Abstract
The bacterium Escherichia coli is used as a model cellular system to test and validate a new technology called Phenotype MicroArrays (PMs). PM technology is a high-throughput technology for simultaneous testing of a large number of cellular phenotypes. It consists of preconfigured well arrays in which each well tests a different cellular phenotype and an automated instrument that continuously monitors and records the response of the cells in all wells of the arrays. For example, nearly 700 phenotypes of E. coli can be assayed by merely pipetting a cell suspension into seven microplate arrays. PMs can be used to directly assay the effects of genetic changes on cells, especially gene knock-outs. Here, we provide data on phenotypic analysis of six strains and show that we can detect expected phenotypes as well as, in some cases, unexpected phenotypes.
Technologies that can provide a cell-wide perspective are very useful. Important “global” technologies using two-dimensional methods were pioneered by O'Farrell and coworkers (O'Farrell 1975) for protein analysis (proteomics), and by Fodor and coworkers (Fodor et al. 1993) for nucleic acid analysis (genomics). These technologies and subsequent refinements allow for global analysis of the important macromolecules of cells that convey the information flow from DNA to RNA to protein. However, the information initially encoded in the genome is ultimately displayed at the cellular level as cellular traits or phenotypes. This paper describes a new technology called Phenotype MicroArrays (PM) that provides an analogous two-dimensional array technology for analysis of live cells (phenomics) to measure hundreds or thousands of cellular properties simultaneously.
A technology for global analysis of cellular phenotypes was first proposed by Bochner (1989b) using microplates for high-throughput assays. Two groups working in genomics of Saccharomyces cerevisiae as a model system recently have tested a large number of strains against 96 (Ross-Macdonald et al. 1999) or 288 (Reiger et al. 1997, 1999) growth phenotypes. These groups used microplate technology to test the growth of yeast strains on the surface of agar. A problem with this approach is that it is difficult to scale it efficiently for high-throughput testing. Agar media have a short shelf life and must be prepared freshly. Scoring of growth is rather subjectively and inefficiently performed with daily visual or photographic records. Estimation of apparent cell mass on a surface is difficult and can be misleading. For example, cellular changes can cause colonies to spread or secrete capsular polysaccharide, thereby appearing much larger than the actual cell number. An ideal high-throughput system would allow for automated, kinetic reading and storing of quantitative phenotypic data directly into computer databases amenable to bioinformatics analyses.
At Biolog, we have employed testing of cellular phenotypes using cell respiration as a reporter system since 1984 (Bochner 1988, 1989a,b). The assay chemistry uses a tetrazolium dye, usually tetrazolium violet, to colorimetrically detect the respiration of cells. Reduction of this dye results in formation of a purple color and because the dye reduction is essentially irreversible under physiological conditions, it accumulates in the well over a period of hours, amplifying the signal and integrating the amount of breathing over time. This provides several major benefits: (1) The color change is easy to monitor; (2) the color change is easy to quantitate; (3) the color change is very sensitive and highly reproducible; and (4) cell respiration can occur independent of cell growth and, in some cases, can measure phenotypes that do not lead to growth.
As part of this technology, the OmniLog instrument has been developed for the purpose of reading and recording the color change in PM assays. The instrument cycles microplates in front of a color CCD camera to read 50 in as little as 5 min and provides quantitative and kinetic information about the response of cells in the PMs. Data are stored directly into computer files and can be recalled and compared with other data at any time.
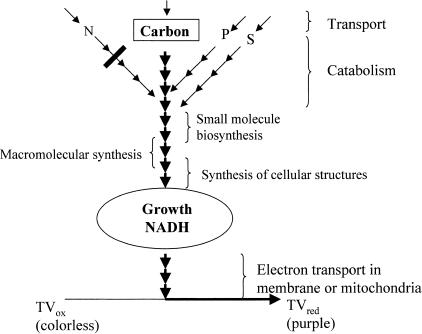
Figure 1 shows how cell respiration can be coupled to a large number and a wide range of cellular phenotypes. In a normal growth situation a coordinated sequence of events must occur. Cells must transport nutrients, catabolize and reform them, produce essential small molecule components, polymerize these into macromolecules, create and assemble subcellular structures, etc. If all of these processes are working normally, the cell can grow and there will be an actual physical flow of electrons from the carbon source to NADH, down the electron transport chain of the cell, and, ultimately, onto the tetrazolium dye to produce the purple color. If one of these processes is working at a subnormal rate it will become a pinchpoint, restricting this flow and resulting in a decrease in purple color. The severity of the restriction is reflected in the degree of loss of purple color. Total loss of function will result in no growth and no purple color. Therefore, colorimetric assay of respiration can provide a virtually universal reporter system for phenomic testing.
Figure 1.
Respiration pathways coupled to cell physiology.
About half of the genes from genomic sequencing efforts have no ascribed function, and even genes with ascribed functions are based primarily on DNA sequence homology, with little or no direct experimental data. Large-scale gene knock-out projects for S. cerevisiae (Burns et al. 1994) have been essentially completed and other projects from Bacillus subtilis (Vagner et al. 1998) to E. coli (Link et al. 1997; Datsenko and Wanner 2000; F. Blattner, pers. comm.) to mouse (Zambrowicz et al. 1998) are at various stages. We developed the PM technology anticipating that it can be used to analyze the effects of loss of gene function, providing a direct experimental linkage between genotype and phenotype. The idea is simply to compare isogenic pairs of strains in PMs to directly assay for the cellular effects of loss of gene function.
RESULTS
First Demonstration of PM Technology and Discovery of Undetected, Accidental Genetic Changes in Cells
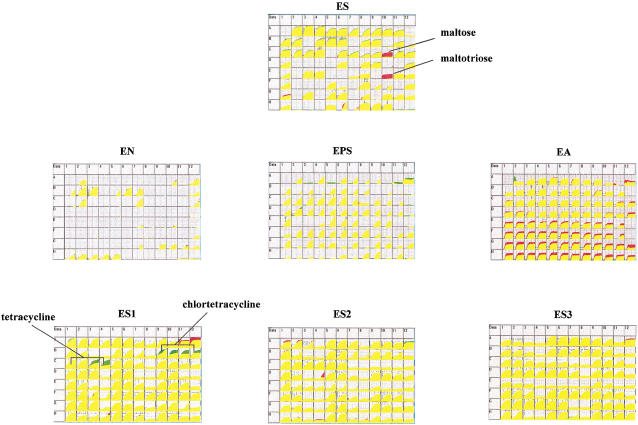
In the first test of the technology we used a PM that measures the carbon catabolic functions of E. coli. We obtained the E. coli strain that was genomically sequenced, MG1655 (Blattner et al. 1997), from Blattner's laboratory, along with a strain (fb753) in which an amber mutation was inserted to inactivate a xylose catabolic gene (xylA). The results of the PM assay of the parental and derived strains are shown in Figure 2. To our surprise, the derived strain had lost metabolism of maltose and maltotriose (circled in red) in addition to xylose (circled in green). The maltose-negative phenotype, which was confirmed in Blattner's laboratory, was due to a second genetic change in the cell line, and the occurrence of the genetic change was traced to a step in the genetic construction where a batch of electrocompetent cells was prepared.
Figure 2.
(a, b) PM analysis of xylA amber mutant strain. (c, d) PM analysis of ynjB::tn10 mutant strain.
A second example of an undetected genetic change occurred in our laboratory several months later. An E. coli strain with only one known lesion in a gene of unknown function (ynjB::tn10, formerly zdj-276::tn10) (Nichols et al. 1998) was examined. The ynjB lesion was moved into a clean genetic background (MG1655) by P1 transduction, selecting for tetracycline resistance. The transductant, strain EP014, was then tested in a series of PMs and found to have a specific defect in nitrogen catabolism. It lost the ability to use L-arginine and L-ornithine as nitrogen sources as shown in Figure 2, c and d.
Closer inspection, however, revealed that this phenotype was due to a mutation close to, but not in, the ynjB gene. The phenotype was ∼5% separable from ynjB::tn10 by P1 transduction (5 of 100 tetracycline-resistant transductants could use L-arginine and L-ornithine as nitrogen sources) and is almost certainly in the astC gene, a recently described neighboring gene coding for the enzyme succinylornithine transaminase (Schneider et al. 1998). Our conclusion is supported by the following evidence. Loss of catabolism of L-arginine and L-ornithine could be due to loss of shared transport functions (Kustu et al. 1979; Wissenbach et al. 1995) or loss of shared catabolic enzymes. We tested the mutant and parental strains for growth in liquid minimal medium and confirmed that the mutant was defective in using L-arginyl-L-arginine as well as L-arginine as nitrogen sources with pyruvate as a carbon source. This confirms that the defect is in a catabolic function, as the arginyl peptide is transported by an oligopeptide system. The astC gene codes for the only enzyme in the AST pathway affecting L-ornithine utilization (Schneider et al. 1998).
Testing Gene Knockouts with Previously Documented Phenotypes
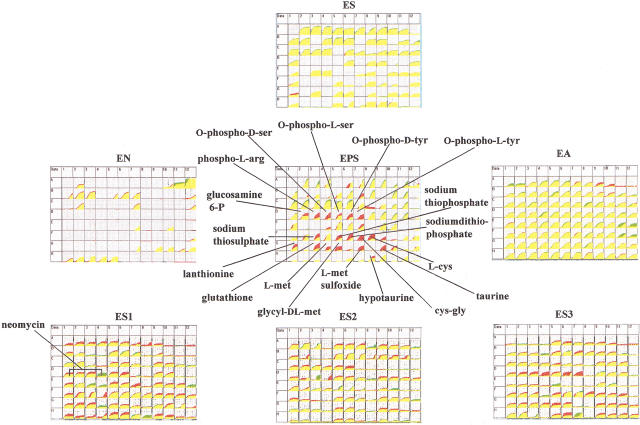
To further validate PM technology, we wanted to test a diverse set of known mutants to see whether it could accurately measure their phenotypes. At that time we had expanded the set of PMs to seven including one measuring carbon catabolism (ES), one measuring nitrogen catabolism (EN), one measuring phosphorus and sulfur catabolism (EPS), one measuring growth stimulation by nutrients (EA, for assay of auxotrophy), and three measuring alterations in sensitivity or resistance to a wide range of toxic chemicals (ES1, ES2, ES3) as probes to various cellular pathways and physiologies. The specific tests are listed in the Methods section. Mutant strains were constructed by P1 transduction of the mutation into MG1655 using a linked drug resistance marker (Table 1) and were tested as isogenic pairs in the OmniLog instrument. The OmniLog records a 96-box chart corresponding to the 96 wells in the PM for each strain and for each PM. The cellular response is recorded showing the amount of purple color in each well (vertical axis) throughout the time course (horizontal axis, usually 24–48 h) of the assay. The mutant strain is recorded as a green tracing and the isogenic MG1655 control strain is recorded as a red tracing. To look for differences, these tracings are graphically overlaid and areas of overlap of green and red are colored yellow (Figure 3). For most phenotypes, the mutant and parental strain give virtually identical phenotypes resulting in yellow color. If the mutant is respiring more slowly in one well its corresponding box in the chart will be red, whereas if the mutant is respiring more rapidly the color will be green.
Table 1.
Strain List
| Strain | Genotype and description | Source |
|---|---|---|
| MG1655 (FB426) | Wild-type Escherichia coli | Jeremy Glasner, Fred Blattner |
| fb753 | MG1655 xylA amber | Jeremy Glasner, Fred Blattner |
| fb757 | MG1655 phoP amber | Jeremy Glasner, Fred Blattner |
| CAG12164 | MG1655 malF3089∷tn10 | Nancy McFarland |
| SP850 (CGSC #7200) | CA8000 ΔcyaA1400∷kan | E. coli genetic stock center |
| MG1655 fruR∷tn5 | Milton Saier | |
| CAG18464 | MG1655 ynjB276∷tn10 | Ken Rudd |
| EP005 | MG1655 malF3089∷tn10 | This study |
| EP007 | MG1655 ΔcyaA1400∷kan | This study |
| EP012 | MG1655 fruR∷tn5 | This study |
| EP014 | MG1655 ynjB276∷tn10 | This study |
Figure 3.
PM analysis of malF::tn10 mutant strain.
First we performed a phenotypic analysis of a strain expected to have a limited and specific defect in carbon metabolism. Strain EP005 has a Tn10 insertion in malF, a gene coding for a maltose transport protein. The result is shown in Figure 3. As expected the only phenotypes observed in this strain were no growth on maltose and maltotriose (red color) and better growth in the presence of tetracycline and chlortetracycline (green color) because of the Tn10 drug resistance element. No other phenotypic changes were detected.
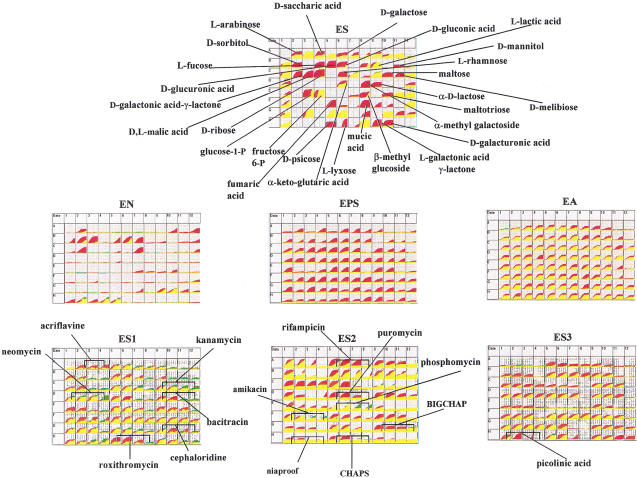
Next we examined knock-outs of some global regulatory genes involved in carbon metabolism. The results obtained with the cya and fruR gene knockouts (EP007 and EP012) are shown in Figures 4 and 5. The cya gene is the structural gene for adenylate cyclase and this mutant cannot make cAMP. Cyclic 3′,5′ AMP is a signal molecule that must bind to the CRP protein to induce expression of genes for secondary carbon sources used by E. coli (Pastan and Adhya 1976). As expected, strain EP007 has defects in using many secondary carbon sources as seen by all of the red areas in the ES carbon array. It also shows the expected resistance to kanamycin and neomycin because of the presence of the kan cassette used to disrupt the cya gene. Because the strain does not grow on pyruvate, it gave all negative phenotypes in the EN, EPS, and EA arrays, which use a pyruvate minimal medium. The cya strain also exhibits numerous phenotypes with regard to sensitivity to toxic chemicals. This is also expected based on previous reports from Ames and coworkers who have shown that cAMP regulates many cellular transport functions and that cya mutants have altered sensitivity to metabolite analogs and antibiotics (Alper and Ames 1978). The drug phosphomycin has been studied most intensively. It is taken up by two independent transport systems in E. coli and Salmonella typhimurium for α-glycerolphosphate and hexose phosphate. These sugar phosphates are secondary carbon sources and their uptake is regulated by cAMP. Cells unable to make cAMP are therefore unable to elevate these transport functions, making them relatively resistant to phosphomycin. Figure 4 shows clearly that the anticipated phosphomycin resistance phenotype is present in EP007.
Figure 4.
PM analysis of cya::kan mutant strain.
Figure 5.
PM analysis of fruR::tn5 mutant strain.
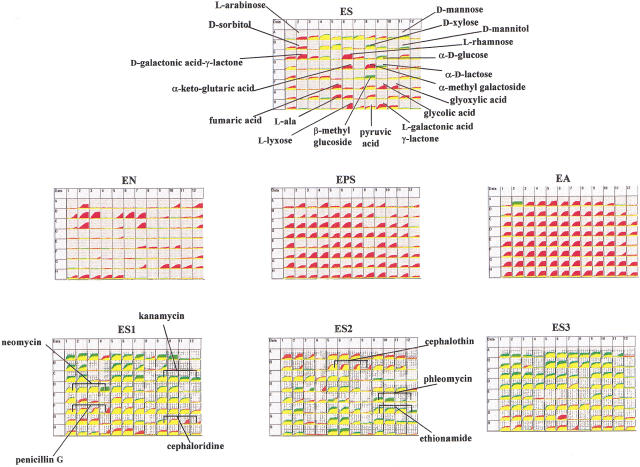
The FruR (also called Cra) protein is another global regulatory element affecting carbon metabolism, but this protein is both a positive and a negative regulator of gene expression (Ramseier et al. 1995). From previous data obtained by Saier and coworkers it is expected to show phenotypes of faster growth with carbon sources such as D-glucose, D-mannose, D-mannitol, D-xylose, and β-methyl glucoside, but slower growth with α-methyl galactoside, L-rhamnose, L-galactonic acid lactone, D-galactonic acid lactone, fumaric acid, glycolic acid, and glyoxylic acid. These phenotypes are all observed in the PM results shown in Figure 5 with both red and green areas in the ES carbon array. Like the cya mutant, this strain will not grow on pyruvate and gave all negative phenotypes in the EN, EPS, and EA arrays. The fruR mutant is also readily distinguished from cya because only a few changes are seen in the ES1, ES2, and ES3 toxicity arrays, and many of these (kanamycin, neomycin, and phleomycin) are due to the Tn5 drug resistance element (Beck et al. 1982) in this strain.
Finally we have tested a phoP mutant. The PhoP protein is a member of a two-component regulatory system (Wanner 1996) controlling many genes in E. coli involved in phosphate utilization, virulence, and survival within macrophages at acidic pH (Groisman et al. 1992; Kasahara et al. 1992). A phoP mutant of E. coli previously constructed and tested for phenotypes was found to have increased sensitivity to magainin 2 but not to other cationic-peptide antibiotics. It had normal growth rates on glucose, glycerol, L-alanine, succinate, and acetate as carbon sources (Groisman et al. 1992). Our results of the PM assay of the phoP mutant strain fb757 are shown in Figure 6, which clearly localizes the physiological changes in this strain. The phoP mutant exhibits normal phenotypes on six of the PMs tested. Nearly all of the phenotypic changes are seen on the EPS array where it shows defects in using several amino acid phosphates and glucosamine-6P as phosphate sources. Interestingly, this strain also exhibits numerous defects in using various sulfur sources.
Figure 6.
PM analysis of phoP amber mutant strain.
DISCUSSION
Phenotypes are the last major area of analysis to become amenable to efficient global analysis. Cellular phenotypes have traditionally been analyzed one at a time and, because phenotypes are often vaguely defined and qualitative, it has not been obvious how one could devise an efficient method with adequate scope and sensitivity for global analysis. We have overcome these difficulties by developing a technology using a colorimetric means for detecting cellular respiration. Respiration is obligatorily coupled to cell growth. One can then devise hundreds or thousands of culture conditions in which specific biological pathways or physiological functions are linked to cell growth. Nearly 700 different assays are presented in this paper, but we are now far along in developing our target number of 2000 assays. With a set of 2000 phenotype assays we expect that we can at least indirectly measure a significant fraction of the functions that cells can perform.
The OmniLog instrument also represents an important technological advance in phenotypic testing. It relieves another technological bottleneck in the monitoring and recording of phenotypic data by allowing for simultaneous data recording on 50 microplates. In a 96-well format this is nearly 5000 wells. It should be straightforward over time to miniaturize the testing further to 384-well microplates providing a 20,000 well capacity and even further beyond that.
Many recently-developed cell-based assay systems employ fluorimetic dyes or fluorescent proteins. Although colorimetric testing is generally viewed as being less sensitive than fluorimetric testing, in this case the color accumulates irreversibly, serving to greatly amplify the signal. Colorimetric measuring instrumentation is much simpler, less costly, and less susceptible to artifacts compared with fluorimetric instruments. Colorimetric assays can also be read by eye (e.g., Fig. 2) to provide visual verification of all results. Two especially nice features of the respiration-based assays are that they are kinetic and quantitative. These features allow for greater sensitivity in detecting more subtle or partial phenotypes.
In this paper we have presented evidence of the high level of accuracy and the sensitivity of PM technology in detecting phenotypes. The ES array, designed to detect changes in carbon metabolism, accurately detected phenotypic changes in xylA, malF, cyaA, and fruR mutant strains. The EN array, designed to detect changes in nitrogen metabolism, accurately detected phenotypic changes in the astC mutant strain. The EPS array, designed to detect changes in phosphorus and sulfur metabolism, detected expected changes in phosphorus metabolism and unexpected changes in sulfur metabolism in the phoP mutant strain. The ES1, ES2, and ES3 sensitivity arrays accurately detected resistance to tetracycline, chlortetracycline, kanamycin, neomycin, bleomycin, and phosphomycin in the strains used in this study. We have also used the EA array to accurately detect phenotypes of auxotrophy for histidine, tryptophan, and pyrimidines in knockout mutants of E. coli and S. typhimurium (data not shown).
We employed E. coli to validate PM technology because it is the most studied and understood cell model and because it is generally assumed that one can do clean genetic constructions for isogenic cell line comparisons. Yet two of our very first experiments with PMs have allowed us to discover unexpected and undetected genetic changes in the strains with which we worked. In one case we found that a strain with a known mutation in ynjB also had a second unknown mutation a few genes away in astC. In another case a stock of electrocompetent cells was prepared using standard protocols to prepare the cells for electroporation. There is evidence that this treatment can induce mutations. Nancy McFarland (pers. comm.) has replica-plated E. coli cells coming out of a frozen state from rich to minimal medium and finds typically about 5% of the colonies are defective in growing in minimal medium. The stock of frozen cells prepared in Blattner's laboratory that picked up a maltose-negative mutation presumably because of this treatment, would have been used in other genetic constructions, inadvertently passing along the unwanted maltose defect into every new strain created. Recently, Miller and coworkers have also used the ES arrays to detect phenotypes resulting from random mutations in mutator strains of E. coli (Funchain et al. 2000).
These observations highlight two important issues that underscore the potential utility of PM technology. In working with cells, we can never know for certain whether they are picking up mutations unless we are willing to sequence the entire genome of each new cell we create. Once a cell line is created, as long as we continue to work with a population regrown from an original stock, we can be reasonably confident that the population will mask genetic defects picked up by single cells. It is important to be aware, in doing cell-based experiments, that cells can and do change constantly in the artificial environments in which we handle them. In the past, where it was impractical to assay more than a few phenotypes, one could not attempt to measure genetic and phenotypic drift. An important use of PM technology is that it will allow people a practical means to characterize cell lines. Although not surveying every gene, it can still provide an important tool in basic biological research as well as in maintenance of cell lines used to produce everything from beer to protein pharmaceuticals.
Even more important is that PMs will allow biologists to detect phenotypes other than those specifically targeted. Many researchers have isolated interesting mutant cell lines that they would like to characterize in greater detail. By analyzing these cell lines in a broad and unbiased way, they may find additional phenotypes that are more interesting and revealing than the phenotypes that were already known.
PM technology will allow broad phenotypic testing to become a simple standard practice. Without a tool of this type, the researcher must guess at expected phenotypes and test these one at a time. PMs allow a simple and unbiased scan of a mutant cell to see how it has changed. Properties instilling better growth such as resistance phenotypes are particularly useful when they can be used for positive selections in genetic constructions. Phosphomycin resistance was used by Ames' group to easily obtain additional mutant alleles in the cya gene (Alper and Ames 1978); however, they had to screen many compounds before finding one that provided a positive selection. With PM technology we made the same observation on phosphomycin in one simple experiment.
Even at a gross level, PM analysis can provide an immediate sense of the phenotypic range and domain of a mutation. Comparing Figures 3 and 4 one can see a dramatic difference between the physiological consequences of a gene affecting carbon metabolism in a specific (malF) versus a global (cyaA) way. Comparing Figures 5 and 6 one can clearly differentiate a global regulatory gene with a domain affecting carbon metabolism (fruR) from one affecting phosphorus and sulfur metabolism (phoP).
In this paper, we have made a casual observation that the E. coli phoP mutant appears to have altered sulfur metabolism. We have also tested an isogenic pair of E. coli strains (from C. Fraley and A. Kornberg, Stanford University) with a deletion of the ppk-ppx genes involved in polyphosphate synthesis. This mutant strain also showed defects in both phosphorus and sulfur metabolism. As far as we are aware, coordinated regulation of phosphorus and sulfur genes and pathways has not been generally recognized by others working in these fields. The one report we are aware of was made in 1963 when Harold and Sylvan, studying polyphosphate metabolism, observed that an intracellular sulfur compound, perhaps glutathione, was a key regulator of polyphosphate accumulation.
PM technology provides another means of global cell analysis and it is instructive to compare the type of information it provides to the information provided by gene expression analysis as well as to consider its overall strengths and limitations. There are important similarities and important differences between global analyses based on gene expression versus those based on phenotypes. In terms of similarities, the quantitative output of a PM assay can be enhanced with color (red for loss of phenotype, yellow for unchanged phenotype, green for gain of phenotype) in a manner analogous to colorization of gene expression data (gene levels going down, staying unchanged, or going up). Also analogous to gene expression data, PM data can be inputted to bioinformatics software and can provide a basis for clustering of genes showing most similar patterns.
One major difference is that PM analysis is much easier to perform than gene expression analysis because it involves simply preparing a standardized cell suspension and inoculating the microwells of the PM. However, to get useful data on genes, one does need to perform genetic work to create the strains to be analyzed. Another major difference is that gene expression analysis provides simultaneous data on thousands of genes, but only under one condition of the cell, whereas PM analysis is normally done on only one gene at a time, but under thousands of conditions of the cell. The technology of PM analysis provides an important complement of gene expression studies and allows one to directly observe the consequences of a genetic change. Extrapolation from gene expression data alone typically depends on complex computer models and may lead to incorrect or incomplete conclusions.
The customization aspects of gene arrays is often more straightforward than with PMs because the chemistry of DNA–RNA hybridization spans all domains of life. We are trying, as much as possible, to design PMs with widely relevant phenotypes such as carbon, nitrogen, phosphorus, and sulfur metabolism. This will allow us to use PMs for a wide range of cell types including aerobic and anaerobic bacteria, yeast, and filamentous fungi. For example, we have already been successful in adapting some of the PMs for use with Pseudomonas aeruginosa, Ralstonia solanacearum, Burkholderia cepacia, Vibrio fluvialis, Sinorhizobium meliloti, Listeria innocua, Staphylococcus aureus, Enterococcus faecalis, Candida albicans, Saccharomyces cerevisiae, and Aspergillus nidulans. It will be important to extend the technology to work with mouse and human cells. We fully expect that this can be accomplished as tetrazolium-based respiration assays of mammalian cells have been used since the advent of the Mossman assay (Mossman 1983).
METHODS
Strains
The E. coli strains used in this study are listed in Table 1. Strains generated in this study were constructed by P1vir-mediated transduction of the desired drug resistance-linked markers present from the appropriate donor strains, into the recipient strain MG1655, using the protocols outlined by Silhavy et al. (1984).
Pregrowth Media
Before inoculating into the EN, EPS, and EA microplates, MG1655 was grown overnight on plates containing the nutrient limited medium, R2A agar (Reasoner and Geldreich 1985). We found that pregrowth of cells on R2A was sufficient to deplete the nutrient reserves of the cells so that subsequent growth in the microplates was entirely dependent on the nutritional supplements provided in each of the wells. R2A was chosen after careful examination of a number of pregrowth media and was the only medium that resulted in no growth and therefore no purple color in the negative control wells for nitrogen source (N-free well), phosphorus source (P-free well), and sulfur source (S-free well). Prior to inoculating into the ES1, ES2, and ES3 microplates, cells were grown overnight on agar plates containing Luria Bertani (LB) nutrient-rich medium.
Defined Minimal Medium
The complete minimal medium used in the EN, EPS, and EA microplates contained 100 mM NaCl, 30 mM triethanolamine HCl (pH 7.1), 25 mM sodium pyruvate, 5.0 mM NH4Cl, 2.0 mM NaH2PO4, 0.25 mM Na2SO4, 0.05 mM MgCl2, 1.0mM KCl, 1.0 μM ferric chloride, and 0.01% tetrazolium violet. Nitrogen-free, phosphorus-free, and sulfur-free versions of this medium were used in the EN and EPS microplates keeping the same concentrations for nitrogen, phosphorus, and sulfur sources.
Nutrient-Rich Medium
Sensitivity/resistance testing in the ES1, ES2, and ES3 microplates was performed in 1:5 diluted LB medium. Each well contained a final concentration of 2.0 g tryptone, 1.0 g yeast extract, and 1.0 g NaCl per liter. Inhibitors and other chemicals were added to this medium at the concentrations listed below. The positive control well of the sensitivity testing panels (well A1 of the ES1 microplate) contains LB without any inhibitor.
Phenotypic Tests in the PMs
MG1655 cells were streaked onto either R2A agar (Acumedia, Baltimore, MD; for EN, EPS, and EA microplates) or LB agar (for ES1, ES2, and ES3 microplates) and were grown overnight at 35°C. Individual colonies were picked up from the surface of the R2A or LB plates using a cotton swab. These cells were suspended in GN/GP-IF (inoculating fluid) at a density corresponding to 85% transmittance in the Biolog turbidimeter using a 20 mm diameter tube. The suspensions were then inoculated into the appropriate microplates at a volume of 100 μl/well. The microplates were placed in the OmniLog instrument for either 48 h (EN, EPS, EA) or 24 h (ES, ES1, ES2, ES3), at which time sufficient purple color had developed in the positive control wells, while the negative control wells remained colorless.
OmniLog Readings
All assays and data recordings were performed using the OmniLog reader with 24- or 48-h incubation at 35° C. Each strain was tested at least twice and the PMs were also examined visually at the end of each incubation for independent verification of results. All phenotypic changes that were consistent and substantial are indicated in Figures 3–6 by black lines.
Establishing a Range of Inhibitor Concentrations
In devising the tests presented in the ES1, ES2, and ES3 microplates, we first needed to establish an optimum concentration range for each of the inhibitory agents used. We chose a concentration range which spanned the sensitivity/resistance threshold for the reference E. coli strain MG1655, to allow for easy discrimination of both resistance and hypersensitivity mutants. We found that this worked best if we used four concentrations (twofold dilutions) for each of the chemicals that were tested.
Preparation of Chemical Stock Solutions and Arrays
Stock solutions of each of the following compounds were prepared in dimethylsulfoxide (DMSO), acetic acid, ethanol, acetone, or water. The stock solutions were then filter-sterilized and stored according to the supplier's instructions. When appropriate, the pH of the stock solutions was adjusted to ∼7. To ensure that the solvents did not affect the growth/respiration of the cells or purple color formation resulting from growth and respiration, we assayed the growth of MG1655 as well as resulting color formation in the presence of each of the solvents. Similar growth patterns in the presence and absence of solvent indicated that the presence of solvent did not adversely affect the cells. The order of the chemicals in the arrays is as listed below, left to right and top to bottom, beginning in the A1 well.
Carbon Sources in the ES Array
Water (negative control), L-arabinose, N-acetyl-D-glucosamine, D-saccharic acid, succinic acid, D-galactose, L-aspartic acid, L-proline, D-alanine, D-trehalose, D-mannose, dulcitol, D-serine, D-sorbitol, glycerol, L-fucose, D-glucuronic acid, D-gluconic acid, D,L-α-glycerol-phosphate, D-xylose, L-lactic acid, formic acid, D-mannitol, L-glutamic acid, glucose 6-phosphate, D-galactonic acid γ-lactone, D,L-malic acid, D-ribose, tween 20, L-rhamnose, D-fructose, acetic acid, α-D-glucose, maltose, D-melibiose, thymidine, L-asparagine, D-aspartic acid, D-glucosaminic acid, 1,2-propanediol, tween 40, α-keto-glutaric acid, α-keto-butyric, α-methyl-D-galactoside, α-D-lactose, lactulose, sucrose, uridine, L-glutamine, m-tartaric acid, glucose 1-phosphate, fructose 6-phosphate, tween 80, α-hydroxy-glutaric acid γ-lactone, α-hydroxy-butyric acid, β-methyl-D-glucoside, adonitol, maltotriose, 2‘-deoxyadenosine, adenosine, glycyl-L-aspartic acid, citric acid, m-inositol, D-threonine, fumaric acid, bromo-succinic acid, propionic acid, mucic acid, glycolic acid, glyoxylic acid, cellobiose, inosine, glycyl-L-glutamic acid, tricarballylic acid, L-serine, L-threonine, L-alanine, L-alanyl-glycine, acetoacetic acid, N-acetyl-β-D-mannosamine, mono-methyl succinate, methyl pyruvate, D-malic acid, L-malic acid, glycyl-L-proline, p-hydroxy phenylacetic acid, m-hydroxy phenylacetic acid, tyramine, D-psicose, L-lyxose, glucuronamide, pyruvic acid, L-galactonic acid γ-lactone, D-galacturonic acid, phenylethylamine, 2-amino ethanol.
Nitrogen Sources in the EN Array
Water (negative control), ammonia, nitrite, nitrate, urea, glutathione (reduced form), alloxan, L-citrulline, putrescine, L-ornithine, agmatine, L-alanine, L-arginine, L-asparagine, L-aspartic acid, L-cysteine, L-glutamic acid, L-glutamine, glycine, L-histidine, L-isoleucine, L-leucine, L-lysine, L-methionine, L-phenylalanine, L-proline, L-serine, L-tyrosine, L-threonine, L-valine, D-alanine, D-asparagine, D-aspartic acid, D-glutamic acid, D-lysine, D-serine, D-valine, N-acetylglycine, L-pyroglutamic acid, L-homoserine, met-ala, n-amylamine, n-butylamine, ethylamine, ethanolamine, ethylene diamine, histamine, (R)-(+)-α phenylethylamine, β-phenylethylamine, tyramine, acetamide, formamide, glucuronamide, lactamide, D(+)-glucosamine, D(+)-galactosamine, D-mannosamine, N-acetyl-D-glucosamine, N-acetyl-D-galactosamine, N-acetyl-D-mannosamine, adenine, adenosine, cytosine, thymine, thymidine, uracil, uridine, xanthine, xanthosine, inosine, DL-α-amino-n-butyric acid, γ-amino-n-butyric acid, ɛ-amino-n-caproic acid, DL-α amino-caprylic acid, hippuric acid, parabanic acid, uric acid, urocanic acid, δ-amino-n-valeric acid, 2-amino-valeric acid, gly-glu, ala-gly, ala-his, ala-thr, gly-met, gly-gln, ala-gln, gly-ala, gly-asn.
Phosphorus Sources in the EPS Array
Water (negative control), phosphate, pyrophosphate, trimetaphosphate, tripolyphosphate, hypophosphite, thiophosphate, adenosine 2‘-monophosphate, adenosine 3′-monophosphate, adenosine 5′-monophosphate, adenosine 2‘:3′-cyclic monophosphate, adenosine 3′:5′-cyclic monophosphate, dithiophosphate, DL-α-glycerophosphate, β-glycerophosphate, phosphatidyl glycerol, phosphoenol pyruvate, phosphocreatine, 2-deoxy-D-glucose 6-phosphate, guanosine 2‘-monophosphate, guanosine 3′-monophosphate, guanosine 5′-monophosphate, guanosine 2‘:3′-cyclic monophosphate, guanosine 3′:5′-cyclic monophosphate, glucose 1-phosphate, glucose 6-phosphate, fructose 1-phosphate, fructose 6-phosphate, mannose 1-phosphate, mannose 6-phosphate, arabinose 5-phosphate, cytidine 2‘-monophosphate, cytidine 3′-monophosphate, cytidine 5′-monophosphate, cytidine 2‘:3′-cyclic monophosphate, cytidine 3′:5′-cyclic monophosphate, glucosamine 1-phosphate, glucosamine 6-phosphate, phospho-L-arginine, O-phospho-D-serine, O-phospho-L-serine, O-phospho-D-tyrosine, O-phospho-L-tyrosine, uridine 2‘-monophosphate, uridine 3′-monophosphate, uridine 5′-monophosphate, uridine 2‘:3′-cyclic monophosphate, uridine 3′:5′-cyclic monophosphate, O-phospho-L-threonine, inositol hexaphosphate, nitrophenyl phosphate, 2-aminoethyl phosphonic acid, 6-phosphogluconic acid, 2-phosphoglyceric acid, phosphoglycolic acid, phosphonoacetic acid, thymidine 3′-monophosphate, thymidine 5′-monophosphate, methylene diphosphonic acid, thymidine 3′:5′-cyclic monophosphate.
Sulfur Sources in the EPS Array
(Beginning with F1 well) Water (negative control), sulfate, thiosulfate, tetrathionate, thiophosphate, dithiophosphate, L-cysteine, cys-gly, L-cysteic acid, cysteamine, L-cysteine-sulphinic acid, cystathionine, lanthionine, D,L-ethionine, glutathione (reduced form), L-methionine, glycyl-D,L-methionine, S-methyl-L-cysteine, L-methionine sulfoxide, L-methionine sulfone, taurine, N-acetyl-D,L-methionine, N-acetyl-cysteine, isethionate, thiourea, thiodiglycol, thioglycolic acid, thiodiglycolic acid, 1-dodecane-sulfonic acid, taurocholic acid, tetramethylene sulfone, hypotaurine, O-acetyl-serine, 3′:3′ thiodipropionic acid, L-djenkolic acid, 2-mercaptoethylamine.
Auxotrophic Supplements in the EA Array
Water (negative control), LB medium (positive control), L-alanine, L-arginine, L-asparagine, L-aspartic acid, adenine, adenosine, 2‘-deoxyadenosine, adenosine 3′:5′-cyclic monophosphate, adenosine 3′-monophosphate, adenosine 5′-monophosphate, L-cysteine, L-glutamic acid, L-glutamine, glycine, L-histidine, L-isoleucine, guanine, guanosine, 2‘-deoxyguanosine, guanosine 3′:5′-cyclic monophosphate, guanosine 3′-monophosphate, guanosine 5′-monophosphate, L-leucine, L-lysine, L-methionine, L-phenylalanine, L-proline, L-serine, cytosine, cytidine, 2‘-deoxycytidine, cytidine 3′:5′-cyclic monophosphate, cytidine 3′-monophosphate, cytidine 5′-monophosphate, L-tryptophan, L-tyrosine, L-threonine, L-valine, D-alanine, D-aspartic acid, thymine, thymidine, thymidine 3′:5′-cyclic monophosphate, thymidine 3′-monophosphate, thymidine 5′-monophosphate, D-glutamic acid, (5)4-amino-imidazole-4(5)-carboxamide, D,L-α,ɛ-diaminopimelic acid, D-biotin, D,L-α-lipoic acid, caprylic acid, uracil, uridine, 2‘-deoxyuridine, uridine 3′:5′-cyclic monophosphate, uridine 3′-monophosphate, uridine 5′-monophosphate, p-amino-benzoic acid, shikimic acid, molybdic acid, folic acid, α-keto-isovaleric acid, D-pantothenic acid, hypoxanthine, inosine, 2‘-deoxyinosine, inosine 3′:5′-cyclic monophosphate, inosine 3′-monophosphate, inosine 5′-monophosphate, thiamine, riboflavin, pyridoxal, pyridoxine, pyridoxamine, quinolinic acid, glutathione (reduced form), L-homoserine lactone, α-ketobutyric acid, β-nicotinamide adenine dinucleotide, nicotinic acid, nicotinamide, N-α-acetyl L-ornithine, L-ornithine, L-citrulline, putrescine, spermidine, spermine, tween 20, tween 40, tween 60, tween 80, δ-amino levulinic acid.
Inhibitory Compounds in the ES1, ES2, and ES3 Arrays
ES1: LB medium (positive control), acriflavine: 4, 8, and 16 μg/mL; ampicillin: 2, 4, 8, and 16 μg/mL; nafcillin: 75, 150, 300, and 600 μg/mL; lincomycin: 50, 100, 200, and 400 μg/mL; chloramphenicol: 0.1, 0.2, 0.4, and 0.8 μg/mL; chlortetracycline: 0.125, 0.25, 0.5, and 1.0 μg/mL; tetracycline: 0.03, 0.07, 0.13, and 0.27 μg/mL; gentamycin: 0.25, 0.5, 1.0, and 2.0 μg/mL; kanamycin: 0.25, 0.5, 1.0, and 2.0 μg/mL; neomycin: 0.75, 1.5, 3.0, and 6.0 μg/mL; vancomycin: 10, 20, 40, and 80 μg/mL; bacitracin: 208, 416, 833, and 1667 μg/mL; clindamycin: 3.3, 6.6, 13, and 26 μg/mL; cloxacillin: 100, 200, 400, and 800 μg/mL; erythromycin: 2.5, 5, 10, and 20 μg/mL; penicillin G: 5, 10, 20, and 40 μg/mL; novobiocin: 33, 66, 133, and 266 μg/mL; spiramycin: 5, 10, 20, and 40 μg/mL; trimethoprim: 0.17, 0.33, 0.67, and 1.3 ng/mL; streptomycin: 0.38, 0.75, 1.5, and 3.0 μg/mL; cephaloridine: 0.75, 1.5, 3.0, and 6.0 μg/mL; cefuroxime: 0.5, 1.0, 2.0, and 4.0 μg/mL; roxithromycin: 10, 20, 40, and 80 μg/mL; piperacillin: 0.5, 1.0, 2.0, and 4.0 μg/mL. ES2: azomycin: 0.2, 0.4, 0.8, and 1.6 μg/mL; rifampicin: 0.25, 0.5, 1.0, and 2.0 μg/mL; tylosin: 25, 50, 100, and 200 μg/ml; cefazolin: 0.5, 1.0, 2.0, and 4.0 μg/mL; cephalothin: 2.5, 5.0, 10, and 20 μg/mL; cefaclor: 0.66, 1.33, 2.7, and 5.3 μg/mL; rifamycin SV: 1.5, 3.0, 5.0, and 10 μg/mL; cefsulodin: 4, 8, 16, and 32 μg/mL; cefotaxime: 0.05, 0.1, 0.2, and 0.4 μg/mL; cefoxitin: 0.75, 1.5, 3.0, and 6.0 μg/mL; puromycin: 12, 24, 48, and 96 μg/mL; spectinomycin: 3.5, 7.0, 14, and 28 μg/mL; fusidic acid: 50, 100, 200, and 400 μg/mL; phosphomycin: 0.2, 0.4, 0.8, and 1.6 μg/mL; phleomycin: 0.25, 0.5, 1.0, and 2.0 μg/mL; amikacin: 0.25, 0.5, 1.0, and 2.0 μg/mL; isoniazid: 300, 600, 1200, and 2400 μg/mL; ethionamide: 25, 50, 100, and 200 μg/mL; sodium dodecyl sulfate: 50, 100, 200, and 400 μg/mL; dodecyltrimethyl ammonium bromide: 10, 20, 40, and 80 μg/mL; BIGCHAP: 2000, 4000, 8000, and 16000 μg/mL; niaproof: 0.08, 0.16, 0.32, and 0.65%, CHAPS: 1500, 3000, 6000, and 12000 μg/mL; N-lauryl sarcosine: 1000, 2000, 4000, and 8000 μg/mL. ES3: nalidixic acid: 0.5, 1.0, 2.0, and 4.0 μg/mL; taurocholic acid: 600, 1200, 2400, and 4800 μg/mL; colistin: 0.25, 0.5, 1.0, and 2.0 μg/mL; procaine: 2500, 5000, 10000, and 20000 μg/mL; diamide: 33, 66, 132, and 264 μg/mL; hydroxylamine: 12, 24, 48, and 96 μg/mL; guanidine: 500, 1000, 2000, and 4000 μg/mL; cupric chloride: 20, 40, 80, and 160 μg/mL; zinc chloride: 10, 20, 40, and 80 μg/mL; cadmium chloride: 5, 10, 20, and 40 μg/mL; nickel chloride: 20, 40, 80, and 160 μg/mL; chromium chloride: 100, 200, 400, and 800 μg/mL; sodium selenite: 100, 200, 300, and 400 μg/mL; potassium tellurite: 0.2, 0.4, 0.8, and 1.6 μg/mL; manganese sulfate: 100, 200, 400, and 800 μg/mL; cobalt chloride: 12, 24, 48, and 96 μg/mL; silver chloride: 2, 4, 8, and 16 μg/mL; potassium chromate: 10, 20, 40, and 80 μg/mL; potassium bromide: 225, 450, 900, and 1800 μg/mL; sodium cyanate: 155, 310, 600, and 1200 μg/mL; sodium azide: 500, 1000, 2000, and 4000 μg/mL; picolinic acid: 50, 100, 200, and 400 μg/mL; potassium superoxide: 100, 200, 400, and 800 μg/mL; menadione: 3.3, 6.6, 13.3, and 26.6 μg/mL.
Acknowledgments
This work is dedicated to the four members of Biolog’s Scientific Advisory Board (Elliot Juni, Bruce Ames, John Ingraham, and Ralph Wolfe) who have been a major influence on and inspiration to the senior author of this paper (B.R.B.). The authors also thank all collaborators who provided strains, as listed in Table 1, especially Jeremy Glasner who created strains specifically for this study. Financial support was from SBIR Grants R43 HG01755–01A1 from NIH-NHGRI (for PMs) and NAS10–12197 from NASA (for OmniLog).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL bbochner@biolog.com; FAX (510) 782-4639.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.186501.
REFERENCES
- Alper MD, Ames BN. Transport of antibiotics and metabolite analogs by systems under cyclic AMP control: Positive selection of Salmonella typhimurium cya and crp mutants. J Bacteriol. 1978;133:149–157. doi: 10.1128/jb.133.1.149-157.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck E, Ludwig G, Auerswald EA, Reiss B, Schaller H. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene. 1982;19:327–336. doi: 10.1016/0378-1119(82)90023-3. [DOI] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Bochner BR. New methods aid microbial identification. Bio/Technology. 1988;6:756. [Google Scholar]
- ————— “Breathprints” at the microbial level. ASM News. 1989a;55:536–539. [Google Scholar]
- ————— Sleuthing out bacterial identities. Nature. 1989b;339:157–158. doi: 10.1038/339157a0. [DOI] [PubMed] [Google Scholar]
- Burns N, Grimwade B, Ross-Macdonald PB, Choi E-T, Finberg K, Roeder GS, Snyder M, et al. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes & Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor SA, Rava RP, Huang XC, Pease AC, Holmes CP, Adams CL. Multiplexed biochemical assays with biological chips. Nature. 1993;364:555–556. doi: 10.1038/364555a0. [DOI] [PubMed] [Google Scholar]
- Funchain P, Yeung A, Stewart JL, Lin R, Slupska MM, Miller JH. The consequences of growth of a mutator strain of Escherichia coli as measured by loss of function among multiple gene targets and loss of fitness. Genetics. 2000;154:959–970. doi: 10.1093/genetics/154.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman EA, Heffron F, Solomon F. Molecular genetic analysis of the Escherichia coli phoP locus. J Bacteriol. 1992;174:486–491. doi: 10.1128/jb.174.2.486-491.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harold FM, Sylvan S. Accumulation of inorganic polyphosphate in Aerobacter Aerogenes. II. Environmental control and the role of sulfur compounds. J Bacteriol. 1963;86:222–231. doi: 10.1128/jb.86.2.222-231.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M, Nakata A, Shinagawa H. Molecular analysis of the Escherichia coli phoP-phoQ operon. J Bacteriol. 1992;174:492–498. doi: 10.1128/jb.174.2.492-498.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustu SG, McFarland NC, Hui SP, Esmon B, Ames GF-L. Nitrogen control in Salmonella typhimurium: Co-regulation of synthesis of glutamine synthetase and amino acid transport systems. J Bacteriol. 1979;138:218–234. doi: 10.1128/jb.138.1.218-234.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link AJ, Phillips D, Church G. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: Application to open reading frame characterization. J Bacteriol. 1997;179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossman T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation in cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Nichols BP, Shafiq O, Meiners V. Sequence analysis of Tn10 insertion sites in a collection of Escherichia coli strains used for genetic mapping and strain construction. J Bacteriol. 1998;180:6408–6411. doi: 10.1128/jb.180.23.6408-6411.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Pastan I, Adhya S. Cyclic adenosine 5′-monophosphate in Escherichia coli. Bacteriol Rev. 1976;40:527–551. doi: 10.1128/br.40.3.527-551.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramseier TM, Bledig S, Michotey V, Feghali R, Saier MH., Jr The global regulatory protein FruR modulates the direction of carbon flow in Escherichia coli. Mol Microbiol. 1995;16:1157–1169. doi: 10.1111/j.1365-2958.1995.tb02339.x. [DOI] [PubMed] [Google Scholar]
- Reasoner DJ, Geldreich EE. A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol. 1985;49:1–7. doi: 10.1128/aem.49.1.1-7.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiger K-J, Kaniak A, Coppee J-Y, Aljinovic G, Baudin-Baillieu A, Orlowska G, Gromadka R, Groudinsky O, di Rago J-P, Slonimski PP. Large-scale phenotypic analysis–the pilot project on yeast chromosome III. Yeast. 1997;13:1547–1562. doi: 10.1002/(SICI)1097-0061(199712)13:16<1547::AID-YEA230>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Reiger K-J, El-Alama M, Stein G, Bradshaw C, Slonimski PP, Maundrell K. Chemotyping of yeast mutants using robotics. Yeast. 1999;15:973–986. doi: 10.1002/(SICI)1097-0061(199907)15:10B<973::AID-YEA402>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Ross-Macdonald P, Coelho PSR, Roemer T, Agarwal S, Kumar A, Jansen R, Cheung K-H, Sheehan A, Symoniatis D, Umansky L, et al. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature. 1999;402:413–418. doi: 10.1038/46558. [DOI] [PubMed] [Google Scholar]
- Schneider BL, Kiupakis AK, Reitzer LJ. Arginine catabolism and the arginine succinyltransferase pathway of Escherichia coli. J Bacteriol. 1998;180:4278–4286. doi: 10.1128/jb.180.16.4278-4286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy TJ, Berman ML, Enquist LW. Experiments with gene fusions. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1984. pp. 111–112. [Google Scholar]
- Vagner V, Dervyn E, Erhlich SD. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- Wanner BL. Phosphorus assimilation and control of the phosphate regulon. In: Neidhardt FC, editor. Escherichia coli and Salmonella typhimurium cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1357–1381. [Google Scholar]
- Wissenbach U, Six S, Bongaerts J, Ternes D, Steinwachs S, Unden G. A third periplasmic transport system for L-arginine in Escherichia coli: Molecular characterization of the artPIQMJ genes, arginine binding and transport. Molec Microbiol. 1995;17:675–686. doi: 10.1111/j.1365-2958.1995.mmi_17040675.x. [DOI] [PubMed] [Google Scholar]
- Zambrowicz BP, Friedrich GA, Buxton EC, Lilleberg SL, Person C, Sands AT. Disruption and sequence identification of 2,000 genes in mouse embryonic stem cells. Nature. 1998;392:608–611. doi: 10.1038/33423. [DOI] [PubMed] [Google Scholar]