.2.1.41 | P07004 GAMMA-GLUTAMYL PHOSPHATE REDUCTASE (GPR) |
| 1.5.1.12 | P09546 BIFUNCTIONAL PUTA PROTEIN |
| 1.5.99.8 | [PROLINE DEHYDROGENASE, DELTA-1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE] |
This family comprises eight enzyme sequences with 11 different enzyme activities. Column descriptions are as listed in Table 1.
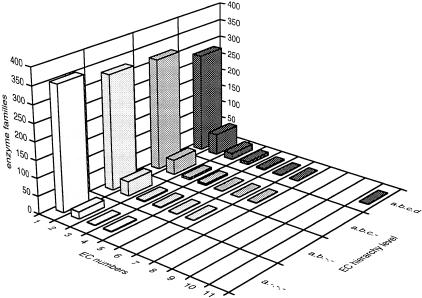
This extreme concentration of unique EC numbers within enzyme families may reflect some strong property of function conservation in the set of known metabolic enzymes of E. coli (Labedan and Riley 1995). There are two potential artifacts, however, that may contribute toward this pattern. First, most of the monofunctional families (93%) correspond to single-member families. In addition, there may be other enzyme homologs in the genome that have not been characterized or included in the EcoCyc database. Second, the EC classification scheme does not always account for the reaction specificity of enzymes. Examples are reaction 1.3.99.1 (Table 1), representing two enzyme complexes catalyzing the reversible interconversion of succinate to fumarate, and reaction 4.1.1.15 (Table 1), representing two subunits of a single enzyme. In fact, when the resolution is reduced (for example by ignoring the fourth, third, or second level of the EC hierarchy), the one-to-one correspondence between families and EC numbers is even more pronounced (Fig. 3).
Figure 3.
Functional versatility of enzyme families at different levels of the EC hierarchy. Frequency distribution of enzyme families (Z-axis) in relation with the number of unique EC numbers (X-axis) these families span and the four levels of the EC hierarchy (Y-axis). The effect observed in Figure 1 is enhanced as the resolution of the EC scheme becomes lower.
Functional Versatility of Enzyme Families in Pathway Space
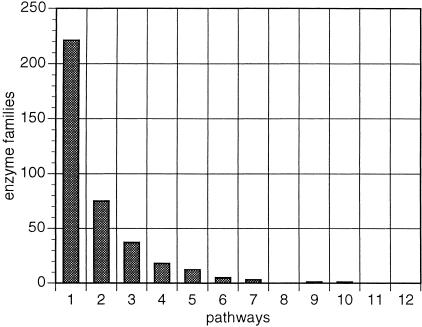
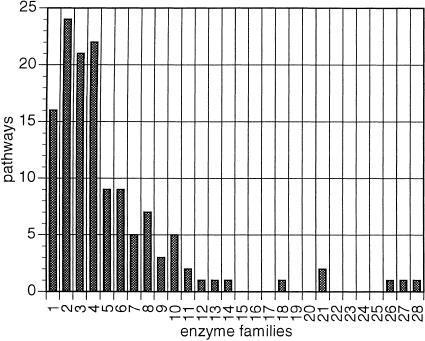
Surprisingly, when the resolution is further reduced by characterizing enzyme function using pathway involvement instead of reaction type, the distribution of enzymes is widened toward multifunctional families (Fig. 4). Still, the majority of enzyme families corresponding to 59% of the total, appear to participate in a single metabolic pathway (Fig. 4). This indicates a sharing of structural types across pathways and may correspond to an evolutionary signature of pathway origins (Jensen 1976). We define enzyme families as “confined” if the number of pathways their members span is less than the number of family members and “promiscuous” if the number of pathways is more than the number of family members.
Figure 4.
Functional versatility of enzyme families as assessed by pathway involvement. Frequency distribution of enzyme families (Y-axis) in relation to the number of unique pathways (X-axis) in which these families appear.
Examples of confined enzyme families include gene products AroG/AroH/AroF (EC 4.1.2.15), involved in the first committed step of the aromatic amino acid biosynthesis pathway, and gene products MurC (EC 6.3.2.8), MurD (EC 6.3.2.9), MurE (EC 6.3.2.13 ), and MurF (EC 6.3.2.15), involved in successive steps of the peptidoglycan biosynthesis pathway. Examples of promiscuous enzyme families include malate dehydrogenase (EC 1.1.1.37), which is known to be involved in six pathways (Ouzounis and Karp 2000), and the cluster of eight members (Table 2) mentioned previously, involved in a total of 10 pathways, including proline biosynthesis, proline utilization, fucose catabolism, methylglyoxal metabolism, rhamnose catabolism, 4-aminobutyrate degradation, and fermentation. A full table of these cases is available on the above-mentioned web site.
A potential artifact in this step may involve the somewhat arbitrary definitions of biochemical pathways (Karp 2000) as well as the threshold values in sequence clustering. We have, however, observed many cases of distinct enzyme families that are known to be involved only in a limited number of pathways, such as the aro gene group mentioned above, suggesting that this pattern is a genuine property of the metabolic map.
Molecular Diversity of Reaction Types in Sequence Space
When the reverse relationship of reaction types to enzyme families was examined, similar patterns are observed. The majority of reaction types, 86% of total, are known to be catalyzed by a single enzyme family (Fig. 5), signifying a low dispersion of catalytic activity across homology groups. Overall, this pattern suggests that sequence largely determines the known functional attributes in term of reaction capability of an enzyme type.
Figure 5.
Molecular diversity of reaction types as assessed by enzyme families. Frequency distribution of unique EC numbers (Y-axis) in relation to the number of enzyme families (X-axis) they span.
The remaining 14% of the EC numbers span more than one enzyme family, which may represent sufficiently divergent sequence clusters or biochemically convergent enzyme types. A well-known case of distinct enzyme families are the class I (FumA and FumB, sharing 80% sequence identity) and class II (FumC) fumarases (Mohrig et al. 1995), all sharing the same EC number (EC 4.2.1.2). Another striking example is a group of four oxidoreductase reactions (EC numbers 1.1.1.−, 1.3.99.1, 1.10.2.−, 1.18.99.1) with members belonging to six different families. A potential explanation is the bias in the EC classification scheme, which provides a high-resolution breakdown of oxidoreductases (EC class 1, with 79 three-level classifications), compared, for instance, with lyases (EC class 4, with 12 three-level classifications) (Bairoch 2000). Another deficiency of the EC classification scheme produces the two extreme examples of a single EC reaction belonging to multiple families (Fig. 5). These reactions correspond to assignments of the same EC number across different (nonhomologous) subunits of enzyme complexes, not necessarily involved in the same catalytic action. The first example is nitrate reductase (EC 1.7.99.4), present in seven families and the second example is NADH dehydrogenase (EC 1.6.5.3), present in 13 different families (Fig. 5). Allowing for these exceptions, the pattern of one reaction–one enzyme family becomes even more pronounced.
Molecular Diversity of Biochemical Pathways in Sequence Space
Finally, and in analogy to the sequence-to-function problem, we examined the extent to which pathways employ members of different enzyme families. The detected enzyme types provide an estimate for the average amount of essential building blocks for each biochemical pathway, in terms of homology groups. Overall, we observe that 63% of the metabolic pathways (83 out of 132) in E. coli employ up to four unique enzyme family types (Fig. 6).
Figure 6.
Molecular diversity of biochemical pathways as assessed by enzyme families. Frequency distribution of small-molecule metabolic pathways (Y-axis) in relation to the number of enzyme family types (X-axis) they use.
In contrast to reaction types, only 12% of pathways (16 out of 132) span a single enzyme family. Some of these pathways contain more than one step, although there is a number of single-step catalytic cascades that are defined as a single pathway (e.g., transaminase reactions) (Ouzounis and Karp 2000).
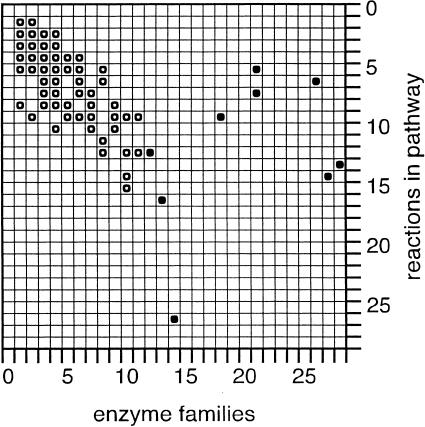
Due to the low number of counts, it is not possible to provide a normalization scheme for this observation. For instance, weighting by the number of steps within pathways does not yield a meaningful pattern (data not shown). With more pathways from different organisms and a deeper understanding of the topological properties of metabolic maps, a re-evaluation of these data may become possible in the future. Currently, we can only describe the number of steps per pathway in relation to the enzyme families these pathways span (Fig. 7).
Figure 7.
The relationship of enzyme families (X-axis) to the number of individual steps per pathway (Y-axis). If there were a one-to-one correspondence of enzyme family types and biochemical reactions, the dominant pattern would be on the diagonal of this matrix. (●) The nine pathways spanning 12 or more enzyme families (mentioned in the Results section); (○) pathway counts (Z-axis, data not shown) and most contain more than one pathway count.
In that respect, it is interesting that 7% of pathways (nine out of 132) span 12 or more enzyme families. These pathways correspond to some of the most complex and extensively studied biochemical cascades (the number of families these pathways span is shown in parentheses): gluconeogenesis (12), purine biosynthesis (13), nucleotide metabolism (14), tricarboxylic acid (TCA) cycle (18), variants of anaerobic and aerobic respiration (21, 26, 27), and fermentation (28). It is worth noting that these pathways appear to span a significant number of enzyme families because of the presence of multiple heteropolymeric enzyme complexes involved in their various catalytic steps (Fig. 7).
DISCUSSION
We have analyzed the sequence-to-function and function-to-sequence problems, using enzyme families. It is instructive to compare these results with a previous study conducted similarly for single enzymes (Ouzounis and Karp 2000). It is striking that the numbers of monofunctional enzymes are comparable, 83% for single enzymes (Ouzounis and Karp 2000) and 75% for enzyme families (this study, as noted above). The reverse relationship also yields very similar percentages for the number of reactions catalyzed by a single enzyme, 91% for single enzymes (Ouzounis and Karp 2000) and 86% for enzyme families (this study, as noted above). These patterns imply that despite currently held views, the catalytic activities of various enzyme types are highly concentrated within enzyme families and there is a dominant one-to-one relationship between sequence and biochemical function.
To characterize biochemical function of metabolic enzymes, we have employed the EC hierarchy assignment and the biochemical pathway involvement. Other, more coarse-grained, functional classification schemes such as the EcoCyc functional classes (Riley 1993) and the Drosophila-derived Gene Ontology scheme (Ashburner et al. 2000) exist. These schemes may be very useful for future analyses of this kind that are not restricted to the metabolic pathway complement but encompass the full spectrum of cellular roles.
Another pattern emerging from the present study is a direct comparison of function divergence within enzyme families (25% of which are multifunctional) to function convergence across families (14% of reactions are catalyzed by more than one family). As pointed out previously, the process of function prediction by homology relies on these principles. Potential errors in this procedure may rise from the class of multifunctional enzyme families. Conversely, the failure to identify specific enzyme types that catalyze a specific reaction may be explained by our inability to detect functionally convergent enzymes.
Using pathways as function descriptors, the one-to-one correspondence between sequence and function is less prominent (see above). A direct comparison of single enzymes (Ouzounis and Karp 2000) and enzyme families with pathway involvement is not possible, because the individual enzymes are highly specific to the corresponding pathways. Our correlation of enzyme families with protein pathways (see above) also sheds some light on the possible mechanisms of pathway evolution.
Early theories for the evolution of biochemical catalysts have suggested that pathways have evolved backward: upstream reactions became possible by the diversification of enzymes that catalyzed the reactions downstream, as compounds became depleted from the environment (Horowitz 1945). This hypothesis allows the development of complex pathways with small modifications of existing ones. It also predicts the accumulation of homologous enzymes within individual pathways, as it would be more probable that similar enzymes are used to handle similar metabolites. Fifty-nine percent of enzyme families are confined within a single metabolic pathway (Fig. 4). Given the significant number of single-member enzyme families, however, the support for the Horowitz hypothesis is rather weak. It appears that only very few pathways exhibit this evolutionary mode.
A competing hypothesis states that biochemical pathways evolve by the recruitment of enzymes in a more opportunistic manner (Jensen 1976). This hypothesis implies that novel biochemical activities have evolved by the use of pre-existing structures (Gerlt and Babbitt 2000). Organisms gained selective advantage first by employing enzymes available through gene duplication to catalyze other reactions and later by further enzyme evolution to fine-tune the enzyme activity. This hypothesis predicts the existence of a high number of homologous enzymes across biochemical pathways. The remaining 41% of enzyme families span multiple pathways, indicating a considerable degree of enzyme recruitment during protein and pathway evolution. The reverse function-to-sequence relationship reveals a high number of pathways (88%) spanning more than one enzyme families, implying that biochemical pathways require a small number of different enzyme types to accomplish the chain of chemical transformations essential for life. This study reflects the knowledge currently available on the metabolic pathways of a single species. It will be interesting to perform similar analyses for other species, when such databases become available, to assess the generality of these observations.
METHODS
First, the entire set of small-molecule metabolism enzymes for the known metabolic complement of E. coli was extracted from the EcoCyc database (Tsoka and Ouzounis 2000). EcoCyc describes the genome and pathways of E. coli solely on the basis of experimental information (Karp et al. 2000). The enzyme set was obtained by formulating a complex query that extracts all proteins catalyzing a reaction whose reactants and products (1) are small molecules and (2) differ. The first condition excludes protein modification and other types of reactions with large molecules, whereas the second condition excludes intracellular transport reactions with no chemical transformation. EcoCyc is currently the only database that allows the extraction of this type of information with such high fidelity. This procedure identified 548 enzymes involved in small-molecule metabolism.
Second, functional descriptions for each enzyme in the form of type of reaction catalyzed (i.e., EC numbers) as well as pathway participation were obtained using EcoCyc (Karp et al. 2000). All sequences of the corresponding entries were extracted using the appropriate pointers in EcoCyc. In addition, protein names and accession numbers were also obtained from SWISS-PROT (Bairoch and Apweiler 2000). Structural descriptions for each enzyme were obtained by first matching each individual sequence to the corresponding PDB entry using BLAST (Altschul et al. 1997) (with an E-value threshold of 10–6) and then extracting protein fold information using SCOP (Lo Conte et al. 2000). Only 39% of these sequences (214 out of 548) have a homolog of known structure. Because of this low coverage, we have based our analyses on sequence similarity alone.
Third, all enzyme sequences were automatically clustered on the basis of sequence similarity, using the GeneRAGE algorithm (Enright and Ouzounis 2000). The algorithm employs a fast sequence similarity search algorithm such as BLAST (Altschul et al. 1997) and represents similarity information between proteins as a binary matrix. Compositionally biased regions are masked with CAST (Promponas et al. 2000). This matrix is subsequently processed through successive rounds of the Smith-Waterman dynamic programming algorithm (Smith and Waterman 1981) to detect inconsistencies, which may represent false-positive or false-negative similarity assignments (Enright and Ouzounis 2000). All parameters for these programs were set to their default values. The resulting clusters comprise protein families with information reflecting the domain structure of proteins. In this analysis, only six two-domain proteins were identified, increasing the effective number of single-domain enzyme entries to 554.
Fourth, the effect of BLAST E-value thresholds for clustering was investigated to identify an optimal threshold value for clustering. Permissive thresholds result in fewer and larger clusters, compared with more stringent values. We have performed full clustering at E-value thresholds ranging from 10−4 to 10−100 and found that the number of enzyme families did not vary significantly (data not shown), indicating that the clustering procedure is quite robust. All results reported here were obtained with the BLAST E-value thresholds of 10−6.
Finally, the analysis involved the detection of functional versatility of the E. coli enzyme families in terms of reaction and pathway properties of the individual family members. This study represents a continuation of previous work, where various metrics for the characterization of an entire metabolic complement were proposed (Ouzounis and Karp 2000). Herein, the additional enzyme family information introduces an evolutionary perspective on the structure and function of biochemical pathways. All of our results are available at http://www.ebi.ac.uk/research/cgg/pathways/families/ and as supplementary data at www.genome.org.
Acknowledgments
The authors thank Anton Enright (Computational Genomics Group) and Peter Karp (SRI International) for comments. This work was supported by the European Molecular Biology Laboratory and the TMR Programme of the European Commission (DG-XII Science, Research and Development). C.O. thanks the UK Medical Research Council and IBM Research for additional support.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
NOTE ADDED IN PROOF
A similar analysis has been performed by Teichmann and colleagues (Teichman et al. 2001, in press).
Footnotes
E-MAIL ouzounis@ebi.ac.uk; FAX 44-1223-494471.
Article published on-line before print: Genome Res., 10.1101/gr.187501.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.187501.
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000;28:304–305. doi: 10.1093/nar/28.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copley RR, Bork P. Homology among (betaalpha)(8) barrels: Implications for the evolution of metabolic pathways. J Mol Biol. 2000;303:627–641. doi: 10.1006/jmbi.2000.4152. [DOI] [PubMed] [Google Scholar]
- desJardins M, Karp PD, Krummenacker M, Lee TJ, Ouzounis CA. Prediction of enzyme classification from protein sequence without the use of sequence similarity. Intell Sys Mol Biol. 1997;5:92–99. [PubMed] [Google Scholar]
- Enright AJ, Ouzounis CA. GeneRAGE: A robust algorithm for sequence clustering and domain detection. Bioinformatics. 2000;16:451–457. doi: 10.1093/bioinformatics/16.5.451. [DOI] [PubMed] [Google Scholar]
- Gerlt JA, Babbitt PC. Can sequence determine function? Genome Biol. 2000;1:r0005.1–r0005.10. doi: 10.1186/gb-2000-1-5-reviews0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegyi H, Gerstein M. The relationship between protein structure and function: A comprehensive survey with application to the yeast genome. J Mol Biol. 1999;288:147–164. doi: 10.1006/jmbi.1999.2661. [DOI] [PubMed] [Google Scholar]
- Horowitz NH. On the evolution of biochemical syntheses. Proc Natl Acad Sci. 1945;31:153–157. doi: 10.1073/pnas.31.6.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos I, Tsoka S, Andrade MA, Janssen P, Audit B, Tramontano A, Valencia A, Leroy C, Sander C, Ouzounis CA. Genome sequences and great expectations. Genome Biol. 2000;2:i0001.1–i0001.3. doi: 10.1186/gb-2000-2-1-interactions0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RA. Enzyme recruitment in evolution of new function. Ann Rev Microbiol. 1976;30:409–425. doi: 10.1146/annurev.mi.30.100176.002205. [DOI] [PubMed] [Google Scholar]
- Jensen RA, Gu W. Evolutionary recruitment of biochemically specialized subdivisions of Family I within the protein superfamily of aminotransferases. J Bacteriol. 1996;178:2161–2171. doi: 10.1128/jb.178.8.2161-2171.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD. What we do not know about sequence analysis and sequence databases. Bioinformatics. 1998;14:753–754. doi: 10.1093/bioinformatics/14.9.753. [DOI] [PubMed] [Google Scholar]
- ————— An ontology for biological function based on molecular interactions. Bioinformatics. 2000;16:269–285. doi: 10.1093/bioinformatics/16.3.269. [DOI] [PubMed] [Google Scholar]
- Karp PD, Riley M, Saier M, Paulsen IT, Paley SM, Pellegrini-Toole A. The EcoCyc and MetaCyc databases. Nucleic Acids Res. 2000;28:56–59. doi: 10.1093/nar/28.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labedan B, Riley M. Gene products of Escherichia coli: Sequence comparisons and common ancestries. Mol Biol Evol. 1995;12:980–987. doi: 10.1093/oxfordjournals.molbev.a040291. [DOI] [PubMed] [Google Scholar]
- Ling M, Allen SW, Wood JM. Sequence analysis identifies the proline dehydrogenase and delta 1-pyrroline-5-carboxylate dehydrogenase domains of the multifunctional Escherichia coliPutA protein. J Mol Biol. 1994;243:950–956. doi: 10.1006/jmbi.1994.1696. [DOI] [PubMed] [Google Scholar]
- Lo Conte L, Ailey B, Hubbard TJ, Brenner SE, Murzin AG, Chothia C. SCOP: A structural classification of proteins database. Nucleic Acids Res. 2000;28:257–259. doi: 10.1093/nar/28.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord JM. Iron- and manganese-containing superoxide dismutases: Structure, distribution, and evolutionary relationships. Adv Exp Med Biol. 1976;74:540–550. doi: 10.1007/978-1-4684-3270-1_45. [DOI] [PubMed] [Google Scholar]
- Mohrig JR, Moerke KA, Cloutier DL, Lane BD, Person EC, Onasch TB. Importance of historical contingency in the stereochemistry of hydratase-dehydratase enzymes. Science. 1995;269:527–529. doi: 10.1126/science.7624773. [DOI] [PubMed] [Google Scholar]
- Ouzounis CA, Karp PD. Global properties of the metabolic map of Escherichia coli. Genome Res. 2000;10:568–576. doi: 10.1101/gr.10.4.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsko GA, Kenyon GL, Gerlt JA, Ringe D, Kozarich JW. On the origin of enzymatic species. Trends Biochem Sci. 1993;18:372–376. doi: 10.1016/0968-0004(93)90091-z. [DOI] [PubMed] [Google Scholar]
- Promponas VJ, Enright AJ, Tsoka S, Kreil DP, Leroy C, Hamodrakas S, Sander C, Ouzounis CA. CAST: An iterative algorithm for the complexity analysis of sequence tracts. Bioinformatics. 2000;16:915–922. doi: 10.1093/bioinformatics/16.10.915. [DOI] [PubMed] [Google Scholar]
- Riley M. Functions of the gene products of Escherichia coli. Microbiol Rev. 1993;57:862–952. doi: 10.1128/mr.57.4.862-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Tsoka S, Ouzounis CA. Prediction of protein interactions: Metabolic enzymes are frequently involved in gene fusion. Nat Genet. 2000;26:141–142. doi: 10.1038/79847. [DOI] [PubMed] [Google Scholar]
- Wilson CA, Kreychman J, Gerstein M. Assessing annotation transfer for genomics: quantifying the relations between protein sequence, structure and function through traditional and probabilistic scores. J Mol Biol. 2000;297:233–249. doi: 10.1006/jmbi.2000.3550. [DOI] [PubMed] [Google Scholar]