Abstract
Although Corynebacterium amycolatum can cause opportunistic infections, it is commonly considered as contaminant. In this report, we present a case of bacteremia caused by C. amycolatum with a novel mutation in the gyrA gene that confers high-level quinolone resistance to the organism.
Keywords: Corynebacterium amycolatum, gyrA mutation, Quinolone resistance
INTRODUCTION
Corynebacterium species except Corynebacterium diphtheriae are part of the normal flora of the skin and mucous membrane and occur as common contaminants in diagnostic specimens. Therefore, physicians often tend to consider these organisms as contaminants. However, these microorganisms have increasingly been recognized as pathogens in immunocompromised patients [1]. Corynebacterium amycolatum, in particular, is a common clinical isolate that often shows multidrug resistance to commonly used antibiotics such as β-lactams, macrolides, and fluoroquinolones [2, 3]. In this report, we present a case of bacteremia caused by C. amycolatum possessing a novel mutation in the gyrA gene, which confers high-level quinolone resistance to the organism.
CASE REPORT
An 88-yr-old man with diabetes mellitus presented to our emergency department with chest pain. There was no definite evidence of acute coronary syndrome, but laboratory examination findings indicated impending end-stage renal disease. Hemodialysis was performed through a subclavian double-lumen catheter on alternate days. After 3 sessions of hemodialysis, on the 11th hospital day, the patient developed fever (temperature, 38.0℃), and the blood profile showed a total white blood cell count of 8,800/µL with 74% neutrophils and an elevated C-reactive protein level of 134.2 mg/L. Hence, samples for blood culture were drawn in triplicate, and empiric antibiotic therapy was initiated with 200 mg of ciprofloxacin administered intravenously every 12 hr. On the 14th hospital day, after 72 hr of incubation, "diphtheroid-like organisms" were detected in all 3 aerobic blood-culture bottles. Although the patient had fever and elevated C-reactive protein level, which do indicate bacteremia, the positive blood culture was considered to show merely contaminants, and therefore, additional blood cultures were performed to identify the causative agent. The repeated blood cultures yielded the same results. Therefore, the antibiotic therapy was changed to vancomycin (1 g/day), and the indwelling double-lumen catheter was removed because of suspected catheter-related infection. Follow-up blood cultures were negative, and the patient recovered fully.
1. Identification
Initially, the "diphtheroid-like organisms" were identified as C. diphtheriae by an API Coryne strip (bioMerieux, Marcy l'Etoile, France). Later, 16S rRNA sequence analysis was performed to confirm the species, as described previously [4]. The isolate exhibited 99.1% identity with C. amycolatum CIP 103452T (GenBank accession no.: X82057), and VITEK 2 Anaerobe and Corynebacterium Identification Card (bioMerieux) analysis confirmed the presence of C. amycolatum with a probability of 99.0%.
2. Susceptibility testing
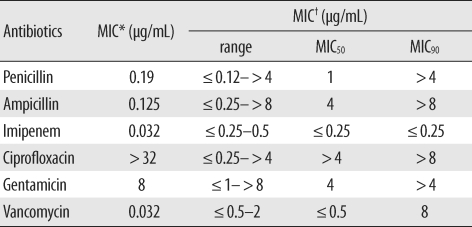
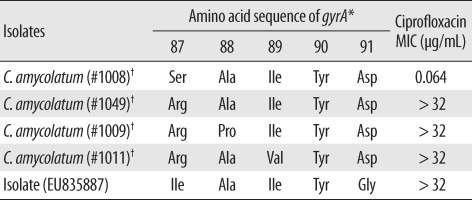
To study the multidrug resistance phenotype of C. amycolatum, we measured the minimum inhibitory concentrations (MICs) of penicillin, ampicillin, imipenem, ciprofloxacin, gentamicin, and vancomycin for this isolate by using Etest (AB bioMerieux, Solna, Sweden). The MIC values are shown in Table 1; ciprofloxacin had the highest MIC value of >32 µg/mL. To elucidate the mechanism underlying the high-level ciprofloxacin resistance, we analyzed the sequence of the quinolone resistance-determining region of the gyrA gene, as described by Sierra et al. [5]. This isolate showed novel mutations in the gyrA gene, wherein the codon for Ser-87 mutated to form an Ile codon and the codon for Asp-91 mutated to a Gly codon (GenBank accession no.:EU835887, Table 2).
Table 1.
MIC values for Corynebacterium amycolatum
*in this case; †MIC values were determined in 60 strains of C. amycolatum by using the method described by Gómez-Garcés et al. [6].
Abbreviation: MIC, minimum inhibitory concentration
Table 2.
Mutations observed in the quinolone resistance-determining regions of the gyrA gene of Corynebacterium amycolatum isolates and their ciprofloxacin MIC values
*The amino acid position number was obtained from the amino acid sequence of the gyrA gene of Corynebacterium glutamicum; †Data reproduced from Sierra et al. [2].
Abbreviations: MIC, minimum inhibitory concentration; Ser, serine; Ala, alanine; Ile, isoleucine; Tyr, tyrosine; Asp, aspartate; Arg, arginine; Pro, proline; Val, valine; Gly, glycine
DISCUSSION
Although its pathogenic role was apparent only in few cases, C. amycolatum, like Corynebacterium striatum, can cause endocarditis, bacteremia, and catheter exit-site infection. In cases wherein clinically significant infection by any of the coryneform bacteria is suspected, species identification and antibiotic susceptibility determination are important to decide and administer the optimal antibiotic and treatment regimens. However, possibilities like misidentification of species or non-availability of adequate data for using automated kits with identification cards should be considered. Genotypic assays such as 16s rRNA sequencing may be required to confirm the identity of species. In this present case, the clinical isolate showed a high level of resistance to ciprofloxacin (MIC value of >32 µg/mL), probably due to the novel mutation in the gyrA gene, wherein the codon for Ser-87 mutated to form an Ile codon and the codon for Asp-91 mutated to a Gly codon [6]. Therefore, in cases of suspected Corynebacterium spp. infection, the physician and the microbiologists in laboratories should confirm the antibiotic susceptibility patterns of the clinical isolates to help decide the optimal antibiotic and treatment regimens.
Footnotes
No potential conflict of interest relevant to this article was reported.
References
- 1.Funke G, von Graevenitz A, Clarridge JE, 3rd, Bernard KA. Clinical microbiology of coryneform bacteria. Clin Microbiol Rev. 1997;10:125–159. doi: 10.1128/cmr.10.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soriano F, Zapardiel J, Nieto E. Antimicrobial susceptibilities of Corynebacterium species and other non-spore-forming gram-positive bacilli to 18 antimicrobial agents. Antimicrob Agents Chemother. 1995;39:208–214. doi: 10.1128/aac.39.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Funke G, Bernard KA. Coryneform gram-positive rods. In: Murray PR, Baron EJ, et al., editors. Manual of clinical microbiology. 9th ed. Washington, DC: ASM Press; 2007. pp. 485–514. [Google Scholar]
- 4.Greisen K, Loeffelholz M, Purohit A, Leong D. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J Clin Microbiol. 1994;32:335–351. doi: 10.1128/jcm.32.2.335-351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sierra JM, Martinez-Martinez L, Vázquez F, Giralt E, Vila J. Relationship between mutations in the gyrA gene and quinolone resistance in clinical isolates of Corynebacterium striatum and Corynebacterium amycolatum. Antimicrob Agents Chemother. 2005;49:1714–1719. doi: 10.1128/AAC.49.5.1714-1719.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gómez-Garcés JL, Alos JI, Tamayo J. In vitro activity of linezolid and 12 other antimicrobials against coryneform bacteria. Int J Antimicrob Agents. 2007;29:688–692. doi: 10.1016/j.ijantimicag.2006.11.032. [DOI] [PubMed] [Google Scholar]