Abstract
Post-transcriptional gene regulation, a regulatory mechanism classically involved in female and male germ cell function has recently been implicated in control of somatic cells of the ovary and testis. Recent advancements in this field may be attributed primarily to the discovery and study of microRNAs (miRNA), small RNA transcripts that can influence mRNA expression via post-transcriptional gene regulatory mechanisms. In the ovary, targeted deletion of Dicer 1, a key enzyme in miRNA biogenesis, provided the first empirical evidence that miRNA/siRNA were critically involved in multiple aspects of ovarian function (folliculogenesis, oocyte maturation, ovulation, and luteal function). Functional studies of miRNA in the ovary have mostly focused on granulosa cells during the critical period of the ovarian cycle surrounding the ovulatory surge of luteinizing hormone (LH). Specific miRNA have been implicated in ovarian responses, due to their transcriptional induction by the LH surge (i.e., miR-21, -132 and -212) or through bioinformatic approaches (miR-224, -17-5p and let-7b). Numerous other miRNA are highly abundant in ovarian somatic tissues, suggesting that we have much to discover with respect to the role of miRNA and regulation of ovarian function. This review will recap the key observations of these early studies and provide insight into future experiments that might further our understanding of ovarian function.
Keywords: microRNA, ovary, oocyte, granulosa
Introduction
Post-transcriptional gene regulation refers to many different regulatory events that can occur following transcription of a nascent messenger RNA transcript. These post-transcriptional processes include: RNA attenuation, alternative splicing, polyadenylation, RNA editing, mRNA transport/localization, mRNA degradation and events associated with the initiation and regulation of translation. Interestingly, many of these post-transcriptional gene regulatory mechanisms have been well studied in both the male and female germ cells, as it was recognized that germ cell transcription and protein synthesis were uncoupled at different periods during gamete development (Racki and Richter 2006). Most recently the identification and discovery of microRNA (miRNA) mediated regulation of gene expression has led to resurgence in the study of post-transcriptional gene regulation (Bartel 2009). MicroRNA function in reproductive tissues and diseases have been covered in depth in previous reviews (Carletti and Christenson 2009; Luense, Carletti et al. 2009). In this mini-review, I will introduce what we have learned about miRNA in the ovary through the use of mouse genetic models and studies of specific miRNA that are differentially expressed in ovarian granulosa cells.
Biosynthesis of small non-coding RNAs
Most miRNA arise from the progressive processing of a large RNA transcript, referred to as primary-miRNA (pri-miRNA) by Drosha and its RNA-binding cofactor DiGeorge syndrome critical region gene 8 (DGCR8) (Bartel 2004). Within the nucleus, these two proteins convert pri-miRNA to ~70-100 base precursor-miRNA (pre-miRNA) that contain a characteristic hairpin loop. The pre-miRNAs are transported to the cytosol where Dicer, a RNAse III endonuclease, removes the hairpin loop to form the ~21 base nucleotide mature miRNA (Bartel 2004). Direct transcription of short hairpin RNAs and mirtrons (i.e., pre-miRNA that arise from intron excision) short circuit the standard miRNA biogenesis process by eliminating the need for Drosha/DGCR8 mediated cleavage (Berezikov, Chung et al. 2007; Babiarz, Ruby et al. 2008). These alternative “pre-miRNA” still require Dicer to remove the hairpin loop and generate the mature miRNA. Thus, a number of laboratories designed floxed Dicer1 (a.k.a. Dicer) deletion constructs to investigate the role of miRNA (Harfe, McManus et al. 2005; Andl, Murchison et al. 2006; Yi, O’Carroll et al. 2006; Mudhasani, Zhu et al. 2008). However, the recent identification of endogenous small interfering RNA (siRNA) that require Dicer for their synthesis and a Dicer independent miRNA biogenic pathway has muddied the simple “paradigm” that deletion of Dicer was synonymous with deletion of all miRNAs.
Endogenous siRNA utilize the well-known RNAi pathway in which Dicer cleaves double stranded RNA (dsRNA) to manipulate gene expression (Babiarz, Ruby et al. 2008; Tam, Aravin et al. 2008; Watanabe, Totoki et al. 2008). In this regulatory system, endogenous dsRNAs are postulated to originate from pseudogenes that encode a complementary mRNA allowing for formation of dsRNA templates needed for Dicer cleavage and formation of endogenous siRNAs (Tam, Aravin et al. 2008; Watanabe, Totoki et al. 2008). To separate the effects of these closely related classes of RNA species (i.e., miRNA and siRNA), laboratories have subsequently targeted the deletion of DGCR8 in tissues, which should then specifically block the miRNA pathway and leave the endogenous siRNA pathway intact (Wang, Medvid et al. 2007).
Lastly, the Dicer independent miRNA biogenesis pathway (Cheloufi, Dos Santos et al. ; Cifuentes, Xue et al.) adds even further complexity to interpretation of the miRNA/siRNA deletion studies. The Dicer independent miRNA synthesis pathway utilizes argonaute 2 (Ago2), a key component of the RNA induced silencing complex (RISC) to catalyze the synthesis of miR-451 from its pre-miRNA form (Cheloufi, Dos Santos et al. ; Cifuentes, Xue et al.). It should be noted that the Dicer independent pathway appears to operate on a specific miRNA (Cheloufi, Dos Santos et al. ; Cifuentes, Xue et al.), ongoing studies will be needed to determine the importance of this biogenic pathway and whether it involves the synthesis of other miRNAs.
Ovarian small non-coding RNAs
MicroRNA, endogenous siRNA and unique Piwi interacting RNA (piRNA) make up the majority of small (< 30 bases) non-coding regulatory RNAs within ovarian tissue (Ahn, Morin et al. ; Tripurani, Xiao et al. ; Choi, Qin et al. 2007; Ro, Song et al. 2007; Mishima, Takizawa et al. 2008; Sirotkin, Ovcharenko et al. 2009; Yao, Lu et al. 2009). Thus far, most of the cloning and next-generation sequencing studies that have identified the small non-coding RNA have utilized whole ovarian tissues, which limit our understanding of the functional role these small RNA play within the ovary. This is largely due to the diverse and heterogeneous nature of ovarian tissue, which contains germ cells and somatic cells (i.e., granulosa, thecal, luteal, endothelial, immune cells, epithelial cells, etc) at varying stages of maturation. While the consensus of the literature is that piRNAs are of germ cell origin (Thomson and Lin 2009), it has not yet been proven that ovarian somatic cells cannot produce these unique RNA transcripts. With respect to endogenous siRNAs, they have been clearly identified in oocytes and in cultured cell lines (Babiarz, Ruby et al. 2008; Tam, Aravin et al. 2008; Watanabe, Totoki et al. 2008), but not yet in ovarian somatic tissue. In the case of miRNA, our laboratory has clearly shown that granulosa cells collected immediately before and 4 h after the ovulatory surge of LH/hCG exhibit differential miRNA gene expression (Fiedler, Carletti et al. 2008). This study indicated that granulosa cells expressed over 212 different miRNA. Interestingly, current studies indicate that while miRNA are present in the oocyte, they appear to have limited or severely suppressed activity (Ma, Flemr et al. ; Suh, Baehner et al.). Lastly, the analysis of miRNA gene expression and function in other ovarian somatic cells such as thecal and luteal cells remains unexplored.
Targeted deletion of Dicer and DGCR8 in ovarian cells
In 2007, Murchison et al. reported the oocyte specific depletion of Dicer in mice using a ZP-3 promoter-Cre (Murchison, Stein et al. 2007). This study revealed that although ZP-3 expression occurs early in the development of the oocyte, the only oocyte defects occurred after the ovulatory surge, with defects in spindle organization and chromatin segregation being observed during meiosis (Murchison, Stein et al. 2007). Moreover, ovarian folliculogenesis appeared normal, indicating that oocyte miRNA and endogenous siRNA post-transcriptional gene regulation did not impact those oocyte genes involved in regulation of the surrounding cumulus cells. The extent of oocyte endogenous siRNA regulation and the specific genes regulated by this mechanism are not well established, although it has been observed that a number of genes involved in microtubule-based processes were upregulated following loss of Dicer (Tam, Aravin et al. 2008). Utilizing two markedly different approaches the laboratories of Schultz/Svoboda and Blelloch (Ma, Flemr et al. 2010; Suh, Baehner et al. 2010) have demonstrated that miRNA function is limited in oocytes and early embryos. Using a series of reporter assays, Ma, Flemr et al. (2010) showed that fully grown GV oocytes and MII eggs exhibit decreased miRNA activity. The targeted deletion of oocyte DGCR8, an enzyme specific for miRNA biosynthesis, had no adverse effect on maturation of oocytes (Suh, Baehner et al. 2010). This finding implicated endogenous siRNA in the developmental abnormalities caused by knocking out Dicer (Suh, Baehner et al. 2010).
In 2008 and 2009, our laboratory and others reported that Amhr2-Cre mediated deletion of Dicer in mice had profound effects on uterine and oviductal development and function as well as ovarian function (Hong, Luense et al. 2008; Nagaraja, Andreu-Vieyra et al. 2008; Gonzalez and Behringer 2009; Pastorelli, Wells et al. 2009). Female infertility was shown to be a result of a primary oviductal defect (Hong, Luense et al. 2008; Gonzalez and Behringer 2009). Additionally, these mice exhibited reduced ovarian function as evidenced by decreased ovulation rates (natural and gonadotropin-induced; Hong, Luense et al. 2008; Nagaraja, Andreu-Vieyra et al. 2008), which was attributed to loss of miRNA within the Amhr-2 expressing granulosa cells. Furthermore, female infertility was observed in another mouse model that exhibits an 80% reduction in DICER expression (Otsuka, Zheng et al. 2008). These female mice exhibited normal ovulation rates but the corpora lutea had reduced progesterone output and as a result the mice were unable to sustain pregnancies. These investigators went on to show that loss of Dicer decreased vascularization of the corpora lutea and that exogenous administration of two miRNA (miR-17-5p and let-7b) could prevent this loss (Otsuka, Zheng et al. 2008). These studies indicate a role for Dicer in ovarian function; however, the role of endogenous siRNA versus miRNA mediated post-transcriptional gene regulation has not been directly addressed in ovarian somatic cells.
MicroRNA Control of Ovarian Function
Our understanding of how specific miRNA regulate developmental and physiological events is in its infancy in most organ systems including the ovary. In addition to the fore mentioned miR-17-5p and let-7b, which were linked to luteal tissue vascularization (Otsuka, Zheng et al. 2008), only 4 other miRNA have been functionally studied in the ovary. Three of these miRNA, miR-21, -132, and -212 increased in granulosa cells following the LH surge (Fiedler, Carletti et al. 2008), while miR-224 expression increased in cultured mouse granulosa cells in response to TGF-β1 exposure (Yao, Yin et al.). The putative functional roles these later four miRNA have on granulosa cell function and ovarian physiology are discussed below.
MicroRNA-224 was identified as one of 16 TGF-β1 regulated miRNA in cultured murine preantral granulosa cells (Yao, Yin et al. 2010). Using bioinformatics these investigators identified Smad4 as a potential target of miR-224 and demonstrated that over expression of miR-224 decreased granulosa cell Smad4 protein levels dramatically. Consistent with a miRNA effect on translation, Smad4 mRNA expression was not significantly changed (although a non-significant 10-15% decrease was observed). The functional aspects of miR-224 over expression and inhibition on granulosa cell proliferation and steroidogenesis were difficult to interpret due to inappropriate statistical analyses and inconsistent induction of miR-224 expression by TGF-β1 (Yao, Yin et al. 2010).
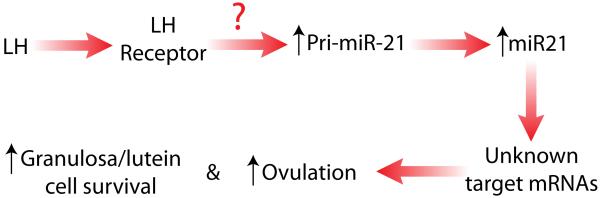
MicroRNA-21, a widely studied miRNA due to its general upregulation in almost all cancers and tumors, was shown to rapidly increase following in vivo LH/hCG administration (Carletti, Fiedler et al. 2010). Expression of mature miR-21 in granulosa cells was preceded by increased in vivo transcription of pri-miR-21. However, when granulosa cells were placed into culture, mature miR-21 expression increased without accompanied changes in pri-miR-21 expression. Furthermore, exogenous administration of cAMP (i.e., 8-bromo-cAMP) had no effect on miR-21 expression in cultured granulosa cells. Interestingly, expression of miR-21 has previously been shown to it self be post-transcriptionally regulated (Davis, Hilyard et al., 2008). Consistent with findings in a number of other cell types, inhibition of miR-21 activity in cultured granulosa cells using a complementary locked nucleic acid (LNA-21) induced apoptosis (Carletti, Fiedler et al. 2010). In other cells a number of miR-21 target genes (>10) have been implicated in miR-21’s anti-apoptotic effects including PTEN, sprouty 2, tropomyocin 1 and programmed cell death 4 (PDCD4) and others see references within (Carletti, Fiedler et al. 2010). Remarkably, none of these known miR-21 target transcript have been shown to change in granulosa cells, therefore the granulosa cell miR-21 target transcript(s) have remained elusive. This observation that targets of miRNA are cell specific is becoming increasing established in the literature (Sood, Krek, et al., 2006). It is noteworthy, that in vivo blockade of miR-21 action following ovarian bursal injection with a blocking LNA-21 decreased the ovulation rate (Carletti, Fiedler et al. 2010). This effect was similar to the effect targeted deletion of Dicer had on ovulation rate (Hong, Luense et al. 2008; Nagaraja, Andreu-Vieyra et al. 2008). These studies indicate a role for miR-21 in preventing apoptosis in the periovulatory granulosa cells as they transition to luteal cells, yet the mechanisms and genes regulated by miR-21 remain to be determined (Figure 1).
Figure 1.
The ovulatory surge of LH signals through the LH receptor on granulosa cells to induce the expression of the pri-miR-21 transcript and ultimately the mature form of the miR-21. MiR-21 in turn regulates the expression of yet unknown gene transcripts to maintain granulosa cell survival and promote ovulation. The ? signifies that we have yet to uncover the transcription factors and intracellular cell signaling cascades that mediate LH-induced increased pri-miR-21 transcription.
In contrast to miR-21, the other two in vivo LH/hCG-induced miRNA (i.e., miR-132 and miR-212) did not increase in granulosa cells upon placement in to culture. Moreover, granulosa cell miR-132 and miR-212, which arise from a single pri-miRNA transcript, exhibited a robust increase in response to 8-Br-cAMP (Fiedler, Carletti et al. 2008). The promoter that drives pri-miR132/212 expression is responsive to cAMP and contains two CREB binding sites (Vo, Klein et al. 2005). Linked to the recognition of the target mRNA transcripts, the seed sequence (i.e., bases 2-8) of these two miRNA share 100% homology suggesting that these miRNA likely regulate a similar set of genes. C-terminal binding protein -1 (CTBP-1) a putative miR-132/212 target gene identified in hippocampus cells was also regulated in murine granulosa cells following blockade of miR-132 and miR-212 action with complementary LNA-oligonucleotides (Vo, Klein et al. 2005; Fiedler, Carletti et al. 2008). However, while a target protein was identified no functional effect on granulosa cell steroidogenesis, cell proliferation, apoptosis was observed following loss of miR-132/212 action (Fiedler, Carletti et al. 2008). Recently, in a study describing cocaine addiction, the phospho-CREB induced expression of miR-212 was shown to regulate an inhibitor of Raf-1 signaling cascade (Hollander, Im et al. 2010). In turn the loss of repression of Raf-1 maintained the increase in phospho-CREB, which could then upregulate miR-132/212 promoter activity and further increase miR-212 expression, setting up a positive feed-forward loop. Following the LH surge the LH receptor is downregulated (Menon, Menon et al. 2010), this type of feed-forward loop could be envisioned to maintain the intracellular signaling required for luteinization.
Conclusions
Insights into ovarian miRNA expression and function are just beginning to be elucidated. Numerous miRNA have been identified in granulosa cells of the ovary with unknown function, several select miRNA have been shown to exhibit differential expression in response to hormonal stimulation and putative functions are being unraveled for these miRNA. Conversely, expression of miRNA by other somatic cells (i.e., thecal and luteal) within the ovary remains unknown. While a role for miRNA in the etiology of diseases in other tissues has been established, whether ovarian miRNA might have a similar role in a reproductive infertility disorders such as polycystic ovarian syndrome (PCOS) has not yet been reported. While much remains to be discovered with respect to miRNA in ovarian tissue, the studies discussed indicate a significant role for miRNA regulation of gene function in the ovary.
Acknowledgements
I would like to thank Dr. Vargheese Chennathukuzhi, Lacey Luense and JB Fitzgerald for their helpful suggestions and critical reading of this review article.
This work was supported in part by funds from NIH grant HD061580 (to L.K.C.) and P20 RR016475 from the National Center for Research Resources. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health.”
References
- Ahn HW, Morin RD, et al. MicroRNA transcriptome in the newborn mouse ovaries determined by massive parallel sequencing. Mol Hum Reprod. 16(7):463–71. doi: 10.1093/molehr/gaq017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andl T, Murchison EP, et al. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol. 2006;16(10):1041–9. doi: 10.1016/j.cub.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, et al. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22(20):2773–85. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Chung WJ, et al. Mammalian mirtron genes. Mol Cell. 2007;28(2):328–36. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carletti MZ, Christenson LK. MicroRNA in the ovary and female reproductive tract. J Anim Sci. 2009;87(14 Suppl):E29–38. doi: 10.2527/jas.2008-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carletti MZ, Fiedler SD, et al. MicroRNA 21 Blocks Apoptosis in Mouse Periovulatory Granulosa Cells. Biol Reprod. doi: 10.1095/biolreprod.109.081448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, et al. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 465(7298):584–9. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Qin Y, et al. Microarray analyses of newborn mouse ovaries lacking Nobox. Biol Reprod. 2007;77(2):312–9. doi: 10.1095/biolreprod.107.060459. [DOI] [PubMed] [Google Scholar]
- Cifuentes D, Xue H, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 328(5986):1694–8. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, et al. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454(7200):56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler SD, Carletti MZ, et al. Hormonal regulation of MicroRNA expression in periovulatory mouse mural granulosa cells. Biol Reprod. 2008;79(6):1030–7. doi: 10.1095/biolreprod.108.069690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez G, Behringer RR. Dicer is required for female reproductive tract development and fertility in the mouse. Mol Reprod Dev. 2009;76(7):678–88. doi: 10.1002/mrd.21010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, McManus MT, et al. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005;102(31):10898–903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander JA, Im HI, et al. Striatal microRNA controls cocaine intake through CREB signalling. Nature. 466(7303):197–202. doi: 10.1038/nature09202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong X, Luense LJ, et al. Dicer1 is essential for female fertility and normal development of the female reproductive system. Endocrinology. 2008;149(12):6207–12. doi: 10.1210/en.2008-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luense LJ, Carletti MZ, et al. Role of Dicer in female fertility. Trends Endocrinol Metab. 2009;20(6):265–72. doi: 10.1016/j.tem.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Flemr M, et al. MicroRNA activity is suppressed in mouse oocytes. Curr Biol. 20(3):265–70. doi: 10.1016/j.cub.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon KM, Menon B, et al. Molecular regulation of gonadotropin receptor expression: Relationship to sterol metabolism. Mol Cell Endocrinol. doi: 10.1016/j.mce.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima T, Takizawa T, et al. MicroRNA (miRNA) cloning analysis reveals sex differences in miRNA expression profiles between adult mouse testis and ovary. Reproduction. 2008;136(6):811–22. doi: 10.1530/REP-08-0349. [DOI] [PubMed] [Google Scholar]
- Mudhasani R, Zhu Z, et al. Loss of miRNA biogenesis induces p19Arf-p53 signaling and senescence in primary cells. J Cell Biol. 2008;181(7):1055–63. doi: 10.1083/jcb.200802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Stein P, et al. Critical roles for Dicer in the female germline. Genes Dev. 2007;21(6):682–93. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraja AK, Andreu-Vieyra C, et al. Deletion of Dicer in somatic cells of the female reproductive tract causes sterility. Mol Endocrinol. 2008;22(10):2336–52. doi: 10.1210/me.2008-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka M, Zheng M, et al. Impaired microRNA processing causes corpus luteum insufficiency and infertility in mice. J Clin Invest. 2008;118(5):1944–54. doi: 10.1172/JCI33680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorelli LM, Wells S, et al. Genetic analyses reveal a requirement for Dicer1 in the mouse urogenital tract. Mamm Genome. 2009;20(3):140–51. doi: 10.1007/s00335-008-9169-y. [DOI] [PubMed] [Google Scholar]
- Racki WJ, Richter JD. CPEB controls oocyte growth and follicle development in the mouse. Development. 2006;133(22):4527–37. doi: 10.1242/dev.02651. [DOI] [PubMed] [Google Scholar]
- Ro S, Song R, et al. Cloning and expression profiling of small RNAs expressed in the mouse ovary. RNA. 2007;13(12):2366–80. doi: 10.1261/rna.754207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirotkin AV, Ovcharenko D, et al. Identification of microRNAs controlling human ovarian cell steroidogenesis via a genome-scale screen. J Cell Physiol. 2009;219(2):415–20. doi: 10.1002/jcp.21689. [DOI] [PubMed] [Google Scholar]
- Sood P, Krek A, et al. Cell type-specific signatures of microRNAs on target mRNA expression. PNAS. 2006;103(8):2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh N, Baehner L, et al. MicroRNA function is globally suppressed in mouse oocytes and early embryos. Curr Biol. 20(3):271–7. doi: 10.1016/j.cub.2009.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453(7194):534–8. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol. 2009;25:355–76. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripurani SK, Xiao C, et al. Cloning and analysis of fetal ovary microRNAs in cattle. Anim Reprod Sci. 120(1-4):16–22. doi: 10.1016/j.anireprosci.2010.03.001. [DOI] [PubMed] [Google Scholar]
- Vo N, Klein ME, et al. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci U S A. 2005;102(45):16426–31. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Medvid R, et al. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39(3):380–5. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453(7194):539–43. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Yao G, Yin M, et al. MicroRNA-224 is involved in transforming growth factor-beta-mediated mouse granulosa cell proliferation and granulosa cell function by targeting Smad4. Mol Endocrinol. 24(3):540–51. doi: 10.1210/me.2009-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao N, Lu CL, et al. A network of miRNAs expressed in the ovary are regulated by FSH. Front Biosci. 2009;14:3239–45. doi: 10.2741/3447. [DOI] [PubMed] [Google Scholar]
- Yi R, O’Carroll D, et al. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet. 2006;38(3):356–62. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]