Abstract
Thioredoxin (Trx) and glutathione (GSH) systems are considered to be two major redox systems in animal cells. They are reduced by NADPH via Trx reductase (TR) or oxidized GSH (GSSG) reductase and further supply electrons for deoxyribonucleotide synthesis, antioxidant defense, and redox regulation of signal transduction, transcription, cell growth, and apoptosis. We cloned and characterized a pyridine nucleotide disulfide oxidoreductase, Trx and GSSG reductase (TGR), that exhibits specificity for both redox systems. This enzyme contains a selenocysteine residue encoded by the TGA codon. TGR can reduce Trx, GSSG, and a GSH-linked disulfide in in vitro assays. This unusual substrate specificity is achieved by an evolutionary conserved fusion of the TR and glutaredoxin domains. These observations, together with the biochemical probing and molecular modeling of the TGR structure, suggest a mechanism whereby the C-terminal selenotetrapeptide serves a role of a protein-linked GSSG and shuttles electrons from the disulfide center within the TR domain to either the glutaredoxin domain or Trx.
In animal cells, redox regulation of many cellular processes is provided by thioredoxin (Trx) and glutathione (GSH; oxidized form of GSH is GSSG) systems (1). NADPH supplies reducing equivalents for these redox systems via pyridine nucleotide disulfide oxidoreductases, which in turn shuttle electrons to downstream proteins and compounds through reversible thiol oxidoreduction. The Trx and GSH systems are involved in a variety of redox-dependent pathways such as providing reducing equivalents for ribonucleotide reductase (the first step in DNA biosynthesis) and peptide methionine sulfoxide reductase, an antioxidant defense and regulation of the cellular redox state (1–3). In addition, the Trx and GSH systems regulate activities of various transcription factors, kinases, and phosphatases, and they were implicated in the redox control of cell growth and death, transcription, cell signaling, and other processes (4, 5).
The animal Trx system is composed of (i) Trx reductase (TR), which is a homodimer of ≈56-kDa subunits and is a member of a pyridine nucleotide disulfide oxidoreductase family; (ii) Trx, which is a ≈12-kDa thiol/disulfide oxidoreductase; and (iii) Trx peroxidase composed of two ≈25-kDa subunits. At least two such Trx systems, cytosolic and mitochondrial, have been identified, and additional homologs of each of these proteins also have been described (6, 7).
The GSH system consists of (i) another member of the pyridine nucleotide disulfide oxidoreductase family, GSSG reductase (GR), a homodimer of ≈55-kDa subunits; (ii) a γ-Glu-Cys-Gly tripeptide, GSH; (iii) an ≈11-kDa thiol/disulfide oxidoreductase, glutaredoxin (Grx); and (iv) a GSH peroxidase (GPx). Although only one form of GR, GSH, and Grx are known in animal cells, five GPxs have been identified. GPxs are either ≈20-kDa monomers or homotetramers of ≈23-kDa subunits and have distinct properties with respect to substrate specificity and tissue and organelle expression (8).
It is noteworthy that among the components of the Trx and GSH systems, several enzymes contain selenocysteine (Sec) residues that are essential for enzyme function. Thus, TR1 (also known as cytosolic TR, TRα, and TrxR1) and TR3 (also known as mitochondrial TR, TRβ, and TrxR2) contain a C-terminal penultimate Sec residue (9–12), and four GPxs contain Sec in their N-terminal active centers (8). Sec residues are generally more reactive and characterized by lower redox potentials than Cys residues, and Sec residues are ionized at physiological pH, whereas Cys residues are typically protonated (10).
In addition to TR1 and TR3, we recently identified a partial cDNA sequence for human TR2 and isolated its mouse ortholog (12). We now describe cloning of the mouse cDNA for this protein (designated TGR for thioredoxin and glutathione reductase) and demonstrate that this enzyme can reduce several components of the Trx and GSH systems. This unusual substrate specificity of TGR is achieved by a natural fusion of TR and Grx domains, which are predicted to communicate through the movement of the C-terminal GSH-like Sec-containing tetrapeptide.
Experimental Procedures
Cloning of Mouse Testes TGR.
A mouse testis expressed sequence tag (EST) sequence (GenBank accession no. AA492975) was identified on the basis of its homology to mouse TRs. We determined the nucleotide sequence of this EST that revealed an incomplete ORF for a new protein. Using primers derived from this partial cDNA sequence, 5′-GTGAACGTAGGCTGTATTCCAA-3′ and 5′-GTTGACATAGGTCACGCCTTT-3′, a mouse testis cDNA library was screened by PCR, and identified clones were further analyzed with respect to insert size by agarose gel electrophoresis (Genome Systems, St. Louis). The longest cDNA identified by this procedure was characterized by nucleotide sequencing.
Isolation of TGR and TR1.
TGR was purified (≈3,000-fold) by using a modified procedure previously developed for the isolation of TR1 (11). Wild-type mouse testes (Pel-Freez Biologicals) (30 g) were homogenized in 5 vol of 25 mM Tris⋅HCl, pH 7.8, containing 1 mM EDTA, 1 mM PMSF, 5 μg/ml aprotinin, 5 μg/ml leupeptin, and 5 μg/ml pepstatin A. After centrifugation at 18,000 g for 40 min, the supernatant was applied onto a DEAE-Sepharose column equilibrated in 25 mM Tris⋅HCl, pH 7.8/1 mM EDTA (buffer A). A gradient of 0–0.5 M NaCl in buffer A was applied, and TGR was detected in fractions from the column by immunoblot assays with antibodies specific for a C-terminal 20-mer peptide of mouse TGR (12). The enzyme eluted at ≈200 mM NaCl, before TR1, which also was assayed by immunoblot assays (with antibodies specific for a C-terminal peptide of TR1; ref. 12) and eluted at ≈250 mM NaCl. Fractions containing TGR were diluted by 2.5-fold in 25 mM Tris⋅HCl, pH 7.5/1 mM EDTA (buffer B) and applied onto an ADP-Sepharose column equilibrated in buffer B. The column was sequentially washed with buffer B, 200 mM NaCl in buffer B, and the enzyme was eluted with 1 M NaCl in buffer B. The fractions containing TGR were made in 0.8 M ammonium sulfate and applied onto a phenyl-HPLC column (TosoHaas, Montgomeryville, PA) equilibrated in 0.8 M ammonium sulfate in buffer B. A gradient of 0.8 M ammonium sulfate in buffer B to buffer B was applied and TGR was eluted at ≈100 mM ammonium sulfate. Fractions containing TGR were collected, concentrated, aliquoted, and stored at −80°C before use. TR1 was isolated (≈2,000-fold) from wild-type mouse livers (Pel-Freez Biologicals) (100 g) according to a previously published procedure (11). TGR and TR1 were purified to homogeneity as assessed by immunoblot assays and Coomassie blue staining of SDS/PAGE gels.
Enzyme Assays.
TR activities of TR1 and TGR were assayed by two methods: (i) NADPH-dependent reduction of 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) determined as the increase in absorbance at 412 nm at 25°C (13) and (ii) NADPH-dependent reduction of 0.5 mg/ml insulin and 3 μM Escherichia coli Trx determined as the decrease in absorbance at 340 nm at 25°C (13). In the TR assays, substrate concentrations were 0.002–0.06 mM NADPH, 0.069–0.69 μM Trx, and 0.00125–0.425 mM DTNB. GSH reductase activity of TGR was assayed as NADPH-dependent reduction of GSSG determined as the decrease in absorbance at 340 nm at 25°C (14). To determine kinetic parameters, substrate concentrations varied from 2.5 to 500 μM GSSG and from 2 to 40 μM NADPH. Grx activity was determined by using 0.01–0.5 mM β-hydroxyethyl disulfide (HED) as a substrate (15), and the activity was assayed as the decrease of absorption at 340 nm at 25°C. Lineweaver–Burk plots were used to determine apparent Km and kcat values (16).
Isolation of Subcellular Fractions.
A 75Se-labeled mouse testis (0.2 g; wild type; 7 months old) was homogenized in 5 vol of ice-cold 0.25 M sucrose/5 mM Tris⋅HCl, pH 7.2/1 mM MgCl2/1 mM PMSF/5 μg/ml aprotinin/5 μg/ml leupeptin/5 μg/ml pepstatin A. The homogenized solution was separated in four steps as follows: (i) The solution was settled by gravity for 20 min to remove unbroken cells. (ii) The liquid fraction was removed and centrifuged at 1,000 g for 20 min. The pellet was considered as the nuclear fraction. (iii) The remaining supernatant was centrifuged at 10,000 g for 20 min. The pellet was considered as the mitochondrial fraction; and (iv) the remaining supernatant was centrifuged at 105,000 g for 2 h. The pellet was considered as the microsomal fraction and the supernatant as the cytosolic fraction. The nuclear, mitochondrial, and microsomal fractions were washed in the homogenization buffer and recentrifuged twice before use (17). Mouse tissues metabolically labeled with 75Se were obtained as described (18).
Gel Filtration.
Mouse testes were homogenized and centrifuged to remove nuclear and mitochondrial fractions as described above. The supernatant containing microsomal and cytosolic fractions (0.8 ml, 4 mg total protein) was applied onto a Superose 6 column (1 × 50 cm, ≈40 ml) that had been equilibrated in 5 mM Tris⋅HCl, pH 7.5/0.25 M sucrose/0.1 M NaCl/1 mM PMSF, followed by elution of proteins in the same solution. TGR was detected in the column fractions by using immunoblot assays. Kav was calculated as Kav = (Velution − Vvoid)/(Vtotal − Vvoid), where V is the volume, and compared with Kav of standards. The standards used were aldolase (158 kDa), catalase (232 kDa), ferritin (440 kDa), and thyroglobulin (669 kDa). Blue Dextran 2000 was used to determine the column void volume.
Immunoprecipitation.
The rat testes microsomal fraction was sonicated in the solution containing 50 mM Tris⋅HCl, 150 mM NaCl, and 1% IGEPAL CA-630, pH 7.4 (immunoprecipitation buffer). The sonicate was divided into five fractions each containing 500 μg of protein, which were supplemented with either 100 μM H2O2, 5 mM DTT, 5 mM NADPH, 5 mM GSH, or water. Each fraction was additionally supplemented with antibodies specific for TGR (4 μg), and the resulting solutions (1 ml each) were rotated for 1.5 h at 4°C. Subsequently, 100 μl of protein A Sepharose was added to each tube and the samples were rotated for an additional hour at 4°C. After a brief centrifugation, the supernatants were removed and concentrated by using Microcon (Millipore) microconcentrators. The pellets were washed four times with 1 ml of the ice-cold immunoprecipitation buffer and used as immunoprecipitate. Both types of samples were subjected to SDS/PAGE analysis under denaturing conditions followed by immunoblot detection with antibodies specific for TGR (19).
Molecular Modeling.
The sybyl 6.6 software package (Tripos Associates, St. Louis) was used to build a molecular model of the TGR homodimer. The homology modeling of the pyridine nucleotide disulfide oxidoreductase and Grx portions of TGR was performed separately by using the composer module of sybyl. The Grx domain was modeled starting with Gly-37. Probable conformations of the nine-residue peptide that linked the TR and Grx portions of TGR were determined by using methods of secondary structure prediction as implemented in sybyl. The conformations were energy-minimized and visually inspected to provide close proximity of the functionally important residues. The homodimer was built from the monomeric chains by superimposing the conservative residues of the active site in the model and the used crystal or NMR structures by using the fit procedure. Energy refinement (500 iterations of energy minimization) was performed with the Kollman all-atom force field and the Kollman charges. The distance-dependent dielectric model (with ɛ equal to 4r) was used for electrostatic energy calculations. The force field parameters and partial atomic charges for the Sec residue were as in ref. 45. The protable module of sybyl was used to assess the quality of the structural model. Approximately 99% of the modeled residues occupied conformationally allowed regions of the Ramachandran plot. Only three cis-peptide bonds were found in the model. Two of them, Val-92–Pro-93 and His-588–Pro-589, were adopted from the template molecules, 1JHB and 1FEC, respectively. The third cis-peptide bond, Gly-612–Cys-613, was necessary to ensure a low-energy conformation of selenosulfide bond formed by the Cys-613–Sec-614 pair in the oxidized state.
Results and Discussion
Cloning and Structural Features of a Mouse Testis TGR.
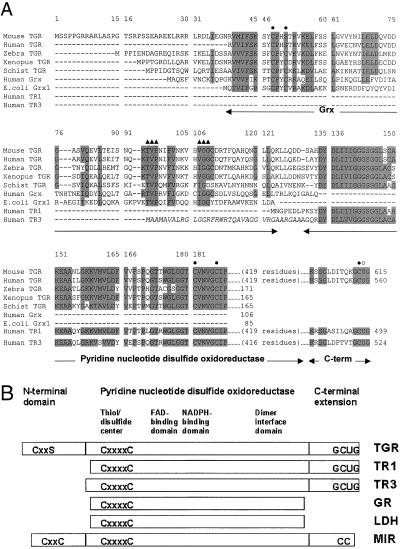
A partial cDNA sequence for human TR2 has been identified (12). We have now cloned mouse cDNA for this protein. The mouse cDNA contained an ORF of 615 aa and was designated TGR. The mouse TGR gene was organized in 16 exons and 15 introns and located on chromosome 6 (GenBank accession number of the bacterial artificial chromosome clone is AC051638) as evidenced by homology analyses using mouse TGR cDNA sequence against sequences generated by the Mouse Genome Project. Mouse TGR exhibited strong sequence homology to a partial sequence of human TGR (formerly TR2, 88% identity), human TR1 (73% identity), and human TR3 (56% identity) (Fig. 1A), and it showed a more distant homology (20–35% identity) to GR, lipoamide dehydrogenase (LDH), and mercuric ion reductase (MIR) from various sources. Like these homologous pyridine nucleotide disulfide oxidoreductases, TGR contained sequences characteristic of NADPH- and FAD-binding domains, a dimer interface domain, and a thiol/disulfide redox active center (Fig. 1B).
Figure 1.
Primary structures of TGR and homologous proteins. (A) Alignment of mouse TGR with partial sequences of human (AF171055, EST), Schistosoma japonicum (AI133811, EST), zebrafish (AW280285, AW778593, and AW153764, ESTs) and Xenopus (AW765014, EST) TGRs, and full-size sequences of human (S47472) and E. coli (P00277) Grxs, human TR1 (S66677), and human TR3 (NP_006431). Conserved residues are shaded. Active site Cys residues (the CxxC motif in the Grx domain, the disulfide center in the pyridine nucleotide disulfide oxidoreductase domain, and the Cys in the C-terminal extension) are shown by closed circles above sequences. The Sec residue (U) is indicated by an open circle above sequences. Residues involved in binding of GSH are shown by closed triangles above sequences. Grx, pyridine nucleotide disulfide oxidoreductase, and C-terminal portions of enzymes are shown by arrows below sequences. Mitochondrial signal peptide in TR3, cleaved in the mature protein, is shown in italics. Residue numbers within the alignment are shown above sequences and within individual sequences on the right. (B) Schematic representation of the domain organization of TGR, TR1, TR3, GR, LDH, and MIR. These six proteins possess active center disulfides (the CxxxxC motif; two cysteines separated by four other residues), FAD- and NADPH-binding domains, dimer interface domain, and other features of the pyridine nucleotide disulfide oxidoreductase family. TGR, TR1, and TR3 also contain an extension with a conserved C-terminal GCUG (U is Sec) tetrapeptide sequence. This tetrapeptide may be viewed as functionally analogous to two molecules of GSH (or to GSSG) in the GSH system. In addition, TGR contains an N-terminal Grx domain, which is structurally and functionally similar to thiol/disulfide oxidoreductases (Grx in the GSH system and Trx in the Trx system). These proteins are substrates for pyridine nucleotide disulfide oxidoreductases and contain an active site CxxC motif (two cysteines separated by two other residues). This motif is replaced by the CxxS motif in mouse and human TGR, but conserved in TGRs from lower eukaryotes. MIR also has a C-terminal extension that is 2 aa shorter than that in TRs and has a CC (two adjacent cysteines) motif. In addition, the majority of (but not all) known MIRs contain one or two copies of an N-terminal metal-binding domain. This domain has the CxxC motif but exhibits a different fold compared with Trx and Grx.
In addition, the TGR contained a C-terminal extension that terminated with Gly-Cys-Sec-Gly, a conserved tetrapeptide found in other animal TRs (Fig. 1 A and B). Sec was encoded by an in-frame TGA codon, whereas the downstream stop signal was TAG.
TGR also contained an N-terminal domain not present in other pyridine nucleotide disulfide oxidoreductases (Fig. 1 A and B). This domain exhibited strong homology to various Grxs and had a characteristic GSH-binding motif. The fusion of TR and Grx or any other two proteins or domains belonging to the Trx and GSH systems had not been previously described. Interestingly, the N-terminal domain of TGR had a single Cys in place of the CxxC redox motif that is involved in a reversible disulfide formation in glutaredoxins. However, a new eukaryotic Grx subfamily was recently characterized, whose members had a single Cys but maintained the Grx function (24). We also detected partial sequences of zebrafish, Xenopus, and the trematode Schistosoma mansoni TGRs, and these proteins contained the CxxC motif within the Grx domain. However, human TGR, like the mouse enzyme, had a single Cys (Fig. 1A). The occurrence of TGR in several nonmammalian eukaryotes suggested an evolutionary advantage of linking the TR and Grx domains.
TGR Exhibits Specificity for GSH and Trx Systems.
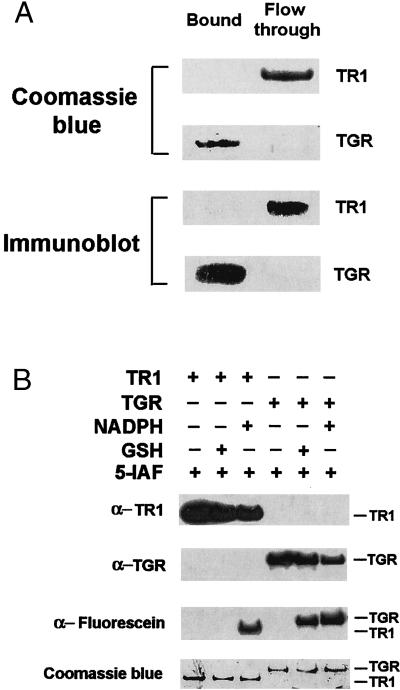
To determine whether TGR had a functional Grx domain, we first directly purified this enzyme to homogeneity from mouse testes and tested its binding to GSH-Sepharose, an affinity resin that had been used previously for isolation of GSH-binding proteins. As a control, we used mouse liver TR1 (11), which is the closest homolog of TGR, but lacks the N-terminal Grx domain. The binding studies demonstrated that TGR bound GSH-Sepharose, whereas TR1 did not (Fig. 2A). In addition, we tested relevant electron donors for their ability to reduce TR1 and TGR. Reduction of enzymes was monitored by alkylation with 5iodoacetamido-fluorescein, the reagent that can covalently modify reactive thiols and selenols (25) including the Sec residue in TR1 (12). The presence of fluorescein was then determined by immunoblot assays with antifluorescein antibodies. We found that NADPH reduced both TR1 and TGR. However, GSH only reduced TGR (Fig. 2B). Thus, TGR exhibited affinity for GSH.
Figure 2.
TGR exhibits affinity for GSH. (A) Binding of TGR to GSH. Purified mouse TR1 and TGR were incubated separately with GSH-Sepharose, after which bound and unbound (flow-through) fractions were analyzed by SDS/PAGE analysis followed by Coomassie blue staining of TR1 (Top) or TGR (second from Top) or subjected to immunoblot analysis with antibodies specific to either TR1 (third from Top) or to TGR (Bottom). (B) Reduction of TGR by GSH or NADPH. TGR or TR1 were incubated with either 100 μM NADPH or 500 μM GSH, alkylated with 20 μM 5-iodoacetamido-fluorescein (5-IAF), and detected by immunoblot analyses with antibodies specific to TR1 (Top), to TGR (second from Top), or to fluorescein (third from Top), or by Coomassie blue staining of the SDS/PAGE gel (Bottom). The positions of TR1 and TGR are shown on the right.
We further examined substrate specificity of TGR by characterizing its reactions with various components of the Trx and GSH systems and other substrates. Table 1 shows that TGR catalyzed NADPH-dependent reductions of DTNB and Trx, which are TR activities (13); NADPH-dependent reduction of GSSG, which is GR activity (14); and NADPH-dependent reduction of a mixed disulfide (β-mercaptoethanol-GSH mixed disulfide) obtained by the reaction of GSH with HED, which is Grx activity (15). In contrast, mouse liver TR1 was only active as TR. Purified mouse testis TGR had lower TR-specific activities than TR1, which could be due to the presence of significant amounts of truncated or inactive forms of TGR in our enzyme preparations. However, kinetic parameters (16) of TGR were similar to those of TR1 for the reduction of Trx (Table 1). Km and kcat of TGR for the reduction of GSSG and HED were somewhat lower than the previously published values for GR and Grx, but catalytic efficiencies of these enzymes could be compared with that of TGR (26–30). Thus, TGR could functionally replace TR, GR, and Grx in in vitro assays. In addition, the NADPH dependence of catalytic activities (Table 1) suggested that the TR and Grx domains of TGR communicated in a redox manner.
Table 1.
Kinetic parameters for TGR catalytic activities
| Substrates | TR1 | TGR | |
|---|---|---|---|
| Specific activity (μmol/min per mg) | NADPH + DTNB (TR activity) | 25 ± 3.0 | 3.1 ± 0.35 |
| NADPH + Trx (TR activity) | 5.9 ± 0.6 | 4.2 ± 0.6 | |
| NADPH + GSSG (GR activity) | ND | 2.0 ± 0.34 | |
| NADPH + HED (Grx activity) | ND | 1.9 ± 0.2 | |
| Apparent Km (μM) | DTNB | 212 | 14.7 |
| NADPH | 19.6 | 10.7 | |
| Trx | 4.73 | 3.0 | |
| GSSG | — | 8.84 | |
| HED | — | 45.2 | |
| Apparent kcat (min−1) | DTNB | 5,180 | 392 |
| Trx | 590 | 340 | |
| GSSG | — | 94 | |
| HED | — | 72 |
Apparent kinetic parameters were determined as described in Experimental Procedures using Lineweaver–Burk plots and varying concentrations of one substrate (2.5–500 μM GSSG, 10–500 μM HED, 0.069–0.69 μM Trx, or 1.25–425 μM DTNB) in a corresponding two-substrate reaction, while maintaining the concentration of a second substrate, NADPH, constant at 0.2 mM. Likewise, NADPH parameters were determined at 2–60 μM NADPH and 5 mM DTNB. ND, not detected.
The fusion of Trx and TR domains was previously reported for an enzyme from the bacterium Mycobacterium leprae (31), and a similar fusion occurs in Arabidopsis thaliana sequences (data not shown). The smaller plant, yeast, and bacterial TRs and larger animal TRs evolved by convergent evolution and independently acquired disulfide reduction function. The smaller enzymes lack the Sec redox center and exhibit a different reaction mechanism and three-dimensional structures compared with animal TRs and homologous pyridine nucleotide disulfide oxidoreductases (32–34). Although the fusion of the Trx and TR domains in bacteria and plants generated enzymes that are components of the same redox system (35), the fusion of the Grx and TR domains in TGR appeared to provide specificity for the GSH system while conserving specificity for the Trx system.
Mammalian TR1s exhibit broad substrate specificity (due to the presence of the highly reactive Sec) and reduce not only Trx and DTNB (Table 1), but also various unrelated redox proteins and compounds, including GPxs, protein disulfide isomerase, NK-lysin, selenite, Sec, vitamin K, alloxan, and other proteins and compounds (13). TR1s exhibit NADPH-dependent peroxidase activity and are capable of reducing H2O2, organic hydroperoxides, and lipid hydroperoxides. Sec is essential for reactions catalyzed by TR1 including peroxidase activities (36–38). In TGR, the presence of the Grx domain, in addition to the sequences highly homologous to TR1, appeared to increase a host of activities by adding those with specificity for the GSH system.
Biochemical Probing of TGR Structure.
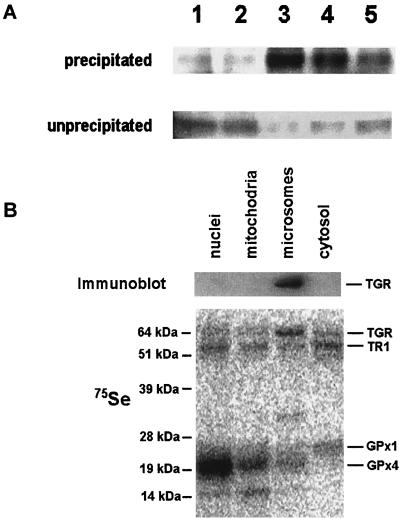
We used polyclonal antibodies specific for the C-terminal peptide of TGR (12) to immunoprecipitate TGR in its reduced and oxidized states from the testis microsomal fraction. Interestingly, TGR was most efficiently precipitated when reducing agents (DTT, NADPH, or GSH) were added to the incubation mixture, whereas the absence of redox agents or the presence of an oxidant H2O2 inhibited the immunoprecipitation reaction (Fig. 3A). These observations suggested that the reduction of TGR caused conformational changes involving the C-terminal peptide and that this peptide was accessible for other proteins when the enzyme was in a reduced state, but buried in the protein globule when the enzyme was oxidized.
Figure 3.
Characterization of TGR. (A) Immunoprecipitation. Microsomal fraction was incubated with purified rabbit polyclonal antibodies specific for the C-terminal peptide of TGR in the presence of 100 μM H2O2 (lane 1), water (lane 2), 5 mM DTT (lane 3), 5 mM NADPH (lane 4), or 5 mM GSH (lane 5). TGR was subsequently precipitated with Protein A-Sepharose, and the pellet (precipitated) and supernatant (unprecipitated) fractions were analyzed by immunoblot assays with antibodies specific for TGR. (B) Localization in subcellular fractions. Homogenate of 75Se-labeled mouse testes was fractionated by differential centrifugation. Nuclear (1,000-g pellet), mitochondrial (10,000-g pellet), microsomal (105,000-g pellet), and cytosolic (supernatant after centrifugation at 105,000 g) fractions were analyzed by immunoblot assays with antibodies specific for TGR (Upper) and by PhosphorImager analyses of 75Se on SDS/PAGE gels (Lower). In addition to the TGR location, the location of major 75Se-labeled selenoproteins, TR1, GPx1, and GPx4, are shown on the right. Masses of standards are shown on the left.
The antibodies specific for the C-terminal peptide also were used to determine the location of TGR in 75Se-labeled subcellular fractions of mouse testis obtained by differential centrifugation. TGR was found in the microsomal fraction (Fig. 3B), the location that distinguished the enzyme from TR1 and Grx, located in the cytosol (39); TR3, located in mitochondria (12, 40–43); and GR, located in both cytosolic and mitochondrial fractions (44). To determine whether the presence of TGR in the microsomal pellet was due to its participation in a large molecular weight complex, the microsomal and cytosolic testis fraction was subjected to chromatography on a calibrated gel-filtration column, followed by the detection of TGR in the column fractions by using immunoblot assays. The calculated molecular mass of the native TGR was ≈130 kDa. Thus, TGR was not a component of a large molecular weight complex, and its experimental mass was consistent with homodimer composition of the enzyme, which is typical of pyridine nucleotide disulfide oxidoreductases.
Molecular Modeling of TGR Structure.
Although three-dimensional structures for animal TRs are not known, structures and reaction mechanisms have been extensively characterized for homologous pyridine nucleotide disulfide oxidoreductases. In these enzymes, the N-terminal thiol/disulfide active center is a key player in the reaction mechanism. During the catalysis, the disulfide in the oxidized enzyme is reduced with electrons provided by NADPH (via enzyme-bound FAD) and further serves as a reductant for specific substrates. Glutaredoxins also have been extensively characterized both structurally and mechanistically (1).
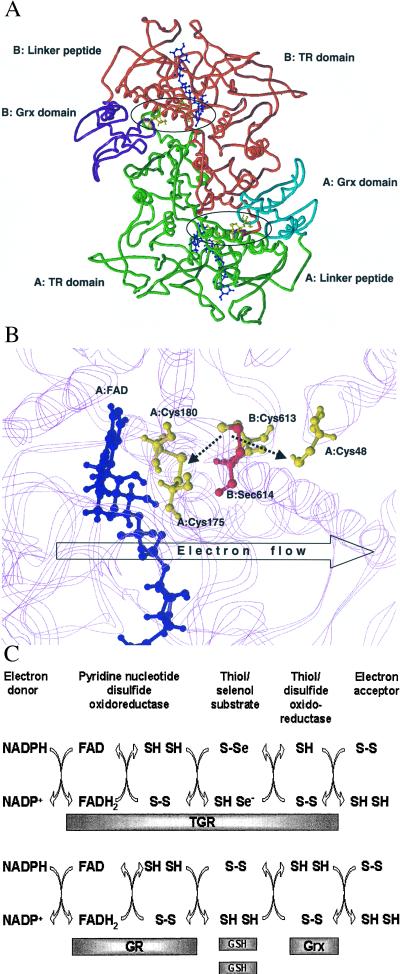
We modeled the pyridine nucleotide disulfide oxidoreductase portion of the TGR homodimer (Fig. 4A) on the basis of crystal structures of three template proteins: GR from Escherichia coli (34.5% identity to TGR; Protein Databank code 1GES), trypanothione reductase from Crithidia fasciculata (34.3% identity; Protein Databank code 1FEC) and MIR from Bacillus sp. strain RC607 (28.4% identity; atomic coordinates kindly provided by Emil Pai, University of Toronto) (45). MIR has a C-terminal extension containing a conserved Cys-Cys dipeptide that delivers Hg2+ to the thiol/disulfide center (46, 47). This extension is homologous to the C-terminal extension of TGR, and the Cys-Cys dipeptide in MIR may be considered as a functional analog of the Cys-Sec pair in TRs (48). Hence, the MIR structure was used to build the C-terminal extension of TGR.
Figure 4.
Molecular modeling and reaction mechanism of TGR. (A) Molecular model of the TGR homodimer. The three-dimensional model was obtained as described under Experimental Procedures. The pyridine nucleotide disulfide oxidoreductase portion and the C-terminal extension (TR domain) of subunit A are shown in green and of subunit B in red. The Grx domain of subunit A is shown in cyan and of subunit B in purple. The location of the TR and Grx domains and the peptide that links these domains (linker peptide) are indicated for both subunits. Active centers are circled. Two FAD molecules are shown in blue, and Cys-48, Cys-175, Cys-180, Cys-613, and Sec-614 in both subunits are shown in yellow. (B) Molecular model of the enzyme active center. FAD molecule is shown in blue. Cys-48, Cys-175, and Cys-180 in subunit A and Cys-613 in subunit B are shown in yellow. Sec-614 in subunit B is shown in red. The location of protein chains that surround the active center are shown in the background in magenta. The Cys-613/Sec-614 and Cys-175/Cys-180 pairs are shown in the oxidized state. Predicted interactions of the Cys-613/Sec-614 dipeptide with the disulfide center (Cys-175/Cys-180) and the active center of the Grx domain (Cys-48) are shown by dashed arrows. In the initial step of catalysis by TGR, NADPH reduces FAD. The predicted direction of electron flow in subsequent steps of catalysis (A: FAD→A: Cys-175/Cys-180→B: Cys-613/Sec-614→A: Cys-48) is shown by the open arrow. (C) Reaction mechanism of TGR. Proposed electron flow involving TGR (Upper) and, for comparison, components of the GSH system (Lower) is shown. TGR is functionally and structurally analogous to a fusion of GR, Grx, and two molecules of GSH and supports five electron transfer events. The roles of redox groups are as follows: the thiol/disulfide redox center initially accepts electrons from NADPH via FAD. In the GSH system, GSSG is then directly reduced by the thiol/disulfide center, whereas in TGR, the C-terminal tetrapeptide serves a role of a protein-linked GSSG and is reduced by the thiol/disulfide center. Electrons are further transferred from GSH to Grx in the GSH system or by a conformational movement of the C-terminal tetrapeptide of TGR from the thiol/disulfide center to the Grx domain. Finally, Grx or the Grx domain interact with an electron acceptor (for example, GSH-mixed disulfide), completing the reaction.
The Grx domain of the TGR molecule was separately modeled by using the NMR-derived structures of human and E. coli Grxs (34.4% identity, Protein Databank code 1JHB; and 36.6% identity, Protein Databank code 3GRX, respectively). The two modeled parts of TGR were linked by a nine-residue peptide.
Analyses of these molecular models (Fig. 4 A and B) revealed predicted structural features that helped in characterizing the reaction mechanism of TGR. First, the thiol/disulfide active center in TGR was located deep in the protein globule and linked to a protein surface through a narrow and long channel, similar to that found in other pyridine nucleotide disulfide oxidoreductases. The thiol/disulfide center did not appear to be accessible for protein substrates (e.g., Trx or the Grx domain) in the TGR model. This is not surprising considering that substrates for homologous pyridine nucleotide disulfide oxidoreductases are small compounds (e.g., GSSG, oxidized lipoamide) whereas Trx and the Grx domain are larger globular structures.
Second, the C-terminal peptide in the TGR model was buried inside the oxidized enzyme with Sec being in the vicinity of the disulfide center. This observation was consistent with our finding that the C-terminal extension of TGR was not accessible in the oxidized state (Fig. 3A). The structure of the C-terminal extension was analogous to the lipoamide arm that interacts with the thiol/disulfide center of LDH within the pyruvate dehydrogenase multienzyme complex (21). Our model (Fig. 4B) revealed that the C-terminal extension of TGR perfectly fits into the channel such that the Cys-Sec motif of one subunit could interact directly with the thiol/disulfide center of another subunit in the TGR homodimer. Whereas the C-terminal extension of MIR is thought to interact with the disulfide center through Hg2+ (46, 47), the C-terminal extension of TGR was 2 aa longer and could make a direct contact with the thiol/disulfide active center.
Third, the predicted binding site for the Grx domain was formed by both subunits of the TGR homodimer (Trx presumably binds in the same place) (Fig. 4A). The nine-residue linker peptide between the Grx domain and the major portion of TGR may ensure the access of Trx in place of the Grx domain. The pullout mechanism for the release of the C-terminal extension from the channel after its reduction by the thiol/disulfide center appeared unlikely. In contrast, the C-terminal extension could reach the surface of the TGR model by a pendulum-like movement within the channel such that the Cys-Sec motif could directly interact with the active site Cys of the Grx domain (Fig. 4B) or be accessible for other targets (e.g., Trx) and such that the C-terminal extension could become accessible for antibodies specific for the C-terminal peptide.
Reaction Mechanism of TGR.
The organization of the TGR homodimer model (Fig. 4A) was such that the Cys-Sec motif of one subunit could transfer electrons from the thiol/disulfide center to the Grx domain of the second subunit and vice versa. The conserved C-terminal Gly-Cys-Sec-Gly motif may be considered as an analog of small disulfide substrates (e.g., GSSG for GR or lipoate for LDH), which is linked to a pyridine nucleotide disulfide oxidoreductase portion of the protein. In other words, TGR may be viewed as a fusion of three components, which correspond to Grx, GR, and GSSG in the GSH system (Figs. 1B and 4C).
These observations suggested a mechanism whereby electrons were transferred from NADPH to downstream electron donors through several redox centers within TGR, NADPH → FAD → thiol/disulfide center → the C-terminal GSSG-like Sec-containing center → the Cys residue within the Grx domain → downstream substrate (Fig. 4 B and C). The key component in this remarkable electron flow was the Gly-Cys-Sec-Gly tetrapeptide that transfers electrons between redox centers through reversible oxidoreduction coupled with conformational changes.
The proposed mechanism is consistent with previous models (9, 20, 22, 23, 34), which proposed, on the basis of kinetics, site-specific mutations and protein microchemistry studies, that the C-terminal redox motif in mammalian TR1 and TR from Plasmodium falciparum can be directly reduced by the N-terminal thiol/disulfide center. Similarity is also evident to LDH, whose substrate, lipoic acid, contains an internal disulfide that transfers electrons and substrate molecules among three different active centers located within three polypeptides (21). Lipoic acid is fused to a protein component of the pyruvate dehydrogenase complex via a long hydrophobic arm that delivers this compound to various reaction centers.
Acknowledgments
We thank Dr. Emil Pai for providing MIR coordinates and Dr. Gautam Sarath for peptide synthesis. This work was supported by National Institutes of Health Grant GM61603 (to V.N.G.) and University of Nebraska Medical Center Cancer Center Grant P30 CA36727 (to S.S.).
Abbreviations
- Trx
thioredoxin
- GSH
glutathione
- GSSG
oxidized GSH
- TR
Trx reductase
- TGR
Trx and GSSG reductase
- GR
GSSG reductase
- Grx
glutaredoxin
- GPx
GSH peroxidase
- Sec
selenocysteine
- EST
expressed sequence tag
- DTNB
5,5′-dithiobis(2-nitrobenzoic acid)
- HED
β-hydroxyethyl disulfide
- LDH
lipoamide dehydrogenase
- MIR
mercuric ion reductase
- GPx4
phospholipid hydroperoxide GSH peroxidase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF349659).
References
- 1.Holmgren A, Arnér E S J, Åslund F, Björnstedt M, Zhong L, Ljung J, Nakamura H, Nikitovic D. In: Oxidative Stress, Cancer, AIDS, and Neurodegenerative Diseases. Montagnier L, Olivier R, Pasquier C, editors. New York: Dekker; 1998. pp. 229–246. [Google Scholar]
- 2.Halliwell B. Free Radical Res. 1999;31:261–272. doi: 10.1080/10715769900300841. [DOI] [PubMed] [Google Scholar]
- 3.Sies H. Free Radical Biol Med. 1999;27:916–921. doi: 10.1016/s0891-5849(99)00177-x. [DOI] [PubMed] [Google Scholar]
- 4.Rhee S G. Exp Mol Med. 1999;31:53–59. doi: 10.1038/emm.1999.9. [DOI] [PubMed] [Google Scholar]
- 5.Finkel T. FEBS Lett. 2000;476:52–54. doi: 10.1016/s0014-5793(00)01669-0. [DOI] [PubMed] [Google Scholar]
- 6.Pedrajas J R, Kosmidou E, Miranda-Vizuete A, Gustafsson J A, Wright A P, Spyrou G. J Biol Chem. 1999;274:6366–6373. doi: 10.1074/jbc.274.10.6366. [DOI] [PubMed] [Google Scholar]
- 7.Seo M S, Kang S W, Kim K, Baines I C, Lee T H, Rhee S G. J Biol Chem. 2000;275:20346–20354. doi: 10.1074/jbc.M001943200. [DOI] [PubMed] [Google Scholar]
- 8.Brigelius-Flohe R. Free Radical Biol Med. 1999;27:951–965. doi: 10.1016/s0891-5849(99)00173-2. [DOI] [PubMed] [Google Scholar]
- 9.Mustacich D, Powis G. Biochem J. 2000;346:1–8. [PMC free article] [PubMed] [Google Scholar]
- 10.Stadtman T C. Annu Rev Biochem. 1996;65:83–100. doi: 10.1146/annurev.bi.65.070196.000503. [DOI] [PubMed] [Google Scholar]
- 11.Gladyshev V N, Jeang K-T, Stadtman T C. Proc Natl Acad Sci USA. 1996;93:6146–6151. doi: 10.1073/pnas.93.12.6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Q-A, Wu Y, Zappacosta F, Jeang K T, Lee B J, Hatfield D L, Gladyshev V N. J Biol Chem. 1999;274:24522–24530. doi: 10.1074/jbc.274.35.24522. [DOI] [PubMed] [Google Scholar]
- 13.Arner E S J, Zhong L, Holmgren A. Methods Enzymol. 1999;300:226–239. doi: 10.1016/s0076-6879(99)00129-9. [DOI] [PubMed] [Google Scholar]
- 14.Smith I K, Vierheller T L, Thorne C A. Anal Biochem. 1988;175:408–413. doi: 10.1016/0003-2697(88)90564-7. [DOI] [PubMed] [Google Scholar]
- 15.Holmgren A, Aslund F. Methods Enzymol. 1995;252:283–292. doi: 10.1016/0076-6879(95)52031-7. [DOI] [PubMed] [Google Scholar]
- 16.Rudolph F B, Fromm H J. Methods Enzymol. 1979;63:138–159. doi: 10.1016/0076-6879(79)63009-4. [DOI] [PubMed] [Google Scholar]
- 17.Ozols J. Methods Enzymol. 1990;182:225–235. doi: 10.1016/0076-6879(90)82019-x. [DOI] [PubMed] [Google Scholar]
- 18.Gladyshev V N, Factor V M, Housseau F, Hatfield D L. Biochem Biophys Res Commun. 1998;251:488–493. doi: 10.1006/bbrc.1998.9495. [DOI] [PubMed] [Google Scholar]
- 19.Kessler S W. Methods Enzymol. 1981;73:442–459. doi: 10.1016/0076-6879(81)73084-2. [DOI] [PubMed] [Google Scholar]
- 20.Gromer S, Wissing J, Behne D, Ashman K, Schirmer R H, Flohe L, Becker K. Biochem J. 1998;332:591–592. doi: 10.1042/bj3320591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voet D, Voet J G. Biochemistry. New York: Wiley; 1995. pp. 541–548. [Google Scholar]
- 22.Wang P F, Arscott L D, Gilberger T W, Muller S, Williams C H., Jr Biochemistry. 1999;38:3187–3196. doi: 10.1021/bi982674g. [DOI] [PubMed] [Google Scholar]
- 23.Zhong L, Arner E S, Holmgren A. Proc Natl Acad Sci USA. 2000;97:5854–5859. doi: 10.1073/pnas.100114897. . (First Published May 9, 2000; 10.1073/pnas.100114897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez-Manzaneque M T, Ros J, Cabiscol E, Sorribas A, Herrero E. Mol Cell Biol. 1999;19:8180–8190. doi: 10.1128/mcb.19.12.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Y, Kwon K S, Rhee S G. FEBS Lett. 1998;440:111–115. doi: 10.1016/s0014-5793(98)01415-x. [DOI] [PubMed] [Google Scholar]
- 26.Carlberg I, Mannervik B. Methods Enzymol. 1985;113:484–490. doi: 10.1016/s0076-6879(85)13062-4. [DOI] [PubMed] [Google Scholar]
- 27.Libreros-Minotta C A, Pardo J P, Mendoza-Hernandez G, Rendon J L. Arch Biochem Biophys. 1992;298:247–253. doi: 10.1016/0003-9861(92)90119-h. [DOI] [PubMed] [Google Scholar]
- 28.Mannervik B, Axelsson K, Larson K. Methods Enzymol. 1981;77:281–285. doi: 10.1016/s0076-6879(81)77038-1. [DOI] [PubMed] [Google Scholar]
- 29.Mieyal J J, Starke D W, Gravina S A, Hocevar B A. Biochemistry. 1991;30:8883–88891. doi: 10.1021/bi00100a023. [DOI] [PubMed] [Google Scholar]
- 30.Luthman M, Holmgren A. J Biol Chem. 1982;257:6686–6690. [PubMed] [Google Scholar]
- 31.Wieles B, van Agterveld M, Janson A, Clark-Curtiss J, Rinke de Wit T, Harboe M, Thole J. Mol Microbiol. 1995;16:921–929. [Google Scholar]
- 32.Kuriyan J, Krishna T S, Wong L, Guenther B, Pahler A, Williams C H, Jr, Model P. Nature (London) 1991;352:172–174. doi: 10.1038/352172a0. [DOI] [PubMed] [Google Scholar]
- 33.Williams CH., Jr FASEB J. 1995;9:1267–1276. doi: 10.1096/fasebj.9.13.7557016. [DOI] [PubMed] [Google Scholar]
- 34.Arscott L C, Gromer S, Schirmer R H, Becker K, Williams C H., Jr Proc Natl Acad Sci USA. 1997;94:3621–3626. doi: 10.1073/pnas.94.8.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang P F, Marcinkeviciene J, Williams C H, Jr, Blanchard J S. Biochemistry. 1998;37:16378–16389. doi: 10.1021/bi980754e. [DOI] [PubMed] [Google Scholar]
- 36.Gasdaska J R, Harney J W, Gasdaska P Y, Powis G, Berry M J. J Biol Chem. 1999;274:25379–25385. doi: 10.1074/jbc.274.36.25379. [DOI] [PubMed] [Google Scholar]
- 37.Lee S R, Bar-Noy S, Kwon J, Levine R L, Stadtman T C, Rhee S G. Proc Natl Acad Sci USA. 2000;97:2521–2526. doi: 10.1073/pnas.050579797. . (First Published February 25, 2000; 10.1073/pnas.050579797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhong L, Holmgren A. J Biol Chem. 2000;275:18121–18128. doi: 10.1074/jbc.M000690200. [DOI] [PubMed] [Google Scholar]
- 39.Rozell B, Holmgren A, Hansson H A. Eur J Cell Biol. 1988;46:470–477. [PubMed] [Google Scholar]
- 40.Gasdaska P Y, Berggren M M, Berry M J, Powis G. FEBS Lett. 1999;442:105–111. doi: 10.1016/s0014-5793(98)01638-x. [DOI] [PubMed] [Google Scholar]
- 41.Lee S R, Kim J R, Kwon K S, Yoon H W, Levine R L, Ginsburg A, Rhee S G. J Biol Chem. 1999;274:4722–4734. doi: 10.1074/jbc.274.8.4722. [DOI] [PubMed] [Google Scholar]
- 42.Miranda-Vizuete A, Damdimopoulos A E, Pedrajas J R, Gustafsson J A, Spyrou G. Eur J Biochem. 1999;261:405–412. doi: 10.1046/j.1432-1327.1999.00286.x. [DOI] [PubMed] [Google Scholar]
- 43.Watabe S, Makino Y, Ogawa K, Hiroi T, Yamamoto Y, Takahashi S Y. Eur J Biochem. 1999;264:74–84. doi: 10.1046/j.1432-1327.1999.00578.x. [DOI] [PubMed] [Google Scholar]
- 44.Kelner M J, Montoya M A. Biochem Biophys Res Commun. 2000;269:366–368. doi: 10.1006/bbrc.2000.2267. [DOI] [PubMed] [Google Scholar]
- 45.Zhao L, Cox A G, Ruzicka J A, Bhat A A, Zhang W, Taylor E W. Proc Natl Acad Sci USA. 2000;97:6356–6361. doi: 10.1073/pnas.97.12.6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schiering N, Kabsch W, Moore M J, Distefano M D, Walsh C T, Pai E F. Nature (London) 1991;352:168–172. doi: 10.1038/352168a0. [DOI] [PubMed] [Google Scholar]
- 47.Engst S, Miller S M. Biochemistry. 1999;38:3519–3529. doi: 10.1021/bi982680c. [DOI] [PubMed] [Google Scholar]