Abstract
A method for identifying and validating single nucleotide polymorphisms (SNPs) with high-density oligonucleotide arrays without the need for locus-specific polymerase chain reactions (PCR) is described in this report. Genomic DNAs were divided into subsets with complexity of ∼10 Mb by restriction enzyme digestion and gel-based fragment size resolution, ligated to a common adaptor, and amplified with one primer in a single PCR reaction. As a demonstration of this approach, a total of 124 SNPs were located in 190 kb of genomic sequences distributed across the entire human genome by hybridizing to high-density variant detection arrays (VDA). A set of independent validation experiments was conducted for these SNPs employing bead-based affinity selection followed by hybridization of the affinity-selected SNP-containing fragments to the same VDA that was used to identify the SNPs. A total of 98.7% (74/75) of these SNPs were confirmed using both DNA dideoxynucleotide sequencing and the VDA methodologies. With flexible sample preparation, high-density oligonucleotide arrays can be tailored for even larger scale genome-wide SNP discovery as well as validation.
With the completion of the first draft of human genomic consensus sequence, there is a predicable increase in interest concerning the types and amount of genetic diversity in the human population. The most abundant type of variations are single nucleotide polymorphisms (SNPs) (Collins et al. 1997). Some of these genetic variations underpin the genetic basis for disease susceptibility and are used in genetic studies as markers for complex traits and diseases.
High-density variation detection arrays (VDAs) have been successfully used for large-scale SNP screening (Chee et al. 1996; Wang et al. 1998; Cargill et al. 1999; Hacia et al. 1999; Halushka et al. 1999). In these experiments, predetermined DNA targets within the genome were amplified with specific PCR reactions. To discover SNPs in this way, each locus in the genome needs to be amplified individually. Although multiple loci can be amplified in a multiplex PCR, this multiplexing approach is costly and labor intensive in the development stages and difficult to scale.
In this study, high-density oligonucleotide arrays were used to screen for SNPs from a subset of human genomic DNA. Each subset of genomic DNAs contained a complexity up to 10 Mb and was amplified with one primer in a single PCR reaction. Sequence variations were screened with the same efficiency as individually amplified targets by hybridizing the same complex genomic sample to multiple designs of VDA. In addition, the SNP candidates were validated by using dideoxynucleotide sequencing and by using a bead-based affinity method to enrich fragments that contained candidate SNPs followed by reanalyzing these enriched fragments in a second round of VDA analysis.
RESULTS
Genome-Wide SNP Discovery
Virtual Restriction Enzyme Digestion and Size Selection
Armed with a working knowledge of the nucleotide sequence of the human genome, restriction enzyme digestion reactions can be carried out in silico. A straightforward computer program to search and “cleave” at designated restriction enzyme recognition sequences and organize the generated fragments according to their size was employed. Genomic sequences were downloaded from Baylor College of Medicine Human Genome Sequencing Center (http://www.hgsc.bcm.tmc.edu/), Lawrence Berkeley Laboratory Human Genome Center (http://www-hgc.lbl.gov/GenomeHome.html) (now a part of Joint Genome Institute, Department of Energy), Stanford Human Genome Center (http://www-shgc.stanford.edu/), The Sanger Centre (http://www.sanger.ac.uk/), The Institute for Genome Research (http://www.tigr.org/), Washington University Genome Sequencing Center (http://genome.wustl.edu/gsc), and Whitehead Institute for Biomedical Research/MIT (http://www.genome.wi.mit.edu/).They were used as input data for this computer program. Using the recognition sequence of EcoRI (5′-GAATTC-3′), and defining the desired size range of identified fragments as 244–344 bp, the program generated a collection of sequence fragments accounting for 0.3% of the input human genome sequences (250 Mb).
Generic PCR Amplification of Complex DNA
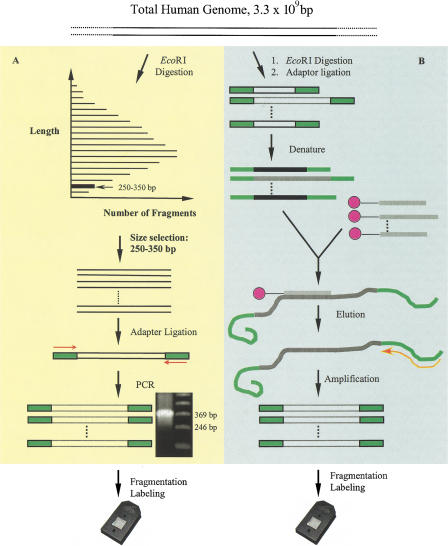
A total of 5 μg of human genomic DNA was digested with EcoRI and the digestion products were resolved using agarose gel electrophoresis (Fig. 1). The DNA fragments of desired size range were excised from the gel and purified. The purified restriction fragments were ligated to a single adaptor. PCR amplification with one primer that hybridized to the adaptor sequences generated the expected fragments (Fig. 1A).
Figure 1.
Schemes of (A) single nucleotide polymorphism (SNP) discovery and (B) validation. (A) Genomic DNA was digested with EcoRI and separated by size on agarose gel. A 250–350 bp fraction was cut from the gel, extracted, and ligated to an adaptor. The ligated DNA was amplified with one primer that interrogated the adaptor sequence. SNPs were screened by hybridizins to VDA. (B) Designated targets were enriched from total genomic DNA by oligonucleotides bound to magnetic beads that interrogated the SNP sites. The genomic DNA was digested with EcoRI and ligated with an adaptor. The enriched targets were amplified with the same primer and screened similarly as in A.
The expected 244–344 bp amplification products were observed after electrophoretic separation (Fig. 1A). The length of PCR product is slightly longer than the excised fragments because of the sequences added by the ligation of the adaptor oligonucleotides. The fold of amplification was estimated to be >2500 by dividing the final yield of DNA after purification by the amount of starting template.
The concordance between the virtual and experimental selections was revealed by the hybridization results of the amplified subset to the VDA. To determine if any particular EcoRI fragment was present in the amplified pool, the empirically derived sequence for each fragment was required to achieve a 50% concordance with the predicted sequence encoded on the array. Thus, the fragments with accordance rate above 50% were considered as being present and amplified from the subset excised from the gel. According to these criteria, 98% of the fragments encoded on the arrays were successfully selected from the total genome and amplified.
Genome-Wide SNP Screening
The nucleotide sequences of 713 EcoRI fragments of the human genome with lengths from 244 to 344 bp (total of 190 kb) were randomly selected from all possible EcoRI fragments generated by virtual restriction enzyme digestion. These sequences were then represented onto a set of seven arrays by the synthesis of eight oligonucleotide probes interrogating each base and each strand of the 190,676 selected bases. The same EcoRI-generated subset was selected and amplified for each of eight unrelated individuals. The RepeatMasker program (A.F.A. Smit and P. Green, unpubl.; http://repeatmasker.genome.washington.edu/cgi-bin/RM2_req.pl) was used to search and detect sequences that are homologous to repetitive sequences found in the human genome. Approximately 33.4% of the selected 190 kb encoded on the array consisted of repeat sequences (e.g., Alus, MIRs, LINEs, LTR elements, MERs, etc).
A total of 124 SNP candidates across the whole genome in eight different individuals were identified (Fig. 2). A total of 88 SNPs were present within unique sequences and 36 in repeat regions. Their distribution among different chromosomes is listed in Table 1. Their allele distribution was as follows: 31 A↔G, 10 A↔T, 41 C↔T, 11 C↔G, and 17 G↔T. The ratio of transitions to transversions was close to 1.5 with the most frequent substitution being C↔T. Of the 124 candidate SNPs that were identified, 92 were successfully amplified and used for concordance studies. Using a dideoxynucleotide sequencing method, 77 of the 92 SNPs (84%) were concordant between the two methods. Discordant SNP candidates were predominantly variations detected only in one chromosome.
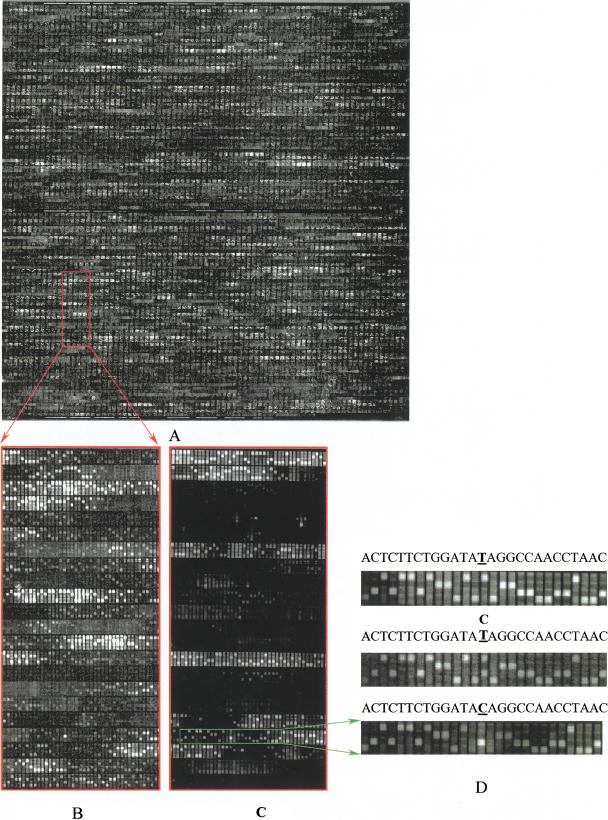
Figure 2.
SNP screening with variant detection arrays (VDAs). (A) The whole image of 40 pM targets hybridized with VDA. (B) An expanded view of a portion of A. (C) The same region of image as in B except that the chip was hybridized with enriched targets. (D) A single nucleotide polymorphism (SNP) detected and confirmed by alternative sample preparation. From top to bottom, the wild type (base T at the center), heterozygous SNP (bases C and T at the center), and homozygous mutation (base C at the center) are shown.
Table 1.
Summary of SNP Screening and Validation by Gel-Based Sequencing
| Chromosome | Number of selected fragments | Number of SNPs | Number of SNPs tested by gel sequencing | Number of SNPs confirmed by gel sequencing |
|---|---|---|---|---|
| 1 | 43 | 8 | 6 | 4 |
| 2 | 3 | 0 | 0 | 0 |
| 3 | 0 | 0 | 0 | 0 |
| 4 | 20 | 2 | 2 | 2 |
| 5 | 56 | 9 | 6 | 4 |
| 6 | 79 | 19 | 15 | 14 |
| 7 | 152 | 27 | 19 | 18 |
| 8 | 7 | 1 | 1 | 1 |
| 9 | 2 | 1 | 0 | 0 |
| 10 | 3 | 1 | 1 | 1 |
| 11 | 4 | 0 | 0 | 0 |
| 12 | 29 | 2 | 2 | 2 |
| 13 | 7 | 0 | 0 | 0 |
| 14 | 6 | 1 | 1 | 1 |
| 15 | 0 | 0 | 0 | 0 |
| 16 | 29 | 5 | 4 | 2 |
| 17 | 40 | 5 | 4 | 3 |
| 18 | 1 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 |
| 20 | 13 | 5 | 5 | 4 |
| 21 | 4 | 0 | 0 | 0 |
| 22 | 34 | 10 | 8 | 6 |
| X | 174 | 27 | 17 | 14 |
| Y | 0 | 0 | 0 | 0 |
| Unknown origin | 7 | 1 | 1 | 1 |
| Total | 713 | 124 | 92 | 88 |
Hybridization of Repetitive Sequences
Generically amplified genome subsets generated by size-selecting restriction enzyme digestion products contain significant amounts of repetitive sequences. Because of their higher abundance in the genome than unique sequences, hybridization intensities of probes that are homologous to repetitive sequences, especially Alu's, are disproportionately high. Such repeat sequences increase misleading cross-hybridization signals.
This cross-hybridization was reduced by adding unlabeled competitive human Cot-1 fraction DNA (GIBCO-BRL). In the presence of Cot-1 DNA, the signal intensities attributable to cross-hybridization were greatly suppressed or diminished whereas those of unique sequences were little affected (Fig. 3). By maintaining an appropriate amount of the Cot-1 DNA in the hybridization solution (see below), the intensities of probes interrogating repetitive sequences were reduced to a level such that no bases were called by the analysis software for these regions. Maintaining the ratio of human Cot-1 fraction to the target at 2:1 effectively suppressed the signals of Alu homologous regions while increasing the ratio to 5:1 suppressed the signals of some L1 homologous sequences. Other sequences that are homologous to less abundant repeat elements, e.g., MER and LTR, did not cause significant cross-hybridization and were little affected by adding Cot-1 fraction.
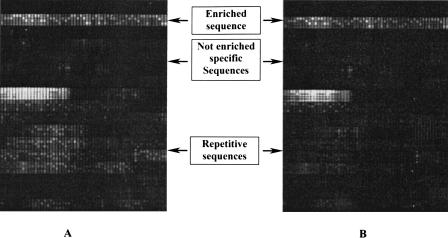
Figure 3.
Suppression of signals of repetitve sequences with human Cot-1 fraction. The variant detection arrays (VDAs) were hybridized with enriched targets without adding Cot-1 (A) and with 30 ng/μL human Cot-1 fraction (B). The bright signals at the top of the image are attributable to unique sequences that were enriched. The dark region below had little intensity because the sequences were not enriched. The lower portion shows the cross-hybridization from repetitive sequences (A) that was diminished by adding human Cot-1 DNA.
SNP Validation with Bead-Selected Targets
In comparison to gel-based sequencing methods used to validate candidate SNPs, an alternative VDA-based method was employed (Fig. 1B). A total of 109 5′-biotinylated oligonucleotides of 25 bases in length were synthesized with possible biallelic variations encoded at the 13th position. These oligonucleotides were pooled according to chip designs and then bound to streptavidin magnetic beads. The sequences selected for the bead-bound oligonucleotides corresponded to 109 of the 713 EcoRI restriction fragments observed during the SNP discovery phase of the project. The 124 candidate SNPs were identified in these 109 EcoRI fragments.
The immobilized oligonucleotides were hybridized to the generically amplified EcoRI genome subset to select those fragments that have the designated candidate SNPs. The bead-selected EcoRI fragments were reamplified in a single PCR reaction as they all had a common adaptor. The amplification products were hybridized to the same VDA that was used to identify the original collection of 124 candidate SNPs. Because the resulting pool of targets was significantly enriched for the few targets, the hybridization signal was substantially improved (Fig. 3). A total of 99 of the 124 SNPs were definitively identified with high confidence, 78 concordant and 21 discordant with the initial screening. Among the 21 discordant candidates, 15 occurred in repeat sequences that were identified by RepeatMasker, nine of the 15 were from three fragments with multiple variations. Among the 25 fragments that were not definitively identified, eight were absent, 17 were either not called by the analysis program although the fragments were present, or possessed signals that resulted in a low confidence score. Reanalysis of the original VDA for all fragments that were absent after affinity selection and hybridization indicated that these fragments possessed low hybridization signals in the region that were recognized by the affinity oligonucleotides.
The chip-based validation was compared to the results of gel-based sequencing. Of the 124 candidate SNPs, 75 SNP candidates were analyzed by both sequencing and bead-selection/VDA validation methods. Both the sequencing and bead-based affinity/VDA validation methods were in agreement for 74 of the 75 candidate SNPs (98.7%). Results by both methods were in agreement concerning the identity of 64 SNPs as well as the fact that 10 of the sites did not contain candidate SNPs. A discordant result was observed at only one site.
DISCUSSION
Genome-Wide SNP Discovery
SNPs are the most abundant form of human genetic variation (Cooper et al. 1985). It is estimated that there are >106 SNPs loci in each human genome. High-density oligonucleotide arrays have been used for large-scale high throughput screening for many of these SNPs (Wang et al 1998; Cargill et al 1999; Hacia et al 1999; Halushka et al 1999; Lindblad-Toh et al. 2000). However, as a prelude to these screening experiments, each of the targeted genomic regions to be screened required individual amplification. To monitor 10% of the proposed SNPs in a copy of the human genome will require 2 × 105 primers. Such a sample preparation approach will be considerably costly and labor intensive given that a significant number of the amplification reactions are likely to fail in the initial attempts. As an assembled final draft of the human genome sequence approaches, an efficient, scalable, and robust sample preparation strategy is needed.
In this study, previously characterized human genomic sequences were initially analyzed by computer to identify the sites where the recognition sequence for the EcoRI restriction enzyme occur. A virtual restriction digestion of 250 Mb of human sequence genomic was carried out and the sequences from 713 EcoRI fragments (190 kb ) contained within the size-selected fragments were used to assist in the design of a SNP screening array. After that, actual EcoRI digestion and size selection were carried out and all the fragments present in the size-selected fragment pool were effectively amplified with overall amplification >2500-fold in 35 cycles of PCR. Although an EcoRI digestion and size selection of the resulting products allowed for identification of SNPs from across the human genome, the use of additional restriction enzymes followed by size selection would allow for greater uniform coverage. In subsequent experiments, fragments with lengths up to 1600 bp were amplified to a similar level of success using a similar strategy (data not shown).
The observed rate of SNP discovery in these experiments was one SNP per 1500 kb genome sequence (124 SNPs/190 kb). However, because ∼33% of the EcoRI fragment sequences are repetitive sequences, the corrected SNP rate for unique sequences was about one SNP per 1000 bp. Additionally, some regions of nonrepetitive sequences were sufficiently homologous to repetitive sequences to hybridize to the human Cot-1 DNA fraction. Because of the high concentration of the Cot-1 DNA used in the experiments, non repetitive sequence regions with up to five base mismatches with the repetitive sequences, especially Alu's, were blocked by the Cot-1 DNA. The real SNP rate was therefore close to previous results, about one SNP per 700 bp in a survey of eight individuals. The estimated occurrence of SNPs in unique sequences in this study was similar to previously reported locus-specific screening results (Wang et al. 1998). For future experiments, omission of repetitive sequences from the chip design is advisable.
Given the availability of >1 × 106 SNPs in the National Center for Biotechnology Information (NCBI) public databases, it is perhaps timely to note that the same strategy can be employed to genotype these newly identified SNPs. By use of precharacterized restriction enzyme digestion and size selection, a collection of hundreds of thousands of SNPs can be included in a restriction enzyme fragment subset, interrogated in one high-density oligonucleotide array, and assayed with one single-tube sample preparation.
Large-Scale Hybridization-Based SNP Validation
An alternative sample preparation has been employed to validate the SNPs discovered with the same chip design. Specific DNA fragment targets were selectively enriched from the total pool by use of oligonucleotides bound to magnetic beads. These affinity oligonucleotides hybridize near the SNP sites. Similar enrichment was obtained when using EcoRI-fragmented total genomic DNA as the target in place of restriction enzyme generated fragments (data not shown).
In a single-pass experiment, the VDA validation gave rise to a similar rate of validation with the gel-sequencing validation and highly concordant results. Eighty percent (99 out of 124) of the tested SNP candidates were definitively identified by hybridization validation whereas 74% (92 out of 124) were identified by gel sequencing. Among the 75 SNP candidates that were definitively determined, 74 were concordant between the two validations. The hybridization-based validation is comparable to the gel-sequencing but uses only one oligonucleotide instead of two primers in PCR amplification. Such a strategy can be used to validate the SNPs that will be generated by the SNP consortium (Marshall 1999; http://snp.cshl.org/) as well as provide a general method to select SNPs of choice for genotyping.
METHODS
Subset Selection and Chip Design
Available human genomic sequences were obtained from the following databases of genome centers: Baylor College of Medicine Human Genome Sequencing Center (http://www.hgsc.bcm.tmc.edu/), Lawrence Berkeley Laboratory Human Genome Center (http://www-hgc.lbl.gov/GenomeHome.html)(now a part of Joint Genome Institute, Department of Energy), Stanford Human Genome Center (http://www-shgc.stanford.edu/), The Sanger Centre (http://www.sanger.ac.uk/), The Institute for Genome Research (http://www.tigr.org/), Washington University Genome Sequencing Center (http://genome.wustl.edu/gsc), and Whitehead Institute for Biomedical Research/MIT(http://www.genome.wi.mit.edu/)] in FASTA format. The version of these sequences was as of February 1999. These sequences were analyzed for the EcoRI restriction recognition sequences and a list of fragments was generated. The size of the fragments was calculated and fragments of the designated size range were output to a .txt file. The sequences were not filtered for repetitive sequences.
A total of 713 fragments of 244–344 bp were selected based on their distribution among 23 chromosomes from the output sequences. The nucleotide sequences within these fragments were built on seven chip designs to interrogate 190,000 bp. These fragments were further divided into 244–269 bp, 270–294 bp, 295–319 bp, and 320–344 bp ranges. The fragments within each size range were split into two sets according to the first base 3′ to the recognition site. The 117 fragments that have an A or T base right 3′ to the recognition sites at both ends in the 244–268 bp range were put in chip design A. Similarly, chip design B had 114 fragments that have a C or G base right 3′ to the recognition sites at both ends in the 244–268 bp range. Design C had 103 fragments that have an A or T base right 3′ to the recognition sites at both ends in the 270–294 bp range. Design D had 103 fragments that have an A or T base right 3′ to the recognition sites at both ends in the 270–294 bp range. Design E had 95 fragments that have an A or T base right 3′ to the recognition sites at both ends in the 295–319 bp range. Design F had 95 fragments that have a C or G base right 3′ to the recognition sites at both ends in the 295–319 bp range. Finally, design G had 88 fragments that have an A or T base right 3′ to the recognition sites at both ends in the 320–344 bp range.
DNA Samples Screened
The eight unrelated individuals (five females and three males) were chosen from Centre d'Etude du Polymorphisme Human (CEPH) pedigrees from Amish (K104-1, -16), Venezuelan (K884-2, -15, -16) and Utah population (K1331-12, -13) as well as an apparently normal nonfetal tissue (NA14672). The genomic DNA samples were either purchased (NA05963B, NA07340A, and NA14672) or isolated from cell lines (GM11036, GM13057, GM05961, GM05995, and GM07007) ordered from the Coriell Cell Repository.
Size Selection and Amplification
A total of 5 μg human genomic DNA ordered from Coriell or isolated from cell lines was digested with 50 units of EcoRI in a final volume of 40 μL solution at 37°C overnight. The digestion mixtures were mixed with 10 μL 6× loading buffer and loaded to 2% agarose gel and separated by electrophoresis in 1× TBE buffer at 4 V/cm. Fragments between 244–344 bp were excised from the gel, isolated with QIAGEN gel extraction kit as per the manufacturer's instructions, ligated to adaptors at 16°C overnight with T4 DNA ligase, and amplified by PCR. The adaptor (5′-GAT CCG AAG GGG TTC G-3′ and 5′-phosphate-AAT TCG AAC CCC TTC GGA TC-3′) and primer (5′-GAT CCG AAG GGG TTC GAA TT-3′) were synthesized by Operon. The PCR reaction contained 15 mM Tris-HCl (pH 8.0), 10 mM KCl, 5 mM MgCl2, 200 μM dNTPs, 5 μM primer, 1 ng ligated DNA, and 5 units of AmpliTaq Gold polymerase (Perkin Elmer). The PCR was performed in a PE-9600 thermocylcer (Perkin Elmer) with 35 cycles of 94°C (30 sec) → 57°C (30 sec) →72°C (2 min).
Target Fragmentation, Labeling and Hybridization, Scanning, and Analysis
PCR products for the eight individuals were purified with QIAGEN kits and fragmented with DNase I. In each 45 μL reaction, 5 μg DNA were digested with 0.6 units of DNase I (Promega) at 37°C for 15 min in 10 mM Tris-acetate (pH 7.5), 10 mM magnesium acetate, and 50 mM potassium acetate. After inactivation of DNase I at 95°C for 15 min, the mixtures were added 2 pmole biotin-N6-ddATP (NEN Life Science Products) and 45 units rTdT (GIBCO BRL), and incubated at 37°C for 1.5 h. 80 μg-Labeled DNA samples were pooled and reduced to 50 μL final volume, heat-denatured by boiling for 30 min, and immediately put on ice. Denatured enzymes were removed by centrifugation. The hybridization was carried out in hybridization solution (3.0 M tetramethylamonium chloride, 10 mM MES, 0.01% Triton-100, 0.5 μg BSA, 0.8 μg/μl human Cot-1 fraction) at 44°C for 40 h on a rotisserie.
Array Washing, Staining, and Scanning
The arrays were washed with 10 mM MES at pH 6.5, with 0.1 M NaCl and 0.01% Triton-100 at 44°C for 30 min, and stained with staining solution (0.01 μg/μL streptavidin-R-phycoerythrin conjugate in 100 mM MES at pH 6.5, with 1 M NaCl, 2.5 μg/μL BSA and 0.01% Triton-100) at 40°C for 15 min and washed on a fluidics station (Affymetrix). The arrays were stained by goat antistrepavidin antibody at 40°C for 30 min and stained by the staining solution before being scanned at 3.4 micron resolution on a GALVO scanner (Wang et al. 1998).
Data Analysis
Candidate SNPs were identified using the same algorithms and were visually reviewed as described previously (Chee et al 1996; Wang et al. 1998). On the array, four probe sequences were synthesized for each base position in each strand with each one at a distinguishable feature. One feature contains the expected base (the reference base) whereas the other three contain a mismatch at the center of the probe sequences. Together, these oligonucleotides test for all four possible bases at the interrogated position. Candidate SNPs were identified by a combination of three algorithms. The first one, base-calling, looked for positions at which hybridization to a substitution base gives a stronger signal than the reference base. “Accordance rate” was defined as the percentage of positions where the reference sequence has the highest intensities. A variant was called when a substitution base had 20% higher intensity than the reference base in any one individual. The second algorithm, mutant fraction, examined the reference base and each of the substitution bases in turn and calculated the fraction of signal present in the nonreference base. The mutant fractions for all eight samples were clustered together to identify possible homozygous and heterozygous individuals. The final algorithm, footprint detection, detected signal loss at the reference positions surrounding a nucleotide substitution by examining all individuals. Subsequently, a reviewer visually inspected the candidate SNPs. An empirical confidence score, “certain,” “likely,” and “possible,” was assigned to each candidate SNP based on the reviewer's judgment. Twenty six candidates were scored as “certain” and 98 as “likely.” Only those that were called with a high-confidence score of “certain” or “likely” were selected for validation and reported here.
Bead Enrichment of Target
We enriched 109 fragments on all chip designs with bead-assisted SNP-specific target selection. A 25-mer oligonucleotide was designed around one SNP in every fragment and synthesized with a 5′ biotin. The oligonucleotides were first bound to streptavidin-magnetic beads pretreated with 100 mM Na2P2O7 and then hybridized to 30 ng denatured PCR products of the genome subset or 3 μg genomic DNA (106copies). The mole ratio between the oligonucleotide and the target fragment was 10,000 : 1. The bead was hybridized to the target in 1× B&W buffer (5 mM Tris-HCl at pH 7.5, 0.5 mM EDTA, 2.0 M NaCl) at 50°C for 16 h on a rotisserie. The beads were washed twice with 1× B&W buffer and transferred to a new tube, and washed 6 times with 1× B&W buffer. The beads were finally washed once with water. DNA fragments were eluted in 30 μL water by incubation at 80°C for 2 min. The targets were amplified with PCR using the same conditions mentioned above. Approximately 2–3 μg purified DNA were fragmented, labeled, and hybridized to arrays. SNPs were called and compared to the initial results.
DNA Sequencing
We used gel-based nucleotide sequencing to confirm the majority of the identified SNPs. Primers were designed with Primer3 (http://www.genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi) for 106 fragments identified with the 121 SNPs classified as “certain” or “likely.” The other three fragments did not have enough flanking sequences for primer design. PCR products were obtained for 109 SNP candidates using genomic DNA as templates for the individuals that were classified as homozygous or heterozygous for an alternative base. The DNA samples were outsourced to Lark Technologies, Inc. for sequencing using dye terminator chemistry on ABI sequencers. The sequence traces were obtained for 92 SNP candidates (23 scored as “certain” and 69 as “likely”) and input into the same program used for SNP detection, and compared to the screening results by visual inspection. Seventy seven of the 92 SNPs (23 “certain,” 54 “likely”) were concordant with the initial screening results.
Acknowledgments
We thank Anthony Berno for the assistance with data analysis, Michael Mittmann and Earl Hubbell for VDA chip design, and Jian-Bing Fan and David J. Lockhart for early involvement in this work.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Shoulian_Dong@affymetrix.com; FAX (408) 481–0422.
Article published on-line before print: Genome Res., 10.1101/gr. 171101.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.171101.
REFERENCES
- Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Lane CR, Lim EP, Kalyanaraman N, Nemesh J, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- Chee M, Yang R, Hubbell E, Berno A, Huang XC, Stern D, Winkler J, Lockhart DJ, Morris MS, Fodor SPS. Accessing genetic information with high-density DNA arrays. Science. 1996;274:610–614. doi: 10.1126/science.274.5287.610. [DOI] [PubMed] [Google Scholar]
- Collins FS, Guyer MS, Charkravarti A. Variations in a theme: Cataloging human DNA sequence variation. Science. 1997;278:1580–1581. doi: 10.1126/science.278.5343.1580. [DOI] [PubMed] [Google Scholar]
- Cooper DN, Smith BA, Cooke HJ, Niemann S, Schmidtke J. An estimate of unique DNA sequence heterozygosity in the human genomes. Hum Genet. 1985;69:201–205. doi: 10.1007/BF00293024. [DOI] [PubMed] [Google Scholar]
- Hacia JG, Fan J-B, Ryder O, Jin L, Edgemon K, Ghandour G, Mayer RA, Sun B, Hsie L, Robins CM, et al. Determination of ancestral alleles for human single-nucleotide polymorphisms using high-density oligonucleotide arrays. Nat Genet. 1999;22:164–167. doi: 10.1038/9674. [DOI] [PubMed] [Google Scholar]
- Halushka MK, Fan J-B, Bentley K, Hsie L, Shen N, Weder A, Cooper R, Lipshutz R, Chakravarti A. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat Genet. 1999;22:239–247. doi: 10.1038/10297. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Winchester E, Daly MJ, Wang DG, Hirschhorn JN, Laviolette J-P, Ardlie K, Reich DE, Robinson E, Sklar P, et al. Large-scale discovery and genotyping of single-nucleotide polymorphisms in the mouse. Nat Genet. 2000;24:381–386. doi: 10.1038/74215. [DOI] [PubMed] [Google Scholar]
- Marshall E. Drug firms to create public database of genetic mutations. Science. 1999;284:406–407. doi: 10.1126/science.284.5413.406. [DOI] [PubMed] [Google Scholar]
- Wang DG, Fan J-B, Siao C-J, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]