Abstract
Background
Various virulence factors and superantigens are encoded by mobile genetic elements. The relationship between clonal background and virulence factors differs in different geographic regions. We compared the distribution and relationship of spa types and virulence genes among methicillin-resistant Staphylococcus aureus (MRSA) strains isolated from a tertiary hospital in 2000-01 and 2007-08.
Methods
In 2000-01 and 2007-08, 94 MRSA strains were collected from 3 intensive care units at a Korean tertiary hospital. We performed spa typing and multiplex PCR for 19 superantigen genes.
Results
Relatively frequent spa types were t037 (40.5%), t002, t601, and t2138 in 2000-01, and t2460 (43.9%), t002, t037, t601, t324, and t2139 in 2007-08. We identified 4 novel spa types, 2 of which were designated as t5076 and t5079. Superantigen profiles were closely linked to spa types. For example, sea, sek, and seq superantigen genes were mainly detected in t037 strains.
Conclusions
Major spa types differed depending on study periods, and the distribution of superantigen genes correlated with spa type.
Keywords: Methicillin-resistant Staphylococcus aureus, spa Typing, Superantigen, Virulence factor
INTRODUCTION
Staphylococcus aureus is the causal pathogen in a wide range of infectious diseases ranging from skin and soft tissue infections to the toxin-mediated diseases pneumonia and bacteremia. S. aureus may also be found in the normal flora of up to 50% of the global population [1]. Since the introduction of penicillin in the 1950s, the control and treatment of S. aureus infection has improved dramatically, but the emergence of methicillin-resistant S. aureus (MRSA) has become a serious threat [2].
Methicillin resistance is encoded by the mecA gene carried by a mobile genetic element, namely, staphylococcal cassette chromosome mec (SCCmec) [3], and in many strains of S. aureus, including MRSA, characteristic superantigens are produced. Initially, 5 types (SEA-SEE) of superantigens were identified, and a total of 19 types have been reported thus far [1].
S. aureus is a polymorphic species with a clonal population structure that is not significantly affected by genetic recombination and mutation; hence, the species exhibits a relatively high degree of genetic linkage disequilibrium [4]. Because of these features, highly discriminating genetic markers are needed to support epidemiological research and prevalence surveys.
Pulsed-field gel electrophoresis (PFGE) is commonly used to determine genetic background, but it is unsuitable for long-term epidemiological studies. Multilocus enzyme electrophoresis and multilocus sequence typing (MLST) have begun to replace PFGE; however, these methods have the disadvantage of yielding data of relatively low resolution. Frénay et al. [5] introduced spa typing, a method by which the polymorphic staphylococcal protein A (spa) coding region is used to identify the genetic background of S. aureus with discriminatory power similar to that of PFGE.
Whole genome analysis has revealed that 75% of the S. aureus genome is comprised of core sequences that represent the clonal background; 10% comprises core variable sequences, and 15% comprises mobile genetic elements [6]. Mobile genetic elements such as plasmids, phages, pathogenicity islands, and genomic islands encode resistance and virulence genes; intra-species or inter-species propagation occurs through horizontal or vertical transfer of these mobile elements. Various virulence factors, including 19 superantigens, are encoded on mobile genetic elements, and the relationship between clonal backgrounds and virulence factors differs in different geographic regions.
We compared the distribution and relationship of spa types and virulence genes among MRSA strains isolated from the intensive care units of a tertiary hospital in 2000-01 and 2007-08. Additionally, periodic changes in spa type prevalence and the frequency of mobile element transfer are identified.
MATERIALS AND METHODS
1. Bacterial strains
Ninety-four strains of S. aureus were isolated from patients suffering from bacteremia in 3 intensive care units (ICUs) (medical ICU [MICU], surgical ICU [SICU], respiratory ICU [RICU]) of a tertiary hospital (Seoul National University Hospital) between January 2000 and December 2001 and between January 2007 and December 2008. A total of 37 S. aureus strains were isolated from the 2000-01 clinical samples. PCR amplification of the spa gene failed in 1 isolate in the year 2000 and in 3 isolates in the year 2001. Therefore, 33 spa-positive isolates (18 MICU strains, 10 SICU strains, and 5 RICU strains) were analyzed. Between January 2007 and December 2008, 57 strains were investigated, and all tested positive for the spa gene. In 2007, 17 strains were isolated from MICU patients; 9, from SICU patients; and 4, from RICU patients. In 2008, 16 strains were isolated from MICU; 10, from SICU; and 1, from RICU. The strains were stored at -70℃.
2. Antibiotic susceptibility tests
Antibiotic susceptibility test profiles for 12 antimicrobial agents (oxacillin, penicillin, gentamicin, ciprofloxacin, clindamycin, chloramphenicol, erythromycin, rifampin, tetracycline, trimethoprim-sulfamethoxazole, vancomycin, and teicoplanin) were determined by the disc diffusion method for the 2000-01 strains. For the 2007-08 strains, the antimicrobial susceptibility tests were performed with disk diffusion method or automated systems such as MicroScan Walk-Away (Dade Behring Inc., West Sacramento, Canada) or VITEK2 (bioMerieux, Marcy l'Etoile, France). Standards for antimicrobial susceptibility testing and interpretation were based on the CLSI standard M100-S10 [7] for 2000-01 strains, and CLSI standard M100-S17 [8] for 2007-08 strains.
3. Nucleic acid extraction
The strains were thawed and inoculated onto blood agar plates using a 10-µL disposable loop, then incubated for 24 hr at 35℃. Colonies were suspended and collected in 1.5-mL tubes with 200 µL distilled water. DNA was extracted using the QIAamp® DNA mini kit (Qiagen GmbH, Hilden, Germany). Nucleic acid extracts were centrifuged at 8,000 rpm for 1 min and stored at -20℃.
4. PCR amplification of mecA
PCR amplification of the mecA gene was performed to confirm MRSA. PCR was performed with 2 µL extracted template DNA, 2 µL mecA primers (GMECAR-1: 5'-ACT GCT ATC CAC CCT CAA AC-3', GMECAR-2: 5'-CTG GTG AAG TTG TAA TCT GG-3') [9], and 10 µL 2× Multiplex master mix (Seegene, Seoul, Korea) in a final volume of 20 µL. The thermal cycling program was as follows: initial denaturation (5 min at 94℃); followed by 30 cycles of denaturation (60 sec at 94℃), annealing (60 sec at 53℃), and extension (60 sec at 72℃); and a single extension (7 min at 72℃).
5. Multiplex PCR amplification of superantigen genes
According to the method described by Hwang et al. [10], multiplex PCR amplification of 20 target genes, including 19 superantigen genes and femA, was performed with 2 µL extracted template DNA, 4 µL 5× primer mixture (Seegene), and 10 µL 2× Multiplex master mix (Seegene) in a final volume of 20 µL. The thermal cycling program was as follows: 15 min at 94℃; 30 cycles of 30 sec at 94℃, 30 sec at 55℃, and 120 sec at 72℃; followed by 10 min at 72℃. A mixture of 7 reference strains (FRI137, FRI361, FRI472, FRI569, FRI913, MNHOCH, and RN4220) was used as the positive control [10].
6. spa typing
spa gene typing was conducted according to the procedure described by Ridom GmbH (http://www.ridom.de). Each reaction mixture contained 5 µL DNA, 1 µL of each primer (spa-1113f: 5'-TAA AGA CGA TCC TTC GGT GAG C-3', spa-1514r: 5'-CAG CAG TAG TGC CGT TTG CTT-3'), and 25 µL Takara premix EX-Taq (Takara Bio Inc., Shiga, Japan) in a final volume of 50 µL. The thermal cycling program was as follows: 94℃ for 5 min; 35 cycles of 94℃ for 30 sec, 55℃ for 45 sec, and 72℃ for 90 sec; followed by 72℃ for 10 min. Amplified PCR products were visualized by agarose gel electrophoresis in a 2% agarose gel at 100 V for 35 min and sequenced at Macrogen Inc. (Seoul, Korea). Data were analyzed using the SpaServer database (http://SpaServer.ridom.de).
RESULTS
1. Antibiotic susceptibility tests
In over 90% of 90 strains, resistance to gentamicin, ciprofloxacin, clindamycin, and erythromycin was observed, and resistance to tetracycline was detected in over 80% of the strains. For chloramphenicol and trimethoprim-sulfamethoxazole, resistance rates were 23.3% and 25.6%, respectively. No resistance to vancomycin and teicoplanin was detected.
2. Distributions of spa types and superantigens in 2000-01 and 2007-08 isolates
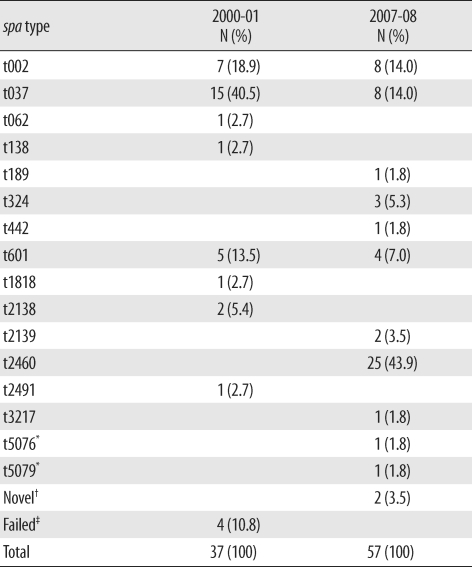
The distribution of spa types in isolates from 2000-01 (N = 37) and 2007-08 (N=57) is shown in Table 1. In 2000-01, 15 isolates (40.5%) were identified as t037; 7 (18.9%), as t002; 5 (13.5%), as t601; and 2 (5.4%), as t2138. One isolate each of t062, t138, t1818, and t2491 was identified, and no spa type was identified in 4 isolates. In 2007-08, 25 isolates (43.9%) were identified as t2460, and 8 isolates were identified as t002 and t037 (14.0%). One isolate each of t189, t442, t3217, t5076, and t5079 was also identified. Among these, t5076 and t5079 were newly registered as novel types, and another 2 types are pending registration in the Ridom spa type database.
Table 1.
Distribution of spa types in 2000-01 and 2007-08
*Novel spa types; †No matched spa type in SpaServer (http://SpaServer.ridom.de); ‡PCR failed.
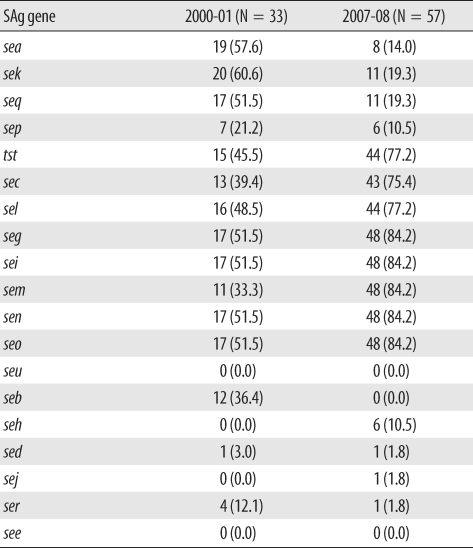
Multiplex PCR for superantigen genes was also performed, and the distribution is shown in Table 2. In the study periods the seg, sei, sem, sen, seo, tst, sec, sel, sea, sek, and seq genes were frequently identified in 14.0-84.2% of the strains. The genes seg, sei, sem, sen, and seo were more frequently identified in 2007-08 than in 2000-01. The genes tst, sec, and sel were also more frequently detected in 2007-08, and the frequency of sea, sek, and seq genes decreased in 2007-08. In addition, seb, seh, sed, sej, and ser genes were relatively uncommon.
Table 2.
Distribution of superantigen (SAg) genes in 2000-01 and 2007-08
Data are N (%) of total strains.
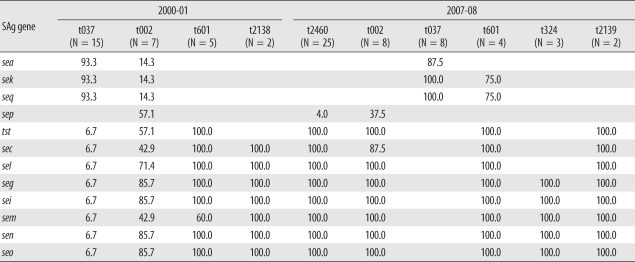
3. Distribution of superantigens relative to major spa types
Superantigen profiles and their relationship to major spa types (≥2 cases) are shown in Table 3. t037 was the most frequent spa type in 2000-01; sea, sek, and seq genes were identified in 14 out of 15 isolates, and t037 in 2007-08 exhibited a similar pattern. In contrast, t601 was associated with tst, sec, sel, seg, sei, sem, sen, and seo gene profiles in 2000-01, and in addition to these genes, sek and seq were also identified in 75% of 2007-08 t601 isolates. Types 324 and 2139 were identified only in 2007-08 and carried seg, sei, sem, sen, and seo (t324) and tst, sec, sel, seg, sei, sem, sen, and seo (t2139) genes. t2138, identified only in 2000-01, carried sec, sel, seg, sei, sem, sen, and seo.
Table 3.
Distribution of superantigen (SAg) genes related to frequently identified spa types in 2000-01 and 2007-08
Data are represented as the percentage of each spa type.
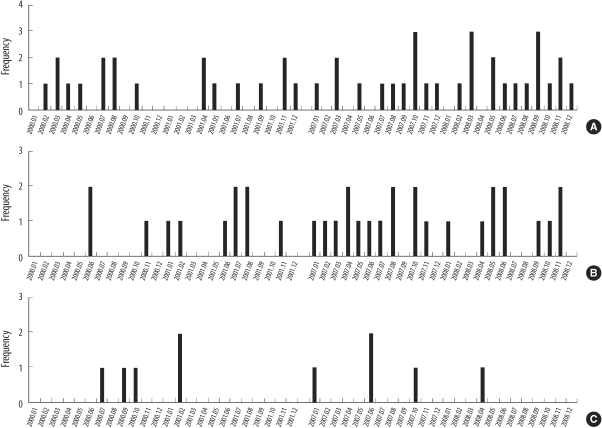
4. Monthly and locational distributions of all identified spa types and type t2460
The monthly and locational distributions of all spa types are shown in Fig. 1. The distribution of MRSA bacteremia was not associated with any particular time of the year. Among the 3 ICUs, MRSA bacteremia was most frequent in MICU (46 isolates, 51.1%). Thirty-four cases of bacteremia occurred in SICU (37.8%) and 10 cases in RICU (10.1%).
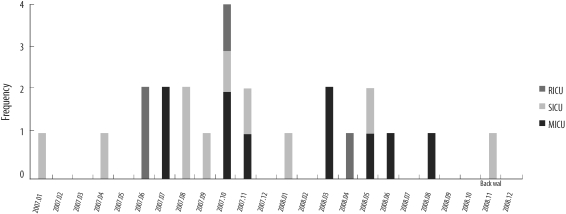
Fig. 1.
Monthly distribution of methicillin-resistant Staphylococcus aureus strains at intensive care units (ICUs) in 2000-01 and 2007-08. Medical ICU (A), surgical ICU (B), and respiratory ICU (C).
t2460, the most prevalent spa type in 2007-08 but not detected in 2000-01, was distributed as shown in Fig. 2. The prevalence of t2460 in 2007-08 was thought to be independent of the temporary outbreak of t2460.
Fig. 2.
Monthly distribution of t2460 methicillin-resistant Staphylococcus aureus strains at intensive care units (ICUs) in 2007-08.
Abbreviations: RICU, respiratory ICU; SICU, surgical ICU; MICU, medical ICU.
DISCUSSION
We evaluated the distribution of superantigen genes among the major spa types in clinical MRSA bacteremia isolates. Mobile genetic elements carrying superantigens are reportedly linked to genetic background, and horizontal transfer of superantigen genes is infrequent in Portugal [11] and Germany [12]; however, a report from USA has indicated that horizontal transfer is frequent [13]. Considering the distribution pattern of superantigen genes in our hospital, vertical transfer seems to be the predominant transmission mode of mobile genetic elements, as is observed in European countries. However, as shown in a few isolates of t002, t037, and t2460, the horizontal transfer of superantigen genes cannot be excluded.
In Tables 2 and 3, the presence of seg, sei, sem, sen, and seo genes, which are encoded within the enterotoxin gene cluster, tended to exclude sea, sek, and seq genes, a pattern that has been described previously [14]. The genes tst, sec, and sel are located in the same pathogenicity island, and sea, sek, and seq are located within a single phage. These are inclined to be closely associated with specific superantigen genes. Further, according a study in Japan, sec, seg, sei, sel, sem, sen, seo, and tst genes are associated with type I γSa4 and type I γSaβ, which are kinds of genomic islands. These features are similar to the findings of the present study, especially for the 2007-08 isolates. Although the proportion of MRSA was relatively higher than that of the Japanese sample (100% vs. 44%) and the population was different, the superantigen genes mentioned above (sec, seg, sei, sel, sem, sen, seo, and tst) seemed to be predominant in both cases. Isolates with sea, sek, and seq genes were confined to methicillin-susceptible S. aureus in Japan, suggesting the existence of some regional differences.
As mentioned earlier, the genetic background is closely related to virulence factors. The enterotoxin gene cluster is always present in MLST CC5, CC22, and CC45 strains but not in CC8, CC12, CC15, and CC395 [1]. It was previously reported that t002, t601, and t2460 are linked to MLST CC5, and t037 is associated with CC8 [15]. Therefore, the distribution of the enterotoxin gene cluster in our study is similar to that of previous findings.
In t002, 3 out of 7 isolates in 2000 and 2001 carried seg, sei, sem, sen, and seo and the remainder carried seg, sei, sen, and seo but not sem. In 2007 and 2008, seg, sei, sem, sen, and seo genes were identified in all t002 strains (8 isolates). Most t037 isolates (95.5%) did not carry seg, sei, sem, sen, and seo genes at any time during the study period. This finding corresponds with that of a European report in which superantigens were closely linked to genetic background. However, considering the frequency of the sep gene in spa-CC002, there remains the possibility of horizontal transfer of mobile genetic elements.
Similarly, t002, t062, t601, and t2460 exhibit spa-CC002 [16] and 13 of 33 isolates (39.3%) in 2000-01 and 37 of 57 isolates (64.9%) in 2007-08 account for spa-CC002. Therefore, the proportion of spa-CC002 showed a tendency to increase over time. Additionally, t037, t324, and t189 belong to spa-CC037 (t037) and spa-CC324 (t324, t189) and their proportions changed from 45.4% (15/33 isolates) and 0% (0/33 isolates) in 2000-01 to 14.0% (8/57 isolates) and 7.0% (4/57 isolates), respectively, in 2007-08.
The numbers of t037, the most frequent spa type in 2000-01, were reduced in 2007-08. In contrast, t2460 was the most frequent spa type in 2007-08 as shown in Table 2. The increase of t2460 strain in 2007-08 was not related to the outbreak, as shown in Figs. 1 and 2. Therefore, t2460 is thought to be one of the major spa types in Korea on the basis of the results of this and a previous study [16]. Additionally, the introduction of community-associated strains cannot be excluded by the presence of t324, which is a new spa type in 2007-08, despite the limited numbers of isolates.
In t2138 and t2139, which were detected in different study periods, the repeat numbers assigned by SpaServer were 26-17-34-34-17-20-17-12-17-16 and 26-23-17-34-130-20-17-12-17-16. Because only 6 nucleotide differences are shown in 3 repeat sequences and similar superantigen profiles, the strains t2138 and t2139 are thought to be closely linked.
In summary, we demonstrated differences in the regional spa types of Korea in comparison to those of Europe and North America, a temporal change of spa types in a single hospital, and a close relationship between spa types and superantigen profiles.
Acknowledgments
We are grateful to Sunhee Lee from the Clinical Research Institute, Seoul National University Hospital, for providing assistance in the laboratory.
This work was supported by grant no. 04-2008-032-0 from the Seoul National University Hospital Research Fund.
Footnotes
No potential conflict of interest relevant to this article was reported.
References
- 1.Holtfreter S, Grumann D, Schmudde M, Nguyen HT, Eichler P, Strommenger B, et al. Clonal distribution of superantigen genes in clinical Staphylococcus aureus isolates. J Clin Microbiol. 2007;45:2669–2680. doi: 10.1128/JCM.00204-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diep BA, Carleton HA, Chang RF, Sensabaugh GF, Perdreau-Remington F. Roles of 34 virulence genes in the evolution of hospital- and community-associated strains of methicillin-resistant Staphylococcus aureus. J Infect Dis. 2006;193:1495–1503. doi: 10.1086/503777. [DOI] [PubMed] [Google Scholar]
- 3.Niemeyer DM, Pucci MJ, Thanassi JA, Sharma VK, Archer GL. Role of mecA transcriptional regulation in the phenotypic expression of methicillin resistance in Staphylococcus aureus. J Bacteriol. 1996;178:5464–5471. doi: 10.1128/jb.178.18.5464-5471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koreen L, Ramaswamy SV, Graviss EA, Naidich S, Musser JM, Kreiswirth BN. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J Clin Microbiol. 2004;42:792–799. doi: 10.1128/JCM.42.2.792-799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frénay HM, Bunschoten AE, Schouls LM, van Leeuwen WJ, Vandenbroucke-Grauls CM, Verhoef J, et al. Molecular typing of methicillin-resistant Staphylococcus aureus on the basis of protein A gene polymorphism. Eur J Clin Microbiol Infect Dis. 1996;15:60–64. doi: 10.1007/BF01586186. [DOI] [PubMed] [Google Scholar]
- 6.Lindsay JA, Moore CE, Day NP, Peacock SJ, Witney AA, Stabler RA, et al. Microarrays reveal that each of the ten dominant lineages of Staphylococcus aureus has a unique combination of surface-associated and regulatory genes. J Bacteriol. 2006;188:669–676. doi: 10.1128/JB.188.2.669-676.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing. Twelfth informational supplement, M100-S12. Wayne, PA: National Committee for Clinical Laboratory Standards; 2002. [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. Nineteenth informational supplement, M100-S19. Wayne, PA: Clinical and Laboratory Standards Institute; 2009. [Google Scholar]
- 9.Mehrotra M, Wang G, Johnson WM. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J Clin Microbiol. 2000;38:1032–1035. doi: 10.1128/jcm.38.3.1032-1035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hwang SY, Kim SH, Jang EJ, Kwon NH, Park YK, Koo HC, et al. Novel multiplex PCR for the detection of the Staphylococcus aureus superantigen and its application to raw meat isolates in Korea. Int J Food Microbiol. 2007;117:99–105. doi: 10.1016/j.ijfoodmicro.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Aires de Sousa M, Conceição T, Simas C, de Lencastre H. Comparison of genetic backgrounds of methicillin-resistant and -susceptible Staphylococcus aureus isolates from Portuguese hospitals and the community. J Clin Microbiol. 2005;43:5150–5157. doi: 10.1128/JCM.43.10.5150-5157.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Witte W, Strommenger B, Cuny C, Heuck D, Nuebel U. Methicillin-resistant Staphylococcus aureus containing the Panton-Valentine leucocidin gene in Germany in 2005 and 2006. J Antimicrob Chemother. 2007;60:1258–1263. doi: 10.1093/jac/dkm384. [DOI] [PubMed] [Google Scholar]
- 13.Varshney AK, Mediavilla JR, Robiou N, Guh A, Wang X, Gialanella P, et al. Diverse enterotoxin gene profiles among clonal complexes of Staphylococcus aureus isolates from the Bronx, New York. Appl Environ Microbiol. 2009;75:6839–6849. doi: 10.1128/AEM.00272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jarraud S, Peyrat MA, Lim A, Tristan A, Bes M, Mougel C, et al. egc, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J Immunol. 2001;166:669–677. doi: 10.4049/jimmunol.166.1.669. [DOI] [PubMed] [Google Scholar]
- 15.Hu DL, Omoe K, Inoue F, Kasai T, Yasujima M, Shinagawa K, et al. Comparative prevalence of superantigenic toxin genes in meticillin-resistant and meticillin-susceptible Staphylococcus aureus isolates. J Med Microbiol. 2008;57:1106–1112. doi: 10.1099/jmm.0.2008/002790-0. [DOI] [PubMed] [Google Scholar]
- 16.Peck KR, Baek JY, Song JH, Ko KS. Comparison of genotypes and enterotoxin genes between Staphylococcus aureus isolates from blood and nasal colonizers in a Korean hospital. J Korean Med Sci. 2009;24:585–591. doi: 10.3346/jkms.2009.24.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]