Abstract
A medium-density linkage map of the ovine genome has been developed. Marker data for 550 new loci were generated and merged with the previous sheep linkage map. The new map comprises 1093 markers representing 1062 unique loci (941 anonymous loci, 121 genes) and spans 3500 cM (sex-averaged) for the autosomes and 132 cM (female) on the X chromosome. There is an average spacing of 3.4 cM between autosomal loci and 8.3 cM between highly polymorphic [polymorphic information content (PIC) ≥ 0.7] autosomal loci. The largest gap between markers is 32.5 cM, and the number of gaps of >20 cM between loci, or regions where loci are missing from chromosome ends, has been reduced from 40 in the previous map to 6. Five hundred and seventy-three of the loci can be ordered on a framework map with odds of >1000 : 1. The sheep linkage map contains strong links to both the cattle and goat maps. Five hundred and seventy-two of the loci positioned on the sheep linkage map have also been mapped by linkage analysis in cattle, and 209 of the loci mapped on the sheep linkage map have also been placed on the goat linkage map. Inspection of ruminant linkage maps indicates that the genomic coverage by the current sheep linkage map is comparable to that of the available cattle maps. The sheep map provides a valuable resource to the international sheep, cattle, and goat gene mapping community.
Sheep play an important role in modern agriculture and possess many heritable traits that are of economic importance. Additionally, the size, physiology, temperament, and lifespan of sheep make them an appropriate model for studying a variety of mammalian biological functions including lung physiology, immunology, endocrinology, reproduction, embryology, and fetal development. Sheep are also useful as disease models for inherited diseases, such as asthma (Wright et al. 1999), muscular dystrophy (McGavin 1974), McArdle's disease (Tan et al. 1997), and Batten's disease (neuronal ceroid lipofuscinosis; Broom et al. 1998), as well as for a variety of infectious diseases. However, although extensive efforts have been devoted to studying the genomes of humans and rodents to gain a better understanding of the role genes play in biological functions, much less effort has been devoted to studying the sheep genome. To improve our understanding of the genetic components of sheep production and biology, there is a need to develop a detailed sheep genome map.
Over the past decade, extensive efforts have been made by the international sheep gene mapping community to develop a useful sheep linkage map (Montgomery et al. 1994; Crawford et al. 1995; de Gortari et al. 1998). Several mapping flocks comprising three-generation full-sibling families have been produced (Crawford et al. 1995; Galloway et al. 1996; de Gortari et al. 1998). The use of these flocks has lead to the construction of low-density autosomal and X chromosome linkage maps (Crawford et al. 1995; Broom et al. 1996; Galloway et al. 1996; Lord et al. 1996; McLaren et al. 1997; de Gortari et al. 1998). For livestock species, such as sheep and cattle, where the costs of rearing and trait evaluation are high relative to the costs of typing a marker (100–1000 : 1), it has been estimated for quantitative trait loci (QTL) studies and genome scans that the optimum spacing between markers lies between 10 and 30 cM (Darvasi and Soller 1994). Furthermore, as studies move from identifying QTLs in populations by genome screens to utilizing and cloning QTLs, it is important to have dense marker maps for livestock. The most recently published sheep linkage map, termed the second generation map, comprised 512 loci with an average spacing of 6 cM (de Gortari et al. 1998). However, many of the markers used in the construction of the second generation map have few alleles or a low polymorphic information content (PIC). The average spacing between markers with a PIC of at least 0.7, or no PIC value and at least eight alleles, was 14.3 cM, and there were a number of regions where there were gaps of >20 cM between markers. In addition, comparisons between the second generation sheep map and recent cattle linkage maps revealed that the second generation sheep map does not span the whole sheep genome as centromeric and/or telomeric regions of a number of sheep chromosomes (1, 2, 9, 10, 14, 17, 18, 23, and 25) lack markers. Consequently there is a need to develop a more comprehensive sheep linkage map containing a larger number of highly polymorphic markers to enable efficient mapping of single gene and polygenic traits in sheep. In this report, we present a medium-resolution linkage map comprising 1062 loci (941 anonymous markers, 121 genes) spanning 3500 cM of the 26 autosomes (sex averaged) and 132 cM of the X chromosome.
RESULTS
Genotyping was performed on the International Mapping Flock (IMF) for 1106 markers resulting in the placement of 1093 markers representing 1062 unique loci on the sheep linkage map (Fig. 1; Table 1). Five hundred and fifty-seven of the mapped loci could be positioned with odds of >1000 : 1 onto an autosomal framework map, and 16 loci were positioned with odds of >1000 : 1 onto the X chromosome framework map. Six markers (AGLA232, OARJL3, UCD022, UCD030, UCD036, and UCD038) were unplaced, six markers (INRA194, TGLA141, BM148, CSIROEST3–38, LSCV18D, and LSCV19) were monomorphic for the IMF, and one marker (TGLA182) was too difficult to score. Twenty-one loci were typed on the IMF by use of more than one marker [CAT, DRB1, DYA, HBB, HRH1, NCAM, IFNG, IGF1, IL3, MAPT, PRF1, RNASE5, SCYA, IL2RB, LAMG, MTNR1A, KAP7, \CSS0M46(CSSM039), \INRA026(MB116), \MCM512 (MCM554), and \TGLA58(BMS710); anonymous loci are prefaced with a backslash]. In addition, it is also likely that the CSRD247 and CSRD270 primers amplify the same microsatellite as there is complete linkage disequilibrium and size correlation between the scoring for these two microsatellites.
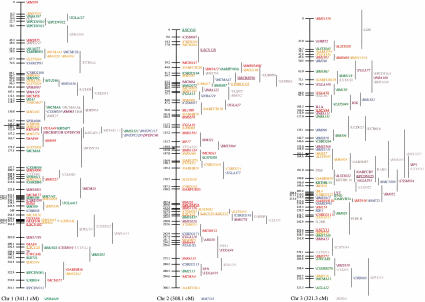
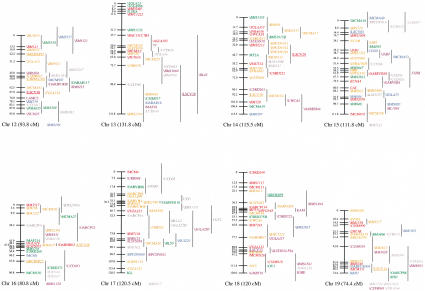
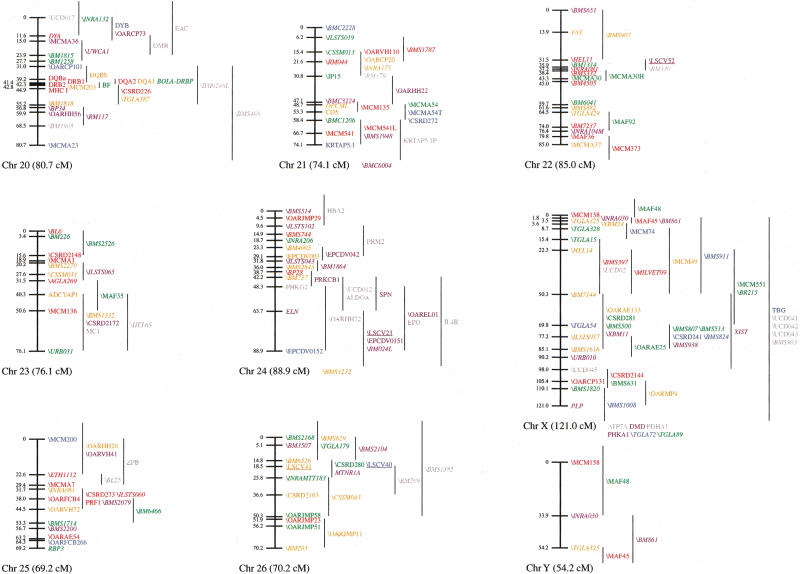
Figure 1.
An updated genetic linkage map for the sheep. Loci shown on the main framework map were positioned with odds of >1000 : 1. Loci to the right of the main framework map were significantly linked to the region indicated by the vertical lines, but could not be positioned on the framework at odds of >1000 : 1. All the autosomal linkage maps are sex-averaged and map distances are estimated by use of the Kosambi mapping function. The X chromosome map shows female map distances, whereas the Y chromosome map shows male map distances. (Horizontal arrows) Likely positions of the centromeres for chromosomes 1, 2, and 3. The remaining chromosomes are oriented with the centromeres at the top. Loci present on the second-generation or X chromosome sheep linkage maps that have not been mapped on the IMF are indicated at the bottom of the relevant chromosomes. The species of origin of the marker(s) used to position a locus is indicated by plain text for sheep markers, italics for cattle markers, and underlined for goat markers. Anonymous loci are prefaced by a backslash. Loci are color-coded by the following system: (red) PIC 0.8 or higher, if no PIC, then 11 or more alleles; (orange) PIC between 0.7 and 0.79, if no PIC, then 8–10 alleles; (green) PIC between 0.6 and 0.69, if no PIC, then 6–7 alleles; (blue) PIC between 0.5 and 0.59, if no PIC, then 5 alleles; (purple) PIC between 0.3 and 0.49, if no PIC then 3–4 alleles; (dark grey) PIC below 0.3, if no PIC, then 2 alleles; (light grey) gene in close proximity to a mapped marker that is within, or closer to, another gene. (*) Although there are at least two DQB genes in sheep, the relationship between the ovine DQB PCR products and DQB genes is unresolved (van Oorschot et al. 1994).
Table 1.
Characteristics of the Ovine Linkage Map
| Chr | Framework map (cM) | Best positions map (cM) | ||||||
|---|---|---|---|---|---|---|---|---|
| sex av | female | male | number of loci | sex av | female | male | number of loci | |
| 1 | 341.1 | 317.6 | 365.6 | 62 | 346.4 | 322.4 | 372.9 | 109 |
| 2 | 308.1 | 288.4 | 328.8 | 51 | 306.3 | 287.4 | 325.9 | 90 |
| 3 | 321.3 | 327.3 | 316.1 | 51 | 326.5 | 331.2 | 322.5 | 93 |
| 4 | 129.0 | 115.6 | 140.4 | 20 | 134.7 | 127.8 | 141.5 | 35 |
| 5 | 152.7 | 153.5 | 189.1 | 23 | 150.4 | 150.0 | 159.2 | 41 |
| 6 | 154.5 | 127.2 | 179.2 | 31 | 157.8 | 132.8 | 181.1 | 57 |
| 7 | 134.3 | 125.7 | 235.0 | 19 | 145.0 | 141.5 | 250.1 | 44 |
| 8 | 124.5 | 117.7 | 129.9 | 19 | 126.7 | 122.5 | 130.1 | 35 |
| 9 | 126.3 | 115.3 | 135.8 | 22 | 126.9 | 119.3 | 133.9 | 38 |
| 10 | 100.4 | 95.4 | 105.0 | 20 | 100.4 | 96.6 | 103.8 | 29 |
| 11 | 118.6 | 105.7 | 132.1 | 21 | 127.1 | 114.3 | 143.6 | 33 |
| 12 | 93.8 | 82.0 | 104.1 | 15 | 102.4 | 86.4 | 107.1 | 28 |
| 13 | 131.8 | 153.1 | 128.8 | 15 | 137.3 | 169.2 | 128.8 | 30 |
| 14 | 115.5 | 93.8 | 137.3 | 17 | 118.4 | 98.6 | 137.8 | 32 |
| 15 | 111.8 | 99.8 | 123.6 | 20 | 124.7 | 101.5 | 146.3 | 41 |
| 16 | 80.8 | 79.4 | 82.6 | 13 | 87.8 | 81.2 | 93.9 | 26 |
| 17 | 120.5 | 96.0 | 143.2 | 19 | 127.3 | 110.0 | 141.3 | 34 |
| 18 | 120.0 | 109.3 | 133.8 | 19 | 123.9 | 108.6 | 145.1 | 34 |
| 19 | 74.4 | 63.1 | 87.3 | 13 | 72.6 | 66.3 | 86.3 | 25 |
| 20 | 80.7 | 71.9 | 93.3 | 16 | 87.0 | 80.3 | 99.7 | 31 |
| 21 | 74.1 | 58.1 | 87.8 | 11 | 75.4 | 58.9 | 88.5 | 23 |
| 22 | 85.0 | 65.9 | 100.0 | 15 | 83.7 | 62.7 | 98.7 | 21 |
| 23 | 76.1 | 65.3 | 87.8 | 10 | 73.3 | 62.8 | 84.3 | 17 |
| 24 | 88.9 | 102.9 | 79.9 | 14 | 97.3 | 106.8 | 90.3 | 29 |
| 25 | 69.2 | 58.7 | 76.4 | 11 | 69.9 | 56.2 | 78.8 | 20 |
| 26 | 70.2 | 56.5 | 84.3 | 10 | 70.7 | 58.1 | 84.3 | 20 |
| Total autosomal | 3403.6 | 3145.2 | 3807.2 | 557 | 3499.9 | 3253.4 | 3875.8 | 1015 |
| X | 80.7 | 121.0 | 55.8 | 16 | 88.3 | 132.1 | 58.6 | 47 |
Fifty-three of the microsatellite primer sets that were used to construct the map amplify more than one set of polymorphic bands in sheep (AGLA22, BM1861, BM1862, BM2023, BM2814, BM3020, BM3212, BM4005, BM6438, BM6507, BM7144, BM8118, BM856, BM9202, BM9248, BMS1915, BMS2742, BMS629, CSN3, CSRD223, CSRD249, CSRD275, CSRD281, CSSME070, ILSTS008, ILSTS010, INRA104, INRA122, LSCV010, LSCV014, LSCV021, LSCV038, LSCV043, MCM63, MCM148, MCM197, MCM541, MCMA6, MCMA19, MCMA30, MCMA54, MILVET09, RM024, RME025, SRCRSP23, SRCRSP24, SRCRSP25, SRCRSP27, TGLA10, TGLA182, URB014, UWCA1, and UWCA28). Generally sheep were genotyped for only one set of bands for each of these primer pairs, with the band set genotyped being selected on the basis of degree of polymorphism and ease of scoring. In some cases, there appeared to be a one to one correspondence between an allele in one band set and an allele in the other band set. Only one set of bands was genotyped for most of these markers. However, where there was no obvious direct correspondence between alleles in different band sets and more than one band set could be easily scored, sheep were genotyped for more than one band set. This process resulted in sheep being genotyped for multiple band sets for products amplified with the AGLA22, BM1862, BM9248, BMS1915, BMS2742, CSRD275, CSRD281, ILSTS010, LSCV010, MCM541, MCMA6, MCMA30, MCMA54, RM024, and URB014 primers. Although most of these band sets had stutter patterns consistent with microsatellites, some lacked stutter bands. The AGLA22, BM1862, BM9248, BMS1915, CSRD275, CSRD281, ILSTS010, LSCV010, and RM024 primers amplified band sets that mapped to different chromosomes, whereas the remainder mapped to the same chromosome. No recombinations were found between the multiple band sets amplified by the BMS2742, MCMA6, MCMA30, and MCMA54 primers, one recombination was found between the band sets amplified by the MCM541 primers, and, although all sets of bands amplified by the URB014 primers mapped to chromosome 1, the positions of the two scored band sets differed by 70 cM.
The average number of informative meioses for mapped markers was 128 (maximum 209: BMS2721, ETH225, ILSTS017, OarCP125, OarCP131, OarCP134, TGLA176, and URB037), with an average of 98 phase known informative meioses (maximum 196: CSRD219, ETH225, ILSTS017, MAF70, MCM140, OarCP131, and TGLA176). The six unmapped markers had between 15 and 43 informative meioses.
The 1093 markers comprised 945 microsatellites, 57 RFLPs, 49 RAPDs, 30 SSCPs, 7 protein polymorphisms and 5 VNTRs. Sixty-three percent of the microsatellites were of cattle origin, thirty-two percent were of sheep origin and five percent were of goat origin. Forty-one percent (448) of the markers are highly polymorphic (PIC of at least 0.7 or no PIC and at least eight alleles).
The map contains 935 anonymous loci (represented by 944 markers: 1 RFLP, 1 SSCP, 3 VNTRs, 890 microsatellites, and 49 RAPDs); 121 genes and 6 ESTs (represented by 149 markers; 56 RFLPs, 29 SSCPs, 7 protein polymorphisms, 2 VNTRs, and 55 microsatellites). In addition, the positions of the CSF2, IRF1, ADH2, and MTP genes are indicated on the map on the basis of their presence within clones containing other linkage-mapped markers (Hawken et al. 1996; Lumsden et al. 1999).
An additional 17 anonymous loci (\BM1520, \BM6404, \BM7165, \BM7213, \BM8217, \BMC6004, \BMS109, \BMS1120, \BMS1232, \BMS1318, \BMS1932, \BMS345, \BMS66, \BP7, \CSSM041, \INRA049, and \INRA194) have been mapped on the MARC sheep flock (de Gortari et al. 1998) but not on the IMF, and an additional 6 markers (ATP7A, DMD, PDHA1, PHKA1, TGLA72 and TGLA89) have been mapped on the sheep X Mapping Flock (XMF; Galloway et al. 1996) but not on the IMF. The chromosomal assignments of these markers are also indicated in Figure 1.
An extensive effort was made to check the veracity of double crossover events for single markers identified by chrompic. This effort resulted in the retyping of sheep by the Centre for Animal Biotechnology for 220 markers. Although the majority of the putative double crossovers turned out to be a consequence of genotyping errors, there was also confirmation of 12 double crossover events represented by a single marker within regions of <20 cM, 6 of which were within regions of <10 cM. There were a further 65 markers for which double crossover events were found within a 20 cM region that were not checked.
The new map contains approximately twice as many loci as the previous sheep linkage map (de Gortari et al. 1998) with twice as many of the markers used being highly polymorphic. There is an average spacing of 3.5 cM between autosomal loci and 8.3 cM between highly polymorphic (PIC ≥ 0.7) autosomal loci and the largest gap between loci is ∼33 cM. The placement of the new markers has extended the coverage of the centromeric (chromosomes 5, 6, 10, 15, 17, 18, 20, and 25) and telomeric (chromosomes 1p, 2p, 3q, 8, 9, 10, 11, 14, 15, 17, 18, 20, 22, 23, 24, and X) regions of many chromosomes. As compared with the second generation ovine linkage map (de Gortari et al. 1998), the number of marker intervals >20 cM and 10 cM in the best positions map has reduced from 40 and 126 to 6 and 88, respectively. There are 11 gaps of >20 cM, and 102 of >10 cM, on the framework map. The maximum distance between adjacent highly polymorphic markers, or a highly polymorphic marker and the end of a linkage group, has reduced from >100 cM to 60 cM. The number of regions of >20 cM between adjacent highly polymorphic markers, or a highly polymorphic marker and the end of a linkage group, has been reduced from 66 to 38.
Overall the present sheep male map is significantly larger than the sheep female map with male sizes being larger than female for all chromosomes except chromosomes 3, 13, and 24 (Table 1). As expected, there is a considerable difference in male and female recombination rates for the pseudoautosomal region of the X chromosome, with the male distance between –CM158 and –TGLA325 being 54.2 cM compared with the female distance of 3.5 cM.
The sheep linkage map contains strong links to both the cattle and goat maps. Approximately 54% of the loci on the sheep linkage map have also been mapped on at least one of the cattle linkage maps, and 20% of the loci have been mapped on the goat linkage map (Fig. 2; Table 2). Six hundred and eight (56%) of the markers on the sheep map are cattle markers, and forty-five (4%) are goat markers. Comparisons of the ends of the sheep and MARC97 linkage groups reveal that the sheep map has about 160 cM at the end of linkage groups that is not represented on the MARC97 map, and the MARC97 map has about 60 cM that is not represented on the sheep map. The sheep linkage map contains 79 markers of bovine origin that have not been mapped in cattle, and 16 markers of caprine origin that have not been mapped in the goat. In addition, the current sheep map contains positions for some cattle and goat markers that have been physically mapped in cattle and goats but have not been placed on a cattle or goat linkage map. The sheep linkage map is useful for these markers as it can be used to predict the positions of these loci on the cattle and goat maps. Furthermore, the sheep linkage map can be used to resolve discrepancies in positions of loci between the various cattle linkage maps. An example of this is given by the mapping of \CSSM004 to sheep chromosome 1. This mapping agrees with its localization to BTA 1 on the MARC97 map (Kappes et al. 1997) in contrast to its positioning on BTA 2 on the IBRP97 map (Barendse et al. 1997).
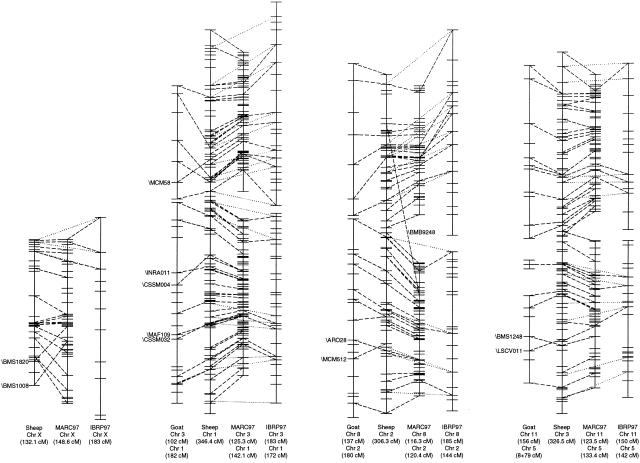
Figure 2.
Comparisons between positions of loci mapped in sheep on ruminant maps. (Tick marks) Positions of all loci that have been mapped in each species. Dashed and dotted lines are drawn between the same locus on different chromosomes: (dashed) goat–sheep, sheep–MARC97, and MARC97–IBRP97; (dotted) sheep–IBRP97. Lines are drawn only for those loci that have been mapped in sheep. (Although many more links exist between the cattle maps, and additional links can be found when comparing the goat and cattle maps, these are not shown.) The only loci identified by name are those whose relative position differs significantly between maps. All chromosomes are depicted with the centromere at the top except for cattle and goat chromosomes 3, 8, 11, and X for which the order has been inverted. (Box) Region of cattle chromosome 9 that is equivalent to the centomeric portion of sheep and goat chromosomes 14.
Table 2.
Comparison of the Number of Loci Shared by the Various Sheep and Cattle Maps
| Linkage mapa | MARC97 | IBRP97b | IRRF95 | MARC97+ IBRP97+ IRRF95 | Goatb |
|---|---|---|---|---|---|
| Number of loci | 1341 | 686 (435) | 226 | 1738 | 305 (240) |
| Number of loci mapped on sheep linkage map | 505 | 206 (145) | 99 | 572 | 209 (172) |
| Number of loci mapped on sheep framework linkage map | 290 | 127 (87) | 63 | 336 | 129 (106) |
Details of linkage maps can be found in the following papers: MARC97, Kappes et al. 1997; IBRP97, Barendse et al. 1997; IRRF95, Ma et al. 1996; and goat, Schibler et al. 1998.
For the comparisons using the IBRP97 or goat map, two sets of numbers are given. The first pertains to the overall number of markers on the IBRP97 or goat linkage maps, while the number in parentheses refers only to the number of markers on the IBRP97 or goat framework maps.
Comparisons between the sheep and cattle autosomal linkage maps reveal that there is generally good agreement between the maps in terms of chromosomal assignments and locus order (Fig. 2). The 17 discrepancies in chromosomal assignment are detailed in Table 3 and comprise 3 sheep microsatellites, 3 sheep protein polymorphisms, 10 cattle microsatellites, and 1 goat microsatellite. The cattle and sheep genotyping for three of the cattle markers (BM4439, BMS2840, and HEL6) that map to nonhomologous chromosomes were performed in the same laboratory. Sheep PCR products were sequenced for five of the cattle markers (BM8115, BM9248, HEL12, ILSTS010, and INRA104) that mapped to nonhomologous chromosomes in sheep, where different laboratories were used for the sheep and cattle genotyping. This sequencing confirmed that the correct forward and reverse primers had been used to amplify the sheep products. However, no homology apart from the primers was found between the sheep and cattle sequences amplified with these primers, with the exceptions of the sequence between the forward primer and the (CA)n repeat for BM9248, and the (TG)n repeat and the reverse primer for ILSTS010. GenBank searches were conducted for all microsatellite primers that belonged to markers that mapped to nonhomologous chromosomes in ruminants. These searches revealed that there were no matches of 100% between the primers and ruminant sequences other than that of the microsatellite for which the primer had been designed. However, several of the primers (BM9248F, BM9248R, HEL6F, HEL6R, ILSTS10R, INRA104F, and MAF48R) were highly similar to other ruminant sequences having only one to four mismatches in sequence.
Table 3.
Loci that Map to Nonhomologous Chromosomes on Sheep, Cattle, and Goat Linkage Maps
| Locus | Sheep | Predicted cattle/goat | Cattleb | Goat | Comment |
|---|---|---|---|---|---|
| \LSCV12 | 2 | 8 | 16 | Primers amplify the same size microsatellite in sheep and goats; primers contain some microsatellite sequence; goat position from Web site as noted in Schibler et al. (1998) map paper. | |
| \BM9248c | 2, 20 | 8, 23 | 2, 12, 13M | Primers amplify multiple microsatellite band sets of varying sizes in sheep and cattle. | |
| \BMS1915a | 3, 14 | 11, 18 | 16M | Primers amplify two microsatellite band sets in sheep of similar size to cattle bands; microsatellite band sets overlap. | |
| \HEL12c | 4 | 4 | 20M,I | Lots of background; microsatellite band set is faint; sheep bands slightly larger than cattle bands. | |
| EAAd | 6 | 6 | 15M,R | Protein polymorphism. | |
| \BM4439 | 8 | 9 | 15M | Cattle has only 19 informative meioses; sheep bands much larger than cattle bands. | |
| \BM8115ac | 13 | 13 | 21M | A low annealing temp (48°C/50°C) was used to amplify sheep DNA. Amplified a constant product as well as a polymorphic band set. Sequencing of both products revealed that the sequence of the constant product was similar to the bovine BM8115 sequence but that the (TG)n repeat had been replaced by a (T)n repeat. The other sequence was unrelated to bovine BM8115. | |
| \ILSTS010c | 5, 14 | 7, 18 | 12M | No stutter pattern associated with sheep bands; probably not a microsatellite. | |
| \BMS2840 | 16 | 20 | 7M | Similar sizes for sheep and cattle bands. | |
| \GMBT11 | 17 | 17 | 26I | VNTR | |
| EAMd | 18 | 21 | 23M,I | Sheep protein typing; cattle typed by PCR; cattle has null allele and only 21 informative meioses. | |
| EACd | 20 | 23 | 18M,R | Protein polymorphism; only 18 informative meioses in sheep but shows no linkage to any chromosome 14 marker. | |
| \OARHH22 | 21 | 29 | 29M | 16 | Reverse primer for goat differs from sheep primer. |
| \MCM373 | 22 | 26 | 2M | Large size difference between sheep and cattle microsatellite bands; may not be the same marker. | |
| \INRA104c | 15, 22 | 15, 26 | 5I | Primers amplify multiple microsatellite band sets in sheep. | |
| \HEL6 | 23 | 24 | 1M,I | Products of similar size in sheep and cattle. | |
| \MAF48 | X | X | 5M | 19 | Sheep bands slightly larger than cattle bands. |
Alternate primer sets used in sheep derived from cattle sequences in GenBank.
M, MARC97; I, IBRP97; R, IRRF95.
The sequence of the primers to amplify sheep DNA has been confirmed by sequencing PCR products.
Sheep and cattle erythrocyte antigens may not be homologous.
The majority of inconsistencies in locus order between the sheep and cattle linkage maps appear as relative marker inversions within regions of <5 cM. These probably relate to the positioning of a number of loci to regions on the best position maps with odds of <1000 : 1. A major difference between the sheep and cattle linkage maps pertains to the organization of the X chromosome. Both the orientation of the X chromosome and the relative positions of the region flanked by \BMS1820 and \BMS1008 differ between sheep and cattle.
In contrast with the good agreement in locus order between the sheep and cattle linkage maps, there are many rearrangements in order between these maps and the goat linkage map (Fig. 2). Of the 121 genes mapped on the sheep linkage map, 105 have homologs that have been mapped on the human map and entered in either the GDB (http://gdbwww.gdb.org/gdb/) or UniGene (http://www.ncbi.nlm.nih.gov/UniGene/) databases, and 92 have homologs that have been positioned on the mouse map and are entered in the MGD (http://www.informatics.jax.org/) database. The genes for which human or murine homologs have not been mapped include members of the keratin and keratin-associated protein families and blood group antigens that have been typed only by protein polymorphisms in sheep. In addition, there are no human or murine homologs of sheep MHC class II DYA and DYB genes.
DISCUSSION
The new sheep linkage map contains twice as many loci as did the previous linkage map (de Gortari et al. 1998), which in turn contained twice as many loci as its predecessor (Crawford et al. 1995). The current map spans ∼3600 cM, and from comparisons with the ends of the MARC97 linkage groups it is estimated to span >98% of the sheep genome. The number of highly polymorphic markers (PIC ≥ 0.7) that have been mapped in sheep has doubled in this map, and 89% of the regions spanned by this map have a highly polymorphic marker within 10 cM. The increased density of highly polymorphic markers will be extremely useful for both genome scans and linkage disequilibrium studies.
Comparisons between the sheep and cattle linkage maps reveal that there are still some sheep chromosomes where the ends of the linkage groups are poorly characterized (Fig. 2). The converse is also true, with some cattle chromosomes lacking regions at the ends of their linkage groups as compared with the linkage groups for the homologous sheep chromosomes. However, the differences between individual linkage group ends are all <20 cM. The EPCDV markers represent an attempt to improve the coverage of markers near the ends of sheep chromosomes (Cribiu et al. 2000).
Despite fewer loci being mapped by linkage analysis in sheep, the current situation in sheep linkage mapping has some advantages when compared with the situation in cattle. More than 98% of the markers that have been mapped by linkage analysis in sheep have been mapped on a common three-generation set of full-sibling sheep families, and it is the policy of the sheep gene mapping community to map all markers on the IMF. In contrast, a number of cattle linkage maps exist that have been developed with different sets of families and different sets of loci (Ma et al. 1996; Barendse et al. 1997; Kappes et al. 1997). As a consequence, some difficulties are experienced in moving between the cattle maps to predict relative positions of loci, and several cattle mapping workshops have been held to address these issues (Taylor et al. 1998; Casas et al. 1999; Gu et al. 2000).
Furthermore, the use of three-generation full-sibling families with typing of all members of the three generations provides phase information that is lacking for some of the cattle maps. This phase information is useful especially for identifying suspect genotypes for retesting. Retesting has revealed that a high proportion of the initially identified double crossover events within small regions involving single markers on the sheep linkage map were due to typing errors. A similar situation was found for the MARC97 cattle map in which 0.33% of initial genotypes were subsequently found to be erroneous (Kappes et al. 1997). Correction of the genotyping errors reduced the overall size of the sheep map, and, in some instances, had a substantial impact on the ordering and position of loci.
Considerable size differences have been reported between the IBRP97 (Barendse et al. 1997) and MARC97 (Kappes et al. 1997) cattle maps with the MARC97 map (2990 cM) being significantly smaller than the IBRP97 map (3680 cM) as demonstrated in Figure 2. Possible explanations for the large discrepancy in the sizes of the cattle maps include: the relative frequencies of erroneous genotypes in the data used to construct the maps; the nature of the crosses used and the number of offspring typed; the completeness of the maps; and differences in recombination rates between the parents and grandparents used to generate the mapping population. Although extensive retesting of suspect genotypes occurred during the construction of the MARC97 map, resulting in a reduction in size of the MARC97 map by 23% from 3680 to 2990 cM (Kappes et al. 1997), there is no indication that this level of error checking occurred when the IBRP map was being prepared (Barendse et al. 1997). It is of interest that the size of 3680 cM for the MARC97 map prior to error corrections is similar to the overall size of the IBRP97 map of 3710 cM. Given the level of error correction undertaken for the sheep map, it appears possible that the actual size of the sheep linkage map may be slightly larger than that of the cattle linkage map.
Overall, the sheep male map is significantly larger than the sheep female map with male sizes being larger than female for all chromosomes except chromosomes 3, 13, and 24. This observation contrasts with the findings for most other mammals in which the female autosomal maps are either considerably larger than the male maps, as is the case with the human, pig, and dog maps (Archibald et al. 1995; Mellersh et al. 1997; Li et al. 1998) or the maps are of a similar size, as with cattle (Barendse et al. 1997; Kappes et al. 1997). One factor likely to contribute to the current difference in size between the male and female sheep linkage maps is the smaller number of male parents and grandparents (8) used to construct the IMF pedigree, as compared with the number of female parents and grandparents (20). The greater number of female parents and grandparents means that this population is likely to contain a greater number of heterozygous individuals that can generate informative meioses information. This difference can be shown to have a greater effect with markers that possess low levels of polymorphism, in which there are often many fewer male informative meioses than female informative meioses. In some instances in which there are only a small number of male informative meioses, this number has lead to an artificial inflation of male chromosomal sizes. An example of this inflation is shown by the intervals between \BMS2614–\BL1134 or \ILSTS020–\BL1134 loci at the end of the linkage map on chromosome 7 in which the female, male, and sex-averaged recombination distances are 0–3.4, 100, and 0–5.0 cM, respectively. This distortion is a consequence of the \BL1134 locus being at the end of a linkage group, having only two alleles, and with only one male grandparent (with only two progeny) and no male parents being informative for the locus as compared with six female grandparents and five female parents.
Differences exist between cattle maps as to whether individual male or female chromosomal maps are larger. Chromosomal workshops have revealed that the IBRP female maps are larger than the male maps for chromosomes 1, 10, and 23, whereas the male maps are larger than the female MARC and TAMU maps for these chromosomes (Beever et al. 1996; Taylor et al. 1997, 1998). Some of these differences are likely to be due to variation in recombination rates between individuals. This phenomenon has been demonstrated for male cattle, humans, and sheep (Yu et al. 1996; Simianer et al. 1997; Lien et al. 1999), and it is likely that recombination rates will also vary amongst females. In addition, as shown by a recent study of human chromosome 19, sex differences in recombination rates also occur along individual chromosomes (Mohrenweiser et al. 1998). The human chromosome 19 study found that whereas the male recombination rate was approximately twice that of females in telomeric regions, it was only 10%–25% of the female rate in more centromeric regions. Whether more densely populated linkage maps will reveal that there are real differences in ruminants between the size of male and female linkage maps, as is the case with humans, remains to be seen.
Although most markers of cattle or goat origin map to the expected chromosomes in the sheep genome, there are a small number of discrepancies (Table 2). It is most likely that the majority of these are due to the comparison not being between homologous loci, to genotyping errors, or to insufficient informative meioses for a marker in one or more species. Possible sources of error for PCR-based markers include the use of primers that do not amplify the locus that they are supposed to as a consequence of either tube mislabeling or an error in the initial categorization of a marker, or primers amplifying different microsatellites in different species. The possibility of primer error was ruled out for all five of the markers that were sequenced. The amplification of different microsatellites in different species with the same primer set is more likely to occur when the stringencies of the annealing conditions differ significantly between the species as more cross-priming events are likely to occur with less stringent conditions. This cross-priming often happens when markers developed in one species are used in another species and was the case for at least six of the primer sets (BM4439, BM8115, HEL12, ILSTS010, LSCV12, and MAF48) that amplified loci mapping to nonhomologous chromosomes in sheep and cattle. In addition, about 50 of the primer sets used in this study amplify more than one set of microsatellite bands in sheep. Although many of the multiple band sets amplified by a single set of primers map to similar regions on a chromosome, and possibly represent duplication events, many map to different chromosomes.
In general, as shown in Figure 2, there is good agreement between locus orders on a chromosome for cattle and sheep maps. The exceptions can probably be explained by locus positions in one or both species being supported by odds of <1000 : 1. However, there are more differences in locus order when comparisons are made between either the sheep or the cattle map and the goat map. One possible reason for these differences is that the goat map is less robust as a consequence of the smaller number of markers that have been placed on it (Schibler et al. 1998).
The recent ruminant mapping studies are able to shed new light on the evolution of chromosomes in Bovidae. Despite the fact that cattle and goats both have 29 acrocentric autosomes as opposed to the 23 acrocentric and 3 metacentric autosomes of sheep, goat chromosomes share some features with sheep chromosomes that are not shared with cattle chromosomes. These include the translocation of a region equivalent to the centromeric end of OAR 9 and CHI 14 to the centromeric region of BTA 9 (Crawford et al. 1995; Vaiman et al. 1996) and the structure of the X chromosome (Galloway et al. 1996; Ponce de Leon et al. 1996). It has been proposed that caprids evolved from a common ancestor with a 2n = 60 karyotype and no metacentric chromosomes (Bunch et al. 1976). However, evidence from the mapping of the trichohyalin (THH) gene in sheep, cattle, and goats suggests that the common ancestor may have had at least one metacentric chromosome equivalent to OAR1. In cattle, the THH gene maps to BTA 1q4 (Schmutz et al. 1998), in goats it maps to CHI 3q2.1 (Schibler et al. 1998), and in sheep it maps to chromosome 1 in the region corresponding to CHI 3 and BTA 3. It seems more likely that the ancestor of OAR1 split in separate places to generate chromosomes 1 and 3 of cattle and goat, than that the ancestral chromosomes for CHI/BTA 1 and 3 would have fused to form OAR 1, and that the region containing the THH gene would then have moved from BTA 3 to 1.
The findings in this study extend those of Galloway and colleagues (1996) and suggest that the organization of the sheep X chromosome is more like that of the goat X chromosome than that of the cattle X chromosome (Ponce de Leon et al. 1996; Hassanane et al. 1998; Piumi et al. 1998). Overall, the cattle X chromosome map appears inverted relative to the sheep and goat maps, with the pseudoautosomal region of sheep and goat X chromosomes being found at the tip of the short arm whereas that of cattle is found at the tip of the long arm. Other differences between ruminant X chromosomes include the relative positions of the centromeres, and the rearrangement of the bovine Xp24–Xq12 and Xq21–Xq24 regions in goats (Piumi et al. 1998). Comparison of the ovine and bovine X chromosome linkage maps reveals that the segment between \BMS1820 (BTA Xq1–Xq2; Sonstegard et al. 1997) and \BMS1008 (BTA Xp11; Sonstegard et al. 1997) has moved from an interstitial position within the bovine X chromosome to the bottom of the sheep X chromosome. It is likely that this rearrangement is equivalent to that found between goat and cattle X chromosomes. In addition, chromosomal painting shows that the cattle Xq41–Xq43 region maps to sheep and goat Xp12–Xq12 (Hassanane et al. 1998).
Even by use of the previous less-complete sheep linkage maps, successes in identifying ovine genomic regions influencing single locus traits based on genome scanning have already been reported. These include the localization of the chromosomal regions containing the genes for Booroola fecundity, callipyge, and Horns on chromosomes 6, 18, and 10, respectively (Montgomery et al. 1993, 1994, 1996; Cockett et al. 1994) and the identification of the fibroblast growth factor receptor 3 (FGFR3) gene as the causative gene for Spider Lamb Syndrome (Cockett et al. 1999) and the bone morphogenetic protein (BMP15) gene as the causative gene for Inverdale fecundity (Galloway et al. 2000). Recently, data have also been presented for the localization of a number of QTLs, including ones for wool quality (Robinson et al. 1997), parasite resistance (Beh et al. 1998; Crawford 1998), facial eczema (Phua et al. 1998), and dagginess (MacDonald et al. 1998).
In conclusion, the current sheep linkage map is a useful medium-density linkage map. The inclusion of 446 highly informative markers will make it of great use to the various international sheep gene mapping and gene discovery projects.
METHODS
Genetic Markers and Genotyping
All markers were genotyped on the IMF. The IMF consists of nine three-generation full-sibling families comprising a total of 127 sheep with 98 F2 progeny and a maximum of 222 informative meioses (Crawford et al. 1995). All F2 progeny share a paternal grandsire, and the founding sheep were derived from the Texel, Coopworth, Perendale, Romney, and Merino breeds. All grandparents were typed for all markers except for some of the markers typed by the USDA on the IMF (de Gortari et al. 1998) for which often only the male grandparents were typed.
To determine the number of alleles and informativeness of markers, new markers typed by the Centre for Animal Biotechnology were also typed on a panel of 40–50 unrelated sheep derived from the Merino, Border Leicester, Suffolk, Romney, Karakul, Finnish Landrace, Poll Dorset, Dorset, Carpet Master, and Texel breeds. PIC values were calculated from the genotyping data by use of the PIC v1.51 program (Ott 1992). This new information was combined with previously reported information on the number of alleles and PIC data for other sheep markers resulting in 600 markers for which both sets of information were available. The number of alleles and PIC values were compared for this data set, and results from this comparison lead to the informativeness relationship, between PIC and number of alleles for markers without PIC data, that was used in Figure 1.
Genotypic data were exchanged between groups by e-mail, and data for an additional 554 markers were merged by CAB with previous autosomal and X chromosome mapping data obtained from IMF typing (Galloway et al. 1996; de Gortari et al. 1998) for linkage analysis. Fifty-two markers were typed on the IMF by more than one group. Scoring discrepancies between groups, which ranged from 0% to 11% (average 2%), were resolved by retyping by CAB.
Marker information for most markers (including primers, product sizes, annealing temperature, Mg2+ ion concentration, and ease of scoring for PCR markers and probes and restriction enzymes for RFLP markers) are detailed at the following Web sites: http://rubens.its.unimelb.edu.au/∼jillm/jill.htm; and http://www.thearkdb.org/browser?species=sheep. Information on hitherto unpublished markers can be obtained from the laboratories detailed in Table 4. The general procedure for PCR reactions was: 20 ng of genomic DNA was amplified in a 5-μL reaction volume consisting of 67 mM Tris-HCl (pH 8.8), 16.6 mM (NH4)2SO4, 0.2 mg/mL gelatin, 0.45% Triton X-100, 50 μM dNTPs, 12.5 ng of both the forward and reverse primers, 0.125 units of Amplitaq DNA polymerase (Perkin Elmer), 27.5 ng of Taq Start Antibody (Clontech), and 0.25 μCi [α33P]d-ATP (AMRAD). Reactions were set up in a 96-well plate and run on a DNA thermal cycler (PTC-100, MJ Research), with the following conditions: an initial denaturation step of 2.5 min at 95°C; followed by 30 cycles of 30-sec steps of denaturation at 95°C, annealing at the appropriate temperature, and extension at 72°C; and a final extension of 2.5 min at 72°C. Reaction products were electrophoresed on denaturing polyacrylamide gels (microsatellites) or non-denaturing MDE (FMC Bioproducts) gels (SSCPs) and visualized by autoradiography.
Table 4.
Sources for Additional Information on Sheep and Goat Markers
| Marker prefix | Laboratory | Contact |
|---|---|---|
| DVEPC, EPCDV, LSCV | Department de Génétique Animale, INRA, France | Daniel Vaiman (vaiman@biotec.jouy.inra.fr) |
| MCM | CSIRO Animal Production, Australia | Ken Beh (K.Beh@prospect.anprod.csiro.au) |
| MAF, OAR | AgResearch Molecular Biology Unit, New Zealand | Allan Crawford (allan.crawford@agresearch.co.nz) |
| CSRD | CSIRO Tropical Agriculture, Australia CAB, University of Melbourne | Blair Harrison (Blair.Harrison@tag.csiro.au) Jill Maddox (jillm@rubens.its.unimelb.edu.au) |
| CSIROEST | Dave Adelson (Dave.Adelson@li.csiro.au) |
Nomenclature
The COGNOSAG nomenclature guidelines have been followed throughout this paper for naming loci (Andresen et al. 1995). These guidelines stipulate that anonymous loci be identified by prefacing them with a backslash.
Linkage Analysis
New genotypes were scored and entered into a database independently by two individuals. The two sets of scoring were compared, and disputed genotypes were either retyped or discarded before generation of CriMap gen files for new markers (Lander and Green 1987). For seven of the loci that had multiple markers systems with no recombinations between the markers representing a locus (CAT, HBB, IGF1, IL2RB, LAMG, MTNR1A, and PRF1), the individual marker scoring was compared, and a haplotype coding system was developed for each locus. The haplotype gen information was then used in the linkage analysis instead of the individual marker information. The “prepare” option of CriMap was used to check for Mendelian segregation, and new markers were assigned to chromosomal groups by use of the “twopoint” option of CriMap with a skeleton map of previously mapped markers. Chromosomal maps were generated by MultiMap v2.3 (Matise et al. 1994). As the IMF pedigree structure differs from that of the CEPH pedigrees, programs were developed to automatically generate suitable ordh, ordj, hash, and names files for use with MultiMap (programs developed by Ian R.W. Evans). Ordh files were created on the basis of the ranking of the loci in order of numbers of phase-known informative meioses. The chrompic option of CriMap was used to identify potential genotyping errors, and double recombinants were retyped where possible. Framework maps were constructed for each chromosome with construction beginning with the pair of markers with the highest number of informative meioses (phase-known) for which a recombination fraction of 0.05–0.35 was supported with a lod score of ≥3.0. For best-position maps, additional markers were placed in the most likely positions on the basis of likelihood data generated by MultiMap. Where several positions of equal likelihood existed, markers were positioned manually to minimize the number of recombinations on a chromosome.
Comparison of Ruminant Maps
Map positions for cattle IBRP97 and MARC97 maps were obtained from ArkDB (http://www.ri.bbsrc.ac.uk/bovmap/arkbov/).
Acknowledgments
This work was supported by a grant from Meat and Livestock Australia. We thank J.G.H. Hickford (CAST), R.A. McGraw (RM44, RM095, RM117), M. Zanotti (MILVET7, MILVET8, MILVET9), D. Groth (BF), P. Schmidt (IFNG), P. Roche (RDA33GH), and B. Kirkpatrick (UWCA1) for supplying primers for mapping, J. Ott for supplying the program PIC 1.51, and I. Evans for developing programs to assist with linkage analysis.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jillm@rubens.its.unimelb.edu.au; FAX 61-3-9347-4083.
Article published on-line before print: Genome Res., 10.1101/gr. 135001. Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.135001.
REFERENCES
- Andresen E, Broad TE, Brown S, Cooper DW, DiStasio L, Dolling CHS, Fleet M, Hill DF, Lauvergne JJ, Lundie RS, et al. Revised guidelines for gene nomenclature in ruminants 1993. Genet Select Evol. 1995;27:89–93. [Google Scholar]
- Archibald AL, Haley CS, Brown JF, Couperwhite S, McQueen HA, Nicholson D, Coppieters W, Van de Weghe A, Stratil A, Winterø AK, et al. The PiGMaP consortium linkage map of the pig (Sus scrofa) Mammal Genome. 1995;6:157–175. doi: 10.1007/BF00293008. [DOI] [PubMed] [Google Scholar]
- Barendse W, Vaiman D, Kemp SJ, Sugimoto Y, Armitage SM, Williams JL, Sun HS, Eggen A, Agaba M, Aleyasin SA, et al. A medium-density genetic linkage map of the bovine genome. Mammal Genome. 1997;8:21–28. doi: 10.1007/s003359900340. [DOI] [PubMed] [Google Scholar]
- Beever JE, Lewin HA, Barendse W, Andersson L, Armitage SM, Beattie CW, Burns BM, Davis SK, Kappes SM, Kirkpatrick BW, et al. Report of the first workshop on the genetic map of bovine chromosome 23. Animal Genet. 1996;27:69–75. doi: 10.1111/j.1365-2052.1996.tb00470.x. [DOI] [PubMed] [Google Scholar]
- Beh KJ, Callaghan MJ, Hulme DJ, Leish Z, DiIenno K, Lenane I. A search for genes affecting gastrointestinal parasite resistance in sheep. Animal Genet. 1998;29S1:67. [Google Scholar]
- Broom JE, Tate ML, Dodds KG. Linkage mapping in sheep and deer identifies a conserved pecora ruminant linkage group orthologous to two regions of HSA16 and a portion of HSA7Q. Genomics. 1996;33:358–364. doi: 10.1006/geno.1996.0211. [DOI] [PubMed] [Google Scholar]
- Broom MF, Zhou C, Broom JE, Barwell KJ, Jolly RD, Hill DF. Ovine neuronal ceroid lipofuscinosis: A large animal model syntenic with the human neuronal ceroid lipofuscinosis variant CLN6. J Med Genet. 1998;35:717–721. doi: 10.1136/jmg.35.9.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunch TD, Foote WC, Spillett JJ. Translocations of acrocentric chromosomes and their implications in the evolution of sheep (Ovis) Cytogenet Cell Genet. 1976;17:122–136. doi: 10.1159/000130704. [DOI] [PubMed] [Google Scholar]
- Casas E, Barendse W, Beever JE, Burns BM, Davis SK, Erhardt G, Forster M, Gomez-Raya L, Kalm E, Kappes SM, et al. Bovine chromosome 4 workshop: Consensus and comprehensive linkage maps. Animal Genet. 1999;30:375–377. doi: 10.1046/j.1365-2052.1999.00529.x. [DOI] [PubMed] [Google Scholar]
- Cockett NE, Jackson SP, Shay TL, Nielsen D, Moore SS, Steele MR, Barendse W, Green RD, Georges M. Chromosomal localization of the callipyge gene in sheep (Ovis aries) using bovine DNA markers. Proc Natl Acad Sci. 1994;91:3019–3023. doi: 10.1073/pnas.91.8.3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockett NE, Shay TL, Beever JE, Nielsen D, Albretsen J, Georges M, Peterson K, Stephens A, Vernon W, Timofeevskaia O, et al. Localization of the locus causing Spider Lamb Syndrome to the distal end of ovine chromosome 6. Mammal Genome. 1999;10:35–38. doi: 10.1007/s003359900938. [DOI] [PubMed] [Google Scholar]
- Crawford AM. Searching for disease resistance QTL. Animal Genet. 1998;29S1:5–6. [Google Scholar]
- Crawford AM, Dodds KG, Ede AJ, Pierson CA, Montgomery GW, Garmonsway HG, Beattie AE, Davies K, Maddox JF, Kappes SW, et al. An autosomal genetic linkage map of the sheep genome. Genetics. 1995;140:703–724. doi: 10.1093/genetics/140.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvasi A, Soller M. Optimum spacing of genetic markers for determining linkage between marker loci and quantitative trait loci. Theor Appl Genet. 1994;89:351–357. doi: 10.1007/BF00225166. [DOI] [PubMed] [Google Scholar]
- de Gortari MJ, Freking BA, Cuthbertson RP, Kappes SM, Keele JW, Stone RT, Leymaster KA, Dodds KG, Crawford AM, Beattie CW. A second-generation linkage map of the sheep genome. Mammal Genome. 1998;9:204–209. doi: 10.1007/s003359900726. [DOI] [PubMed] [Google Scholar]
- Galloway SM, Hanrahan V, Dodds KG, Potts MD, Crawford AM, Hill DF. A linkage map of the ovine X chromosome. Genome Res. 1996;6:667–677. doi: 10.1101/gr.6.8.667. [DOI] [PubMed] [Google Scholar]
- Galloway SM, McNatty KP, Cambridge LM, Laitinen MPE, Juengel JL, Jokiranta TS, McLaren RJ, Luiro K, Dodds KG, Montgomery GW, et al. Mutations in an oocyte-derived growth factor gene (BMP15) cause increased ovulation rate and infertility in a dosage-sensitive manner. Nature Genet. 2000;25:279–283. doi: 10.1038/77033. [DOI] [PubMed] [Google Scholar]
- Gu Z, Gomez-Raya L, Vage DI, Elo K, Barendse W, Davis G, Grosz M, Erhardt G, Kalm E, Reinsch N, et al. Consensus and comprehensive linkage maps of bovine chromosome 7. Animal Genet. 2000;31:206–209. doi: 10.1046/j.1365-2052.2000.00627.x. [DOI] [PubMed] [Google Scholar]
- Hassanane MS, Chaudhary R, Chowdhary BP. Microdissected bovine X chromosome segment delineates homoeologous chromosomal regions in sheep, goat and buffalo. Chromosome Res. 1998;6:213–217. doi: 10.1023/a:1009263718667. [DOI] [PubMed] [Google Scholar]
- Hawken RJ, Broom MF, van Stijn TC, Lumsden JM, Broad TE, Maddox JF. Mapping the ovine genes encoding IL3, IL4, IL5, and CSF2 to sheep chromosome 5q13-q15 by FISH. Mammal Genome. 1996;7:858–859. doi: 10.1007/s003359900252. [DOI] [PubMed] [Google Scholar]
- Kappes SM, Keele JW, Stone RT, McGraw RA, Sonstegard TS, Smith TPL, Lopez-Corrales NL, Beattie CW. A second-generation linkage map of the bovine genome. Genome Res. 1997;7:235–249. doi: 10.1101/gr.7.3.235. [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P. Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci. 1987;84:2363–2367. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WT, Fann CSJ, Ott J. Low-order polynomial trends of female-to-male map distance ratios along human chromosomes. Human Hered. 1998;48:266–270. doi: 10.1159/000022814. [DOI] [PubMed] [Google Scholar]
- Lien S, Cockett NE, Klungland H, Arnheim N, Georges M, Gomez-Raya L. High-resolution gametic map of the sheep callipyge region: Linkage heterogeneity among rams detected by sperm typing. Animal Genet. 1999;30:42–46. doi: 10.1046/j.1365-2052.1999.00430.x. [DOI] [PubMed] [Google Scholar]
- Lord EA, Lumsden JM, Dodds KG, Henry HM, Crawford AM, Ansari HA, Pearce PD, Maher DW, Stone RT, Kappes SM, et al. The linkage map of sheep chromosome 6 compared with orthologous regions in other species. Mammal Genome. 1996;7:373–376. doi: 10.1007/s003359900107. [DOI] [PubMed] [Google Scholar]
- Lumsden JM, Lord EA, Cato SA, Richardson TE, van Stijn TC, Broom MF, Patel K, Montgomery GW. The application of AFLP fingerprinting to construct a YAC contig containing ADH2 and MTP on sheep chromosome 6. Cytogenet Cell Genet. 1999;84:225–229. doi: 10.1159/000015264. [DOI] [PubMed] [Google Scholar]
- Ma RZ, Beever JE, Da Y, Green CA, Russ I, Park C, Heyen DW, Everts RE, Fisher SR, Overton KM, et al. A male linkage map of the cattle (Bos taurus) genome. J Hered. 1996;87:261–271. doi: 10.1093/oxfordjournals.jhered.a022999. [DOI] [PubMed] [Google Scholar]
- MacDonald PA, McEwan JC, Goosen GJ, Dodds KG, Green RS, Wheeler RW, Knowler KJ, Greer GJ, Crawford AM. A “dagginess” QTL in sheep. Animal Genet. 1998;29S1:67. [Google Scholar]
- McGavin MD. An animal model of human disease: Progressive ovine muscular dystrophy. Comp Pathol Bullet. 1974;6:3–4. [Google Scholar]
- McLaren RJ, Rogers GR, Davies KP, Maddox JF, Montgomery GW. Linkage mapping of wool keratin and keratin-associated protein genes in sheep. Mammal Genome. 1997;8:938–940. doi: 10.1007/s003359900616. [DOI] [PubMed] [Google Scholar]
- Matise TC, Perlin M, Chakravarti A. Automated construction of genetic linkage maps using an expert system (MultiMap): A human genome linkage map. Nature Genet. 1994;6:384–390. doi: 10.1038/ng0494-384. [DOI] [PubMed] [Google Scholar]
- Mellersh CS, Langston AA, Acland GM, Fleming MA, Ray K, Wiegand NA, Francisco LV, Gibbs M, Aguirre GD, Ostrander EA. A linkage map of the canine genome. Genomics. 1997;46:326–336. doi: 10.1006/geno.1997.5098. [DOI] [PubMed] [Google Scholar]
- Mohrenweiser HW, Tsujimoto S, Gordon L, Olsen AS. Regions of sex-specific hypo- and hyper-recombination identified through integration of 180 genetic markers into the metric physical map of human chromosome 19. Genomics. 1998;47:153–162. doi: 10.1006/geno.1997.5097. [DOI] [PubMed] [Google Scholar]
- Montgomery GW, Crawford AM, Penty JM, Dodds KG, Ede AJ, Henry HM, Pierson CA, Lord EA, Galloway SM, Schmack AE, et al. The ovine Booroola fecundity gene (FecB) is linked to markers from a region of human chromosome 4q. Nature Genet. 1993;4:410–414. doi: 10.1038/ng0893-410. [DOI] [PubMed] [Google Scholar]
- Montgomery GW, Lord EA, Penty JM, Dodds KG, Broad TE, Cambridge L, Sunden SLF, Stone RT, Crawford AM. The Booroola fecundity (FecB) gene maps to sheep chromosome 6. Genomics. 1994;22:148–153. doi: 10.1006/geno.1994.1355. [DOI] [PubMed] [Google Scholar]
- Montgomery GW, Henry HM, Dodds KG, Beattie AE, Wuliji T, Crawford AM. Mapping of the Horns (Ho) locus in sheep: A further locus controlling horn development in domestic animals. J Hered. 1996;87:358–363. doi: 10.1093/oxfordjournals.jhered.a023014. [DOI] [PubMed] [Google Scholar]
- Ott J. Strategies for characterizing highly polymorphic markers in human gene mapping. Am J Hum Genet. 1992;51:283–290. [PMC free article] [PubMed] [Google Scholar]
- Phua SH, Garmonsway HG, Dodds KG, McEwan JC, Morris CA, Towers NR, Hyndman DL, Pitchford WS, Crawford AM. Detection of a QTL for resistance to facial eczema disease in sheep. Animal Genet. 1998;29S1:74. [Google Scholar]
- Piumi F, Schibler L, Vaiman D, Oustry A, Cribiu EP. Comparative cytogenetic mapping reveals chromosome rearrangements between the X chromosomes of two closely related mammalian species (cattle and goats) Cytogenet Cell Genet. 1998;81:36–41. doi: 10.1159/000015004. [DOI] [PubMed] [Google Scholar]
- Ponce de Leon FA, Ambady S, Hawkins GA, Kappes SM, Bishop MD, Robl JM, Beattie CW. Development of a bovine X chromosome linkage group and painting probes to assess cattle, sheep, and goat X chromosome segment homologies. Proc Natl Acad Sci. 1996;93:3450–3454. doi: 10.1073/pnas.93.8.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NA, McPartlan H, Ward W, Matthews ME. DNA testing for breeding better sheep. Wool Technol Sheep Breeding. 1997;45:147–156. [Google Scholar]
- Schibler L, Vaiman D, Oustry A, Giraud-Delville C, Cribiu EP. Comparative gene mapping: A fine-scale survey of chromosome rearrangements between ruminants and humans. Genome Res. 1998;8:901–915. doi: 10.1101/gr.8.9.901. [DOI] [PubMed] [Google Scholar]
- Schmutz SM, Moker JS, Berryere TG. In situ hybridization of five loci to cattle chromosome 1. Cytogenet Cell Genet. 1998;81:51–53. doi: 10.1159/000015007. [DOI] [PubMed] [Google Scholar]
- Simianer H, Szyda J, Ramon G, Lien S. Evidence for individual and between-family variability of the recombination rate in cattle. Mammal Genome. 1997;8:830–835. doi: 10.1007/s003359900587. [DOI] [PubMed] [Google Scholar]
- Sonstegard TS, Lopez-Corrales NL, Kappes SM, Stone RT, Ambady S, Ponce de León FA, Beattie CW. An integrated genetic and physical map of the bovine X chromosome. Mammal Genome. 1997;8:16–20. doi: 10.1007/s003359900339. [DOI] [PubMed] [Google Scholar]
- Tan P, Allen JG, Wilton SD, Akkari PA, Huxtable CR, Laing NG. A splice-site mutation causing ovine McArdle's disease. Neuromuscular Disorders. 1997;7:336–342. doi: 10.1016/s0960-8966(97)00062-x. [DOI] [PubMed] [Google Scholar]
- Taylor JF, Lutaaya E, Sanders JO, Turner JW, Davis SK. A medium density microsatellite map of BTA10: Reassignment of INRA69. Animal Genet. 1997;28:360–362. doi: 10.1111/j.1365-2052.1997.00177.x. [DOI] [PubMed] [Google Scholar]
- Taylor JF, Eggen A, Aleyasin A, Armitage SM, Barendse W, Beever JE, Bishop MD, Brenneman RA, Burns BM, Davis SK, et al. Report of the first workshop on the genetic map of bovine chromosome 1. Animal Genet. 1998;29:228–235. doi: 10.1111/j.1365-2052.1998.00287.x. [DOI] [PubMed] [Google Scholar]
- Vaiman D, Brunalfi A, Bensaada M, Derbois C, Vaiman A, Crawford A, Metezeau P, Cribiu EP. Isolation of subtelomeric DNA sequences labelling sheep and goat chromosome ends. Genet Selec Evol. 2000;32:599–619. doi: 10.1186/1297-9686-32-6-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaiman D, Schibler L, Bourgeois F, Oustry A, Amigues Y, Cribiu EP. A genetic linkage map of the male goat genome. Genetics. 1996;144:279–305. doi: 10.1093/genetics/144.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oorschot RA, Maddox JF, Adams LJ, Fabb SA. Characterization and evolution of ovine MHC class II DQB sequence polymorphism. Animal Genet. 1994;25:417–424. doi: 10.1111/j.1365-2052.1994.tb00532.x. [DOI] [PubMed] [Google Scholar]
- Wright CD, Havill AM, Middleton SC, Kashem MA, Lee PA, Dripps DJ, O'Riordan TG, Bevilacqua MP, Abraham WM. Secretory leukocyte protease inhibitor prevents allergen-induced pulmonary responses in animal models of asthma. J Pharmacol Exp Therapeut. 1999;289:1007–1014. [PubMed] [Google Scholar]
- Yu J, Lazzeroni L, Qin J, Huang MM, Navidi W, Erlich H, Arnheim N. Individual variation in recombination among human males. Am J Hum Genet. 1996;59:1186–1192. [PMC free article] [PubMed] [Google Scholar]